Abstract
Long intergenic noncoding RNAs (lincRNAs) are increasingly recognized as important mediators of many biological processes relevant to human pathophysiologies, including cardiovascular diseases. In vitro studies have provided important knowledge of cellular functions and mechanisms for an increasing number of lincRNAs. Dysregulated lncRNAs have been associated with cell fate programming and development, vascular diseases, atherosclerosis, dyslipidemia and metabolic syndrome, and cardiac pathological hypertrophy. However, functional interrogation of individual lincRNAs in physiological and disease states is largely limited. The complex nature of lincRNA actions and poor species conservation of human lincRNAs pose substantial challenges to physiological studies in animal model systems and in clinical translation. This review summarizes recent findings of specific lincRNA physiological studies, including MALAT1, MeXis, Lnc-DC and others, in the context of cardiovascular diseases, examines complex mechanisms of lincRNA actions, reviews in vivo research strategies to delineate lincRNA functions and highlights challenges and approaches for physiological studies of primate-specific lincRNAs.
Keywords: Long intergenic noncoding RNAs, Cardiovascular diseases, Animal models
1. Introduction
Pervasive transcription in mammalian genome generate tens of thousands RNA transcripts that do not encode proteins. Long intergenic noncoding RNAs (lincRNAs) are defined as over 200 nucleotides in length and, like protein-coding mRNAs, most are spliced and 3’ polyadenylated [2]. Compared to mRNAs, lincRNAs are poorly conserved across species, and their expression is lower and more tissue specific [2–4]. Although the biological roles of most lincRNAs remain to be elucidated, they are emerging as key mediators in a vast variety of cellular functions, such as cell proliferation and differentiation, and have been implicated in many human pathologies, including cancer [5,6], cardiovascular diseases (CVD) [7–9] and metabolic disorders [10,11]. In addition, recent functional genomic studies suggest that long noncoding RNAs, not the nearby protein-coding genes, are the likely causal element driving human disease association at some genome wide association study (GWAS) loci. These include candidates like ANRIL at the 9p21 locus for CVD [12–15], CCAT2 for colon cancer [16], and lnc13 for celiac disease association [17]. Indeed, multiple functional variants at lncRNA loci have been identified for CVD traits (Table 1) [18–22] including ANRIL variants that regulate the expression of CDKN2A/B and atherogenic pathways [18,19], in cis SNPs related to the expression level of Myocardial Infarction Associated Transcript (MIAT) and corresponding susceptibility of myocardial infarction [20], and H19 variants associated with increased IGF2 expression increased CVD risk [21]. These findings suggest that studying functional roles of lincRNAs in pathophysiology may provide new insights into causal pathogenic mechanism and opportunities for novel diagnostic and therapeutic strategies in human diseases, including CVD.
Table 1.
Examples of lncRNA loci that harbor SNPs associated with cardiovascular diseases.a.
| lncRNA | SNPs | Functional effects | Related disease | References |
|---|---|---|---|---|
| ANRIL | rs1333049, rs10757278, rs2383206 | Suppress expression of CDKN2A/B and regulate atherogenic pathways | Atherosclerosis | [18,19] |
| MIAT | Intron 1: 5338C > T (rs2331291), exon 3: 8813G > A, exon 3: 9186G > A (rs2301523), exon5: 11093 G > A, exon5: 11741 G > A, exon5: 12311 C > T | Regulate expression level of MIAT | Myocardial infarction | [20] |
| H19 | rs217727, rs2067051 | Increase expression level of IGF2 | Coronary artery disease | [21] |
| Linc-VWF | rs1558324 | Regulate expression level of VWF | Inflammation | [22] |
lncRNA, long noncoding RNA; ANRIL, antisense noncoding RNA in the INK4 locus; CDKN2A/B, cyclin-dependent kinase Inhibitor 2A/B; IGF2, insulin-like growth factor 2; MIAT, myocardial infarction associated transcript; SNP, single-nucleotide polymorphism; VWF, Von Willebrand factor.
Modified from Dechamethakun, J Hum Genet. 2017 [110].
LincRNAs mediate gene expression or function through transcriptional and post-transcription regulation. Through their primary sequence motifs or secondary structure, lncRNAs can interact with specific proteins or RNA partners, such as transcriptional regulators and histone modifying enzymes in the nucleus [23,24], or RNA-binding proteins and microRNA in the cytoplasm [25,26]. Some lincRNAs encode microRNAs within their gene loci and are precursors for functional microRNAs [27]. Independent of RNA-dependent mechanisms, lincRNAs can act in cis through regulatory DNA elements or transcription activities at lincRNA genomic loci [28,29]. Individual lincRNAs can regulate diverse gene pathways or distinct cellular processes through multiple distinct molecular mechanisms. For example, our group has shown that adipose linc-ADAL binds with distinct protein partners in adipocyte nucleus and cytoplasm to regulate adipocyte differentiation and de novo lipogenesis respectively [30]. LincRNA-p21 regulates the expression of nearby gene CDKN1A through in cis-acting DNA elements while also modulating a larger set of genes in trans through RNA-dependent mechanism [31–33]. The complex nature of lncRNA actions underscores the importance of studying their physiological roles in tissues and whole organisms. This review summarizes examples of CVD-related lincRNAs (Fig. 1) and their physiological impacts in animal models (Table 2), discusses in vivo research strategies for interrogating lincRNA function (Table 3) and highlights challenges and approaches for in vivo studies of primate-specific lincRNAs (Fig. 2). The primary focus of this review is on lincRNAs i.e., lncRNAs that are intergenic, the genomic location of the vast majority of lncRNAs and GWAS loci for CVD, and for that reason we do not discuss in detail lncRNAs that are antisense or overlap protein coding genes, including ANRIL the well-studied lncRNA at the 9p21 locus for CVD [12–15].
Fig. 1.
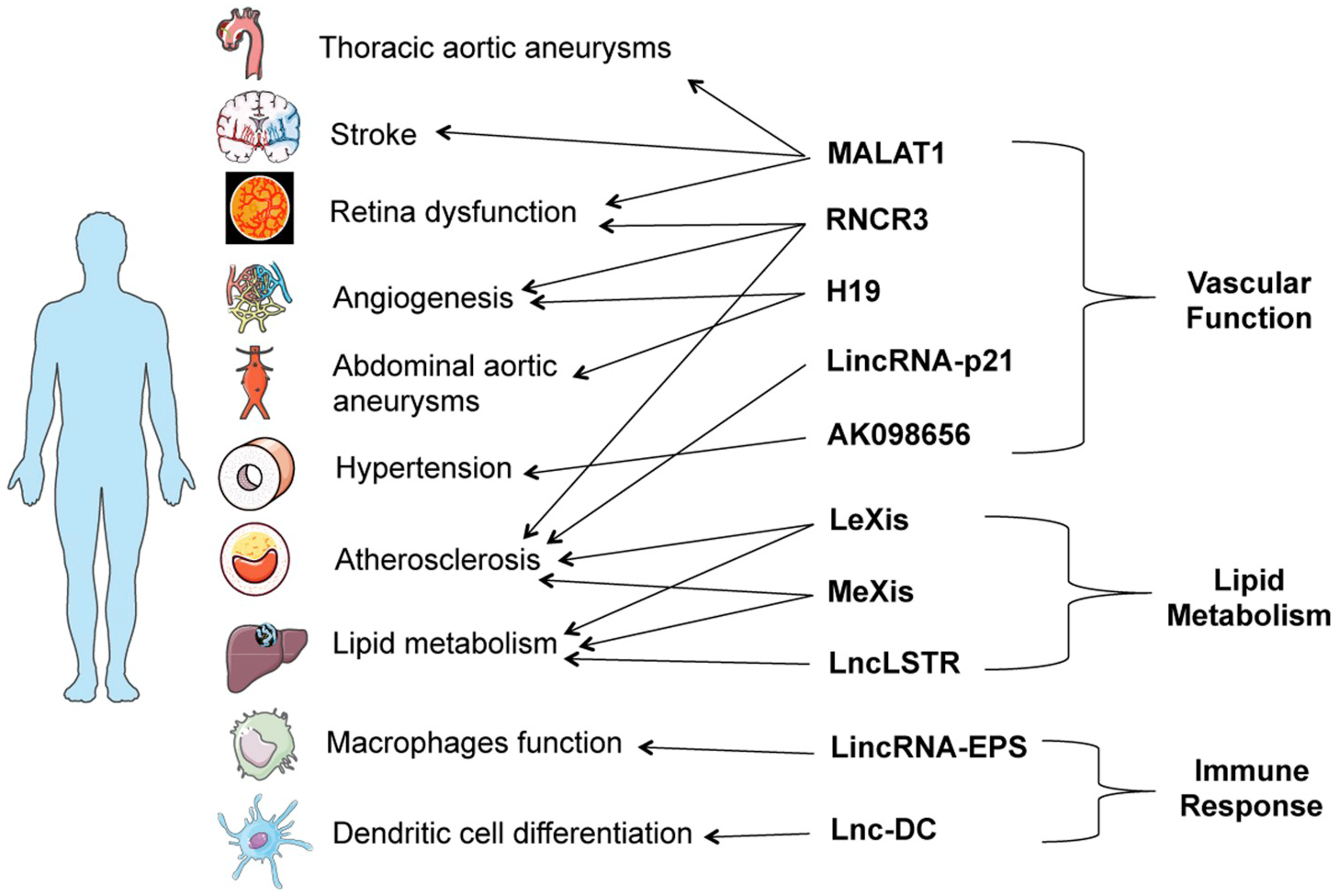
Established physiological and pathophysiological roles of lincRNAs in cardiovascular diseases.
Table 2.
List of CVD-related lincRNA examples and their physiological impacts in animal models. Human (H) and mouse (M) genomic locations of these lincRNAs are based on human hg19 and mouse MM10 genome build.
| Biological process | LincRNA | Conservation | Genomic location in human and mouse | In vivo perturbation | Physiological impact | Ref. |
|---|---|---|---|---|---|---|
| Vascular function | MALAT1 | Yes | H-chr11 (+): 65265224–65273940 | Genetic deletion of gene locus | Reduced endothelial cell proliferation and angiogenesis | [51] |
| M-chr19(−): 5795690–5802671 | Enhanced SMC contractile function and reduced thoracic aneurysm growth | [56] | ||||
| H19 | Yes | H-chr11 (−): 2016406–2019065 | RNA delivery by nano-vesicles | Improved wound healing in diabetic rats | [71] | |
| M-chr7(−): 142575530–142578146 | Genetic deletion of gene locus | Reduced SMC apoptosis and development of abdominal aneurysms | [72] | |||
| lincRNA-p21 | Yes | H-chr6(−): 36631169–36635073 | RNAi-mediated knockdown | Increased neointima formation in the carotid artery injury model | [75] | |
| M-chr17(−): 29057474–29079126 | ||||||
| RNCR3 | Yes | H-chr8(−): 9757574–9760839 | RNAi-mediated knockdown | Increased formation of atherosclerotic lesion in mouse aortas | [78] | |
| M-chr14(+): 64588115–64593961 | ||||||
| AK098656 | Unknown | H-chr16(−):80601000–80606705 | Transgenic rat model | Spontaneous hypertension, smaller diameters and reduced contractile protein expression of resistant arteries | [79] | |
| Lipid metabolism | IncLSTR LeXis | No | M-chr1(+):151138034 −151144095 | RNAi-mediated knockdown in liver | Reduced plasma triglyceride levels, increased hepatic expression of lipoprotein ApoC2 | [80] |
| Yes | H-chr9(−):107752367–107754061 | Virus-mediated RNA overexpression | Reduced hepatic cholesterol biosynthesis, serum cholesterol and atherosclerotic lesion | [81,82] | ||
| M-chr4(−): 53201519–53220013 | Genetic deletion and lincRNA knockdown in liver | Increased lipid accumulation and expression of lipogenesis is genes in the liver | [81] | |||
| MeXis | Yes | M-chr4(−):53261356–53265492 | RNAi-mediated knockdown in liver | Increased hepatic ABCA1 expression and serum cholesterol level | [83] | |
| H-chr9(+):107854176–107881755 | Genetic deletion of gene locus | Decreased cholesterol efflux in macrophages; increased aortic atherosclerotic lesions | [83] | |||
| Immune response | lincRNA-EPS | unknown | M-chr4(−): 109402279–109406257 | Genetic deletion of gene locus | Enhanced basal and TLR4 induced expression of immune response genes in macrophages; decreased survival rate following endotoxemia challenge | [38] |
| Lnc-DC | Yes | H-chr17(−): 58160927–58165828 | RNAi-mediated knockdown in bone marrow | Impaired mouse dendritic cell differentiation | [84] | |
| M-chr11(+)83746940–83752646 |
Table 3.
Different strategies to perturb lincRNAs in vivo and their impacts on the endogenous lincRNA genetic loci.
| Method | Perturbation of endogenous lincRNA gene | |||
|---|---|---|---|---|
| RNA | DNA | Transcription | ||
| RNA-based approach | RNAi or ASO | + | − | − |
| Transgene for overexpression | + | − | − | |
| DNA-based approach | Promoter deletion | + | (−)a | + |
| Gene body deletion | + | + | + | |
| Transcription termination signal insertion | + | (−)a | + | |
| CRISPRi or CRISPR activation | + | − | + | |
+ = disrupts; − = no effect.
No major disruption of DNA sequences in the lincRNA gene body.
Fig. 2.
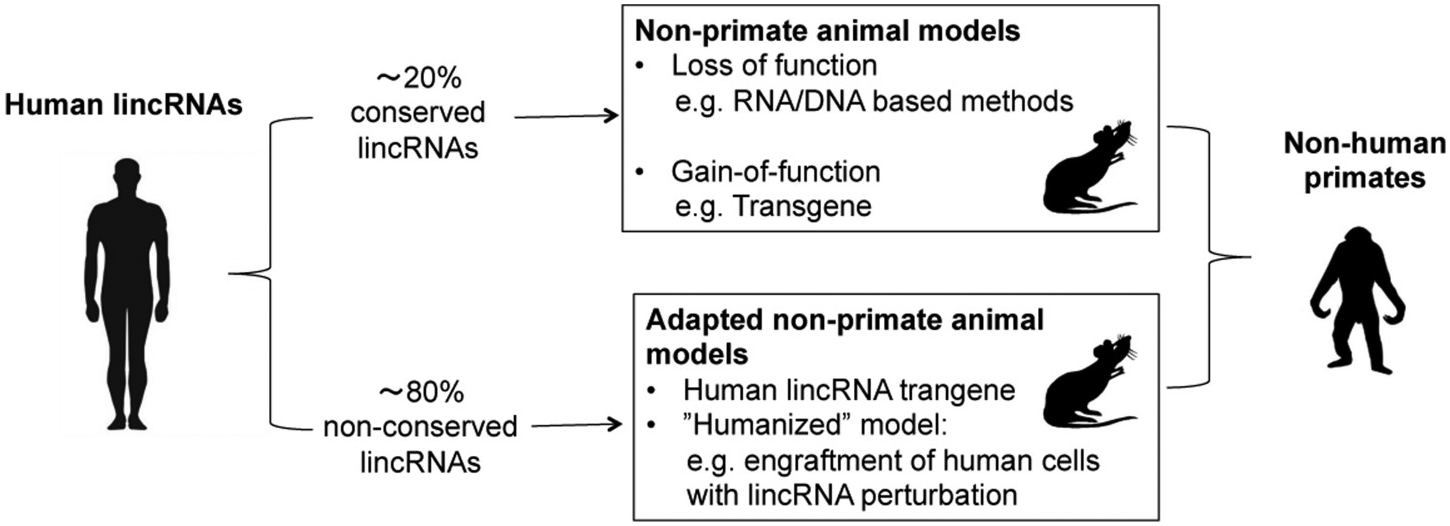
Strategies for interrogation of conserved and non-conserved human lincRNAs in vivo.
2. Importance of physiological studies in lincRNA research
Over the past decade, lincRNA studies have flourished in many research fields, from systematic identification and categorization of lincRNAs in an increasing number of species to in-depth functional and mechanistic interrogation of individual lincRNAs. Although the functional significance of most lincRNAs remain unclear, in vitro loss-of-function and gain-of-function studies have provided valuable insights into the potential biological roles of lincRNAs, at least in cellular context. However, emerging evidence has suggested that knowledge of lincRNA functions gleaned from in vitro systems may poorly inform or match the physiological impact of these lincRNAs in animal studies. The limitation of in vitro findings has been observed for both well-characterized lincRNAs (e.g. NEAT1) and newly identified lincRNAs. For example, NEAT1 knockdown in cell lines markedly impaired the formation of papaspeckles [34,35], nuclear bodies important for gene regulation and mRNA processing. Mice with NEAT1 genetic deficiency also had impaired papaspeckles in specific tissues yet exhibited no apparent physiological abnormalities [36]. LincRNA-EPS, an erythroid lincRNA, was recently found to regulate mouse erythroid differentiation [37] yet genetic ablation of this lincRNA showed no effects on erythropoiesis or hemoglobin levels in mice [38]. Conflicting findings from cell and animal studies are not specific to lincRNA research and can be attributed to various factors, such as biological difference between cellular and physiological conditions, the interactions of target genes with other genetic and physiological factors in animal models. For example, lincRNA HOTAIR regulates gene silencing of homeotic genes in HoxD cluster [39] and Hotair−/− mice on C57B6 background demonstrated impaired skeleton development consistent across independent studies [40,41]. However, Hotair−/− mice on a mixed ground showed minimal impairment [42], suggesting that mouse genetic background may have a significant impact on lincRNA knockout phenotypes in vivo.
3. Physiological roles of lincRNAs in cardiovascular diseases
3.1. LincRNAs in vascular function
3.1.1. MALAT1
MALAT1 (metastasis-associated lung adenocarcinoma transcript 1) is a conserved lincRNA ubiquitously expressed in a number of cells and tissues. Its expression is frequently elevated in many types of cancers [43,44] and other pathological conditions, such as hypoxia and hyperglycemia [45–47]. As a nuclear lincRNA, MALAT1 plays important roles in alternative splicing by modulating the distribution and activities of splicing factors in nuclear speckles [48,49]. In addition, MALAT1 is involved in epigenetic gene regulation by interacting with polycomb protein CBX4 [50]. Transcriptome analyses from MALAT1 knockdown studies suggest MALAT1 regulates gene expression of cell cycle regulators such as CCNA2, CCNB1, and CCNB2 in endothelial cells [51]. Consistent with these findings, mouse xenograft studies demonstrate that both genetic deficiency and knockdown of MALAT1 significantly reduce tumor development and metastasis [52–54]. Notably, recently studies support an emerging regulatory role of MALAT1 in the pathogenesis of cardiovascular diseases. Michalik et al. demonstrated that MALATs1−/− mice showed a delayed vessel extension in the retina revascularization of neonatal mice while in vivo inhibition of MALAT1 by antisense oligonucleotides suppressed blood flow recovery in the ischemic hindlimb muscle of adult mice, consistent with impaired proliferation of MALAT1-knockdown endothelial cells in vitro [51]. Similarly, MALAT1−/− mice exhibit higher expression of pro-apoptotic and pro-inflammatory genes in the cerebral cortex, larger brain infarct size, worsened neurological scores, and reduced sensorimotor functions after transient focal cerebral ischemia [55]. Together, these in vivo studies support an important role of MALAT1 in promoting endothelial cell proliferation and blood vessel growth [51,55]. Interestingly, Cardenas et al. recently revealed that MALAT1 form a ternary complex with histone deacetylase HDAC9 and the chromatin-remodeling enzyme BRG1 to recruit polycomb repressive complex 2 (PRC2) and inhibit gene expression of contractile proteins in vascular smooth muscle cells [56]. Importantly, in vivo genetic ablation of MALAT1 restored contractile protein expression in mouse aortas and reduced the development of thoracic aortic aneurysms [56]. The distinct regulatory roles of MALAT1 in different vascular cells highlight the complexity of lincRNA action in diverse pathophysiological conditions and the challenge in therapeutic targeting of this ubiquitously expressed lincRNA.
3.1.2. H19
H19 (H19 imprinted maternally expressed transcript) is one of the first identified imprinted lncRNAs and is only transcribed from the maternal allele [57]. Both the primary sequences and the secondary structures of H19 are well conserved across many mammals [58,59]. H19 expression is highly induced during embryogenesis, reduced after birth and only retained in a few adult tissues, such as skeleton muscles, adipose and adrenals [60,61]. H19 is often abnormally activated in many types of cancers and has been increasingly recognized as an oncogene [62,63]. Besides serving as a precursor of microRNA miR-675 [64], H19 has been shown to interact with a variety of proteins (such as RNA-binding proteins HuR [65], KSRP [66] and hnRNPU [67]) and microRNAs (such as Let-7 [68] and miR-106 [69]) to regulate genes involved in cell proliferation, migration, differentiation and tumor-igenesis.
For endothelial cells, increased H19 expression in microvessels has been associated with increased angiogenesis in glioma tissues [70]. Hyperglycemia-induced reduction of H19 expression in endothelial cells was linked to impaired angiogenesis in diabetes [71] and Tao et al. showed that in vivo delivery of exogenous H19 by extracellular vesicle-mimetic nano-vesicles markedly improve wound healing in diabetic rats, supporting a pro-angiogenic role of H19 [71]. For SMCs, recent conflicting studies reported opposing effects of H19 on in vitro proliferation and apoptosis: Li et al. showed H19 knockdown significantly inhibited apoptosis and increased proliferation of human aortic SMCs [72] while Zhang el al. observed the opposite phenotype in human aortic SMCs stimulated by oxidized LDL (ox-LDL) [73]. Importantly, Li et al. demonstrated that H19 expression in aortic media SMCs was elevated in mouse models of abdominal aortic aneurysms (AAAs) and in vivo H19 knockdown limited AAA growth, consistent with the pro-apoptotic effect of H19 in their in vitro SMC studies [72]. The exact regulatory roles of H19 in SMC function and vascular disorders may dependent on different H19-interacting partners in distinct pathophysiological and SMC phenotype contexts. For example, Li et al. demonstrated H19 promote SMC apoptosis via HIF1α [73] while Zhang et al. showed H19 inhibited apoptosis of ox-LDL stimulated SMCs via miR-148b/WNT/β-catenin [73]. Further studies are required to resolve these apparently conflicting data.
3.1.3. lincRNA-p21
lincRNA-p21 is a conserved and p53-regulated lincRNAs. It has been shown to act in trans by forming a repressive complex with hnRNPK to inhibit many gene in p53 transcriptional network [31] or binding directing with target mRNAs to suppress their translation [32]. In addition, lincRNA-p21 appears to regulate its nearby protein-coding gene Cdkn1a (p21) through cis-regulatory DNA elements in the locus [29,33]. As a p53-target lincRNA, the expression of lincRNA-p21 was decreased in many cancer types [74]. Many studies on lincRNA-p21 focused on p53-dependent apoptosis, cell cycle regulation and cancer research [74]. Wu et al. demonstrated that the expression level of lincRNA-p21 was reduced in mouse aortas with atherosclerotic plaques and in human coronary artery tissues of CAD patients [75]. Consistent with previous finding on the effects of lincRNA-p21 in cell cycle, in vitro knockdown of lincRNA-p21 promoted proliferation and reduced apoptosis of vascular SMCs [75]. In vivo lincRNA-p21 knockdown in mouse carotid arteries by lentiviral-based shRNA markedly increased neointima formation in the carotid artery injury model [75]. Furthermore, Wu et al. present a novel mechanism of lincRNA-p21 to promote p53 activity in SMCs. LincRNA-p21 modulates the dynamic interaction between p53 and its post-transcriptional regulators p300 and MDM2 by directly binding MDM2, reducing p53-MDM2 complex while promoting p53-p300 interaction. This results in a marked increase in P53 activities [75].
3.1.4. RNCR3
RNCR3 (Retinal non-coding RNA 3), is a conserved lincRNAs highly expressed in brain tissues [27]. Its expression is actively regulated during mouse retina development [76] and the differentiation of neurons and oligodendrocytes [77]. Similar to H19, RNCR3 also serves as the precursor for a microRNA, miR-124a. Genetic ablation of RNCR3 resulted in several defects in mouse brain development, including smaller brain size and aberrant growth of dentate granule cell axons [27]. These phenotypes were rescued by overexpressing miR-124a [27], suggesting RNCR3 act through miR-124a to regulate central nervous system development. Recently Shan et al. demonstrated that RNCR3 was expressed in vascular smooth muscle and endothelial cells and was increased by hypercholesterolemia stress (oxidized LDL) in vitro [78]. RNCR3 expression was marked increased in mouse aortas with atherosclerotic lesions [78]. Importantly, in vivo knockdown of RNCR3 by viral-based short hairpin RNA (shRNA) accelerated development of atherosclerosis, supporting a protective role of RNCR3 in atherosclerosis [78].
3.1.5. AK098656
AK098656 is a human lincRNA that are highly expressed in human testis [2] and appears not conserved in rodents [79]. Its expression is also predominantly detected in smooth muscle cells among 15 human cells derived from various tissues [79]. Jin et al. found that the level of AK098656 was significantly elevated in the plasma of hypertensive patients [79]. In vitro knockdown of AK098656 inhibited, whereas overexpression increased, SMC proliferation, suggesting a regulatory role of AK098656 in promoting SMC synthetic phenotype [79]. A transgenic rat model overexpressing AK098656 had spontaneous hypertension as well as narrowed diameter and reduced contractile protein expression of resistant arteries [79]. Mechanistically, AK098656 binds with SMC contractile protein myosin heavy chain-11 and fibronectin-1 to promote their degradation, possibly by interacting with 26S proteasome non-ATPase regulatory subunit 11 [79].
3.2. lincRNAs in lipid metabolism
3.2.1. lncLSTR
lncLSTR (liver-specific triglyceride regulator) is a mouse lincRNA predominantly expressed in liver [80]. Its liver expression fluctuates during fasting and refeeding, suggesting a regulatory role in energy metabolism [80]. In vivo lncLSTR knockdown in mouse liver significantly reduced plasma triglyceride levels in normal and hyperlipidemic mice by increasing expression of liver-secreted lipoprotein ApoC2, a potent activator of lipoprotein lipase (LPL) and enhancing LPL-mediated triglyceride clearance in peripheral tissues [80]. Interestingly, in vitro lncLSTR knockdown in hepatocytes showed no effects on ApoC2 expression, suggesting lncLSTR does not directly regulate apoC2 through a cell-autonomous mechanism [80]. In vivo lncLSTR knockdown reduced liver expression of Cyp8b1, a key enzyme determining the ratio of two most abundant bile acids in mouse, cholic acid and muricholic acid, thus resulting in increased muricholic acid in bile pool composition [80]. Muricholic acid exhibited greater induction of apoC2 genes after binding with bile acid receptor FXR in liver [80]. In summary, lncLSTR modulates hepatic and plasma apoC2 levels through liver Cyp8b1-FXR pathway to regulate LPL-mediated triglyceride clearance in peripheral tissues, highlighting a novel lincRNA mechanism that operates in vivo in both physiological and pathophysiological contexts. Notably, Li et al. failed to detect a human ortholog for lncLSTR in annotated human lincRNA catalogs [80]. Further studies are required to determine if lncLSRT is encoded in human or has any functional impact on lipid metabolism in primates.
3.2.2. LeXis
LeXis (liver-expressed LXR induced sequence) is a conserved, liver-enriched lincRNA that is robustly induced by high cholesterol diet feeding and activation of liver x receptor (LXR) [81]. Recent studies demonstrated that in vivo LeXis overexpression in mouse liver significantly reduced expression of hepatic cholesterol biosynthesis genes, total serum cholesterol and triglyceride levels and atherosclerotic lesion in aortas [81,82]. In addition, genetic deficiency and in vivo knockdown of LeXis increased hepatic lipid accumulation and elevated gene expression of lipogenesis genes [81].
3.2.3. MeXis
MeXis (macrophage-expressed LXR-induced sequence) is a conserved and macrophage-enriched lincRNAs that is encoded near protein-coding gene ABCA1, an important regulator of cholesterol efflux. MeXis expression in mouse macrophage was markedly induced by physiologic lipid signals, such as oxidized or acetylated LDL, and LXR activation [83]. Sallam et al. demonstrated that MeXis regulated the expression of nearby gene ABCA1 by interacting with transcriptional coactivator DDX17 in macrophages [83]. In addition, overexpressing MeXis in liver by adenovirus significantly increased hepatic ABCA1 expression and serum cholesterol level [83]. MeXis−/− mice exhibited decreased ApoA-I-dependent cholesterol efflux capacity and increased cholesterol content in macrophages after Western diet feeding [83]. Importantly, atherosclerotic plaque in aortas was significantly increased in hyperlipidemic mice transplanted with MeXis−/− bone marrows [83], supporting macrophage MeXis upregulation of ABCA1-mediated cholesterol efflux and protection from atherosclerosis development in mice.
3.3. lincRNAs in immune response
3.3.1. LincRNA-EPS
lincRNA-EPS is a conserved lincRNAs, whose expression is high in resting macrophages and markedly reduced by toll like receptor (TLR) activation [38]. Atianand et al. recently demonstrated that lincRNA-EPS suppresses transcription of immune response genes in resting macrophages by associating with chromatin at regulatory regions of these genes through hnRNPL nuclear protein and controlling nucleosome positioning [38]. LincRNA-EPS−/− mice show enhanced basal and TLR4 induced expression of immune response genes in macrophages while the numbers of macrophages, dendritic cells, lymphocytes, natural killer cells and red blood cells were unaffected [38]. Furthermore, lincRNA-EPS−/− showed elevated levels of inflammatory cytokines in serum and peritoneal fluid as well as decreased survival rate following LPS challenges [38]. These findings support that lincRNA-EPS plays an important role in controlling both the homeostatic and TLR-inducible immune gene responses in vivo.
3.3.2. Lnc-DC
Lnc-DC is a conserved lincRNA that is detected in many immune cells, including macrophages, monocytes and lymphocytes [84]. Lnc-DC expression is highly enriched in dendritic cells and actively regulated by PU.1 transcription factor [84]. Wang et al. recently demonstrated that lnc-DC regulates dentritic cell differentiation by interacting with STAT3 [84], a transcription factor important to dendritic cell function. Lnc-DC bound directly to STAT3 in the cytoplasm and promoted STAT3 phosphorylation on tyrosine-705 by preventing STAT3 dephosphorylation by SHP1 [84]. Knockdown of lnc-DC in mouse bone marrow cells resulted in impaired mouse dendritic cell differentiation after bone marrow transplantation [84], supporting a regulatory role of lnc-DC in dendritic cell function in vivo.
4. Application and limitation of animal models in lincRNA research
Loss-of-function and gain-of-function approaches are widely used to study physiologies of protein-coding and non-coding genes in animal models. Unlike coding genes exerting their biological functions through proteins, a lincRNA gene can act through multiple elements, including diverse functions of the mature lincRNA transcript itself, the DNA sequences within the lincRNA gene loci, and transcription activity of the lincRNA. For example, lincRNA-p21 has been identified to regulate its neighboring protein-coding gene Cdkn1a in cis [33]. Recent studies demonstrate that deletion of lincRNA-p21 locus markedly reduced Cdkn1a gene expression even in mouse tissues not expressing lincRNA-p21, suggesting lincRNA-p21 gene acts through functional enhancer DNA elements at the locus, i.e., in a RNA-independent manner [29]. Indeed, many in cis-acting lincRNAs appear to share the same promoter or upstream enhancer regions with nearby protein coding genes. Engreitz et al. recently found that multiple lincRNAs (e.g. Blustr and Bendr) regulated nearby gene expression by the transcriptional activities at these lincRNA loci, not by lincRNA transcripts themselves [28]. Genetic ablation of lincRNA transcription, by deleting promoter regions, inserting premature transcription termination signals or perturbing promoter-proximal splice sites, significantly reduced expression of the lincRNA neighboring protein genes [28]. Whereas sequential deletions of Blustr exons and introns had no effect on nearby gene expression, suggesting that Blustr in cis regulation is independent of specific sequences [28]. Similarly, Anderson et al. demonstrate that inhibiting transcription of Upperhand, a lincRNA sharing a bi-directional promoter with nearby gene Hand2, markedly reduced Hand2 expression while knockdown of the mature Upperhand transcript by antisense oligonucleotides (ASOs) had no effect on Hand2 level [85]. Since RNA molecules, genomic regulatory DNA and gene transcription can all contribute to lincRNA molecular mechanisms, it is critical to carefully design experimental strategies for animal models that address each possible mechanism and perturb these individual potential actions of lincRNA. Interpreting in vivo phenotypes using complementary approaches, including recently developed CRISPRi or CRISPR activation, which allow robust transcriptional modulation without altering genetic sequences (Table 3), are often necessary to dissect regulatory mechanisms and physiological functions of lincRNA.
In vivo loss-of-function approaches targeting lincRNA transcript molecules include RNA interference (RNAi, e.g. siRNA and shRNA) and antisense RNA analogues (e.g. locked nucleic acids ASO). ASOs bind with RNA transcripts and activate RNase H mediated RNA degradation while siRNA and shRNA activate RNA-induced silencing complex to degrade targets [86]. Systemic administration of siRNA or ASO by tail vein injection or osmotic pump delivery can achieve sustained knockdown of lincRNA abundance in many tissues in rodents [87,88]. Viral-based shRNAs (e.g. adeno-associated virus) can be applied locally or systemically, providing opportunities to examine lincRNA functions in tissue-specific manner. However, there are still concerns regarding efficiency and off-target effects of in vivo lincRNA knockdown by RNAi. RNAi methods often results in inefficient knockdown of nuclear RNAs as RNAi-mediated RNA destabilization predominantly occurs in the cytoplasm. Importantly, these RNA-targeting strategies preclude interrogation of lincRNAs acting through the transcription or DNA elements at their genomic loci, but can be a useful complement when applied with other genetic strategies (Table 3).
For lincRNA knockout animal models, several strategies can be used to target lincRNA genetic loci and prevent lincRNA transcription, including deleting promoter regions or whole gene bodies of lincRNAs and inserting transcription termination signals (e.g. polyadenylation). While these approaches are very effective to ablate lincRNA expression in vivo, there are several caveats to take into account. For the lincRNAs sharing bi-directional promoters or upstream DNA elements with nearly protein-coding genes, genetic ablation of transcription activities at these loci would likely inhibit the expression of both lincRNAs and local genes [28]. While transcription-mediated in cis regulation of nearby genes might be the mechanism for some lincRNAs, it is possible that they also may act through lincRNA transcripts to regulate genes in trans. In these cases, the unintended knockdown of nearby protein coding genes would confound interrogation of the function of an active transcribed lincRNA molecule. For lincRNAs acting through regulatory DNA element at lincRNA loci, inserting polyadenylation signals after the lincRNA transcription start sites may cause no significant perturbation of regulatory DNA sequences and this potential lincRNA function, even if lincRNA transcription is ablated. In addition, it should be noted that any one lincRNA can function through multiple mechanisms, e.g. DNA-dependent in cis regulation and RNA-dependent in trans action. For example, genetic deletion of lincRNA Fendrr locus impaired lung maturation and mesenchymal differentiation [89] while polyadenylation signal insertion resulted in heart and body wall defects in mouse models [90], suggesting distinct physiological roles of Fendrr in development through independent mechanisms. These findings highlight that lincRNA transgenic animal models and rescue studies are important to delineate specific RNA-dependent lincRNA actions.
5. Challenges to and strategies for the study of non-conserved lincRNAs in vivo
LincRNAs have evolved more recently and more rapidly than protein coding genes and microRNAs [91]. It has been estimated that the majority of primate lincRNAs (~80%) are not conserved in mouse [92,93]. Conservation has to be considered at several levels and requires a systematic comparison of lincRNAs across species in terms of genomic (DNA) positional conservation, primary sequence, transcription status and splicing patterns, as well as secondary structure [94]. For example, most primate and human lincRNAs are not “syntenic” (positionally conserved) in rodents, i.e., at the genomic level the 5′ and/or 3′ neighboring protein coding genes are not the same. In this context, conservation of cis-regulatory functions cannot be considered or studied in mice models. Furthermore, a large percentage of human lincRNAs that are positionally syntenic between human and mice have not been found to be expressed (transcribed) in mouse tissues suggesting that these molecules and functions evolved later and only in primate species. Importantly, sequence similarity does strongly hint at functional conservation but there haven’t been enough studies to suggest that lack of sequence similarity implies lack of functional conservation. For example, FENDRR (FOXF1 adjacent non-coding developmental regulatory RNA) represents one example of a functional lncRNA with a conserved position yet very limited sequence conservation between human and mouse [95].
An increasing amount of data support the concept that many primate-specific lincRNAs, not found in rodents or other organisms, play important regulatory roles in cellular processes, such as pluripotency and differentiation, and have been implicated in human cancer and cardiometabolic disorders [96–98]. While in vitro studies provide useful insights on biological roles of primate-specific lincRNAs in cells, it is crucial to study the physiological impacts of these non-conserved lincRNAs. Yet, interrogation of these non-conserved lincRNAs in animal models is particularly challenging as most model system, such as rodents, often do not encode them. Perturbation of protein coding genes and microRNAs by RNAi or transgene have been applied to non-human primates in translational or pre-clinical studies. However, RNA-based strategies may not always capture lincRNA actions as discussed above. Additionally, non-human primates are scarce and far more costly than other animal models, thus limiting their use and feasible application for most investigators. Recently, however, non-primate models have been adapted and can be very useful tools for examination of these nonconserved lincRNAs In vivo (Fig. 2).
Transgene approach can be used to express primate-specific lincRNAs in non-primate animal models. It is possible, although not widely demonstrated, that the protein or RNA partners of these lincRNAs are conserved and can interact with primate-specific lincRNAs in non-primate animal models. For example, transient overexpression of a primate-specific lincRNA LncND in mouse brains elicited a phenotype on neuron development consistent with that found in vitro in human cell studies [99], supporting the plausibility of the transgene approach for some non-conserved lincRNAs. And bacterial artificial chromosome (BAC) transgene mouse models can utilize the gene body and genomic regulatory DNAs of non-conserved lincRNA loci to drive lincRNA expression at physiological levels and with tissue expression patterns as endogenous lincRNAs in vivo [100]. For lincRNAs that require for function a significant portion of the transcript to be produced, the transgene approach is important to examine non-conserved lincRNAs with some RNA-dependent mechanisms.
Another in vivo approach is to engraft human cells expressing primate-specific lincRNAs in rodent models with immune deficiency. Combined with RNA-based or genetic perturbation of lincRNAs in human cells, these “humanized” animal models permit study of primate-specific lincRNAs acting in their native cellular context and the examination of their functional impacts under physiological conditions. As an example, Xenograft model of human cancer cells in immunodeficient rodents (e.g. nude and NSG mice) have been widely used to study the roles of human lincRNA in tumor development and metastasis [101,102]. Similarly, implantation of human hepatocytes and adipocytes derived from primary cells or IPSC (induced pluripotent stem cells) in immunodeficient mice has been used to study protein-coding genes in hepatic and adipose function in vivo [103,104]. Such models can be adapted to interrogate primate-specific lincRNAs in lipid metabolism homeostasis and cardiometabolic disorders. Indeed, our group is pursuing such strategies for the study of several non-conserved, human adipose and macrophage lincRNAs for which we have already demonstrated functions in in vitro models [30,105,106].
Although not an in vivo approach per se, it’s worth noting that recent technical advances in tissue engineering have established stem cell-based organoids as a powerful experimental method to study human physiology and diseases [107,108]. Such organoid models are in vitro 3D cell clusters derived from embryonic cells or IPSs. In the presences of artificial extracellular matrices and suitable induction exogenous factors, these pluripotent cells form organized cell clusters that exhibit similar functionalities as many in vivo tissues, such as liver, kidney, brain and intestine [107,108]. These organoid systems recapitulate important biological processes involved in tissue physiological functions, including cellular mechanisms, cell-cell interactions and cell-matrix interactions. Organoids are more physiologically relevant than in vitro 2D cell systems and more easily adaptable to biological or genetic manipulation than in vivo animal models [109]. As a near-physiological model system, organoids can serve a complementary approach to study physiological roles of non-conserved lincRNAs.
For clinical translation and study designs to study human lincRNAs, we face unavoidable challenges and uncertainties, including that (a) RNA sequencing (RNA-seq) of human material is required to identify novel non-coding RNA that are not conserved outside primates (i.e., most human lincRNAs), (b) lincRNAs have lower expression and greater tissue specificity than protein coding mRNAs thus, relative to mRNAs, they require more complete tissue profiling and deeper RNA-seq for comprehensive and reproducible annotation in humans, and (c) low expression and RNA degradation pose difficulties for detection and measurement of lincRNA levels in body fluids such as blood and plasma and thus for querying the potential roles of secreted lincRNAs in physiology and disease.
6. Concluding remark
LincRNAs have been increasingly identified as key mediators in many CVD-related cellular and biological processes. Recent advances in gene editing technologies, such as CRISPR/Cas, are greatly facilitating research on novel genes and will enhance efforts to reveal new knowledge of lincRNA in normal physiology and disease. While different animal models of lincRNA perturbation have provided solid evidence to support important physiological roles of lincRNAs in cardiovascular health, their complex biological functions and regulatory mechanisms as well as the marked primate-specificity (> 80%) of most human lincRNAs pose great challenges yet even greater opportunities to advance lincRNA research and to reveal novel mechanisms of cardiometabolic pathophysiology and disease. Determining the physiological impact of lincRNA using in vivo models is crucial to understanding basic lincRNA functions and translating lincRNAs into novel diagnostic and therapeutic strategies in CVD.
HIGHLIGHTS.
Long intergenic noncoding RNAs (lincRNA) play regulatory roles in cardiovascular health.
Physiological interrogation of lincRNAs is challenging.
Complementary approaches are often needed in the examination of lincRNA functions in cardiovascular diseases.
Acknowledgements
X.Z. tragically and suddenly passed away during the final revisions and writing of this manuscript. We dedicate this paper to her memory and to the gentle soul and talented scientist that our community has lost.
Financial support
This work was supported by grants from National Institutes of Health, R01-HL-132561 (M.P.R.) R01-HL-113147 (M.P.R.), K24-HL-107643 (M.P.R.), and the American Diabetes Association 1-16-PDF-137(X.Z). M.P.R. is also supported by R01-HL-111694 and UL1-TR-001873. (M.P.R.).
Footnotes
Conflicts of interest
The authors declared they do not have anything to disclose regarding conflict of interest with respect to this manuscript.
References
- [2].Cabili MN, Trapnell C, Goff L, et al. , Integrative annotation of human large intergenic noncoding RNAs reveals global properties and specific subclasses, Genes Dev. 25 (2011) 1915–1927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Derrien T, Johnson R, Bussotti G, et al. , The GENCODE v7 catalog of human long noncoding RNAs: analysis of their gene structure, evolution, and expression, Genome Res. 22 (2012) 1775–1789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Ulitsky I, Bartel DP, lincRNAs: genomics, evolution, and mechanisms, Cell 154 (2013) 26–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Huarte M, The emerging role of lncRNAs in cancer, Nat. Med 21 (2015) 1253–1261. [DOI] [PubMed] [Google Scholar]
- [6].Schmitt AM, Chang HY, Long noncoding RNAs in cancer pathways, Cancer Cell 29 (2016) 452–463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Lorenzen JM, Thum T, Long noncoding RNAs in kidney and cardiovascular diseases, Nat. Rev. Nephrol 12 (2016) 360–373. [DOI] [PubMed] [Google Scholar]
- [8].Uchida S, Dimmeler S, Long noncoding RNAs in cardiovascular diseases, Circ. Res 116 (2015) 737–750. [DOI] [PubMed] [Google Scholar]
- [9].Simion V, Haemmig S, Feinberg MW, LncRNAs in Vascular Biology and Disease, Vascul. Pharmacol, 2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Giroud M, Scheideler M, Long non-coding RNAs in metabolic organs and energy homeostasis, Int. J. Mol. Sci 18 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Leti F, DiStefano JK, Long Noncoding RNAs as Diagnostic and Therapeutic Targets in Type 2 Diabetes and Related Complications, Genes (Basel) vol. 8, (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Holdt LM, Beutner F, Scholz M, et al. , ANRIL expression is associated with atherosclerosis risk at chromosome 9p21, Arterioscler. Thromb. Vasc. Biol 30 (2010) 620–627. [DOI] [PubMed] [Google Scholar]
- [13].Pasmant E, Sabbagh A, Vidaud M, et al. , ANRIL, a long, noncoding RNA, is an unexpected major hotspot in GWAS, Faseb. J. : Off. Publ. Fed. Am. Soc. Exp. Biol 25 (2011) 444–448. [DOI] [PubMed] [Google Scholar]
- [14].Holdt LM, Hoffmann S, Sass K, et al. , Alu elements in ANRIL non-coding RNA at chromosome 9p21 modulate atherogenic cell functions through trans-regulation of gene networks, PLoS Genet. 9 (2013) e1003588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Holdt LM, Stahringer A, Sass K, et al. , Circular non-coding RNA ANRIL modulates ribosomal RNA maturation and atherosclerosis in humans, Nat. Commun 7 (2016) 12429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Ling H, Spizzo R, Atlasi Y, et al. , CCAT2, a novel noncoding RNA mapping to 8q24, underlies metastatic progression and chromosomal instability in colon cancer, Genome Res. 23 (2013) 1446–1461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Castellanos-Rubio A, Fernandez-Jimenez N, Kratchmarov R, et al. , A long non-coding RNA associated with susceptibility to celiac disease, Science 352 (2016) 91–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Congrains A, Kamide K, Oguro R, et al. , Genetic variants at the 9p21 locus contribute to atherosclerosis through modulation of ANRIL and CDKN2A/B, Atherosclerosis 220 (2012) 449–455. [DOI] [PubMed] [Google Scholar]
- [19].Congrains A, Kamide K, Katsuya T, et al. , CVD-associated non-coding RNA, ANRIL, modulates expression of atherogenic pathways in VSMC, Biochem. Biophys. Res. Commun 419 (2012) 612–616. [DOI] [PubMed] [Google Scholar]
- [20].Ishii N, Ozaki K, Sato H, et al. , Identification of a novel non-coding RNA, MIAT, that confers risk of myocardial infarction, J. Hum. Genet 51 (2006) 1087–1099. [DOI] [PubMed] [Google Scholar]
- [21].Gao W, Zhu M, Wang H, et al. , Association of polymorphisms in long non-coding RNA H19 with coronary artery disease risk in a Chinese population, Mutat. Res 772 (2015) 15–22. [DOI] [PubMed] [Google Scholar]
- [22].Liu Y, Ferguson JF, Xue C, et al. , Tissue-specific RNA-Seq in human evoked inflammation identifies blood and adipose LincRNA signatures of cardiometabolic diseases, Arterioscler. Thromb. Vasc. Biol 34 (2014) 902–912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Cao J, The functional role of long non-coding RNAs and epigenetics, Biol. Proced. Online 16 (2014) 11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Vance KW, Ponting CP, Transcriptional regulatory functions of nuclear long noncoding RNAs, Trends Genet. 30 (2014) 348–355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Rashid F, Shah A, Shan G, Long non-coding RNAs in the cytoplasm, Dev. Reprod. Biol 14 (2016) 73–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Bayoumi AS, Sayed A, Broskova Z, et al. , Crosstalk between long noncoding RNAs and MicroRNAs in health and disease, Int. J. Mol. Sci 17 (2016) 356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Sanuki R, Onishi A, Koike C, et al. , miR-124a is required for hippocampal axogenesis and retinal cone survival through Lhx2 suppression, Nat. Neurosci 14 (2011) 1125–1134. [DOI] [PubMed] [Google Scholar]
- [28].Engreitz JM, Haines JE, Perez EM, et al. , Local regulation of gene expression by lncRNA promoters, transcription and splicing, Nature 539 (2016) 452–455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Groff AF, Sanchez-Gomez DB, Soruco MML, et al. , In vivo characterization of linc-p21 reveals functional cis-regulatory DNA elements, Cell Rep. 16 (2016) 2178–2186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Zhang X, Xue C, Lin J, et al. , Interrogation of nonconserved human adipose lincRNAs identifies a regulatory role of linc-ADAL in adipocyte metabolism, Sci. Transl. Med 10 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Huarte M, Guttman M, Feldser D, et al. , A large intergenic noncoding RNA induced by p53 mediates global gene repression in the p53 response, Cell 142 (2010) 409–419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Yoon JH, Abdelmohsen K, Srikantan S, et al. , LincRNA-p21 suppresses target mRNA translation, Mol. Cell 47 (2012) 648–655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Dimitrova N, Zamudio JR, Jong RM, et al. , LincRNA-p21 activates p21 in cis to promote Polycomb target gene expression and to enforce the G1/S checkpoint, Mol. Cell 54 (2014) 777–790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Sunwoo H, Dinger ME, Wilusz JE, et al. , MEN epsilon/beta nuclear-retained non-coding RNAs are up-regulated upon muscle differentiation and are essential components of paraspeckles, Genome Res. 19 (2009) 347–359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Sasaki YT, Ideue T, Sano M, et al. , MENepsilon/beta noncoding RNAs are essential for structural integrity of nuclear paraspeckles, Proc. Natl. Acad. Sci. U. S. A 106 (2009) 2525–2530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Nakagawa S, Naganuma T, Shioi G, et al. , Paraspeckles are subpopulation-specific nuclear bodies that are not essential in mice, J. Cell Biol 193 (2011) 31–39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Hu W, Yuan B, Flygare J, et al. , Long noncoding RNA-mediated anti-apoptotic activity in murine erythroid terminal differentiation, Genes Dev. 25 (2011) 2573–2578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Atianand MK, Hu W, Satpathy AT, et al. , A long noncoding RNA lincRNA-EPS acts as a transcriptional brake to restrain inflammation, Cell 165 (2016) 1672–1685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Rinn JL, Kertesz M, Wang JK, et al. , Functional demarcation of active and silent chromatin domains in human HOX loci by noncoding RNAs, Cell 129 (2007) 1311–1323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Li L, Liu B, Wapinski OL, et al. , Targeted disruption of Hotair leads to homeotic transformation and gene derepression, Cell Rep. 5 (2013) 3–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Lai KM, Gong G, Atanasio A, et al. , Diverse phenotypes and specific transcription patterns in twenty mouse lines with ablated LincRNAs, PloS One 10 (2015) e0125522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Amandio AR, Necsulea A, Joye E, et al. , Hotair is dispensible for mouse development, PLoS Genet. 12 (2016) e1006232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Tian X, Xu G, Clinical value of lncRNA MALAT1 as a prognostic marker in human cancer: systematic review and meta-analysis, BMJ Open 5 (2015) e008653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Wei Y, Niu B, Role of MALAT1 as a Prognostic Factor for Survival in Various Cancers: a Systematic Review of the Literature with Meta-analysis vol. 2015, Dis. Markers, 2015, p. 164635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Lelli A, Nolan KA, Santambrogio S, et al. , Induction of long noncoding RNA MALAT1 in hypoxic mice, Hypoxia 3 (2015) 45–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].Puthanveetil P, Chen S, Feng B, et al. , Long non-coding RNA MALAT1 regulates hyperglycaemia induced inflammatory process in the endothelial cells, J. Cell Mol. Med 19 (2015) 1418–1425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].Biswas S, Thomas AA, Chen S, et al. , MALAT1: an epigenetic regulator of inflammation in diabetic retinopathy, Sci. Rep 8 (2018) 6526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [48].Tripathi V, Ellis JD, Shen Z, et al. , The nuclear-retained noncoding RNA MALAT1 regulates alternative splicing by modulating SR splicing factor phosphorylation, Mol. Cell 39 (2010) 925–938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [49].Tripathi V, Shen Z, Chakraborty A, et al. , Long noncoding RNA MALAT1 controls cell cycle progression by regulating the expression of oncogenic transcription factor B-MYB, PLoS Genet. 9 (2013) e1003368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [50].Yang L, Lin C, Liu W, et al. , ncRNA- and Pc2 methylation-dependent gene relocation between nuclear structures mediates gene activation programs, Cell 147 (2011) 773–788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [51].Michalik KM, You X, Manavski Y, et al. , Long noncoding RNA MALAT1 regulates endothelial cell function and vessel growth, Circ. Res 114 (2014) 1389–1397. [DOI] [PubMed] [Google Scholar]
- [52].Zhou X, Liu S, Cai G, et al. , Long non coding RNA MALAT1 promotes tumor growth and metastasis by inducing epithelial-mesenchymal transition in oral squamous cell carcinoma, Sci. Rep 5 (2015) 15972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [53].Gutschner T, Hammerle M, Eissmann M, et al. , The noncoding RNA MALAT1 is a critical regulator of the metastasis phenotype of lung cancer cells, Cancer Res. 73 (2013) 1180–1189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [54].Hu Y, Lin J, Fang H, et al. , Targeting the MALAT1/PARP1/LIG3 Complex Induces DNA Damage and Apoptosis in Multiple Myeloma, Leukemia, (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [55].Zhang X, Tang X, Liu K, et al. , Long noncoding RNA Malat1 regulates cerebrovascular pathologies in ischemic stroke, J. Neurosci 37 (2017) 1797–1806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [56].Lino Cardenas CL, Kessinger CW, Cheng Y, et al. , An HDAC9-MALAT1-BRG1 complex mediates smooth muscle dysfunction in thoracic aortic aneurysm, Nat. Commun 9 (2018) 1009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [57].Gabory A, Jammes H, Dandolo L, The H19 locus: role of an imprinted non-coding RNA in growth and development, Bioessays 32 (2010) 473–480. [DOI] [PubMed] [Google Scholar]
- [58].Juan V, Crain C, Wilson C, Evidence for evolutionarily conserved secondary structure in the H19 tumor suppressor RNA, Nucleic Acids Res. 28 (2000) 1221–1227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [59].Smits G, Mungall AJ, Griffiths-Jones S, et al. , Conservation of the H19 non-coding RNA and H19-IGF2 imprinting mechanism in therians, Nat. Genet 40 (2008) 971–976. [DOI] [PubMed] [Google Scholar]
- [60].Lustig O, Ariel I, Ilan J, et al. , Expression of the imprinted gene H19 in the human fetus, Mol. Reprod. Dev 38 (1994) 239–246. [DOI] [PubMed] [Google Scholar]
- [61].Han DK, Liau G, Identification and characterization of developmentally regulated genes in vascular smooth muscle cells, Circ. Res 71 (1992) 711–719. [DOI] [PubMed] [Google Scholar]
- [62].Matouk IJ, Halle D, Gilon M, et al. , The non-coding RNAs of the H19-IGF2 imprinted loci: a focus on biological roles and therapeutic potential in Lung Cancer, J. Transl. Med 13 (2015) 113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [63].Raveh E, Matouk IJ, Gilon M, et al. , The H19 Long non-coding RNA in cancer initiation, progression and metastasis - a proposed unifying theory, Mol. Canc 14 (2015) 184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [64].Cai X, Cullen BR, The imprinted H19 noncoding RNA is a primary microRNA precursor, RNA 13 (2007) 313–316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [65].Keniry A, Oxley D, Monnier P, et al. , The H19 lincRNA is a developmental reservoir of miR-675 that suppresses growth and Igf1r, Nat. Cell Biol 14 (2012) 659–665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [66].Giovarelli M, Bucci G, Ramos A, et al. , H19 long noncoding RNA controls the mRNA decay promoting function of KSRP, Proc. Natl. Acad. Sci. U. S. A 111 (2014) E5023–E5028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [67].Wang S, Wu X, Liu Y, et al. , Long noncoding RNA H19 inhibits the proliferation of fetal liver cells and the Wnt signaling pathway, FEBS Lett. 590 (2016) 559–570. [DOI] [PubMed] [Google Scholar]
- [68].Kallen AN, Zhou XB, Xu J, et al. , The imprinted H19 lncRNA antagonizes let-7 microRNAs, Mol. Cell 52 (2013) 101–112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [69].Imig J, Brunschweiger A, Brummer A, et al. , miR-CLIP capture of a miRNA targetome uncovers a lincRNA H19-miR-106a interaction, Nat. Chem. Biol 11 (2015) 107–114. [DOI] [PubMed] [Google Scholar]
- [70].Jia P, Cai H, Liu X, et al. , Long non-coding RNA H19 regulates glioma angiogenesis and the biological behavior of glioma-associated endothelial cells by inhibiting microRNA-29a, Cancer Lett. 381 (2016) 359–369. [DOI] [PubMed] [Google Scholar]
- [71].Tao SC, Rui BY, Wang QY, et al. , Extracellular vesicle-mimetic nanovesicles transport LncRNA-H19 as competing endogenous RNA for the treatment of diabetic wounds, Drug Deliv. 25 (2018) 241–255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [72].Li DY, Busch A, Jin H, et al. , H19 Induces Abdominal Aortic Aneurysm Development and Progression, Circulation, 2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [73].Zhang L, Cheng H, Yue Y, et al. , H19 knockdown suppresses proliferation and induces apoptosis by regulating miR-148b/WNT/beta-catenin in ox-LDL -stimulated vascular smooth muscle cells, J. Biomed. Sci 25 (2018) 11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [74].Chen S, Liang H, Yang H, et al. , LincRNa-p21: function and mechanism in cancer, Med. Oncol 34 (2017) 98. [DOI] [PubMed] [Google Scholar]
- [75].Wu G, Cai J, Han Y, et al. , LincRNA-p21 regulates neointima formation, vascular smooth muscle cell proliferation, apoptosis, and atherosclerosis by enhancing p53 activity, Circulation 130 (2014) 1452–1465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [76].Blackshaw S, Harpavat S, Trimarchi J, et al. , Genomic analysis of mouse retinal development, PLoS Biol. 2 (2004) E247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [77].Mercer TR, Qureshi IA, Gokhan S, et al. , Long noncoding RNAs in neuronalglial fate specification and oligodendrocyte lineage maturation, BMC Neurosci. 11 (2010) 14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [78].Shan K, Jiang Q, Wang XQ, et al. , Role of long non-coding RNA-RNCR3 in atherosclerosis-related vascular dysfunction, Cell Death Dis. 7 (2016) e2248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [79].Jin L, Lin X, Yang L, et al. , AK098656, a novel vascular smooth muscle cell-dominant long noncoding RNA, promotes hypertension, Hypertension 71 (2018) 262–272. [DOI] [PubMed] [Google Scholar]
- [80].Li P, Ruan X, Yang L, et al. , A liver-enriched long non-coding RNA, lncLSTR, regulates systemic lipid metabolism in mice, Cell Metabol. 21 (2015) 455–467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [81].Sallam T, Jones MC, Gilliland T, et al. , Feedback modulation of cholesterol metabolism by the lipid-responsive non-coding RNA LeXis, Nature 534 (2016) 124–128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [82].Tontonoz P, Wu X, Jones M, et al. , Long noncoding RNA facilitated gene therapy reduces atherosclerosis in a murine model of familial hypercholesterolemia, Circulation 136 (2017) 776–778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [83].Sallam T, Jones M, Thomas BJ, et al. , Transcriptional regulation of macrophage cholesterol efflux and atherogenesis by a long noncoding RNA, Nat. Med 24 (2018) 304–312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [84].Wang P, Xue Y, Han Y, et al. , The STAT3-binding long noncoding RNA lnc-DC controls human dendritic cell differentiation, Science 344 (2014) 310–313. [DOI] [PubMed] [Google Scholar]
- [85].Anderson KM, Anderson DM, McAnally JR, et al. , Transcription of the non-coding RNA upperhand controls Hand2 expression and heart development, Nature 539 (2016) 433–436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [86].Watts JK, Corey DR, Silencing disease genes in the laboratory and the clinic, J. Pathol 226 (2012) 365–379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [87].Geary RS, Norris D, Yu R, et al. , Pharmacokinetics, biodistribution and cell uptake of antisense oligonucleotides, Adv. Drug Deliv. Rev 87 (2015) 46–51. [DOI] [PubMed] [Google Scholar]
- [88].Gao K, Huang L, Nonviral methods for siRNA delivery, Mol. Pharm 6 (2009) 651–658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [89].Sauvageau M, Goff LA, Lodato S, et al. , Multiple knockout mouse models reveal lincRNAs are required for life and brain development, Elife 2 (2013) e01749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [90].Grote P, Wittler L, Hendrix D, et al. , The tissue-specific lncRNA Fendrr is an essential regulator of heart and body wall development in the mouse, Dev. Cell 24 (2013) 206–214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [91].Kutter C, Watt S, Stefflova K, et al. , Rapid turnover of long noncoding RNAs and the evolution of gene expression, PLoS Genet. 8 (2012) e1002841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [92].Necsulea A, Soumillon M, Warnefors M, et al. , The evolution of lncRNA repertoires and expression patterns in tetrapods, Nature 505 (2014) 635–640. [DOI] [PubMed] [Google Scholar]
- [93].Hezroni H, Koppstein D, Schwartz MG, et al. , Principles of long noncoding RNA evolution derived from direct comparison of transcriptomes in 17 species, Cell Rep. 11 (2015) 1110–1122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [94].Ulitsky I, Evolution to the rescue: using comparative genomics to understand long non-coding RNAs, Nat. Rev. Genet 17 (2016) 601–614. [DOI] [PubMed] [Google Scholar]
- [95].Dey BK, Mueller AC, Dutta A, Long non-coding RNAs as emerging regulators of differentiation, development, and disease, Transcription 5 (2014) e944014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [96].Durruthy-Durruthy J, Sebastiano V, Wossidlo M, et al. , The primate-specific noncoding RNA HPAT5 regulates pluripotency during human preimplantation development and nuclear reprogramming, Nat. Genet 48 (1) (2016. January) 44–52, 10.1038/ng.3449 Epub 2015 Nov 23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [97].Rigoutsos I, Lee SK, Nam SY, et al. , N-BLR, a primate-specific non-coding transcript leads to colorectal cancer invasion and migration, Genome Biol. 18 (2017) 98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [98].Xiao T, Liu L, Li H, et al. , Long noncoding RNA ADINR regulates adipogenesis by transcriptionally activating C/EBPalpha, Stem Cell Reports 5 (2015) 856–865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [99].Rani N, Nowakowski TJ, Zhou H, et al. , A primate lncRNA mediates notch signaling during neuronal development by sequestering miRNA, Neuron 90 (2016) 1174–1188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [100].Van Keuren ML, Gavrilina GB, Filipiak WE, et al. , Generating transgenic mice from bacterial artificial chromosomes: transgenesis efficiency, integration and expression outcomes, Transgenic Res. 18 (2009) 769–785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [101].Wang Y, Zeng X, Wang N, et al. , Long noncoding RNA DANCR, working as a competitive endogenous RNA, promotes ROCK1-mediated proliferation and metastasis via decoying of miR-335–5p and miR-1972 in osteosarcoma, Mol. Canc 17 (2018) 89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [102].Zhang Y, Pitchiaya S, Cieslik M, et al. , Analysis of the androgen receptor-regulated lncRNA landscape identifies a role for ARLNC1 in prostate cancer progression, Nat. Genet 50 (2018) 814–824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [103].Papazyan R, Liu X, Liu J, et al. , FXR activation by obeticholic acid or nonsteroidal agonists induces a human-like lipoprotein cholesterol change in mice with humanized chimeric liver, J. Lipid Res 59 (2018) 982–993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [104].Min SY, Kady J, Nam M, et al. , Human ‘brite/beige’ adipocytes develop from capillary networks, and their implantation improves metabolic homeostasis in mice, Nat. Med 22 (2016) 312–318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [105].Zhang H, Xue C, Wang Y, et al. , Deep RNA sequencing uncovers a repertoire of human macrophage long intergenic noncoding RNAs modulated by macrophage activation and associated with cardiometabolic diseases, Journal of the American Heart Association 6 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [106].Ballantyne RL, Zhang X, Nunez S, et al. , Genome-wide interrogation reveals hundreds of long intergenic noncoding RNAs that associate with cardiometabolic traits, Hum. Mol. Genet 25 (2016) 3125–3141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [107].Yin X, Mead BE, Safaee H, et al. , Engineering stem cell organoids, Cell stem cell 18 (2016) 25–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [108].Fatehullah A, Tan SH, Barker N, Organoids as an in vitro model of human development and disease, Nat. Cell Biol 18 (2016) 246–254. [DOI] [PubMed] [Google Scholar]
- [109].Sun Y, Ding Q, Genome engineering of stem cell organoids for disease modeling, Protein Cell 8 (2017) 315–327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [110].Dechamethakun S, Muramatsu M, Long noncoding RNA variations in cardiometabolic diseases, J. Hum. Genet 62 (2017) 97–104. [DOI] [PubMed] [Google Scholar]


