Highlights
-
•
Knowledge of the extent of COVID-19 epidemic and the level of herd immunity is urgently needed
-
•
We develop a Multiplex bead-based IgG detection tool for pathogenic human coronaviruses.
-
•
The novel assay is highly sensitive and specific.
-
•
Our novel assay is more sensitive than a commercial EIA.
-
•
We show that an immunoassay using a single antigen can lead to false positive results.
Keywords: COVID-19, SARS-CoV2, SARS, Luminex, Serology
Abstract
Background
Knowledge of the COVID-19 epidemic extent and the level of herd immunity is urgently needed to help manage this pandemic.
Methods
We used a panel of 167 samples (77 pre-epidemic and 90 COVID-19 seroconverters) and SARS-CoV1, SARS-CoV2 and MERS-CoV Spike and/or Nucleopcapsid (NC) proteins to develop a high throughput multiplex screening assay to detect IgG antibodies in human plasma. Assay performances were determined by ROC curves analysis. A subset of the COVID-19+ samples (n = 36) were also tested by a commercial NC-based ELISA test and the results compared with those of the novel assay.
Results
On samples collected ≥14 days after symptoms onset, the accuracy of the assay is 100 % (95 % CI: 100−100) for the Spike antigen and 99.9 % (95 % CI:99.7−100) for NC. By logistic regression, we estimated that 50 % of the patients have seroconverted at 5.7 ± 1.6; 5.7 ± 1.8 and 7.9 ± 1.0 days after symptoms onset against Spike, NC or both antigens, respectively and all have seroconverted two weeks after symptoms onset. IgG titration in a subset of samples showed that early phase samples present lower IgG titers than those from later phase. IgG to SARS-CoV2 NC cross-reacted at 100 % with SARS-CoV1 NC. Twenty-nine of the 36 (80.5 %) samples tested were positive by the commercial ELISA while 31/36 (86.1 %) were positive by the novel assay.
Conclusions
Our assay is highly sensitive and specific for the detection of IgG antibodies to SARS-CoV2 proteins, suitable for high throughput epidemiological surveys. The novel assay is more sensitive than a commercial ELISA.
1. Introduction
In December 31th, 2019, WHO was informed on cases of pneumonia with unknown etiology in Wuhan City, Hubei Province of China and, in January 30th, the new disease was declared a public health emergency of international concern. The virus causing this severe acute respiratory syndrome (SARS) was rapidly identified as a betacoronavirus named SARS-CoV2 [1]. This new coronavirus disease, now called COVID-19, has spread globally in six months, locking down the world, infecting millions of people and killing 0.4 million of them as of June, 7th 2020. To date, there is no effective specific treatment nor prophylactic vaccine. Most countries worldwide took restrictive measures including lockdown and social distancing to flatten the epidemic curve and limit virus transmission. The novel SARS-CoV2 coronavirus induces a large spectrum of disease from asymptomatic infections to severe pneumonia and death. Thus, while expecting rapid development of effective vaccines and treatments, it is urgently needed to know the extent of the epidemic, to estimate the level of persons who have been in contact with the virus and recovered from it and the level of herd immunity [2,3].
Studies from around the world [[4], [5], [6], [7], [8], [9], [10]] reported on immune responses to SARS-CoV2 in the early weeks of the infection using ELISA, plaque reduction neutralization tests (PRNT), chemiluminescence or a combination of these methods. The antigens most commonly used were the spike glycoprotein S1 with the receptor binding domain [8,11], the nucleocapsid protein or both [7,9]. Assays such as PRNT and neutralization are not suited for large scale high throughput surveys as it is currently needed for SARS-CoV2 serology because they are time- and bench work-demanding, especially if two antigens are used. There is thus a need for alternative methods for screening in the context of epidemiological surveys. In earlier works on other viral infections, we have developed highly sensitive and specific microspheres bead-based tests to detect antibodies in human and wildlife samples to identify antibodies to a wide diversity of HIV/SIV and Ebola viruses [12,13]. Here, we developed an assay using the same technology to simultaneously detect IgG antibodies to the highly pathogenic human coronaviruses, SARS-CoV1, SARS-CoV2 and MERS-CoV, using two viral antigens for each of the SARS viruses. The assay presented an accuracy of 100 % and 99.9 % to detect SARS-CoV2 spike and nucleocapsid, respectively.
2. Material and methods
2.1. Human plasma
We used a panel of 167 samples (Table 1 ) to validate our assay. Of these, 77 were COVID-19 negative and were collected in 2015 as described eralier [13]. The remaining 90 samples were from consenting COVID-19 patients hospitalized in Montpellier University hospitals and included in the “COVIDOtheque cohort” (ClinicalTrials.gov Identifier: NCT04347850). The cohort received an institutional ethics committee approval (CPP Ile de France III, n°2020-A00935−34). They were collected between March, 26th and April, 25th, 2020 from RT-qPCR confirmed COVID-19 cases as described earlier [14].
Table 1.
Characteristics of convalescent and negative control samples used in the study.
| Convalescents | |
|---|---|
| Sample collection date | 26/03/2020−25/04/2020 |
| Number included | 90 |
| Gender | |
| Male | 55 |
| Female | 22 |
| Unspecified | 13 |
| Age (years) median |
72 |
| Range | 33−99 |
| Days since symptoms onset (days) | |
| Median Range 3−47 |
19 |
| Negative controls | |
| Collection date | December 2015 |
| Number included | 77 |
2.2. Recombinant proteins
We used commercially available recombinant Nucleocapsid and/or Spike (S1) proteins derived from SARS-CoV1, SARS-CoV2 and MERS-CoV. The proteins were from Sinobiologicals and purchased as lyophilized powders from Interchim (Montluçon, France) and resuspended in a buffer and at concentration as per manufacturer’s instructions, aliquoted and stored until use.
2.3. Protein coupling to Luminex beads and multiplex screening for IgG antibodies to SARS-CoV1, SARS-CoV2 and MERS-CoV in plasma
We described in detail in our previous works the protocol for coupling proteins and peptides to Luminex microsphere beads [12,13]. In brief, recombinant spike proteins (1 μg/1.25 × 106 beads) and nucleocapsid (2 μg/1.25 × 106 beads) were covalently coupled on carboxyl functionalized fluorescent magnetic beads (Luminex Corp., Austin, TX) with the BioPlex amine coupling kit (Bio-Rad Laboratories, Marnes-la-Coquette, France) according to the manufacturer’s instructions. For each recombinant protein-coupled bead set, we used 2000 beads/μl of assay buffer. Preliminary experiments on different plasma dilutions (1/100−1/1000) showed that the dilution 1/200 gave the best signal to noise ratio. Diluted samples were incubated with coupled beads for 16 h at 4 °C. Reactions were revealed after incubation with a biotin-labeled anti-human IgG and streptavidin-R-phycoerythrin conjugate. Antigen-antibody reactions were read on BioPlex-200 equipment (Bio-Rad, Marnes-la-Coquette. France) and the results were expressed as median fluorescence intensity (MFI) per 100 beads. To determine IgG titers of a subset of samples against the different antigens tested, we performed a 2-fold serial dilution of these samples from 1/100 to 1/12,800 and tested them as described above. The titer was the highest value of reciprocal dilution factor given a signal above the cut-off.
2.4. Calculation of cut-off, sensitivity, specificity and accuracy
To calculate the cut-off, we used the (Mean+3xSD) formula by calculating the mean of MFI of the 77 COVID-19 negative samples for each of the recombinant proteins tested. We added to the value obtained three times the standard deviation. The result obtained was considered as the cut-off for each antigen. We also used receiver operating characteristics (ROC) curve analysis to determine the cut-off values for SARS-CoV2 antigens (because we only had convalescent samples from SARS-CoV2 patients), their sensitivity, specificity and accuracy. The ROC curve and other statistical analysis were performed with Graphpad Prism8 (San Diego, CA, USA).
2.5. Nucleocapsid based ELISA test
We used Abbott SARS-CoV-2 IgG for Alinity EIA as per manufacturer instructions to test a subset of 36 samples of the COVID-19+ samples.
3. Results
3.1. Sensitivity, specificity and accuracy to detect COVID-19 IgG in convalescent plasma
To evaluate the performance of our Luminex-based COVID-19 IgG antibody detection assay, we tested a panel of 167 samples (Table 1). The majority (71.4 %) of convalescent patients for whom the gender was specified were males and the median age was 72 years. The median duration between COVID-19 symptoms onset and sample collection was 19 days spanning from 3 to 47 days.
Mean signal intensities in the COVID-19 group were 7475 ± 3576 and 7692 ± 3864 for Spike and NC, respectively. In the negative control group, these values were 233 ± 197 and 95 ± 129 for the Spike and NC proteins, respectively.
We first calculated the cut-off values for positivity for both SARS-CoV2 Spike and Nucleocapsid recombinant proteins using the two methods described in the methods section above. For Spike recombinant protein, the cut-off values were 832 and 1030 MFI by Mean+3xSD and ROC curve analysis methods, respectively. For the nucleocapsid recombinant protein, these values were respectively 482 and 491 with Mean+3xSD and ROC curve analysis methods.
Because previous reports showed that a steady state of IgG response to a viral infection is reached at 2 weeks after exposure [5,[15], [16], [17]], we selected a subset of samples collected 14 days or more after onset of COVID-19 symptoms to determine the clinical performance of our assay for the detection of IgG antibodies to COVID-19. Results from that analysis (Table 2 ) showed that sensitivity of both recombinant proteins was 100 %. The specificity of the Spike protein was also 100 % while that of Nucleocapsid was 98.7 %. The overall accuracy of both antigens taken individually was 100 % for Spike and 99.9 % for Nucleocapsid.
Table 2.
Sensitivity, specificity and accuracy of the xMAP assay to detect IgG to SARS-CoV2 antigens in 138 samples; 77 negative control samples and 61 samples from COVID-19 patients ≥ days after onset of symptoms.
| Spike | Nucleocapsid (NC) | Spike + and NC+ | ||||
|---|---|---|---|---|---|---|
| ≥ Day14 (n = 138) | 95 % CI | ≥ Day14 (n = 138) | 95 % CI | ≥ Day14 (n = 138) | 95 % CI | |
| Sensitivity (%) | 100.0 | 92.7−100 | 100.0 | 92.7−100 | 100.0 | 94.−100 |
| Specificity (%) | 100.0 | 94.2−100 | 98.7 | 92.2−100 | 100.0 | 95.2−100 |
| Accuracy (%) | 100.0 | 100.0−100 | 99.9 | 99.7−100 | 100.0 | 97.3−100 |
| PPV (%) | 100.0 | 94.1−100 | 98.4 | 91.4−99.7 | 100.0 | 94.1−100 |
| NPV (%) | 100.0 | 95.2−100 | 100.0 | 95.2−100 | 100.0 | 95.2−100 |
We also combined the results of NC and Spike antigens to evaluate a sample status. By doing so, all the assay parameters (sensitivity, specificity, accuracy and the predictive values) were 100 % (Table 2).
We next stratified the capacity of our novel assay to detect IgG directed against the spike and nucleocapsid by time after symptoms onset. We defined 3 categories: samples collected less than a week, between one week and two weeks and two weeks or more after symptoms onset. Table 3 summarizes these data and show that 50 % of patients seroconverted during the first week for both antigens, 77.7 % and 83.3 % between 1 and 2 weeks after symptoms onset on nucleocapsid and Spike, respectively. Two weeks or more after symptoms onset, 100 % of patients had seroconverted against both antigens.
Table 3.
Sensitivity of the xMAP assay to detect IgG anti SARS-CoV2 antigens stratified by time since symptoms onset.
| Time since symptoms onset | N | IgG anti-NC Positive n positive (%) |
IgG anti-SP n positive (%) |
NC + SP+ n positieve (%) |
|---|---|---|---|---|
| ≤ Day 7 | 10 | 5 (50) | 5 (50) | 3 (30) |
| Day8-Day13 | 19 | 15 (78.9) | 16 (84.2) | 15 (78.9) |
| After Day13 | 61 | 61 (100) | 61 (100) | 61(100) |
| Total | 90 | 81 (90) | 82 (91.1) | 79 (87.7) |
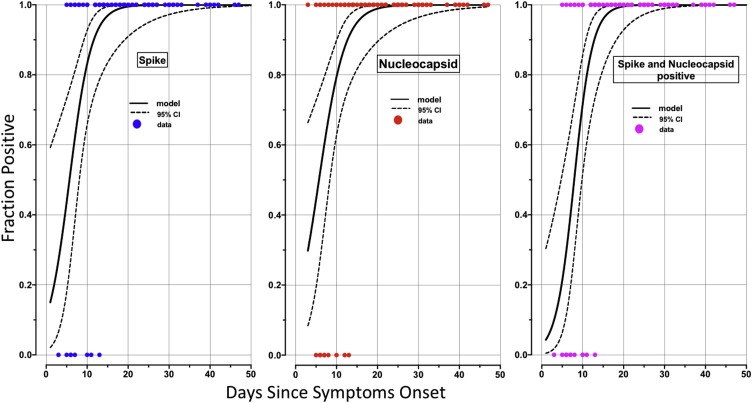
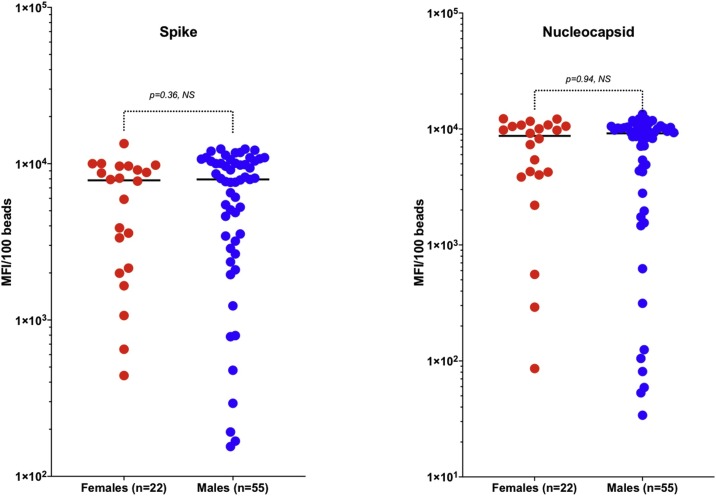
We used logistic regression to model IgG response to Spike, Nucleocapsid or to both antigens simultaneously as a function of time since symptoms onset. Results from this analysis (Fig. 1 A, B &C), showed that 50 % of the patients seroconverted at 5.9 ± 1.6; 5.7 ± 1.8 and 7.9 ± 1.0 days since symptoms onset against Spike, Nucleocapsid or both antigens, respectively. Virtually all the patients have seroconverted by 15–20 days after symptoms of COVID-19 were identified, generalizing and confirming the observation from raw data presented in Table 3. There were no significant differences in distribution of IgG response to Spike and nucleocapsid antigens between males and females (Fig. 2 ).
Fig. 1.
Timing of seroconversion during SARS-CoV2 infection.
Logistic regression was used to represent the dynamics of seroconversion in 90 COVID-19 seroconverters for Spike (left panel), Nucleocapsid (middle panel) or both (right panel). The figures show the fraction of IgG antibody positive samples as a function of time since symptoms onset. The bold curves represent the regression and dashed lines the 95 % confidence interval. All the patients have seroconverted two weeks after the onset of symptoms.
Fig. 2.
Comparison of IgG response to SARS-CoV2 antigens in male and female convalescent COVID 19 patients.
The figures compare IgG response to SARS-CoV2 antigens (Spike and Nucleocapsid) stratified by gender. There was no statistically significant difference between the two genders for both antigens. The groups were compared by the non-parametric Mann-Whitney U test
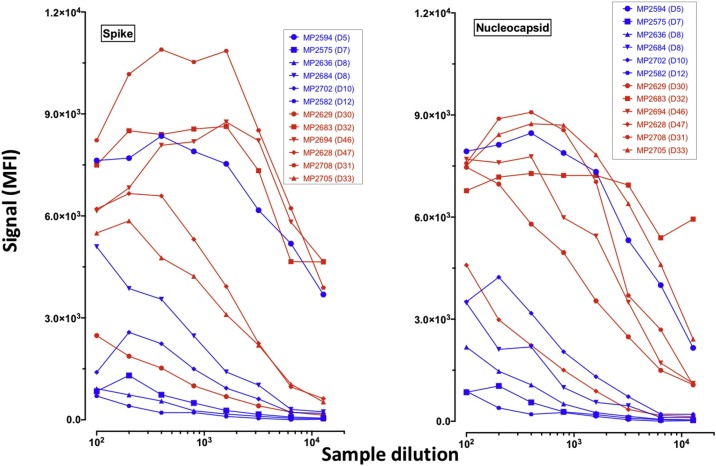
To determine IgG titers to COVID-19 antigens, we selected 6 samples from the early phase of COVID-19 symptoms (<14 days) and 6 others from later stages (>30 days) and tested serial dilutions of these samples until negativation. Results from these titration curves showed that (Table 4 and Fig. 3 ), overall and as expected, IgG titers of samples from later phase were higher than those from earlier phase. This observation stands for both the Spike and the Nucleocapsid proteins. At 4 weeks or later after symptoms onset, 3/6 and 5/6 of the tested samples presented IgG titers above 12,800 against Spike and Nucleocapsid, respectively. This proportion was only 1/6 for both antigens for samples collected before 2 weeks after symptoms onset.
Table 4.
End-point dilution titers of IgG antibodies to SARS-CoV2 Spike and Nucleocapsid recombinant proteins in a subset of early and later phase samples.
| Sample ID | Age | Gender | Time since symptoms onset | IgG titer Spike | IgG titer Nucleocapsid |
|---|---|---|---|---|---|
| MP2594 | 60 | F | 5 | >12,800 | >12,800 |
| MP2575 | 55 | M | 7 | 200 | 400 |
| MP2636 | 75 | F | 8 | <100 | 800 |
| MP2684 | 75 | F | 8 | 3200 | 3200 |
| MP2702 | 74 | F | 10 | 800 | 6400 |
| MP2582 | NA | NA | 12 | <100 | 100 |
| MP2629 | NA | F | 30 | 800 | >12,800 |
| MP2708 | 74 | M | 31 | >12,800 | >12,800 |
| MP2683 | 71 | F | 32 | >12,800 | >12,800 |
| MP2705 | 73 | M | 33 | 3200 | 1600 |
| MP2694 | 75 | F | 46 | >12,800 | >12,800 |
| MP2628 | 68 | M | 47 | 6400 | >12,800 |
Fig. 3.
Titration of IgG antibodies to SARS-CoV2 Spike and Nucleocapsid recombinant proteins.
To determine the titers of IgG antibodies to SARS-CoV2 Spike and Nucleocapsid in a subset of the samples of our panel, we performed 2-fold serial dilutions on six samples from the early phase of symptoms onset and six from later phase (> 30 days). The graphs show the changes of IgG binding intensities to the Spike (left panel) and the Nucleocapsid proteins (right panel) at the different dilutions. Curves in blue are early phase samples and those in red, from later phase.
3.2. Cross-reactions of COVID-19 convalescent samples with SARS-CoV1 and MERS-CoV antigens
Although very diverse, some coronavirus proteins are conserved through the different clades while others, like Spike proteins, are quite species-specific [18]. To estimate the level of antibody cross-reactions induced by SARS-CoV2 in convalescent plasma, we also tested our positive control panel samples on the other highly pathogenic human coronaviruses, namely SARS-CoV1 and MERS-CoV recombinant proteins. Data from Table 5 summarize the results of this comparison. Of the 61 samples of presumably fully seroconverted COVID-19+ patients (i.e. two weeks after symptom onset) tested on the five antigens, 100 % cross-reacted with SARS-CoV1 Nucleocapsid protein and 45.9 % also cross-reacted with SARS-CoV1 Spike protein. Notably, only 2 (3.3 %) of the 61 cross-reacted with MERS-CoV Nucleocapsid. These data are perfectly in line with the phylogenetic proximity of these viruses [19].
Table 5.
Cross-reactions of 61 SARS-CoV2 convalescent samples (> 2weeks after onset of symptoms) with SARS-CoV1 and MERS-CoV antigens.
| N positive/N tested | % | |
|---|---|---|
| SARS-CoV1-NC+ | 61 | 100 |
| SARS-CoV1-SP+ | 28 | 45.9 |
| MERS-CoV-NC+ | 2 | 3.3 |
3.3. Comparison with a commercial EIA assay
To evaluate the performance of our novel assay with a commercially available EIA assay, we tested a subset of 36 samples, collected between 1 and 30 days after symptom onset, from the COVID-19+ panel. The EIA assay, United States FDA approved for emergency access, uses SARS-CoV nucleocapsid as antigen. The commercial EIA identified 29 samples positive of 36 tested (80.5 %) while our novel assay detected 31/36 (86.1 %) tested on the same NC antigen. And additional sample, negative by the commercial and was reactive on the Spike antigen. This sample was collected from a patient at day1 post symptoms onset.
4. Discussion
In most countries of the world affected by the Covid-19 pandemic, the coverage of viral detection by molecular means has been low and thus, the actual epidemic spread of the SARS-CoV2 is unknown. One possibility to fill this gap is to perform serological diagnosis and surveys. This is especially important for patients with mild to moderate illness and who do not refer to medical care, or refer later, after 2 weeks, when the probability of virus detection is low. Serological diagnosis is also an important tool to understand the extent of COVID-19 in the community and to define the level of herd immunity.
We chose the Spike and Nucleocapsid recombinant proteins as antigens because they have been shown to be highly immunogenic during coronavirus infections in humans or non-human primates [20]. Our data showed that for both antigens, the sensitivity was 100 % (Table 2). However, while the specificity of Spike antigen was also 100 %, that of the Nucleocapsid antigen was slightly lower (98.7 %) because one sample from the pre-epidemic panel reacted weakly above the cut-off threshold with that antigen. This could reflect a non-specific binding or a cross-reaction with one of the mild coronaviruses circulating in France in 2015. Overall, the accuracy of both antigens was above 99 % (Table 2). Because a fully established IgG response in a natural infection normally covers all immunogenic antigens, we also analyzed the performance of our assay by combining the two antigens we tested. As expected, this resulted in a highly sensitive and specific assay with 100 % performance for all the parameters evaluated. French as well as international health authorities recommend that serological diagnostic assays should present a clinical specificity of at least 98 % and a clinical sensitivity of 90 % or more [21]. Our assay largely fulfills these criteria.
When we stratified the samples by time since symptoms onset, we observed that 100 % of patients have seroconverted after two weeks (Table 3). A recent work reported that 100 % of patients (n = 125) tested for COVID-19 were IgG positive by day 17 after symptoms onset [6]. Two comprehensive reviews on different aspects of the human immune responses to coronavirus infections, including SARS-CoV2, showed that in most patients, IgG-seroconversion occurs from the second week since symptom onset onwards, with the kinetics and breath depending on the severity or not of the disease [20,22]. However, it is too early to know if antibody response induced by SARS-CoV2 will persist over time and for how long and if they will be protective upon re-exposure to the same or a related virus. For other human coronavirus, including SARS-CoV1 and MERS-CoV, IgG antibodies have been detected up to 2–3 years after infection [23,24].
Another major concern in the antibody response to SARS-CoV2 is the nature, breath and titers of IgGs. Here, we found that in 8/12 samples collected ≥ 2 weeks after symptoms onset, IgG titers above 12,800 were observed. It is not known if these IgGs are neutralizing or not. Ju and colleagues [25] for instance isolated potentially neutralizing monoclonal antibodies with high titers from memory B-cells of SARS-CoV2 seroconverters.
One of the multiple advantages of the Luminex technology is the possibility of multiplexing. As previously reported [25], we observed here high level cross-reactivity between the Nucleocapsid proteins of SARS-CoV1 and SARS-CoV2. Hence, EIA using this antigen for COVID-19 serodetection in areas where SARS-CoV1 circulated might lead to false positive results. This should be especially taken into consideration when performing epidemiological surveys. To limit this peculiarity and significantly increase the specificity of our assay, we considered a sample as positive if it was simultaneously reactive on Nucleocapsid and Spike protein. We successfully applied such an algorithm for the serology of Ebolavirus in human and wildlife samples [13,[26], [27], [28], [29]]. One limitation of such a strategy is the difference in the kinetics of antibody response to these different antigens. It is very likely that surface and internal proteins will induce different kinetics of IgG responses. Hence, an algorithm combining two or more different antigens is most pertinent in the steady phase of the antibody response.
Finally, we compare our novel assay with a commercially available EIA assay. On the same viral antigen, our assay was more sensitive than the reference assay. The observation of Luminex assay being more sensitive than other EIA has already been reported by our group and others for different pathogens [13].
In summary, we have developed a highly sensitive and specific multi-target serological tool for the detection of IgG antibodies to SARS-CoV1, SARS-CoV2 and MERS-CoV infections. The assay is at least as sensitive as a commercial EIA and is fully suited for high throughput sero-epidemiological surveys.
CRediT authorship contribution statement
Ahidjo Ayouba: Conceptualization, Data curation, Formal analysis, Funding acquisition, Writing - original draft, Writing - review & editing. Guillaume Thaurignac: Data curation, Formal analysis, Writing - review & editing. David Morquin: Resources, Writing - review & editing. Edouard Tuaillon: Resources, Writing - original draft, Writing - review & editing. Raisa Raulino: Data curation, Writing - review & editing. Antoine Nkuba: Data curation, Writing - review & editing. Audrey Lacroix: Data curation, Writing - review & editing. Nicole Vidal: Data curation, Writing - review & editing. Vincent Foulongne: Resources, Writing - review & editing. Vincent Le Moing: Resources, Writing - review & editing. Jacques Reynes: Resources, Writing - review & editing. Eric Delaporte: Conceptualization, Resources, Funding acquisition, Writing - review & editing. Martine Peeters: Conceptualization, Formal analysis, Funding acquisition, Writing - original draft, Writing - review & editing.
Declaration of Competing Interest
All the authors declared no conflict of interest.
Acknowledgements
This work was supported by Institut de Recherche pour le Développement (IRD), the French Agence Nationale de la Recherche (ANR; ZOOCOV grant) and Montpellier University of Excellence (MUSE) emergency response funding (PANCOV-S grant). Raisa Raulino was supported by PhD grant from INSERM and University of Montpellier and Antoine Nkuba by a ARTS PhD grant from IRD. The funders had no role in study design, data collection and interpretation, or the decision to submit the work for publication.
References
- 1.Chen L., Liu W., Zhang Q., Xu K., Ye G., Wu W. RNA based mNGS approach identifies a novel human coronavirus from two individual pneumonia cases in 2019 Wuhan outbreak. Emerg. Microbes Infect. 2020;(9):313–319. doi: 10.1080/22221751.2020.1725399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Goudsmit J. The paramount importance of serological surveys of SARS-CoV-2 infection and immunity. Eur. J. Epidemiol. 2020;35:331–333. doi: 10.1007/s10654-020-00635-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Krammer F., Simon V. Serology assays to manage COVID-19. Science. 2020;368:1060. doi: 10.1126/science.abc1227. [DOI] [PubMed] [Google Scholar]
- 4.Fontanet A., Tondeur L., Madec Y., Grant R., Besombes C., Jolly N. Cluster of COVID-19 in northern France: a retrospective closed cohort study. medRxiv. 2020 2020. 04.18.20071134. [Google Scholar]
- 5.Wölfel R., Corman V.M., Guggemos W., Seilmaier M., Zange S., Müller M.A. Virological assessment of hospitalized patients with COVID-2019. Nature. 2020;581:465–469. doi: 10.1038/s41586-020-2196-x. [DOI] [PubMed] [Google Scholar]
- 6.Bryan A., Pepper G., Wener M.H., Fink S.L., Morishima C., Chaudhary A. Performance characteristics of the abbott architect SARS-CoV-2 IgG assay and seroprevalence in Boise. Idaho. J Clin Microbiol. 2020 doi: 10.1128/JCM.00941-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ni L., Ye F., Cheng M.L., Feng Y., Deng Y.Q., Zhao H. Detection of SARS-CoV-2-Specific humoral and cellular immunity in COVID-19 convalescent individuals. Immunity. 2020 doi: 10.1016/j.immuni.2020.04.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Perera R.A., Mok C.K., Tsang O.T., Lv H., Ko R.L., Wu N.C. Serological assays for severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), March 2020. Euro Surveill. 2020;25 doi: 10.2807/1560-7917.ES.2020.25.16.2000421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Qu J., Wu C., Li X., Zhang G., Jiang Z., Li X. Profile of IgG and IgM antibodies against severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) Clin. Infect. Dis. 2020 doi: 10.1093/cid/ciaa489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Team C-I Clinical and virologic characteristics of the first 12 patients with coronavirus disease 2019 (COVID-19) in the United States. Nat. Med. 2020 doi: 10.1038/s41591-020-0877-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Stadlbauer D., Amanat F., Chromikova V., Jiang K., Strohmeier S., Arunkumar G.A. SARS-CoV-2 seroconversion in humans: a detailed protocol for a serological assay, antigen production, and test setup. Curr. Protoc. Microbiol. 2020;57:e100. doi: 10.1002/cpmc.100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ahuka-Mundeke S., Ayouba A., Mbala-Kingebeni P., Liegeois F., Esteban A., Lunguya-Metila O. Novel multiplexed HIV/simian immunodeficiency virus antibody detection assay. Emerg Infect Dis. 2011;17:2277–2286. doi: 10.3201/eid1712.110783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ayouba A., Touré A., Butel C., Keita A.K., Binetruy F., Sow M.S. Development of a sensitive and specific serological assay based on luminex technology for detection of antibodies to Zaire ebola virus. J. Clin. Microbiol. 2017;55:165–176. doi: 10.1128/JCM.01979-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lescure F.X., Bouadma L., Nguyen D., Parisey M., Wicky P.H., Behillil S. Clinical and virological data of the first cases of COVID-19 in Europe: a case series. Lancet Infect. Dis. 2020;20:697–706. doi: 10.1016/S1473-3099(20)30200-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.To K.K., Tsang O.T., Leung W.S., Tam A.R., Wu T.C., Lung D.C. Temporal profiles of viral load in posterior oropharyngeal saliva samples and serum antibody responses during infection by SARS-CoV-2: an observational cohort study. Lancet Infect. Dis. 2020;20:565–574. doi: 10.1016/S1473-3099(20)30196-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Guo L., Ren L., Yang S., Xiao M., Chang D., Yang F. Profiling early humoral response to diagnose novel coronavirus disease (COVID-19) Clin. Infect. Dis. 2020 doi: 10.1093/cid/ciaa310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tuaillon E., Bollore K., Pisoni A., Debiesse S., Renault C., Marie S. Detection of SARS-CoV-2 antibodies using commercial assays and seroconversion patterns in hospitalized patients. medRxiv. 2020 doi: 10.1016/j.jinf.2020.05.077. 2020.05.04.20090027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Agnihothram S., Gopal R., Yount B.L., Jr., Donaldson E.F., Menachery V.D., Graham R.L. Evaluation of serologic and antigenic relationships between middle eastern respiratory syndrome coronavirus and other coronaviruses to develop vaccine platforms for the rapid response to emerging coronaviruses. J. Infect. Dis. 2014;209:995–1006. doi: 10.1093/infdis/jit609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Joffrin L., Goodman S.M., Wilkinson D.A., Ramasindrazana B., Lagadec E., Gomard Y. Bat coronavirus phylogeography in the Western Indian Ocean. Sci. Rep. 2020;10:6873. doi: 10.1038/s41598-020-63799-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Vabret N., Britton G.J., Gruber C., Hegde S., Kim J., Kuksin M., et al. Immunology of COVID-19: current state of the science. Immunity. [DOI] [PMC free article] [PubMed]
- 21.2020. Haute Autorité De Santé HAS. cahier_des_charges_test_serologique_covid19’.https://www.hassante.fr/upload/docs/application/pdf/202004/cahier_des_charges_test_serologique_covid19.pdf [Google Scholar]
- 22.Huang A.T., Garcia-Carreras B., Hitchings M.D.T., Yang B., Katzelnick L., Rattigan S.M. A systematic review of antibody mediated immunity to coronaviruses: antibody kinetics, correlates of protection, and association of antibody responses with severity of disease. medRxiv. 2020 doi: 10.1038/s41467-020-18450-4. 2020.04.14.20065771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Cao W.C., Liu W., Zhang P.H., Zhang F., Richardus J.H. Disappearance of antibodies to SARS-associated coronavirus after recovery. N. Engl. J. Med. 2007;357:1162–1163. doi: 10.1056/NEJMc070348. [DOI] [PubMed] [Google Scholar]
- 24.Liu W., Fontanet A., Zhang P.H., Zhan L., Xin Z.T., Baril L. Two-year prospective study of the humoral immune response of patients with severe acute respiratory syndrome. J. Infect. Dis. 2006;193:792–795. doi: 10.1086/500469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ju B., Zhang Q., Ge X., Wang R., Yu J., Shan S. Potent human neutralizing antibodies elicited by SARS-CoV-2 infection. bioRxiv. 2020 doi: 10.1038/s41586-020-2380-z. 2020.03.21.990770. [DOI] [PubMed] [Google Scholar]
- 26.Ayouba A., Ahuka-Mundeke S., Butel C., Mbala Kingebeni P., Loul S., Tagg N. Extensive serological survey of multiple african nonhuman primate species reveals low prevalence of immunoglobulin g antibodies to 4 ebola virus species. J. Infect. Dis. 2019;220:1599–1608. doi: 10.1093/infdis/jiz006. [DOI] [PubMed] [Google Scholar]
- 27.De Nys H.M., Kingebeni P.M., Keita A.K., Butel C., Thaurignac G., Villabona-Arenas C.J. Survey of ebola viruses in Frugivorous and insectivorous bats in Guinea, Cameroon, and the Democratic Republic of the Congo, 2015-2017. Emerg Infect Dis. 2018;24:2228–2240. doi: 10.3201/eid2412.180740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Diallo M.S.K., Rabilloud M., Ayouba A., Touré A., Thaurignac G., Keita A.K. Prevalence of infection among asymptomatic and paucisymptomatic contact persons exposed to Ebola virus in Guinea: a retrospective, cross-sectional observational study. Lancet Infect. Dis. 2019;19:308–316. doi: 10.1016/S1473-3099(18)30649-2. [DOI] [PubMed] [Google Scholar]
- 29.Keita A.K., Butel C., Thaurignac G., Diallo A., Nioke T., Traoré F. Serological evidence of ebola virus infection in Rural Guinea before the 2014 west african epidemic outbreak. Am. J. Trop. Med. Hyg. 2018;(99):425–427. doi: 10.4269/ajtmh.18-0105. [DOI] [PMC free article] [PubMed] [Google Scholar]