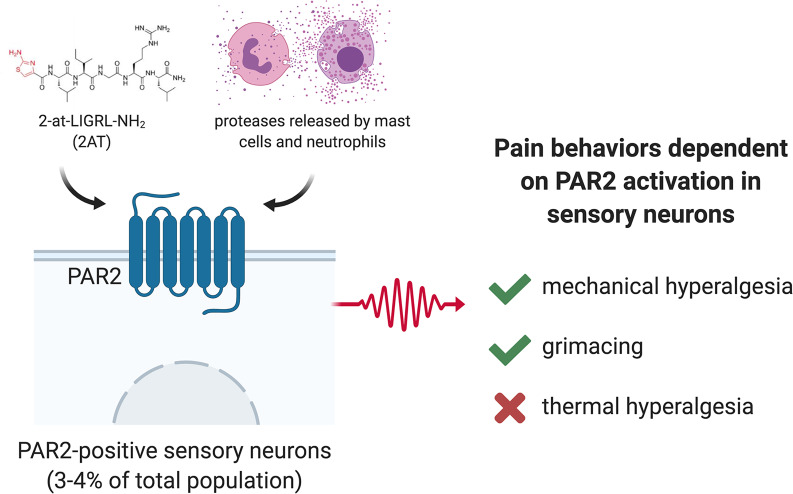
Abstract
Protease-activated receptor 2 (PAR2) has long been implicated in inflammatory and visceral pain, but the cellular basis of PAR2-evoked pain has not been delineated. Although PAR2-evoked pain has been attributed to sensory neuron expression, RNA-sequencing experiments show ambiguous F2rl1 mRNA detection. Moreover, many pharmacological tools for PAR2 are nonspecific, acting also on the Mas-related GPCR family (Mrg) that are highly enriched in sensory neurons. We sought to clarify the cellular basis of PAR2-evoked pain. We developed a PAR2–conditional knockout mouse and specifically deleted PAR2 in all sensory neurons using the PirtCre mouse line. Our behavioral findings show that PAR2 agonist–evoked mechanical hyperalgesia and facial grimacing, but not thermal hyperalgesia, are dependent on PAR2 expression in sensory neurons that project to the hind paw in male and female mice. F2rl1 mRNA is expressed in a discrete population (~4%) of mostly small-diameter sensory neurons that coexpress the Nppb and IL31ra genes. This cell population has been implicated in itch, but our work shows that PAR2 activation in these cells causes clear pain-related behaviors from the skin. Our findings show that a discrete population of DRG sensory neurons mediate PAR2-evoked pain.
Keywords: Neuroscience
Keywords: Anesthesiology, Pain, Proteases
A discreet population of DRG sensory neurons mediate PAR2-evoked pain.
Introduction
Protease-activated receptors (PARs) are G protein–coupled receptors (GPCRs) that are targeted by endogenous proteases. These proteases cleave the extracellular N-terminus of the receptor to reveal a tethered peptide ligand that then induces cellular signaling (1). These receptors can signal at the cell membrane, but they also continue to generate signaling after internalization via endosomal signaling pathways (2). PARs have been implicated in many pathological states (1, 3–8). Research on PAR2 has focused on pain and inflammation due to the long-standing observation of decreased pain sensitization in F2rl1–/– mice (9). Stemming from this original finding, many subsequent studies have focused on how PAR2 signaling occurs in nociceptors, but much of this body of evidence was built using a tool peptide PAR2 agonist, SLIGRL, which is now known to also be an agonist of the Mas-related GPCR, MrgprC11 (encoded by the Mrgprx1 gene) and may also act on other Mrg receptors (10, 11). Given that the Mrg family of receptors is highly enriched in dorsal root ganglion (DRG) neurons (12, 13), this complicates interpretation of some of the pharmacological literature on the topic. Another emerging issue in the field is that coagulation factor II (thrombin) receptor-like 1 (F2rl1) gene expression in many bulk and single-cell DRG-sequencing data sets is either undetectable or on the threshold of detection limits (14, 15). This includes single-cell experiments from visceral afferents (16). These findings are surprising given that PAR2 is widely considered an important therapeutic target for visceral pain with the model that PAR2 in visceral afferents is activated by endogenous proteases released during visceral inflammation (2, 17–20).
These disparate and somewhat controversial findings raise important questions about our understanding of PAR2 in the biology of pain. We sought to elucidate the cellular basis of PAR2-evoked pain by generating a conditional knockout mouse for the F2rl1 gene. Importantly, another group independently generated a similar mouse and crossed it with the Scn10aCre mouse to generate a nociceptor-specific knockout of PAR2. They found decreased mechanical hypersensitivity in response to proteases that are thought to act on PAR2, consistent with the hypothesis that PAR2-evoked pain is mediated specifically by nociceptors (2). This study also substantially advanced the field of PAR2 biology by demonstrating that PAR2 continues to signal once it is internalized via endosomal signaling. However, this study did not address different pain modalities or delineate precisely which populations of nociceptors express the F2rl1 mRNA.
We targeted exon 2 of F2rl1 to create a sensory neuron–specific conditional PAR2-knockout mouse using the PirtCre line (21). Our findings demonstrate that PAR2-evoked mechanical hypersensitivity and affective pain are lost in these mice while thermal hyperalgesia is lost in response to exogenous agonists but intact for endogenous protease-induced activation of PAR2. RNAscope in situ hybridization and cellular signaling assays on cultured mouse DRG neurons show that PAR2 is expressed by a small population of nociceptors that express several markers that identify itch nociceptors. Interestingly, we find that PAR2 activation leads only to pain, and not itch responses, demonstrating that this subpopulation of nociceptors signals pain with an appropriate stimulus.
Results
Evaluation of PAR2 expression in DRG sensory neurons.
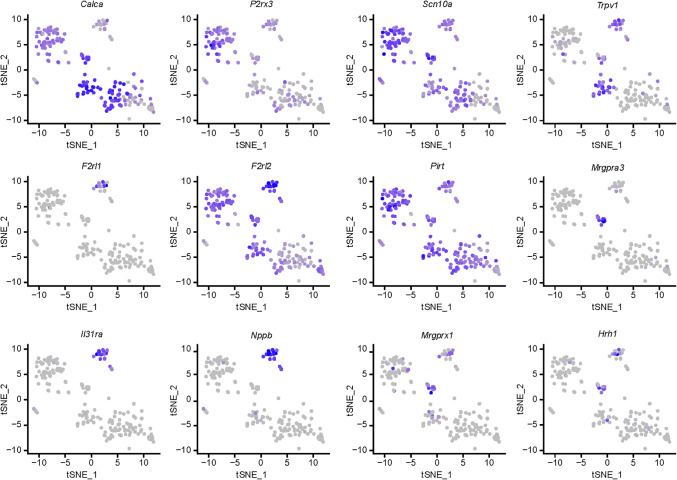
Recent RNA-sequencing studies find very low expression levels for F2rl1 mRNA in DRG neurons (14–16), a surprising finding given the large literature on PAR2 signaling in DRG neurons (1). We reevaluated F2rl1 mRNA expression in a deeply sequenced mouse single-cell RNA-sequencing data set (22). We found that F2rl1 mRNA was detected but only in a small subset of cells (Figure 1). Single-cell expression for F2rl1 was identified in a subpopulation of neurons with gene markers Il31ra and Nppb that coexpress Hrh1 and Mrgprx1, all genes thought to mark a set of sensory neurons that are important for itch sensation (13, 23, 24). These neurons also coexpressed Trpv1.
Figure 1. Delineation of F2rl1-expressing DRG neurons from single-cell RNA-sequencing experiments.
t-Distributed stochastic neighbor embedding (t-SNE) plots show gene expression clustering from 204 single-cell RNA-sequencing profiles of mouse DRG neurons. Gene names are indicated above each t-SNE plot, and color saturation represents normalized gene expression level. Single-cell RNA-sequencing of mouse DRG neurons demonstrates that F2rl1 mRNA is expressed in a discrete population of sensory neurons that also express gene markers implicated in itch, such as Nppb and Il31ra. This population of sensory neurons also expresses Trpv1.
We next evaluated F2rl1 mRNA expression in mouse DRG using RNAscope. We used probes for mRNAs encoding PAR2, calcitonin gene-related peptide (CGRP), and P2X3 receptor (P2X3R) in triple-labeling experiments (Figure 2A). PAR2 expression was found in a small subset (3.4%) of neurons in F2rl1floxPirt+/+ mice. We generated a conditional knockout of PAR2, F2rl1floxPirtCre, and tested for knockout of PAR2 mRNA expression in DRG neurons of F2rl1floxPirt+/+ and F2rl1floxPirtCre mice. We detected PAR2 mRNA expression in F2rl1floxPirt+/+ mice but not conditional knockout mice (Figure 2B). In F2rl1floxPirt+/+ mice, PAR2 expression was rare in cells expressing mRNA encoding CGRP, but most PAR2 mRNA–expressing cells also coexpressed P2X3R (Figure 2, C–E). PAR2 mRNA–expressing cells were almost entirely small diameter (Figure 2D). As an additional control for the specificity of our conditional knockout approach, we did RNAscope experiments on skin sections from F2rl1floxPirt+/+ and F2rl1floxPirtCre mice. We observed PAR2 mRNA expression in populations of skin cells in both genotypes, demonstrating that PAR2 was not knocked out in skin cells using the PirtCre approach (Figure 2F).
Figure 2. F2rl1 is expressed by a small subset of sensory neurons.
DRG neurons (A–E) and hind paw skin (F) from F2rl1floxPirt+/+ and F2rl1floxPirtCre mice were dissected and prepared for RNAscope in situ hybridization. White arrows indicate cells positive for F2rl1 mRNA. (A) Representative original magnification ×20 images of Calca (green), P2rx3 (red), and F2rl1 (white) mRNA signal in the DRG of F2rl1floxPirt+/+ and F2rl1floxPirtCre mice. These images show that the F2rl1floxPirtCre mice do not express F2rl1 mRNA in sensory neurons while the F2rl1floxPirt+/+ mice do. Scale bar: 50 μm. (B) Original magnification ×40 overlay image showing RNAscope signal at the cellular level. Scale bar: 10 μm. (C) Percentage of Calca+ and P2rx3+ neurons that coexpress F2rl1. Around 2% of Calca+ neurons express F2rl1 mRNA while around 6% of P2rx3+ neurons express F2rl1 mRNA. (D) Histogram illustrating the diameter of neurons expressing F2rl1. F2rl1+ neurons are small- to medium-diameter neurons (16–46 μm). (E) Pie chart illustration of the percentage of F2rl1+ cells that colocalize with Calca+ and P2rx3+ neurons. About 3%–4% of DRG neurons are F2rl1+, of which almost all are P2rx3+ neurons. (F) Representative hind paw skin images of F2rl1 (white) and DAPI (blue) signal from a F2rl1floxPirt+/+ mouse and a F2rl1floxPirtCre mouse. The last image shows hind paw skin from a F2rl1floxPirt+/+ mouse stained with a negative probe control (bacterial dapB). These images show the specificity of conditional knockout of F2rl1 expression is restricted to only sensory neurons and not skin cells. Scale bar: 20 μm.
PAR2 agonist–induced signaling occurs exclusively in PAR2-expressing DRG neurons.
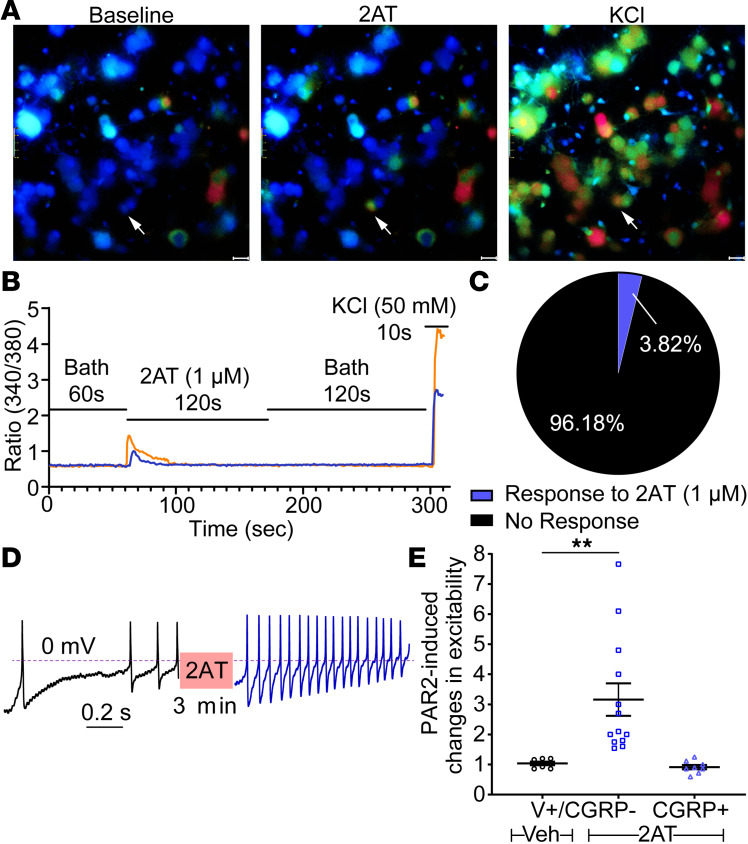
Having established that PAR2 mRNA expression is restricted to a small proportion of DRG neurons, we tested whether this restricted expression pattern would be found in functional assays. We began with Ca2+ imaging of DRG neurons in culture. In previous studies, we observed increased [Ca2+]i in primary trigeminal ganglion neurons after treatment with the specific PAR2 agonist, 2-at-LIGRL-NH2 (2AT, 1 μM) (25–27). We found that 2AT induced Ca2+ signaling in approximately 4% of DRG neurons (Figure 3, A–C). We also assessed whether PAR2-mediated plasticity could be observed in DRG neurons using patch clamp electrophysiology. To do this, we focused on Trpv1-expressing neurons using a genetically tagged line because our analysis of RNA-sequencing data revealed that PAR2 overlaps with a subset of Trpv1-expressing cells. As predicted, we found that ramp-evoked spiking was augmented by 2AT (1 μM, 3 minutes) treatment but only in CGRP–TRPV1+ cells (Figure 3, D and E).
Figure 3. 2AT-evoked Ca2+ signaling is specific for PAR2-expressing neurons.
Primary mouse DRG cultures were prepared for Ca2+ imaging (A–C) or whole-cell current clamp recordings (D and E). (A) Representative original magnification ×40 images of cultured DRG neurons loaded with Fura 2 at baseline and upon treatment with 2AT (1 μM) and KCl (50 mM), a positive control for neuronal Ca2+ signaling. White arrows highlight an individual cell responsive to both 2AT and KCl (i.e., PAR2+ neurons). Scale bar: 20 μm. (B) Representative traces of 2 cultured DRG neurons showing changes in [Ca2+]i (plotted as 340/380 nm ratiometric change) in response to 2AT and KCl. Baseline measures were recorded for 60 seconds in normal bath solution. Cells were then treated with 2AT (1 μM) for 120 seconds, washed in normal bath solution for 120 seconds, and treated with KCl (50 mM) for 10 seconds. Cells with at least 20% ratiometric change in response to KCl treatment were classified as neurons, and out of these, neurons with at least 40% ratiometric change in response to 2AT treatment were classified as PAR2+. (C) Pie chart illustrating the percentage of PAR2+ neurons in culture as characterized by response to 2AT (1 μM). About 3%–4% of primary cultured DRG neurons (KCl responsive) are PAR2+ (2AT responsive). (D) Whole-cell current clamp recordings reveal increased firing of TRPV1+CGRP– cultured DRG neurons from CGRPcre/+-ER Rosa26LSL-tDTomato/+ TRPV1-GFP reporter mice after activation with 2AT (1 μM). A linear ramp (0 to 0.1 nA for 1 second) was applied to the patched neuron to generate an action potential (AP) train before and after 2AT (1 μM) treatment. (E) Electrophysiological experiments demonstrate that 2AT (1 μM) induced hyperexcitability exclusively in TRPV1+CGRP– neurons. Changes in neuronal excitability were based on the ratio of current ramp-generated AP frequencies after and before vehicle/2AT treatment. n = 7 for TRPV1+CGRP– neurons treated with vehicle, n = 13 for TRPV1+CGRP– neurons treated with 2AT (1 μM), and n = 8 for CGRP+ neurons treated with 2AT (1 μM). Data represent mean ± SEM. One-way ANOVA with Tukey’s multiple comparisons (E) **P < 0.01.
PAR2 agonists can cause development of chronic pain via activation of extracellular signal–regulated protein kinase (ERK1/2) signaling (28). We also tested whether 2AT evokes activation of ERK signaling specifically in PAR2 mRNA–expressing cells. First, we evaluated PAR2 mRNA expression in DRG cultures from mice using RNAscope. We observed PAR2 expression in approximately 3% of cells, almost all of which also expressed P2X3R mRNA (Figure 4, A and B). We then exposed cultured mouse DRG neurons to 2AT (1 μM, 10 minutes) and then did RNAscope for PAR2 mRNA and immunocytochemistry (ICC) for p-ERK. Strikingly, we observed increased ERK phosphorylation but only in cells that also expressed PAR2 mRNA (Figure 4, C and D). These experiments demonstrate that 2AT acts specifically on PAR2-expressing cells to induce increased intracellular Ca2+, augmented cellular excitability, and enhanced ERK activity.
Figure 4. 2AT-evoked increased phosphorylated ERK signal intensity is specific for F2rl1-expressing neurons.
Primary mouse DRG cultures were prepared for RNAscope in situ hybridization and immunocytochemistry. (A) Representative original magnification ×40 images of P2rx3 and F2rl1 mRNA and Neurofilament 200 (NF200) protein signal of cultured DRG neurons from WT mice. White arrows indicate a neuron positive for F2rl1 mRNA signal. Smaller image panels display zoomed-in images of the indicated single neuron. Scale bar: 20 μm. Zoomed-in image scale bar: 2 μm. (B) Pie chart illustrating the percentage distribution of neuronal F2rl1 and P2rx3 expression in vitro. About 3%–4% of primary cultured DRG neurons are F2rl1+, almost all of which are also P2rx3+. (C) Representative images of F2rl1 mRNA signal and phosphorylated ERK (p-ERK) immunolabeling in cultured DRG neurons from WT mice after treatment with vehicle or 2AT (1 μM) for 10 minutes. Scale bar: 2 μm. F2rl1+ DRG neurons treated with 2AT show increased p-ERK signal when compared with vehicle treatment. (D) Signal intensity of p-ERK increased markedly in F2rl1+ neurons after treatment with 2AT (1 μM) when compared with the vehicle treatment group. No significant difference in p-ERK signal intensity is seen between vehicle- and 2AT-treated groups in the F2rl1– neurons. p-ERK signal was quantified through mean gray intensity value and normalized to the average p-ERK signal intensity value for the vehicle treatment groups. n = 15 and n = 16 for F2rl1+ neurons treated with vehicle or 2AT, respectively. n = 88 for both vehicle and 2AT treatment groups in F2rl1– neurons. Data represent mean ± SEM. One-way ANOVA with Bonferroni’s multiple comparisons (D) ***P < 0.001.
PAR2-driven mechanical hyperalgesia and grimace are mediated by sensory neurons.
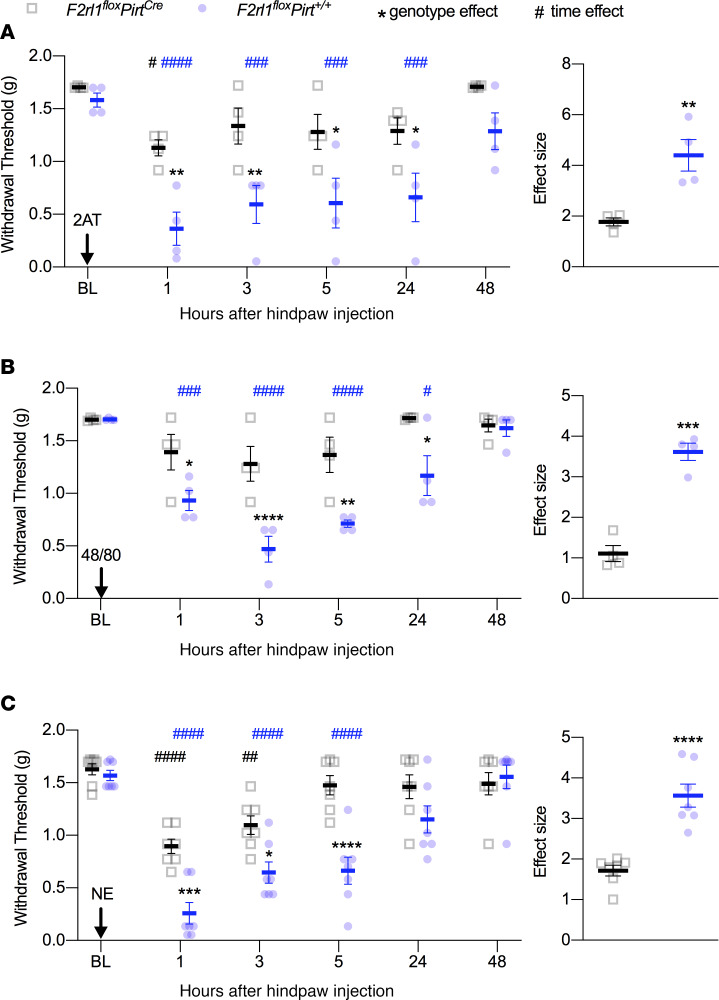
To determine whether PAR2 receptor activity in DRG sensory neurons is responsible for specific types of nociceptive behaviors, we injected the PAR2 agonist, 2AT, the mast cell degranulator 48/80, or neutrophil elastase (NE) into the hind paws of either control F2rl1floxPirt+/+ or PAR2–conditional knockout F2rl1floxPirtCre mice. Using von Frey testing, we found that when 2AT was injected into the hind paws of mice lacking PAR2 in sensory neurons (F2rl1floxPirtCre), the mice showed only a very transient mechanical hypersensitivity (Figure 5A). In contrast, F2rl1floxPirt+/+ mice displayed mechanical hypersensitivity that lasted for at least 24 hours and that was significantly greater than PAR2–conditional knockout mice (Figure 5A). Strikingly similar results were obtained for both 48/80 (Figure 5B) and NE (Figure 5C), demonstrating that PAR2-mediated mechanical hypersensitivity requires sensory neuron PAR2 expression in mice.
Figure 5. PAR2-induced mechanical hypersensitivity is sensory neuron mediated.
Mice were injected with PAR2 agonists before assessing mechanical sensitivity via von Frey testing 1, 3, 5, 24, and 48 hours after hind paw injection. Baseline (BL) measures were obtained before administering 2AT, 48/80, or NE. When compared with F2rl1floxPirt+/+ mice, F2rl1floxPirtCre mice show decreased mechanical hypersensitivity in response to 2AT (30 pmol) (A), 48/80 (6.5 nmol) (B), and NE (10 units) (C). Effect size is determined by calculating the cumulative difference between the baseline value and the value for each time point. *P < 0.05 compared with F2rl1floxPirt+/+ or F2rl1floxPirtCre groups. #P < 0.05 compared with baseline measures. n = 4 for F2rl1floxPirt+/+ and F2rl1floxPirtCre groups treated with 2AT and 48/80, n = 7 for F2rl1floxPirt+/+ and F2rl1floxPirtCre groups treated with NE. Data are expressed as mean ± SEM. Two-way ANOVA used for withdrawal threshold, with Holm-Šidák and Dunnett’s multiple comparisons: *P < 0.05, **P < 0.01, ***P < 0.001, and ****P < 0.0001; ##P < 0.01, ###P < 0.001, and ####P < 0.0001. Unpaired t test used for effect size: **P < 0.01, ***P < 0.001, and ****P < 0.0001. Asterisks denote significant differences between genotypes. Pound signs denote significant differences versus BL as a function of time.
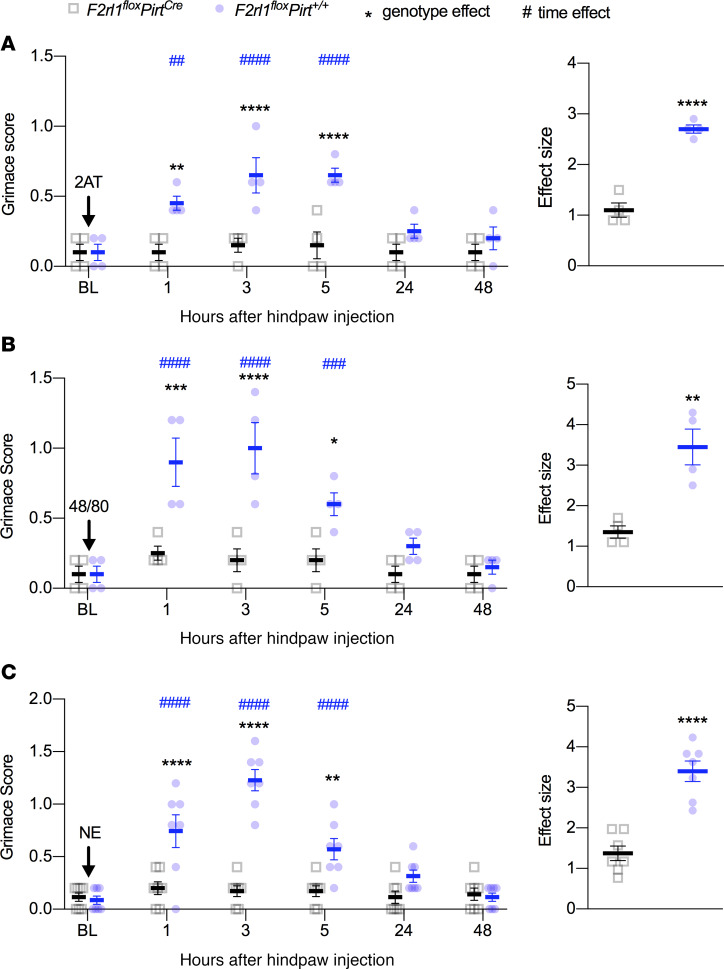
To determine whether PAR2 activation results in changes in affective measures of pain, we injected 2AT, 48/80, and NE into the hind paws of either F2rl1floxPirt+/+ or F2rl1floxPirtCre mice and recorded grimacing behaviors. F2rl1floxPirtCre did not show any signs of grimacing in response to 2AT injection while F2rl1floxPirt+/+ showed a significant increase in mouse grimace scores for up to 5 hours after injection (Figure 6A). Likewise, 48/80 (Figure 6B) and NE (Figure 6C) caused grimacing for about 5 hours in F2rl1floxPirt+/+, but no grimacing responses were noted in F2rl1floxPirtCre mice. Therefore, grimacing in response to PAR2 activation also depends on PAR2 expression in DRG neurons.
Figure 6. PAR2 agonists effectuate facial grimacing via PAR2+ sensory neurons.
Mice were injected with PAR2 agonists and grimacing was subsequently scored 1, 3, 5, 24, and 48 hours after hind paw injection. Baseline (BL) measures were obtained before administering 2AT, 48/80, or NE. When compared with F2rl1floxPirt+/+ mice, F2rl1floxPirtCre mice show decreased facial grimacing in response to 2AT (30 pmol) (A), 48/80 (6.5 nmol) (B), and NE (10 units) (C). Effect size is determined by calculating the cumulative difference between the value for each time point and the baseline value. *P < 0.05 compared with F2rl1floxPirt+/+ or F2rl1floxPirtCre groups. #P < 0.05 compared with baseline measures. n = 4 for F2rl1floxPirt+/+ and F2rl1floxPirtCre groups treated with 2AT and 48/80, n = 7 for F2rl1floxPirt+/+ and F2rl1floxPirtCre groups treated with NE. Data are expressed as mean ± SEM. Two-way ANOVA used for grimace score, with Holm-Šidák and Dunnett’s multiple comparisons: *P < 0.05, **P < 0.01, ***P < 0.001, and ****P < 0.0001; ##P < 0.01, ###P < 0.001, and ####P < 0.0001. Unpaired t test used for effect size: **P < 0.01, and ****P < 0.0001. Asterisks denote significant differences between genotypes. Pound signs denote significant differences versus BL as a function of time.
Thermal hyperalgesia findings were less clear-cut than mechanical sensitivity and grimace. Injection of 2AT into the hind paws of F2rl1floxPirtCre did not cause thermal hyperalgesia when compared with baseline (Figure 7A). F2rl1floxPirt+/+ mice showed thermal hyperalgesia only at the 24-hour time point, and the effect size of thermal hyperalgesia was greater in mice with intact PAR2 expression in DRG neurons (Figure 7A). Injection of 48/80 caused significant thermal hyperalgesia at the 5- and 48-hour time points only in F2rl1floxPirtCre mice, but the effect size did not differ between genotypes (Figure 7B). NE caused robust thermal hyperalgesia in both genotypes that lasted for 48 hours (Figure 7C).
Figure 7. NE- and 48/80-evoked thermal hyperalgesia is not mediated via PAR2+ sensory neurons.
Mice were injected with PAR2 agonists and then latency to paw withdrawal was measured via the Hargreaves test 1, 3, 5, 24, and 48 hours after hind paw injection. Baseline (BL) measures were obtained before administering 2AT, 48/80, or NE. When compared with F2rl1floxPirt+/+ mice, F2rl1floxPirtCre mice showed decreased thermal hyperalgesia in response to only 2AT (30 pmol) (A) but not to 48/80 (6.5 nmol) (B) or NE (10 units) (C). Effect size is determined by calculating the cumulative difference between the baseline value and the value for each time point. *P < 0.05 compared with F2rl1floxPirt+/+ or F2rl1floxPirtCre groups. #P < 0.05 compared with baseline measures. n = 4 for F2rl1floxPirt+/+ and F2rl1floxPirtCre groups treated with 2AT and 48/80, and n = 7 for F2rl1floxPirt+/+ and F2rl1floxPirtCre groups treated with NE. Data are expressed as mean ± SEM. Two-way ANOVA with Holm-Šidák and Dunnett’s multiple comparisons #P < 0.05, ##P < 0.01, and ###P < 0.001. Unpaired t test *P < 0.05.
To determine whether PAR2 receptor activation results in changes in temperature indicative of inflammation of the paw, we used infrared FLIR imaging. With 2AT injection, we did not note any change in hind paw temperature in either genotype at any time point (Figure 8A). On the other hand, 48/80 caused a significant increase in paw temperature but only in F2rl1floxPirt+/+ mice (Figure 8B). Upon injection of NE into the hind paw, we again observed that only F2rl1floxPirt+/+ mice showed a significant increase in paw temperature (Figure 8C).
Figure 8. Effects of 48/80 and NE on paw temperature are sensory neuron PAR2-mediated.
Mice were injected with PAR2 agonists, and then hind paw temperatures were measured via FLIR imaging 1, 3, 5, 24, and 48 hours after hind paw injection. Baseline measures were obtained before administering 2AT, 48/80, or NE. 2AT (30 pmol) (A) did not cause an increase in paw temperature in either genotype. Unlike the F2rl1floxPirt+/+ mice, F2rl1floxPirtCre mice did not show an increase in paw temperature in response to 48/80 (6.5 nmol) (B) and NE (10 units) (C). Effect size is determined by calculating the cumulative difference between the value for each time point and the baseline value. *P < 0.05 compared with F2rl1floxPirt+/+ or F2rl1floxPirtCre groups. #P < 0.05 compared with baseline measures. n = 4 for F2rl1floxPirt+/+ and F2rl1floxPirtCre groups treated with 2AT and 48/80, and n = 7 for F2rl1floxPirt+/+ and F2rl1floxPirtCre groups treated with NE. Data are expressed as mean ± SEM. Two-way ANOVA used for temperature, with Holm-Šidák and Dunnett’s multiple comparisons: *P < 0.05, and **P < 0.01; #P < 0.05, ##P < 0.01, and ###P < 0.001. Unpaired t test used for effect size: *P < 0.05, and **P < 0.01. Asterisks denote significant differences between genotypes. Pound signs denote significant differences versus BL as a function of time.
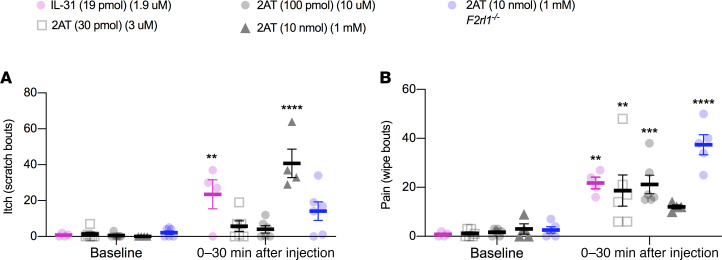
Our data show that F2rl1 mRNA is very specifically expressed in a subset of sensory neurons that are associated with itch behavior. Therefore, we assessed acute itch and pain behaviors using the cheek scratch versus wipe assay (29). We used 3 concentrations of 2AT, 2 of which should be specific for PAR2 and a higher concentration that could potentially activate Mrg receptors (10, 11). We also used interleukin-31 (IL-31, 19 pmol) because the PAR2-expressing population of cells express the IL-31 receptor (IL-31R), and IL-31R signaling is linked to itch behaviors (30). We observed that IL-31 caused an increase in itch bouts, but low doses of 2AT (30 and 100 pmol) did not (Figure 9A). The higher dose of 2AT (10 nmol) did cause itch bouts in WT mice, but this effect was not seen in global F2rl1–/– mice. IL-31 and both low doses of 2AT caused wipes, indicative of pain behaviors, in WT mice (Figure 9B). The higher dose of 2AT did not cause significant wiping behavior in WT mice but did cause wiping in F2rl1–/– mice. These findings show that 2AT, at concentrations that are specific for PAR2 activation (11), only causes acute pain behaviors and not itching.
Figure 9. Activation of PAR2+ sensory neurons via low-dose intradermal administration of 2AT induces pain but not itch bouts.
The subset of sensory neurons that express F2rl1 mRNA also express several markers, such as the IL-31R, implicated in itch. We investigated the role of PAR2 activation in this subset of sensory neurons via the cheek scratch versus wipe assay. This assay differentiates itch and pain behaviors in mice through hind limb scratching and forelimb wiping, respectively. After habituation, baseline behavior was video-taped for 15 minutes before injection. Intradermal injections of IL-31 (19 pmol) and low-dose 2AT (30 pmol and 100 pmol) were administered into the shaved left cheek of WT mice. High-dose 2AT (10 nmol) was administrated to both WT and F2rl1–/– (global PAR2 KO) mice. (A) Itch (scratch bouts) and (B) pain (wipe bouts) were scored up to 30 minutes after injections. IL-31 caused both significant scratching and wiping compared with baseline. However, low doses of 2AT (PAR2-specific concentrations) caused only pain behaviors and not scratching. High-dose 2AT (PAR2-nonspecific concentration) caused scratching but not significant wiping in WT mice. In F2rl1–/– mice, high-dose 2AT caused wiping but not significant scratching. n = 4, n = 6, n = 6, and n = 4 for WT mice treated with IL-31 (30 pmol) and 2AT (30 pmol, 100 pmol, and 10 nmol), respectively; n = 6 for F2rl1–/– mice treated with 2AT (10 nmol). Data are expressed as mean ± SEM. Two-way ANOVA with Bonferroni’s multiple comparisons (baseline versus treatment) **P < 0.01, ***P < 0.001, and ****P < 0.0001.
Discussion
PAR2 was one of the first pain targets identified using knockout mouse technology (9) and has remained a prominent target in the pain field for 2 decades (1, 8). While many aspects of PAR2 physiology and pharmacology have been revealed, the elucidation of other targets, such as the Mrg family of GPCRs for some broadly used PAR2 tools, has complicated interpretation of much of the existing literature (10). Moreover, several recent RNA-sequencing papers have reported surprisingly low levels of F2rl1 gene expression in dorsal root ganglia and/or in single DRG neurons (14, 16, 22). Our experiments were aimed at gaining better clarity on which neurons in the DRG express PAR2 and what aspects of pain behavior are driven by these neurons. Combined with a recent study that independently generated a nociceptor-specific F2rl1-knockout mouse with nearly identical findings (2), our work makes it clear that sensory neuron–expressed PAR2 is required for mechanical hypersensitivity and spontaneous pain behaviors caused by PAR2 activation in the paw, at least in male mice. A unique aspect of our work is that we demonstrate that this effect is driven by a very small population of DRG neurons. We conclude that sensory neuron–expressed PAR2 is a key target for mechanical and spontaneous pain driven by the release of endogenous proteases from many types of immune cells.
Thermal hyperalgesia caused by inflammation is at least partially mediated by PAR2 (9). In our experiments we did not observe any genotype differences in 48/80- or NE-evoked thermal hyperalgesia, indicating that this effect is likely driven by PAR2 expression in nonneuronal cells. Interestingly, PAR2 is expressed by a small subset of DRG nociceptors that also express TRPV1, but not CGRP, a receptor that is required for the generation of thermal hyperalgesia in inflammatory conditions (31–33). It is possible that PAR2 is not expressed by a sufficient proportion of these neurons to cause thermal hyperalgesia. Many previous studies have shown that PAR2 activation sensitizes TRPV1 (34–36), but many of these studies used SLIGRL, a PAR2-activating peptide that also stimulates Mrg receptors (10). The cellular basis of PAR2-mediated thermal hyperalgesia remains unresolved.
PAR2 has previously been implicated in itch, but this literature is also complicated by the nonspecific nature of some widely used PAR2 pharmacological tools. For instance, SLIGRL-induced itch is mediated by an Mrg GPCR, not by PAR2 (10). Nevertheless, some itch-causing agents, such as cowhage, activate PAR2 (37, 38). Interestingly, our RNAscope and analysis of single-cell RNA-sequencing experiments clearly show that F2rl1 mRNA is expressed in a population of DRG neurons that are known to be critical for itch behaviors in mice (23). Importantly, the contribution of this subset of neurons to nociception has not been clear. Because of these previous and current findings, we tested whether 2AT can cause pain or itch behaviors in mice. At 2AT doses that are specific for PAR2 (11), we observed clear pain behaviors, consistent with our grimace findings. We did not observe itch behaviors. IL-31, which acts via a receptor that is expressed by this same population of cells (30), produced both itch and pain behaviors, indicating that activating these neurons is capable of driving both types of behavioral outcomes. These differential outcomes may be mediated by encoding at the level of the spinal cord, as it has recently been shown that burst firing in those itch circuits is required to drive itch behavior (39, 40). At high concentrations of 2AT, we noted both pain and itch behaviors, and the pattern of these behaviors was different in F2rl1–/– and WT mice. Although we did not explore the mechanisms driving this difference experimentally, it may be explained by a complex pattern of recruitment of different populations of nociceptors because of the lack of specificity of the compound at concentrations above ~10 μM (11). This pattern would necessarily be different in mice lacking PAR2. It could also be explained by differential innervation patterns for different types of afferents. In this regard, it has recently been shown that jugular neurons expressing an itch receptor, MrgprC11, signal bronchoconstriction, and airway hyperresponsiveness (41). The physiological outcome of PAR2 activation is likely dependent on the peripheral and central innervation target of the PAR2-expressing neuron.
Previous work from our group has demonstrated that activation of PAR2 leads to the development of a persistent pain state termed hyperalgesic priming (28). Hyperalgesic priming models involve a priming stimulus, such as 2AT, triggering neuronal plasticity that causes long-lasting sensitivity to subthreshold doses of inflammatory cytokines (e.g., prostaglandin E2) after resolution of the initial insult (42, 43). The hyperalgesic priming model serves as an important experimental model for chronic pain conditions in which a transition occurs from an acute to chronic pain state. The priming caused by PAR2 activation indicates a direct role of the receptor in the development of chronic pain disorders where protease release could be involved. The pain plasticity seen after 2AT treatment requires PAR2-mediated activation of ERK and downstream signaling to translation initiation factors that alter gene expression in nociceptors (28, 44, 45). These PAR2-mediated effects could have been due to signaling in sensory neurons or other nonneuronal cells. Our current work demonstrates that this effect is driven mainly by nociceptor-expressed PAR2 because 2AT-evoked mechanical hypersensitivity and grimacing are gone with sensory neuron–specific deletion of the F2rl1 gene and PAR2 activation activates ERK in this specific population of nociceptors. Although we provide compelling evidence that most aspects of PAR2-mediated pain are due to PAR2 in nociceptors, it remains to be seen what tissues these nociceptors innervate. This is an important question to address in future studies. PAR2 has been implicated in gastrointestinal pain (2, 18), but few, if any, colonic sensory neurons express F2rl1 mRNA (16). Discovering the innervation pattern of this population of nociceptors will clarify which pain disorders are likely to benefit from PAR2 antagonist therapy.
Methods
Animals.
To generate F2rl1flox mice, loxP sites were inserted flanking the F2rl1 gene exon 2 on chromosome 13. The F2rl1 gene contains 2 exons, and exon 2 was targeted because it contains the majority of the coding sequence of the PAR2 protein. Mice were generated on a C57BL/6J background through a contract with Cyagen Biosciences (see Supplemental Methods for project report; supplemental material available online with this article; https://doi.org/10.1172/jci.insight.137393DS1). A neomycin selection cassette was inserted and removed through Frt-mediated recombination. Mice were crossed with PirtCre mice (21), provided by Xinzhong Dong at Johns Hopkins University (Baltimore, Maryland, USA) through a material transfer agreement, at University of Texas at Dallas, to generate experimental animals for behavioral experiments. Additional C57BL/6J mice were bred in our colony for cell culture and cellular anatomy studies.
The Rosa26LSL-tDTomato/+ mouse line on B6.129 background was obtained from the Jackson Laboratory (Bar Harbor, Maine, USA). Trpv1GFP mouse lines were purchased from the GENSAT program (Mutant Mouse Resource and Research Center, University of North Carolina School of Medicine, Chapel Hill, North Carolina, USA, and UCD, Davis, California, USA). The Calcacre/+-ER mouse line was provided by Pao-Tien Chuang (UCSF, San Francisco, California, USA) (46). Adult male mice were used in described electrophysiology experiments.
Experimental reagents.
2AT was made as previously described (25, 26). NE was purchased from Elastin Products Company, Inc. (SE563). Compound 48/80 was purchased from MilliporeSigma (C2313).
Behavioral methods.
In behavioral experiments, we used male mice. Behavioral observers were blinded to genotype and treatment in all experiments. Mechanical sensitivity was measured using von Frey filament testing (47). Animals were acclimated to suspended Plexiglas chambers (11.4 × 7.6 × 7.6 cm) with wire mesh bottoms (1 cm2). Withdrawal thresholds to probing the hind paws were determined before experimental treatment and at 1, 3, 5, 24, and 48 hours after administration. Paw withdrawal thresholds were determined by applying von Frey filaments to the nonglabrous plantar aspect of the hind paws, and a response was indicated by a withdrawal of the paw. The withdrawal thresholds were determined by the Dixon up-down method by using blinded observers. The maximum filament strength was 2 g for the experiments.
Mouse grimace scoring was performed as described by Langford et al. (48). The mouse grimace scale is a pain assay used to measure affective pain in response to multiple types of stimuli (49–52). Mice were placed individually in the same suspended Plexiglas chambers with wire mesh bottoms as previously described, allowed to acclimate for 1 hour, and then scored by blinded scorers before experimental treatment and at 1, 3, 5, 24, and 48 hours after administration. The scores of each animal subject were averaged at each time point by group.
Thermal sensitivity was measured using the Hargreaves method (53). Mice were placed on a warmed glass floor (29°C) 20 minutes before each testing, and using a Hargreaves apparatus (IITC Model 390), a focused beam of high-intensity light was aimed at the plantar nonglabrous surface of the hind paws. The intensity of the light was set to 30% of maximum with a cutoff value of 20 seconds. The latency to withdraw the hind paw was measured to the nearest 0.01 seconds. The hind paws were measured before treatment and at 1, 3, 5, 24, and 48 hours after administration.
Paw inflammation was investigated by measuring the temperature of the animal’s hind paws. All testing was performed in a climate-controlled room with an ambient temperature of 21°C ± 2°C. Animals were allowed to acclimate in the testing room for 1 hour before testing. Colorized infrared thermograms that captured the nonglabrous surface of the animal’s hind paws were obtained using a FLIR T-Series Thermal Imaging Camera (FLIR Systems, Inc). The thermograms were captured before experimental treatment and at 1, 3, 5, 24, and 48 hours after administration. Thermogram analysis was performed using the FLIR Thermal Imaging Software. For each thermogram image, a straight line was drawn on the plantar surface of both hind paws, and the mean temperature was recorded from the average of each pixel along the drawn line. The raw temperatures were then plotted for ipsilateral and contralateral hind paws for each individual animal.
To assess cheek itch and wipe bouts, mice were anesthetized briefly with isoflurane, and the left cheek was shaved 2–3 days before intradermal injection. Animals were allowed to adapt to the experimental conditions by placing them in the same suspended Plexiglas chambers with wire mesh bottoms, as previously described, before experiments began. On the day of the experiment, mice were habituated for 1 hour in the acrylic boxes, and then their baseline behavior was recorded for 15 minutes. For each mouse, 2 camcorders (Samsung HMX-F90 and HMX-QF20) were placed in front of and behind the mouse, and recordings were done simultaneously. After 15 minutes of recording baseline behavior, the mice were restrained and received a 10-μL intradermal injection into the cheek of either 2AT (30 pmol, 100 pmol, or 10 nmol) or IL-31 (19 pmol). Injections were done using a Hamilton syringe (catalog 80901) with a 30G needle held parallel to the skin and inserted superficially. Once mice were injected, they were placed back into the acrylic boxes, and their behavior was videotaped for 30 minutes. The video recordings from the 2 camcorders positioned in front of and behind each mouse were edited together into 1 video to give a simultaneous view of 2 angles of the mouse. The behavior in the videos was scored by students who were blinded to the experimental groups. Wiping and scratching behaviors were scored as described previously by Shimada et al. (29).
DRG cultures.
For primary neuronal cultures used in calcium imaging and RNAscope in situ hybridization, dorsal root ganglia were dissected from adult male and female Institute of Cancer Research mice (bred at University of Texas at Dallas and originally obtained from Envigo) and suspended in Hanks’ balanced salt solution without calcium and magnesium before culturing. Ganglia were incubated at 37°C for 25 minutes in 1 mg/mL papain (LS003119; Worthington), followed by 25 minutes of incubation at 37°C in 3 mg/mL collagenase type 2 (LS004176; Worthington) and 2 mg/mL Dispase II (04942078001; MilliporeSigma). Ganglia were then triturated in HBSS with a 1-mL pipette tip. The solution was passed through a 70-μm cell strainer (22363548; Thermo Fisher Scientific), and the cells were resuspended in DMEM/F12/GlutaMAX (Gibco, Thermo Fisher Scientific) culture medium nourished with 10% fetal bovine serum (FBS; SH30088.03; Hyclone) and 1% penicillin/streptomycin (pen-strep; 15070-063; Gibco, Thermo Fisher Scientific). Cells were plated and allowed to adhere for 2 hours, and each well was then flooded with the same supplemented culture medium described previously with an additional 10 ng/mL nerve growth factor (NGF; 01-125; MilliporeSigma) and 3 μg/mL 5-fluoro-2′-deoxyuridine + 7 μg/mL uridine (FRD+U; MilliporeSigma) added. Thereafter, neurons were kept at 37°C and 5% CO2 in an incubator with supplemented culture medium with NGF and FRD+U changed every other day until further experimentation.
DRG cultures for immunocytochemistry and RNAscope experiments were prepared as described above with the neurons plated on 8-well Chamber Slides (154534; Nunc Lab-Tek). The neurons were resuspended in culture medium, plated in 100 μL in each well, and allowed to adhere for 2 hours. Then, the wells were flooded with culture medium supplemented with 10% FBS, 1% pen-strep, 10 ng/mL NGF, and 3 μg/mL + 7 μg/mL FRD+U. Neurons were kept at 37°C and 5% CO2 with medium changed every other day until their use. Protease III (Advanced Cell Diagnostics) treatment concentration for RNAscope-ICC was optimized to 1:30.
For primary DRG neuronal cultures for electrophysiology recordings, L3-L5 DRG neurons were quickly removed from male reporter mice CGRPER-cre/+ Rosa26LSL-tDTomato/+ and CGRPER-cre/+ Rosa26LSL-tDTomato/+ TRPV1-GFP. DRG neurons were dissociated by treatment with a 1 mg/mL Collagenase/Dispase (Roche) solution. Cells were maintained in DMEM supplemented with 2% FBS, 2 mM l-glutamine, 100 U/mL penicillin, and 100 μg/mL streptomycin. The experiments were performed within 6–36 hours after DRG neuron plating.
Calcium imaging.
DRG neurons were dissected and cultured as described before and were plated on poly-d-lysine–coated dishes (P35GC-1.5-10-C; MatTek) with additional laminin coating (L2020; MilliporeSigma). Neurons were used within 24 hours of plating. DRG neurons were loaded with 1 μg/μL Fura 2AM (108964-32-5; Life Technologies) for 1 hour before changing to normal bath solution (135 mM NaCl, 5 mM KCl, 10 mM HEPES, 1 M CaCl2, 1 M MgCl2 and 2 M glucose, adjusted to pH 7.4 with N-methyl-glucamine, osmolarity of 300 ± 5 milliosmoles). The cells were then treated with 1 μM 2AT dissolved in normal bath solution for 120 seconds. Images were acquired on the Olympus IX73 inverted microscope at original magnification ×40. For purposes of analysis, cells that responded with at least 20% ratiometric change (340 nm/380 nm) in extracellular Ca2+ upon treatment of KCl were classified as neurons. Out of this classification, neurons that responded with at least 40% ratiometric change upon treatment of 2AT were classified as PAR2+. The experiment was performed using the MetaFluor Fluorescence Ratio Imaging Software.
Tissue preparation.
Lumbar DRG neurons and hind paw skin were rapidly dissected, embedded in optimal cutting temperature compound, and flash-frozen immediately in dry ice. Tissues were sectioned at 20 μm onto charged slides. Sections were only briefly thawed to adhere to the slide but were immediately returned to the –20°C cryostat chamber until completion of sectioning.
RNAscope in situ hybridization.
RNAscope in situ hybridization multiplex version 1 was performed as instructed by Advanced Cell Diagnostics (ACD). Slides were transferred from the cryostat directly into cold (4°C) 10% formalin for 15 minutes and then dehydrated in 50% ethanol (5 minutes), 70% ethanol (5 minutes), and 100% ethanol (10 minutes) at room temperature. The slides were air-dried briefly, and then boundaries were drawn around each section using a hydrophobic pen (ImmEdge PAP pen; Vector Labs). When the PAP pen boundaries had dried, sections were incubated in protease IV reagent for 2 minutes at room temperature. Slides were washed briefly in 1X phosphate-buffered saline (PBS, pH 7.4) at room temperature. Each slide was then placed in a prewarmed humidity control tray (ACD) containing dampened filter paper. For DRG experiments, F2rl1 (PAR2; 417541; ACD), Calca (CGRP; 417961; ACD), and P2rx3 (P2X3R; 521611; ACD) probes were pipetted onto each section until fully submerged and then incubated for 2 hours at 40°C. For hind paw skin, only the F2rl1 or bacterial dapB (negative control) probes were used. Slides were then washed 2 times in 1X RNAscope wash buffer and returned to the oven for 30 minutes after submersion in AMP-1 reagent. Washes and amplification were repeated using AMP-2, AMP-3, and AMP-4 (ALT-B) reagents with 15-minute, 30-minute, and 15-minute incubation periods, respectively. Slides were then washed 2 times in 0.1 M phosphate buffer (PB, pH 7.4). The DRG slides were then processed for immunohistochemistry, while the hind paw skin slides were incubated for 5 minutes in 1:5000 DAPI (ACD), washed in 0.1 M PB, air-dried, and coverslipped with ProLong Gold mounting medium (Thermo Fisher Scientific).
Immunohistochemistry.
After completion of RNAscope in situ hybridization, DRG slides were incubated in blocking buffer (10% normal goat serum, 0.3% Triton X-100 in 0.1 M PB) for 1 hour at room temperature while being shielded from light. Slides were placed in a light-protected, humidity-controlled tray and incubated in primary antibody (mouse anti–Neurofilament 200; clone N52; MAB5266; MilliporeSigma) at 1:500 in blocking buffer overnight at 4°C. The next day, slides were washed 2 times in 0.1 M PB and then incubated in secondary antibody, goat anti-mouse IgG (H+L) Alexa Fluor 405 (1:2000; A-31553; Invitrogen, Thermo Fisher Scientific) for 1 hour at room temperature. Sections were washed 2 times in 0.1 M PB, air-dried, and coverslipped with ProLong Gold mounting medium.
Image analysis for DRG sections.
Three mice per genotype were imaged on an Olympus FV3000 confocal microscope at original magnification ×20. One image was acquired of each mouse DRG section, and 3 sections were imaged per mouse (total: 9 images). The raw image files were brightened and contrasted equally in Olympus CellSens software and then analyzed manually 1 cell at a time for expression of Calca, P2rx3, and F2rl1. Cell diameters were measured using the polyline tool. NF200 signal (not shown), Calca, P2rx3, and F2rl1 were used to quantify the total neuronal population. Representative images of hind paw skin are shown from F2rl1floxPirt+/+ and F2rl1floxPirtCre mice with a negative control from an F2rl1floxPirt+/+ animal imaged at the same settings.
RNAscope in situ hybridization on DRG cultures.
DRG cultures were prepared as described with the neurons plated on 8-well Chamber Slides (154534; Nunc Lab-Tek) coated with poly-d-lysine (P0899; MilliporeSigma). On day 5, the cultures were treated with 1 μM 2AT or vehicle (culture medium) for 10 minutes in the incubator. The samples were then prepared as instructed by ACD. The chambers were disassembled and the slides submerged in 1X PBS. They were transferred to 10% formalin for 30 minutes at room temperature, followed by 3 washes in 1X PBS. Hydrophobic boundaries were drawn around each well as previously described. Each well was incubated with protease III reagent (1:30 in 1X PBS) for 10 minutes at room temperature. Slides were washed in 1X PBS and then placed in a prewarmed humidity control tray containing dampened filter paper. F2rl1 and P2rx3 probes were pipetted onto each well. Two wells received only control probes, negative (bacterial dapB) or positive (320881; ACD). Slides were incubated in the probes, followed by washes, and amplification as previously described. After completion of RNAscope in situ hybridization, ICC was performed.
Immunocytochemistry.
The following steps were performed in a light-protected humidity control tray. Slides were incubated in blocking buffer (10% normal goat serum in 0.1 M PB) with 0.02% Triton X-100 for 1 hour at room temperature. They were then incubated overnight at 4°C with primary antibody, mouse anti–Neurofilament 200 at 1:500 and rabbit anti–phospho-p44/42 MAPK T202/Y204 (p-ERK; 9101; Cell Signaling Technology), at 1:250 in blocking buffer. The next day, slides were washed twice in 0.1 M PB and incubated for 1 hour at room temperature with secondary antibodies, goat anti-mouse IgG (H+L) Alexa Fluor 405 (A-31553; Invitrogen, Thermo Fisher Scientific) and goat anti-rabbit IgG (H+L) Alexa Fluor 647 (A-21245; Invitrogen, Thermo Fisher Scientific), both at 1:2000 in blocking buffer. Slides were washed twice in 0.1 M PB, air-dried, and coverslipped with ProLong Gold Antifade mounting medium.
Image analysis for DRG cultures.
Using an Olympus confocal microscope (FV1200), 5 wells of each treatment were imaged. Per well, 3–6 images were taken at original magnification ×40 with 9 Z-slices for a total of 22 images per treatment. The raw image files were projected to their maximum Z, brightened and contrasted equally in Olympus CellSens software, and analyzed manually for expression of P2rx3 and F2rl1. NF200 signal was used to verify the neuronal population. Z-slices that did not contain the neuron in focus were excluded from analysis. Regions of interest of each neuron were drawn using the ellipse tool, and p-ERK signal was quantified using mean gray intensity value. Background values taken from the negative control were subtracted before analysis. Percentages of neurons expressing P2rx3 and F2rl1 were summed from both treatment groups, while p-ERK intensity was compared among 2AT- and vehicle-treated F2rl1-positive and -negative neurons. Representative images at original magnification ×40 are shown for P2rx3, F2rl1, and NF200 expression, along with a zoomed-in image of a single neuron. Zoom-in of a single neuron is also shown for p-ERK expression in a representative F2rl1+ neuron.
Electrophysiology.
Recordings were made in whole-cell current clamp configurations at 22°C–24°C. Data were acquired and analyzed using an Axopatch 200B amplifier and pCLAMP10.2 software (Molecular Devices). Recorded data were filtered at 5 kHz and sampled at 30 kHz. Borosilicate pipettes (Sutter) were polished to resistances of 2–3 MΩ. Access resistance (Rs) was compensated (40%–80%) when appropriate up to the value of 6–8 MΩ. Data were rejected when Rs changed more than 20% during recording, leak currents were more than 50 pA, or input resistance was less than 300 MΩ. Standard external solution contained (in mM): 140 NaCl, 5 KCl, 2 CaCl2, 1 MgCl2, 10 d-glucose, and 10 HEPES pH 7.4. The standard pipette (internal) solution contained (in mM): 140 KCl, 1 MgCl2, 1 CaCl2, 10 EGTA, 10 d-glucose, 10 HEPES pH 7.3, 2.5 ATP, and 0.2 GTP. Drugs were applied by a fast, pressure-driven, computer-controlled, 4-channel system (ValveLink8; AutoMate Scientific) with quartz application pipettes.
CGRP+ DRG small (<30 pF) neurons from CGRPcre/+-ER Rosa26LSL-tDTomato/+ mice were randomly selected for recording. TRPV1+CGRP– DRG neurons from CGRPcre/+-ER Rosa26LSL-tDTomato/+ TRPV1-GFP reporter mice were selected for recording because they have PAR2 (15). To characterize modulation of TRPV1+CGRP– or CGRP+ DRG neuron excitation by vehicle (control) or PAR2-activating peptide (2AT), the following sequence of recording protocols was applied: (a) a single AP in current clamp configuration was generated with a 0.5-ms and 1-nA current step to define the type of sensory neurons (54); (b) a linear ramp from 0 to 0.1 nA for 1 second was applied to generate a control AP train; (c) the patched neuron was treated for 2–5 minutes with vehicle or PAR2 activator; and then (d) the ramp as in the step 2 was reapplied. Data were accumulated from 3–5 independent mouse DRG neuronal cultures. Each culture was generated from 1 male mouse. Changes in neuronal excitability were calculated by dividing AP frequency generated by a current ramp after vehicle or drug treatment to AP frequency produced by the ramp before treatment. Excitability was determined to be regulated by 2AT when the drug treatment produced a statistically significant increase in AP frequency versus vehicle treatment (i.e., control).
Bioinformatics.
Read counts for each coding gene for 204 single-cell RNA-sequencing profiles of mouse DRG sensory neurons were obtained from Gene Expression Omnibus (accession number GSE63576) (22). t-SNE–based clustering and visualization of the single-cell data sets were performed using Seurat package 2.2.1 (55, 56).
Statistics.
All statistical tests used GraphPad Prism version 8.4.1 (GraphPad Software, Inc.). Differences between groups were evaluated using 1- and 2-way ANOVAs followed by Bonferroni’s, Tukey’s, Dunnett’s, or Holm-Šídák multiple comparisons for data sets with 3 or more groups. Unpaired 2-tailed t tests were done for data sets with only 2 groups as indicated in the text and figure legends. Outliers were assessed using a Grubb’s test and excluded. Only 1 outlier data point was identified in this study and is noted in the figure legend for that data set. All statistics, including t, q, degrees of freedom, and exact P values, are shown in Supplemental Tables 1 and 2. All data are represented as mean ± SEM with P < 0.05 considered significant.
Study approval.
All animal protocols were approved by the University of Texas at Dallas Institutional Animal Care and Use Committee and were consistent with the NIH’s Guide for the Care and Use of Laboratory Animals (National Academies Press, 2011).
Author contributions
TJP, GD, ANA, SB, and JV designed experiments for the study. SNH, MK, JMM, AA, SS, AW, PRR, SNB, DKN, and MDB performed experiments and analyzed data. TJP, GD, ANA, SB, JV, PRR, MDB, SNH, MK, JMM, AA, SS, and SNB provided commentary and contributed to writing the manuscript.
Supplementary Material
Acknowledgments
This work was supported by NIH grants R01NS065926 (to TJP), R01NS102161 (to TJP and ANA), and R01NS098826 (to TJP, SB, JV, and GD). The authors thank Xinzhong Dong at Johns Hopkins University and Pao-Tien Chuang at UCSF for providing us the PirtCre mouse line and the Calcacre/+-ER mouse lines, respectively.
Version 1. 04/30/2020
In-Press Preview
Version 2. 06/04/2020
Electronic publication
Footnotes
Conflict of interest: The authors have declared that no conflict of interest exists.
Copyright: © 2020, American Society for Clinical Investigation.
Reference information: JCI Insight. 2020;5(11):e137393.https://doi.org/10.1172/jci.insight.137393.
Contributor Information
Shayne N. Hassler, Email: snhassle@central.uh.edu.
Moeno Kume, Email: mxk132430@utdallas.edu.
Ayesha Ahmad, Email: ayesha.ahmad@utdallas.edu.
Stephanie Shiers, Email: sis150030@utdallas.edu.
Andi Wangzhou, Email: axw131330@utdallas.edu.
Dhananjay K. Naik, Email: dhananjay.naik2@utdallas.edu.
Michael D. Burton, Email: michael.burton@utdallas.edu.
Josef Vagner, Email: vagner@email.arizona.edu.
Scott Boitano, Email: sboitano@email.arizona.edu.
Armen N. Akopian, Email: AKOPIAN@UTHSCSA.EDU.
Gregory Dussor, Email: gregory.dussor1@utdallas.edu.
Theodore J. Price, Email: theodore.price@utdallas.edu.
References
- 1.Bunnett NW. Protease-activated receptors: how proteases signal to cells to cause inflammation and pain. Semin Thromb Hemost. 2006;32(suppl 1):39–48. doi: 10.1055/s-2006-939553. [DOI] [PubMed] [Google Scholar]
- 2.Jimenez-Vargas NN, et al. Protease-activated receptor-2 in endosomes signals persistent pain of irritable bowel syndrome. Proc Natl Acad Sci U S A. 2018;115(31):E7438–E7447. doi: 10.1073/pnas.1721891115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cocks TM, Moffatt JD. Protease-activated receptor-2 (PAR2) in the airways. Pulm Pharmacol Ther. 2001;14(3):183–191. doi: 10.1006/pupt.2001.0285. [DOI] [PubMed] [Google Scholar]
- 4.Kunzelmann K, Schreiber R, König J, Mall M. Ion transport induced by proteinase-activated receptors (PAR2) in colon and airways. Cell Biochem Biophys. 2002;36(2-3):209–214. doi: 10.1385/CBB:36:2-3:209. [DOI] [PubMed] [Google Scholar]
- 5.Coelho AM, Ossovskaya V, Bunnett NW. Proteinase-activated receptor-2: physiological and pathophysiological roles. Curr Med Chem Cardiovasc Hematol Agents. 2003;1(1):61–72. doi: 10.2174/1568016033356715. [DOI] [PubMed] [Google Scholar]
- 6.Schaffner F, Ruf W. Tissue factor and PAR2 signaling in the tumor microenvironment. Arterioscler Thromb Vasc Biol. 2009;29(12):1999–2004. doi: 10.1161/ATVBAHA.108.177428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Rothmeier AS, Ruf W. Protease-activated receptor 2 signaling in inflammation. Semin Immunopathol. 2012;34(1):133–149. doi: 10.1007/s00281-011-0289-1. [DOI] [PubMed] [Google Scholar]
- 8.Bao Y, Hou W, Hua B. Protease-activated receptor 2 signalling pathways: a role in pain processing. Expert Opin Ther Targets. 2014;18(1):15–27. doi: 10.1517/14728222.2014.844792. [DOI] [PubMed] [Google Scholar]
- 9.Vergnolle N, et al. Proteinase-activated receptor-2 and hyperalgesia: a novel pain pathway. Nat Med. 2001;7(7):821–826. doi: 10.1038/89945. [DOI] [PubMed] [Google Scholar]
- 10.Liu Q, et al. The distinct roles of two GPCRs, MrgprC11 and PAR2, in itch and hyperalgesia. Sci Signal. 2011;4(181):ra45. doi: 10.1126/scisignal.2001925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Boitano S, et al. Development and evaluation of small peptidomimetic ligands to protease-activated receptor-2 (PAR2) through the use of lipid tethering. PLoS One. 2014;9(6):e99140. doi: 10.1371/journal.pone.0099140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tiwari V, et al. Mas-related G protein-coupled receptors offer potential new targets for pain therapy. Adv Exp Med Biol. 2016;904:87–103. doi: 10.1007/978-94-017-7537-3_7. [DOI] [PubMed] [Google Scholar]
- 13.Meixiong J, Dong X. Mas-related G protein-coupled receptors and the biology of itch sensation. Annu Rev Genet. 2017;51:103–121. doi: 10.1146/annurev-genet-120116-024723. [DOI] [PubMed] [Google Scholar]
- 14.Ray P, et al. Comparative transcriptome profiling of the human and mouse dorsal root ganglia: an RNA-seq-based resource for pain and sensory neuroscience research. Pain. 2018;159(7):1325–1345. doi: 10.1097/j.pain.0000000000001217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Usoskin D, et al. Unbiased classification of sensory neuron types by large-scale single-cell RNA sequencing. Nat Neurosci. 2015;18(1):145–153. doi: 10.1038/nn.3881. [DOI] [PubMed] [Google Scholar]
- 16.Hockley JRF, et al. Single-cell RNAseq reveals seven classes of colonic sensory neuron. Gut. 2019;68(4):633–644. doi: 10.1136/gutjnl-2017-315631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kawabata A, et al. Suppression of pancreatitis-related allodynia/hyperalgesia by proteinase-activated receptor-2 in mice. Br J Pharmacol. 2006;148(1):54–60. doi: 10.1038/sj.bjp.0706708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cenac N, et al. Role for protease activity in visceral pain in irritable bowel syndrome. J Clin Invest. 2007;117(3):636–647. doi: 10.1172/JCI29255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhang W, et al. Proteinase-activated receptor 2 mediates thermal hyperalgesia and is upregulated in a rat model of chronic pancreatitis. Pancreas. 2011;40(2):300–307. doi: 10.1097/MPA.0b013e318201cbc1. [DOI] [PubMed] [Google Scholar]
- 20.Roman K, Done JD, Schaeffer AJ, Murphy SF, Thumbikat P. Tryptase-PAR2 axis in experimental autoimmune prostatitis, a model for chronic pelvic pain syndrome. Pain. 2014;155(7):1328–1338. doi: 10.1016/j.pain.2014.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kim YS, et al. Coupled activation of primary sensory neurons contributes to chronic pain. Neuron. 2016;91(5):1085–1096. doi: 10.1016/j.neuron.2016.07.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Li CL, et al. Somatosensory neuron types identified by high-coverage single-cell RNA-sequencing and functional heterogeneity. Cell Res. 2016;26(1):83–102. doi: 10.1038/cr.2015.149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mishra SK, Hoon MA. The cells and circuitry for itch responses in mice. Science. 2013;340(6135):968–971. doi: 10.1126/science.1233765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.LaMotte RH, Dong X, Ringkamp M. Sensory neurons and circuits mediating itch. Nat Rev Neurosci. 2014;15(1):19–31. doi: 10.1038/nrn3641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Boitano S, Flynn AN, Schulz SM, Hoffman J, Price TJ, Vagner J. Potent agonists of the protease activated receptor 2 (PAR2) J Med Chem. 2011;54(5):1308–1313. doi: 10.1021/jm1013049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Flynn AN, et al. The protease-activated receptor-2-specific agonists 2-aminothiazol-4-yl-LIGRL-NH2 and 6-aminonicotinyl-LIGRL-NH2 stimulate multiple signaling pathways to induce physiological responses in vitro and in vivo. J Biol Chem. 2011;286(21):19076–19088. doi: 10.1074/jbc.M110.185264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hassler SN, et al. Protease activated receptor 2 (PAR2) activation causes migraine-like pain behaviors in mice. Cephalalgia. 2019;39(1):111–122. doi: 10.1177/0333102418779548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tillu DV, et al. Protease-activated receptor 2 activation is sufficient to induce the transition to a chronic pain state. Pain. 2015;156(5):859–867. doi: 10.1097/j.pain.0000000000000125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Shimada SG, LaMotte RH. Behavioral differentiation between itch and pain in mouse. Pain. 2008;139(3):681–687. doi: 10.1016/j.pain.2008.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Cevikbas F, et al. A sensory neuron-expressed IL-31 receptor mediates T helper cell-dependent itch: Involvement of TRPV1 and TRPA1. J Allergy Clin Immunol. 2014;133(2):448–460. doi: 10.1016/j.jaci.2013.10.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Caterina MJ, Schumacher MA, Tominaga M, Rosen TA, Levine JD, Julius D. The capsaicin receptor: a heat-activated ion channel in the pain pathway. Nature. 1997;389(6653):816–824. doi: 10.1038/39807. [DOI] [PubMed] [Google Scholar]
- 32.Tominaga M, et al. The cloned capsaicin receptor integrates multiple pain-producing stimuli. Neuron. 1998;21(3):531–543. doi: 10.1016/s0896-6273(00)80564-4. [DOI] [PubMed] [Google Scholar]
- 33.Caterina MJ, et al. Impaired nociception and pain sensation in mice lacking the capsaicin receptor. Science. 2000;288(5464):306–313. doi: 10.1126/science.288.5464.306. [DOI] [PubMed] [Google Scholar]
- 34.Amadesi S, et al. Protease-activated receptor 2 sensitizes the capsaicin receptor transient receptor potential vanilloid receptor 1 to induce hyperalgesia. J Neurosci. 2004;24(18):4300–4312. doi: 10.1523/JNEUROSCI.5679-03.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Dai Y, et al. Proteinase-activated receptor 2-mediated potentiation of transient receptor potential vanilloid subfamily 1 activity reveals a mechanism for proteinase-induced inflammatory pain. J Neurosci. 2004;24(18):4293–4299. doi: 10.1523/JNEUROSCI.0454-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Amadesi S, et al. Protease-activated receptor 2 sensitizes TRPV1 by protein kinase Cepsilon- and A-dependent mechanisms in rats and mice. J Physiol (Lond) 2006;575(pt 2):555–571. doi: 10.1113/jphysiol.2006.111534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Akiyama T, Merrill AW, Zanotto K, Carstens MI, Carstens E. Scratching behavior and Fos expression in superficial dorsal horn elicited by protease-activated receptor agonists and other itch mediators in mice. J Pharmacol Exp Ther. 2009;329(3):945–951. doi: 10.1124/jpet.109.152256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Akiyama T, Lerner EA, Carstens E. Protease-activated receptors and itch. Handb Exp Pharmacol. 2015;226:219–235. doi: 10.1007/978-3-662-44605-8_13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Pagani M, et al. How gastrin-releasing peptide opens the spinal gate for itch. Neuron. 2019;103(1):102–117.e5. doi: 10.1016/j.neuron.2019.04.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Petitjean H, Séguéla P, Sharif-Naeini R. Bursting enables GRP neurons to engage spinal itch circuits. Neuron. 2019;103(1):5–7. doi: 10.1016/j.neuron.2019.06.009. [DOI] [PubMed] [Google Scholar]
- 41.Han L, et al. Mrgprs on vagal sensory neurons contribute to bronchoconstriction and airway hyper-responsiveness. Nat Neurosci. 2018;21(3):324–328. doi: 10.1038/s41593-018-0074-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Reichling DB, Levine JD. Critical role of nociceptor plasticity in chronic pain. Trends Neurosci. 2009;32(12):611–618. doi: 10.1016/j.tins.2009.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kandasamy R, Price TJ. The pharmacology of nociceptor priming. Handb Exp Pharmacol. 2015;227:15–37. doi: 10.1007/978-3-662-46450-2_2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Moy JK, et al. The MNK-eIF4E signaling axis contributes to injury-induced nociceptive plasticity and the development of chronic pain. J Neurosci. 2017;37(31):7481–7499. doi: 10.1523/JNEUROSCI.0220-17.2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Moy JK, Khoutorsky A, Asiedu MN, Dussor G, Price TJ. eIF4E phosphorylation influences Bdnf mRNA translation in mouse dorsal root ganglion neurons. Front Cell Neurosci. 2018;12:29. doi: 10.3389/fncel.2018.00029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Song H, Yao E, Lin C, Gacayan R, Chen MH, Chuang PT. Functional characterization of pulmonary neuroendocrine cells in lung development, injury, and tumorigenesis. Proc Natl Acad Sci U S A. 2012;109(43):17531–17536. doi: 10.1073/pnas.1207238109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Chaplan SR, Bach FW, Pogrel JW, Chung JM, Yaksh TL. Quantitative assessment of tactile allodynia in the rat paw. J Neurosci Methods. 1994;53(1):55–63. doi: 10.1016/0165-0270(94)90144-9. [DOI] [PubMed] [Google Scholar]
- 48.Langford DJ, et al. Coding of facial expressions of pain in the laboratory mouse. Nat Methods. 2010;7(6):447–449. doi: 10.1038/nmeth.1455. [DOI] [PubMed] [Google Scholar]
- 49.Matsumiya LC, et al. Using the Mouse Grimace Scale to reevaluate the efficacy of postoperative analgesics in laboratory mice. J Am Assoc Lab Anim Sci. 2012;51(1):42–49. [PMC free article] [PubMed] [Google Scholar]
- 50.Leach MC, Klaus K, Miller AL, Scotto di Perrotolo M, Sotocinal SG, Flecknell PA. The assessment of post-vasectomy pain in mice using behaviour and the Mouse Grimace Scale. PLoS One. 2012;7(4):e35656. doi: 10.1371/journal.pone.0035656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Akintola T, Raver C, Studlack P, Uddin O, Masri R, Keller A. The grimace scale reliably assesses chronic pain in a rodent model of trigeminal neuropathic pain. Neurobiol Pain. 2017;2:13–17. doi: 10.1016/j.ynpai.2017.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Tuttle AH, et al. A deep neural network to assess spontaneous pain from mouse facial expressions. Mol Pain. 2018;14:1744806918763658. doi: 10.1177/1744806918763658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Hargreaves K, Dubner R, Brown F, Flores C, Joris J. A new and sensitive method for measuring thermal nociception in cutaneous hyperalgesia. Pain. 1988;32(1):77–88. doi: 10.1016/0304-3959(88)90026-7. [DOI] [PubMed] [Google Scholar]
- 54.Patil MJ, Hovhannisyan AH, Akopian AN. Characteristics of sensory neuronal groups in CGRP-cre-ER reporter mice: comparison to Nav1.8-cre, TRPV1-cre and TRPV1-GFP mouse lines. PLoS One. 2018;13(6):e0198601. doi: 10.1371/journal.pone.0198601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Butler A, Hoffman P, Smibert P, Papalexi E, Satija R. Integrating single-cell transcriptomic data across different conditions, technologies, and species. Nat Biotechnol. 2018;36(5):411–420. doi: 10.1038/nbt.4096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Gribov A, et al. SEURAT: visual analytics for the integrated analysis of microarray data. BMC Med Genomics. 2010;3:21. doi: 10.1186/1755-8794-3-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.