Abstract
BACKGROUND
Epidemiologic studies suggest that metformin has antitumor effects. Laboratory studies indicate metformin impacts cancer stem-like cells (CSCs). As part of a phase II trial, we evaluated the impact of metformin on CSC number and on carcinoma-associated mesenchymal stem cells (CA-MSCs) and clinical outcomes in nondiabetic patients with advanced-stage epithelial ovarian cancer (EOC).
METHODS
Thirty-eight patients with stage IIC (n = 1)/III (n = 25)/IV (n = 12) EOC were treated with either (a) neoadjuvant metformin, debulking surgery, and adjuvant chemotherapy plus metformin or (b) neoadjuvant chemotherapy and metformin, interval debulking surgery, and adjuvant chemotherapy plus metformin. Metformin-treated tumors, compared with historical controls, were evaluated for CSC number and chemotherapy response. Primary endpoints were (a) a 2-fold or greater reduction in aldehyde dehydrogenase–positive (ALDH+) CD133+ CSCs and (b) a relapse-free survival at 18 months of more than 50%.
RESULTS
Metformin was well tolerated. Median progression-free survival was 18.0 months (95% CI 14.0–21.6) with relapse-free survival at 18 months of 59.3% (95% CI 38.6–70.5). Median overall survival was 57.9 months (95% CI 28.0–not estimable). Tumors treated with metformin had a 2.4-fold decrease in ALDH+CD133+ CSCs and increased sensitivity to cisplatin ex vivo. Furthermore, metformin altered the methylation signature in CA-MSCs, which prevented CA-MSC–driven chemoresistance in vitro.
CONCLUSION
Translational studies confirm an impact of metformin on EOC CSCs and suggest epigenetic change in the tumor stroma may drive the platinum sensitivity ex vivo. Consistent with this, metformin therapy was associated with better-than-expected overall survival, supporting the use of metformin in phase III studies.
TRIAL REGISTRATION
ClinicalTrials.gov NCT01579812.
Keywords: Oncology
Keywords: Human stem cells
A phase-II translational study demonstrates that metformin treatment is associated with a reduction in cancer stem-like cells and a maintenance of chemosensitivity.
Introduction
It is estimated that in 2020, 21,750 women will be diagnosed with epithelial ovarian cancer (EOC) and 13,940 will die of ovarian cancer (1). While antiangiogenic therapy has offered improvement in progression-free survival (PFS), a significant overall survival (OS) advantage is unclear (2, 3). Similarly, poly (ADP-ribose) polymerase inhibitors have demonstrated an impressive increase in PFS in BRCA mutation carriers, but OS data are not mature (4). Furthermore, these therapeutic approaches are extremely costly (5–9). Thus new, cost-effective approaches are needed to reduce relapse rates for patients with EOC and improve OS. Metformin, a low-cost modulator of cellular metabolism, represents one potential approach.
Preclinically, metformin has demonstrated antitumor effects in several cancers (10, 11). Epidemiologic studies, while not in complete agreement, have indicated that patients with ovarian cancer taking metformin compared with patients not taking metformin have a significantly longer OS (12–16).
Many mechanisms of metformin’s anticancer activity have been proposed. Several studies have suggested metformin modulates AMPK signaling, AKT activity, and the induction of apoptosis (17, 18). Metabolic actions have been proposed related to gluconeogenesis, mitochondrial function, and cellular metabolism (19, 20). Metformin has been reported to inhibit epithelial-mesenchymal transition (EMT), inhibit IGF signaling, and selectively suppress cancer stem-like cell (CSC) growth (21–25). In ovarian cancer, metformin is reported to reverse chemotherapy resistance, reduce cancer cell migration and metastasis, and prevent EMT (17, 20, 26–28). We reported that metformin targets aldehyde dehydrogenase–positive (ALDH+) ovarian CSCs (29, 30) and enhances response to chemotherapy (31).
Currently, at least 55 clinical trials are evaluating metformin as a cancer treatment (32). Here, we present results of a nonrandomized phase II study of metformin administered in combination with chemotherapy for nondiabetic patients with advanced-stage EOC. The primary objective of this study was a translation endpoint to evaluate the impact of metformin on CSCs and 18-month relapse-free survival (RFS).
Results
Patient population
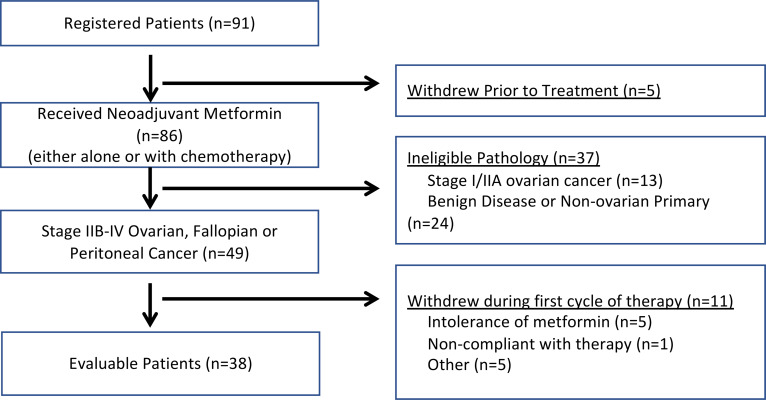
Ninety-one patients were enrolled in the study between October 28, 2011, and March 8, 2016. Study design was as indicated (Figure 1). Five patients withdrew consent before treatment initiation related to timing/location of surgery. Eighty-six patients were treated with metformin before surgery. Of these patients, 37 were excluded due to ineligible pathology (24 had benign disease or nonovarian carcinoma, and 13 had stage I/IIA ovarian carcinoma). An additional 10 were nonevaluable due to withdrawn consent for intolerance of metformin, personal issues (e.g., moving to other cities), or noncompliance (Figure 2). Target accrual was 50 evaluable patients; however — due to the unexpectedly high number of patients with early-stage disease who were enrolled but ultimately not eligible, an unexpectedly high number of nonovarian malignancies, and limited financial support — the trial closed early.
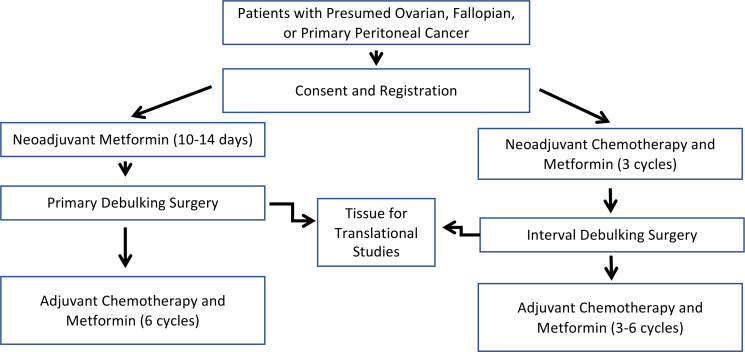
Figure 1. Clinical study trial design.
Patients were treated with metformin before debulking surgery either for 2 weeks or along with 3 cycles of neoadjuvant chemotherapy. Surgical specimens were used for later CSC studies. Metformin was continued along with adjuvant chemotherapy for a total of 6 cycles.
Figure 2. Enrollment and exclusion in the clinical trial.
Of 91 patients enrolled, 38 were evaluable. Excluded patients included those who withdrew before trial initiation, were ineligible due to benign or lower stage pathology, or withdrew after treatment was initiated but before receiving chemotherapy due to intolerance of metformin, noncompliance, or other reasons, such as moving or choosing to receive adjuvant treatment at another institution.
Table 1 summarizes patient characteristics. Twenty-five (65.8%) had stage III disease, and 12 (31.6%) had stage IV disease. Twenty-three patients (60.5%) received neoadjuvant chemotherapy; 15 (39.5%) received adjuvant therapy. Thirty (78.9%) achieved optimal debulking status (<1 cm residual disease). Twenty-seven (71.1%) had platinum-sensitive tumor. Two of the 23 (8.7%) patients receiving neoadjuvant chemotherapy and metformin achieved pathologic complete responses with no evidence of residual disease at interval debulking surgery. Thirty-two patients (84.2%) completed at least 6 cycles of chemotherapy with metformin. Two patients chose to continue metformin therapy (off-label, provided by their internist) after completion of the study.
Table 1. Clinical characteristics of enrolled patients.

Safety
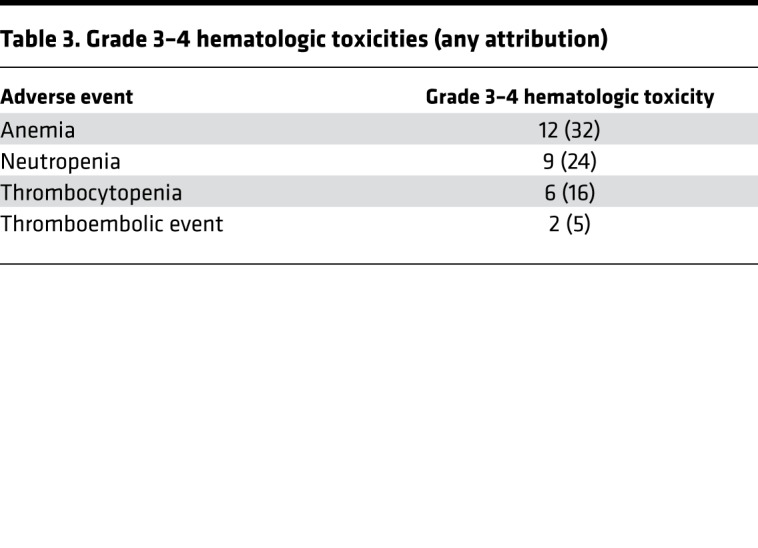
Table 2 and Table 3 detail toxicities potentially related to metformin. A severe dermatologic reaction was the only grade 4, nonhematologic toxicity. One patient had grade 3 diarrhea. The anticipated metformin-associated diarrhea and nausea were the most common side effects. Five of 16 patients treated at 1000 mg BID withdrew from the trial due to metformin-related gastrointestinal (GI) side effects. Subsequently, patients were enrolled with target doses of 500 mg BID. Three patients were dose-reduced to 500 mg/daily secondary to GI side effects. All patients who experienced nausea were taking 1000 mg BID. Hepatologic adverse effects included mild elevation in AST and ALT. Anticipated rates of hematologic and neurologic toxicity were observed.
Table 2. Metformin-attributed adverse events.
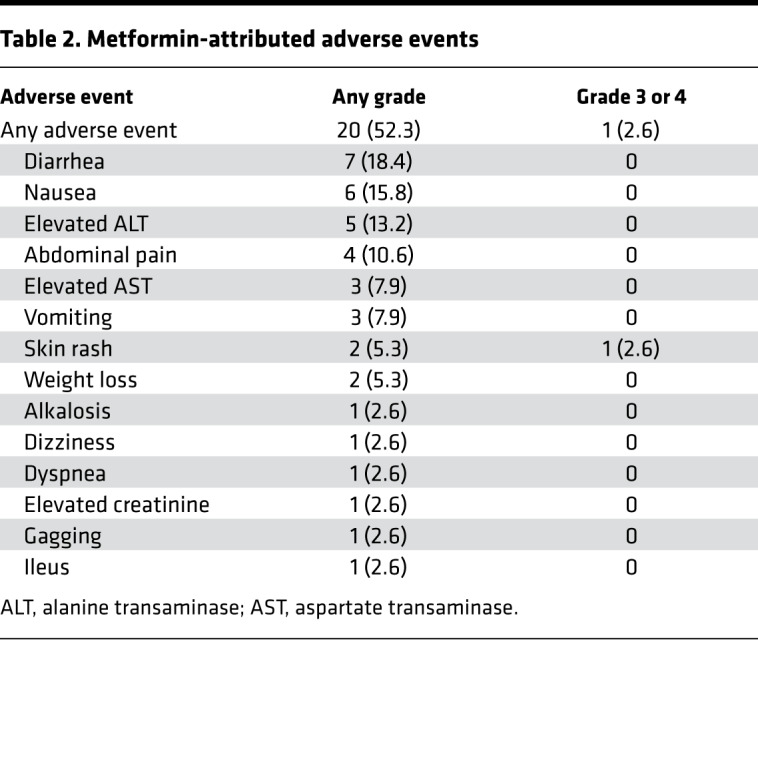
Table 3. Grade 3–4 hematologic toxicities (any attribution).
Efficacy
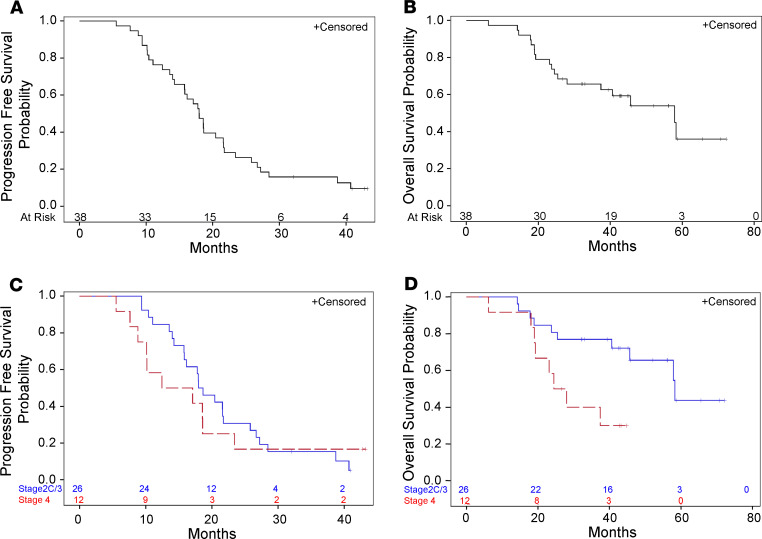
The data cutoff for primary analysis was January 22, 2018. With a median follow-up of 45 months, the median PFS for the entire population was 18.0 months (95% CI 14.0–21.6) (Figure 3A). For the 27 patients with nonpersistent disease after therapy, RFS at 18 months was 59.3% (95% CI 38.6–70.5). Median OS was 57.9 months (95% CI 28.0–not estimable [NE]; Figure 3B), with a 3-year OS of 65.7% (95% CI 48.3–78.4) (Figure 3B). Comparing by stage, those with stage IIC/III disease had a median PFS of 18.3 months (95% CI 5.8–21.7), while patients with stage IV disease had a median PFS of 14.8 months (95% CI 7.6–23.4) (Figure 3C). Median OS by stage was 58.0 months (95% CI 44–NE) for patients with stage IIC/III disease and 22 months (95% CI 7–NE) for patients with stage IV disease (Figure 3D).
Figure 3. Kaplan-Meier estimates of progression-free and overall survival.
(A and B) Kaplan-Meier estimates of PFS and OS for the entire population. Median PFS was 18.0 months (14.0–21.6). Median OS was 57.9 months (28.0–not estimable). (C and D) Kaplan-Meier estimates of PFS and OS by stage.
Translational studies
Metformin treatment reduces CSCs.
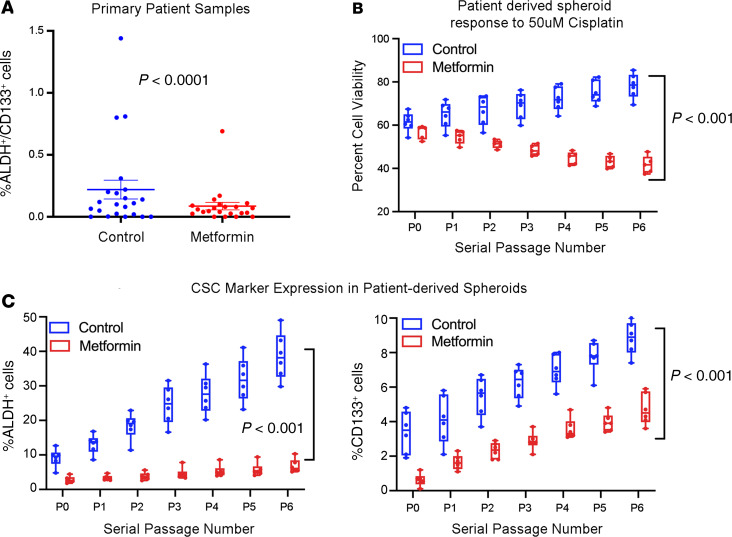
Prior studies suggest metformin can reduce cancer “stemness” (21, 25, 31). We have shown that ALDH+CD133+ ovarian cancer cells are enriched for ovarian CSCs (29). Thus, a primary endpoint of this study was to evaluate ALDH+CD133+ CSCs in the metformin-treated specimens and matched non–metformin-treated control patients. For homogeneity, we only evaluated samples from 22 patients with stage III/IV high-grade serous cancer. Controls met the eligibility criteria set forth for the trial and were consented for tissue collection via an IRB-approved tumor banking protocol. Selecting from a bank of more than 200 patient samples, 22 controls were matched to have identical stage, histology, and chemotherapy (including adjuvant vs. neoadjuvant). Average age of controls at the time of surgery was similar at 61.1 years (range 42–76). Among control patients, 75% underwent optimal debulking (15 of 20; debulking status unavailable for 2). Flow cytometry revealed metformin-treated patients exhibited an average 2.4-fold reduction in percentage of ALDH+CD133+ cells compared with non–metformin-treated ovarian cancer controls (P < 0.0001, Figure 4A).
Figure 4. Tumors treated with metformin have decreased cancer stemness.
(A) Summary of FACS analysis of ALDH+CD133+ CSCs in metformin-treated (n = 22) and matched control ovarian cancers (n = 22) demonstrating a 2.4-fold decrease in CSCs in metformin-treated tumors. (B) Cell viability of tumor cells from metformin-treated patients (n = 6) or control patients (n = 7) grown in suspension, passaged weekly, and treated with cisplatin (5 replicates each). Tumor cells from metformin-treated patients maintain platinum sensitivity with serial passage, while control tumor cells increase platinum resistance over time. (C) Evaluation of ALDH and CD133 expression in metformin-treated and control tumor cells grown in suspension and after serial passages. Metformin-treated samples start at a lower baseline and increase less over time relative to controls. Lines in boxes represent averages. The whiskers depict the minimum and maximum values, and the length of the box represents the interquartile range. Statistical significance between passages was assessed with 2-sided Student’s t tests and comparisons made between metformin and nonmetformin samples at each passage, using 1-/2-way ANOVA and Tukey’s post hoc analysis to determine specific significant differences (P < 0.05). All data are expressed as mean ± SEM.
To further evaluate the stemness of these tumors, tumor cells from 6 metformin-treated patients and 7 controls were grown in hanging-drop suspensions in serum-free media and serially passaged. Spheroids were then analyzed for response to cisplatin therapy over time and expression of ALDH or CD133. In line with a potential reduction in stemness, metformin-treated cells were more sensitive to platinum treatment, unlike controls, and appeared not to develop therapeutic resistance with passaging (P < 0.001, Figure 4B). Consistent with the initial analysis, spheroids from metformin-treated patients initially demonstrated reduced levels of both ALDH and CD133 (Figure 4C). Furthermore, these levels stayed lower over time and, in the case of ALDH, appeared to increase less over time.
Metformin affects DNA methylation of cancer-associated MSCs in the tumor microenvironment.
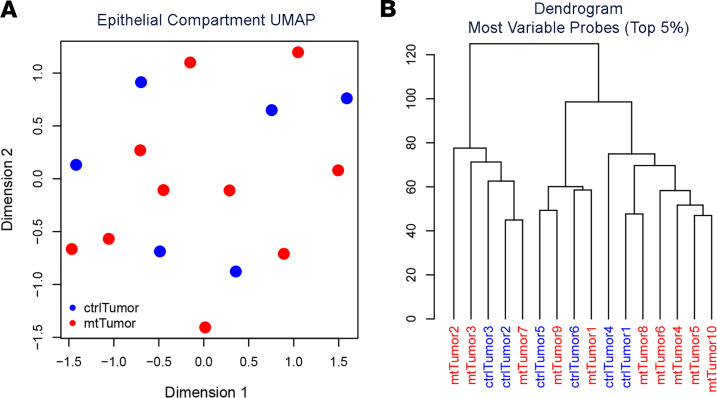
Metformin use has been associated with an immediate impact on blood cell DNA methylation (33). Metformin has also been reported to modify cancer cell DNA methylation (34–36). Given the reduction in CSC numbers and the persistence of platinum response ex vivo, we hypothesized a potential epigenetic change in cancer cells and performed DNA methylation analysis on tumor cells from control and metformin-treated tumors using the Illumina Infinium MethylationEPIC DNA methylation array. To eliminate changes associated with chemotherapy, we only used tumors that were treated with metformin neoadjuvantly in the absence of chemotherapy. Uniform manifold approximation and projection (UMAP), using all CpG sites, did not separate metformin-treated tumor cells and nontreated control tumors (Figure 5A). After correction for multiple comparisons, we were unable to identify statistically significant differentially methylated loci, using either supervised or unsupervised hierarchical clustering with the top 5% most variable sites (Figure 5B).
Figure 5. Metformin does not affect DNA methylation of bulk cancer cells.
(A) Primary 2-dimensional UMAP analysis showing intermixed metformin-treated tumor cells (mtTumor, red, n = 10) and nontreated control tumor cells (ctrlTumor, blue, n = 6). (B) Dendrogram of hierarchical clustering using the top 5% most variable CpGs in tumor cells from control and metformin-treated patients.
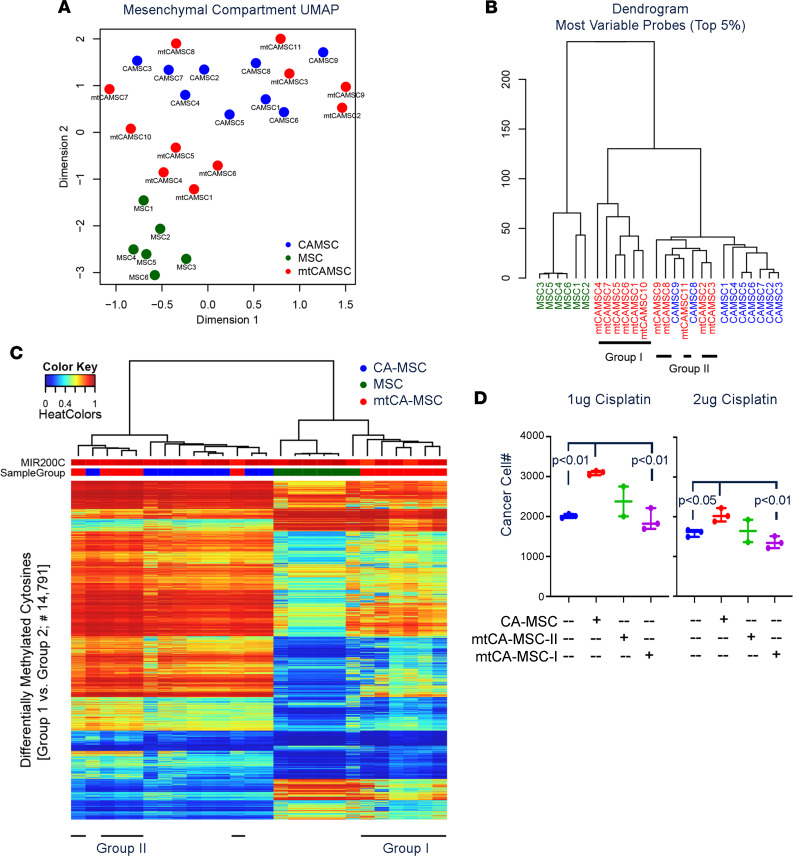
We previously demonstrated that mesenchymal stem cells (MSCs) are important members of the ovarian tumor microenvironment (TME) (37, 38) that assume a unique protumorigenic cancer-associated MSC (CA-MSC) phenotype (39). One of the protumorigenic aspects of the CA-MSCs is to enhance stemness and chemoresistance. We therefore similarly analyzed the impact of metformin on CA-MSC DNA methylation status. We profiled 3 groups of MSC samples, including MSCs derived from normal omental adipose tissue, CA-MSCs derived from high-grade serous ovarian cancer (HGSOC) omental metastasis (control-CA-MSCs), and CA-MSCs derived from metformin-treated HGSOC omental metastasis (metformin-CA-MSCs). A similar UMAP analysis as done with the tumors demonstrated that normal MSCs and control-CA-MSCs segregated into 2 distinct groups. Metformin-CA-MSCs separated into 2 groups, one overlapping control-CA-MSCs and the other creating an intermediate group between CA-MSCs and normal MSCs (Figure 6A). An unsupervised hierarchical clustering analysis on the top 5% CpG sites, selected based on variance, recapitulated the global UMAP comparison (Figure 6B). Five of the 11 metformin-CA-MSCs (group II) clustered with the control-CA-MSCs, while 6 (group I) demonstrated a distinct methylation pattern. Even with multiple-comparison correction and small sample size, we were able to identify 14,791 differentially methylated CpG sites (within 4986 differentially methylated regions) (Figure 6C) at the 5% FDR level. Group I metformin-CA-MSCs segregated with normal MSCs.
Figure 6. Metformin impacts DNA methylation of host CA-MSCs and prevents CA-MSC–induced chemoresistance.
(A) Primary 2-dimensional UMAP analysis for the MSC groups, including metformin-CA-MSCs (mtCAMSC, red, n = 11), control-CA-MSCs (CA-MSC, blue, n = 9), and normal adipose MSCs (MSC, green, n = 6). (B) Dendrogram from hierarchical clustering using the top 5% most variable CpGs for the MSCs. Metformin-CA-MSC samples were split into 2 groups as indicated. (C) DNA methylation heatmap of 14,791 probes differentially methylated between group I and group II. MIR200C promoter DNA methylation level (unmethylated in epithelial cells and methylated in mesenchymal cells) is included as a measure of CA-MSC purity. Group I metformin-CA-MSCs segregate with normal MSCs, distinct from control-CA-MSCs. (D) Viable cancer cell number following coculture with the indicated MSC types and the indicated doses of cisplatin. A Student’s 2-tailed t test was used for comparison.
To determine whether CA-MSC methylation profiles correlated with outcome, we compared the OS of patients whose CA-MSCs sorted into groups I and II. All 5 patients in group II (CA-MSC profile more like non–metformin-treated controls) were deceased. In contrast, only 3 of 6 patients in group I (patients whose CA-MSCs sorted with normal MSCs) were deceased.
To determine whether the metformin response of CA-MSCs could cause better patient outcomes, we performed in vitro chemosensitivity assays with CAOV3 ovarian cancer cell lines cocultured with control CA-MSCs (n = 3), group I metformin-CA-MSCs (n = 3), and group II metformin-CA-MSCs (n = 2). Consistent with prior studies (37), non–metformin-treated CA-MSCs significantly increased chemoresistance of ovarian cancer cells (Figure 6D). However, ovarian cancer cells cultured with group I metformin-treated CA-MSCs demonstrated no increase in chemoresistance (Figure 6D). Group II metformin-CA-MSCs had an intermediate phenotype. Taken together these data suggest metformin’s impact on CA-MSCs may prevent CA-MSC–driven chemoresistance. Supporting the idea that metformin may help maintain platinum sensitivity, of 21 platinum-sensitive patients (patients who experience recurrence at least 6 months after completion of adjuvant chemotherapy) for whom data on response to second-line therapy were available, 18 (82%) demonstrated a response to second-line therapy (11 complete response, 7 partial response, 3 progressive disease). This compares favorably with historical controls, in which a response of 50%–65% has been observed (40, 41).
Discussion
This is the first prospective study to our knowledge to evaluate metformin as a treatment in nondiabetic patients with ovarian cancer. The trial met the primary endpoint of ≥2-fold reduction of ALDH+CD133+ CSC and ≥50% RFS at 18 months; however, the RFS endpoint has a wide CI. The median PFS of 18 months and OS of 57.9 months compare favorably to other clinical trials with similar patient populations and historical expectations (2, 42).
The metformin dose of 500 mg BID was well tolerated. However, doses of 1000 mg BID were not tolerated because of adverse GI effects. This may be unique to the ovarian cancer population because patients found the GI side effects mentally distressing; they were reminiscent of patients’ presenting ovarian cancer symptoms and provoked anxiety. Consequently, many patients would not consider dose reduction of the metformin, withdrawing from the trial.
Outcome results, while encouraging, are limited because this was a nonrandomized trial. While cross-trial comparisons are obviously problematic, median PFS and OS compare favorably to the outcomes in recent landmark clinical trials with similar patient populations, such as GOG218 (the majority of patients were stage III with optimally debulked disease) and GOG262 (2, 42–47). While our study had a higher rate of optimal debulking than these 2 studies, this likely reflects the fact that 60% of the patients in the current study received neoadjuvant chemotherapy, which increases optimal debulking rates but does not affect OS (48). Of note, because bevacizumab was not approved for adjuvant therapy at the time of this study, no patients were treated with bevacizumab.
Selective targeting of CSCs is a proposed mechanism of action of metformin (27, 32, 49–51). We previously found metformin was associated with a ~2-fold reduction in ovarian CSCs in animal studies and increased in chemotherapy response (31). Exactly how metformin reduced CSC numbers was unclear. Translational studies completed as part of this clinical trial showed a very similar 2.4-fold CSC reduction in metformin-treated patients. Consistent with reduced stemness, metformin-treated patient cells demonstrated an increased sensitivity to platinum ex vivo, and unlike controls, resistance to platinum did not increase with passage. Sensitivity to platinum maintained over time could explain the modest PFS yet excellent OS of the metformin-treated population. Our study did not include metformin as a maintenance therapy for financial reasons; however, if metformin works to reduce chemotherapy resistance, continued use may further improve patient outcomes. This is being tested in an ongoing randomized phase II trial of metformin as a chemotherapy adjuvant (ClinicalTrials.gov NCT02122185).
Metformin may also affect stemness indirectly via an impact on the TME. We observed strong metformin-related DNA methylation changes in CA-MSCs of patients with good outcomes. Unlike non–metformin-treated CA-MSCs, CA-MSCs from metformin-treated tumors did not have the ability to drive chemoresistance ex vivo. Because MSCs are well established as regulators of CSCs (37, 52, 53), we speculate metformin may, in part, affect CSCs indirectly via its effect on MSCs. Because MSCs are known to be potent regulators of the immune response, a metformin impact on MSCs would be consistent with its reported immunomodulatory effects (24, 54). Improved OS without significant impact on PFS could also be attributable to an improvement in antitumor immune response.
Although our translational findings are of interest, they are limited in interpretation because this was not a randomized trial. However, control tumors were well-matched contemporaries, from the same institution. Our studies are consistent with a randomized phase II trial in lung cancer of an EGFR inhibitor with or without metformin, which demonstrated statistically significant PFS improvement with an impressive 14-month improvement in OS (1). In contrast, clinical trials of metformin in pancreatic cancer did not show a survival benefit (55). If metformin acts to prevent the development of chemoresistance, it is unlikely to have an impact on pancreatic cancer, which, unlike ovarian and lung cancer, is rather therapy resistant on primary presentation. Further, if metformin acts via conversion of MSCs to CA-MSCs, studies of metformin in pancreatic cancer, where the tumor stroma is well established, are less likely to have positive results. Consistent with this, for patients with stage IV ovarian cancer, we observed OS consistent with historical controls whereas, for stage III patients, we observed a better-than-expected OS.
In conclusion, we demonstrated that metformin is well tolerated in nondiabetic patients. Metformin treatment resulted in a significant reduction in the CSC population and alteration of DNA methylation of CA-MSCs, which eliminated CA-MSC–driven increases in chemoresistance. This was associated with a better-than-expected median OS, particularly for patients with stage II–III disease. This study strongly supports the use of metformin in phase III clinical trials for adjuvant treatment of EOC.
Methods
Study design and eligibility criteria.
This was a single-center, open-label phase II trial of patients with a new diagnosis of confirmed advanced-stage EOC. All patients gave written informed consent before participation in the study. Progression was defined using Gynecologic Cancer InterGroup–Response Evaluation Criteria in Solid Tumors criteria (56). Primary endpoints were an 18-month median RFS and 2-fold reduction in CSCs versus non–metformin-treated historical control samples. PFS and OS were secondary endpoints. Survival intervals were defined from the date of diagnosis to the date of first evidence of progression or death from any cause.
Eligible patients had evidence of malignancy consistent with stage IIC, III, or IV ovarian, fallopian, or primary peritoneal cancer (57); ECOG performance status 0–2; age 18–80 years; and intact renal (serum creatinine ≤ 1.4 mg/dL) and hepatic function (bilirubin ≤ 1.5 times the upper limit of normal and AST and ALT ≤ 2.5 times the upper limit of normal). Patients having a diagnosis of diabetes mellitus, metformin use in the preceding 6 months, hypersensitivity to metformin, or history of metabolic acidosis were excluded, as were those with a history of other active malignancies.
Procedures.
Patients were initially treated with oral metformin 500 mg BID for 7 days and then increased to 1000 mg twice daily. However, after an initial high dropout rate (n = 5 of the first 16) due to metformin-related GI side effects, the remaining patients were enrolled with target dose of 500 mg twice daily. Patients either received 7–10 days of metformin before primary debulking surgery, followed by at least 6 cycles of adjuvant metformin and platinum/taxane chemotherapy or 3 cycles of neoadjuvant metformin plus platinum/taxane chemotherapy, followed by interval debulking and 3–6 cycles of adjuvant platinum/taxane chemotherapy with metformin (Figure 1). Patients were consented and started metformin the same day they were evaluated for surgery to ensure receiving metformin neoadjuvantly did not delay patients undergoing surgery. Patients were treated with either standard Q3 week carboplatin (AUC = 6) and paclitaxel (175 mg/m2) or Q3 week carboplatin (AUC = 6) and weekly taxol (80 mg/m2) per the physician’s preference. The decision to treat patients with initial surgery or neoadjuvant chemotherapy was up to the treating surgeon, based on perceived ability to perform optimal debulking. Pill counts and patient journals were assessed after every cycle to confirm treatment compliance. Metformin was discontinued after completion of the study.
Safety and toxicity monitoring.
Patients were evaluated for toxicity at enrollment and with each therapeutic cycle. Adverse events were graded according to Common Terminology Criteria for Adverse Events (version 4.0) (58). The number and proportion of the highest graded toxicity for each category were reported. Patients were withdrawn from the study for serious metformin-related nonhematologic adverse events (grade 3 or greater). Dose reduction was allowed for low-grade toxicity.
CSC studies.
Tumor samples were processed into live single-cell suspensions (59) and frozen for batch analysis. The proportion of ALDH1+CD133+ CSCs was evaluated via flow cytometry as previously described (60–63).
Hanging-drop spheroids.
Tumor cell suspensions from 6 neoadjuvant metformin-treated patients and 7 control patients were gown as previously described in 5 replicate plates (60). After 7 days, spheroids were harvested and disaggregated and single cells counted and replated on a new hanging-drop plate as drops of 100 cells, to form passage 1. This process was repeated weekly for 6 passages. Live-cell phase microscopy images of the spheroids were collected (Olympus IX81 and CellSens software) on days 1, 3, and 7 of culture for each passage to monitor spheroid formation/proliferation (60–63). At the time of disaggregation, a portion of cells was used for flow cytometry analysis of ALDH and CD133 as above. Seven-day-old spheroids were treated with cisplatin at a concentration of 50 μM. The effect of drug treatment was determined at 72 hours, using the Alamar blue assay as described (60–62).
DNA methylation profiling.
Tumor cell suspensions were washed 3 times to eliminate dead cells, and DNA was collected using the DNeasy column-based purification kit (QIAGEN). Patient-derived CA-MSCs and normal omental MSCs were cultured as described (37–39), then FACS-sorted for the cell surface markers CD105, CD90, and CD73, with genomic DNA isolated as above. DNA methylation profiling was performed using the Infinium MethylationEPIC BeadChip Kit (Illumina). Raw IDAT files were processed using R package SeSAMe (64) with noob background correction (65), nonlinear dye bias correction, and nondetection masking. A beta value was calculated for each locus as the ratio of methylated signal intensity to the sum of unmethylated and methylated signal intensities, with a range from 0 to 1, corresponding to the fraction of methylated allele in the assayed sample at this locus. We masked measurements from suboptimally designed probes due to overlapping with SNPs and mapping issues (66). Dimensionality reduction for visualizing sample similarities was achieved by UMAP, arXiv: 1802.03426, using all CpG sites assayed. Hierarchical clustering was done with the R function hclust (67), based on the top 5% most variable CpG probes, and performed alongside the UMAPs. Differential methylation analysis was conducted using R package DMRcate (68), version 1.8.6, with default significance cutoff settings (FDR controlled at 5% with the Benjamini-Hochberg procedure for multiple corrections).
MSC–cancer cell coculture chemosensitivity assay.
Assays were performed as previously described (37). Briefly, GFP-labeled CAOV3 cells (ATCC) were cocultured with patient-derived MSCs, 1:1 ratio for 24 hours (10,000 cancer cells:10,000 MSCs), in a 1:1 mixture of DMEM–10% FBS and complete MSC media, then treated with 1 μg or 2 μg of cisplatin for 48 hours. Cells were harvested and viable (DAPI–) CAOV3-GFP cells were counted, using flow cytometry with constant time and volume across samples.
Statistics.
The study was powered for 2 objectives, CSC number and PFS at 18 months. Based on prior analysis, we assumed an average CSC number of 3% and hypothesized, from preliminary data, that metformin would reduce CSC number to 1.5%. To calculate power, the scale of the measurements was transformed to angular equivalents, using an adaptation of the arcsine transformation of the square root of the proportion, in order to normalize the otherwise skewed distribution. Assuming mild variability, an assumption formalized by setting the standard deviation equal to half the mean for each group, with 50 treated and 50 control cases, using a 2-sample, 2-sided Student’s t test would yield over 90% power to detect the hypothesized difference significantly, with at most 5% type I error.
PFS at 18 months was powered using a 1-sided, 1-sample exact binomial test (DSTPLAN, Version 4.2). Allowing at most 2.5% type I error, 49 patients were needed to achieve 80% power to detect 20% change in PFS. OS was a secondary endpoint. Time-to-event endpoints, OS and PFS, were estimated using the product-limit method of Kaplan and Meier, with overall median study-wide follow-up time estimated using reverse censoring for survival.
Translational studies statistics.
Spheroid studies were repeated 5 times, with at least 40 spheroids (technical replicates) interrogated for each analysis at each time point and at least 6 patient samples. Spheroid proliferation at day 7 was normalized to day 1 for each specimen. Normalized viability is expressed as percentage of untreated controls. Statistical significance between passages was assessed with 2-sided Student’s t tests and comparisons made between metformin and nonmetformin samples at each passage, using 1-/2-way ANOVA and Tukey’s post hoc analysis to determine specific significant differences (P < 0.05). Three independent flow cytometry analyses were performed to identify an average percentage of ALDH+ or CD133+ populations. All data are expressed as mean ± SEM. Statistical analyses were performed using Prism 7 (GraphPad) and SAS version 9.4.
Study approval.
The study (protocol HUM00047900, ClinicalTrials.gov identifier NCT01579812) received the approval of the University of Michigan Rogel Comprehensive Cancer Center IRB. All patients gave written informed consent before participation in the study.
Author contributions
JRB, JJS, and LC acquired data. RKR, CJ, KM, FM, SU, JRL, BCO, LC, and LGC performed patient enrollment, tissue acquisition, and patient care. KAG, HF, and HS performed biostatistical and bioinformatic analysis. DKC, SET, LGC, RS, KY, PM, MB, GM, and RJB performed laboratory experimentation. JRB, DKC, SET, KAG, LGC, and RJB designed the study and prepared the manuscript.
Supplementary Material
Acknowledgments
This work was supported by the University of Michigan Clinical and Translational Science Award, the A. Alfred Taubman Medical Research Institute, and NIH R01-5R01CA211913. This project used the UPMC Hillman Cancer Center Cancer Genomics Facility, which is supported in part by award P30CA047904.
Version 1. 05/05/2020
In-Press Preview
Version 2. 06/04/2020
Electronic publication
Funding Statement
This work was supported by the University of Michigan CTSA, the Taubman Institute, and the Magee-Womens Research Institute. This project used the UPMC Hillman Cancer Center Genomic Core that is supported in part by award P30CA047904.
Footnotes
Conflict of interest: The authors have declared that no conflict of interest exists.
Copyright: © 2020, American Society for Clinical Investigation.
Reference information: JCI Insight. 2020;5(11):e133247.https://doi.org/10.1172/jci.insight.133247.
Contributor Information
Daniel K. Chan, Email: chandk2@upmc.edu.
Kent A. Griffith, Email: kentg@umich.edu.
Huihui Fan, Email: huihui.fan@vai.org.
Robert Szulawski, Email: rsupmc27@gmail.com.
Kun Yang, Email: kunyang@med.umich.edu.
R. Kevin Reynolds, Email: rkr@med.umich.edu.
Carolyn Johnston, Email: johnstob@med.umich.edu.
Karen McLean, Email: karenmcl@med.umich.edu.
Shitanshu Uppal, Email: uppal@umich.edu.
J. Rebecca Liu, Email: rliu@med.umich.edu.
Lourdes Cabrera, Email: lcabrera@med.umich.edu.
Brian C. Orr, Email: orrbc2@upmc.edu.
Francesmary Modugno, Email: fm@cs.cmu.edu.
Pooja Mehta, Email: mpooja@umich.edu.
Michael Bregenzer, Email: mbregenz@umich.edu.
Geeta Mehta, Email: mehtagee@umich.edu.
Hui Shen, Email: hui.shen@vai.org.
Ronald J. Buckanovich, Email: buckanovichrj@mwri.magee.edu.
References
- 1.Siegel RL, Miller KD, Jemal A. Cancer statistics, 2020. CA Cancer J Clin. 2020;70(1):7–30. doi: 10.3322/caac.21590. [DOI] [PubMed] [Google Scholar]
- 2.Burger RA, et al. Incorporation of bevacizumab in the primary treatment of ovarian cancer. N Engl J Med. 2011;365(26):2473–2483. doi: 10.1056/NEJMoa1104390. [DOI] [PubMed] [Google Scholar]
- 3.Monk BJ, Pujade-Lauraine E, Burger RA. Integrating bevacizumab into the management of epithelial ovarian cancer: the controversy of front-line versus recurrent disease. Ann Oncol. 2013;24(suppl 10):x53–x58. doi: 10.1093/annonc/mdt472. [DOI] [PubMed] [Google Scholar]
- 4.Moore K, et al. Maintenance olaparib in patients with newly diagnosed advanced ovarian cancer. N Engl J Med. 2018;379(26):2495–2505. doi: 10.1056/NEJMoa1810858. [DOI] [PubMed] [Google Scholar]
- 5.Chan JK, et al. Bevacizumab in treatment of high-risk ovarian cancer--a cost-effectiveness analysis. Oncologist. 2014;19(5):523–527. doi: 10.1634/theoncologist.2013-0322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hinde S, Epstein D, Cook A, Embleton A, Perren T, Sculpher M. The cost-effectiveness of bevacizumab in advanced ovarian cancer using evidence from the ICON7 trial. Value Health. 2016;19(4):431–439. doi: 10.1016/j.jval.2016.01.013. [DOI] [PubMed] [Google Scholar]
- 7.Mehta DA, Hay JW. Cost-effectiveness of adding bevacizumab to first line therapy for patients with advanced ovarian cancer. Gynecol Oncol. 2014;132(3):677–683. doi: 10.1016/j.ygyno.2014.01.021. [DOI] [PubMed] [Google Scholar]
- 8.Zhong L, Tran AT, Tomasino T, Nugent E, Smith JA. Cost-effectiveness of niraparib and olaparib as maintenance therapy for patients with platinum-sensitive recurrent ovarian cancer. J Manag Care Spec Pharm. 2018;24(12):1219–1228. doi: 10.18553/jmcp.2018.24.12.1219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Smith HJ, Walters Haygood CL, Arend RC, Leath CA, Straughn JM. PARP inhibitor maintenance therapy for patients with platinum-sensitive recurrent ovarian cancer: a cost-effectiveness analysis. Gynecol Oncol. 2015;139(1):59–62. doi: 10.1016/j.ygyno.2015.08.013. [DOI] [PubMed] [Google Scholar]
- 10.Deng J, et al. Novel application of metformin combined with targeted drugs on anticancer treatment. Cancer Sci. 2019;110(1):23–30. doi: 10.1111/cas.13849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Coyle C, Cafferty FH, Vale C, Langley RE. Metformin as an adjuvant treatment for cancer: a systematic review and meta-analysis. Ann Oncol. 2016;27(12):2184–2195. doi: 10.1093/annonc/mdw410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kumar S, et al. Metformin intake is associated with better survival in ovarian cancer: a case-control study. Cancer. 2013;119(3):555–562. doi: 10.1002/cncr.27706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wang SB, Lei KJ, Liu JP, Jia YM. Continuous use of metformin can improve survival in type 2 diabetic patients with ovarian cancer: a retrospective study. Medicine (Baltimore) 2017;96(29):e7605. doi: 10.1097/MD.0000000000007605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Urpilainen E, et al. Prognosis of ovarian cancer in women with type 2 diabetes using metformin and other forms of antidiabetic medication or statins: a retrospective cohort study. BMC Cancer. 2018;18(1):767. doi: 10.1186/s12885-018-4676-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bodmer M, Becker C, Meier C, Jick SS, Meier CR. Use of metformin and the risk of ovarian cancer: a case-control analysis. Gynecol Oncol. 2011;123(2):200–204. doi: 10.1016/j.ygyno.2011.06.038. [DOI] [PubMed] [Google Scholar]
- 16.Bakhru A, Buckanovich RJ, Griggs JJ. The impact of diabetes on survival in women with ovarian cancer. Gynecol Oncol. 2011;121(1):106–111. doi: 10.1016/j.ygyno.2010.12.329. [DOI] [PubMed] [Google Scholar]
- 17.Galdieri L, Gatla H, Vancurova I, Vancura A. Activation of AMP-activated protein kinase by metformin induces protein acetylation in prostate and ovarian cancer cells. J Biol Chem. 2016;291(48):25154–25166. doi: 10.1074/jbc.M116.742247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gotlieb WH, et al. In vitro metformin anti-neoplastic activity in epithelial ovarian cancer. Gynecol Oncol. 2008;110(2):246–250. doi: 10.1016/j.ygyno.2008.04.008. [DOI] [PubMed] [Google Scholar]
- 19.Lengyel E, et al. Metformin inhibits ovarian cancer growth and increases sensitivity to paclitaxel in mouse models. Am J Obstet Gynecol. 2015;212(4):479.e1–479.e10. doi: 10.1016/j.ajog.2014.10.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Liu X, Romero IL, Litchfield LM, Lengyel E, Locasale JW. Metformin targets central carbon metabolism and reveals mitochondrial requirements in human cancers. Cell Metab. 2016;24(5):728–739. doi: 10.1016/j.cmet.2016.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bao B, Azmi AS, Ali S, Zaiem F, Sarkar FH. Metformin may function as anti-cancer agent via targeting cancer stem cells: the potential biological significance of tumor-associated miRNAs in breast and pancreatic cancers. Ann Transl Med. 2014;2(6):59. doi: 10.3978/j.issn.2305-5839.2014.06.05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gadducci A, Biglia N, Tana R, Cosio S, Gallo M. Metformin use and gynecological cancers: a novel treatment option emerging from drug repositioning. Crit Rev Oncol Hematol. 2016;105:73–83. doi: 10.1016/j.critrevonc.2016.06.006. [DOI] [PubMed] [Google Scholar]
- 23.Sarfstein R, Friedman Y, Attias-Geva Z, Fishman A, Bruchim I, Werner H. Metformin downregulates the insulin/IGF-I signaling pathway and inhibits different uterine serous carcinoma (USC) cells proliferation and migration in p53-dependent or -independent manners. PLoS One. 2013;8(4):e61537. doi: 10.1371/journal.pone.0061537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hirsch HA, Iliopoulos D, Struhl K. Metformin inhibits the inflammatory response associated with cellular transformation and cancer stem cell growth. Proc Natl Acad Sci U S A. 2013;110(3):972–977. doi: 10.1073/pnas.1221055110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hirsch HA, Iliopoulos D, Tsichlis PN, Struhl K. Metformin selectively targets cancer stem cells, and acts together with chemotherapy to block tumor growth and prolong remission. Cancer Res. 2009;2009(19):7507–7511. doi: 10.1158/0008-5472.CAN-09-2994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhang R, et al. Inhibitory effects of metformin at low concentration on epithelial-mesenchymal transition of CD44(+)CD117(+) ovarian cancer stem cells. Stem Cell Res Ther. 2015;6:262. doi: 10.1186/s13287-015-0249-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rattan R, Giri S, Hartmann LC, Shridhar V. Metformin attenuates ovarian cancer cell growth in an AMP-kinase dispensable manner. J Cell Mol Med. 2011;15(1):166–178. doi: 10.1111/j.1582-4934.2009.00954.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Xie Y, Peng Z, Shi M, Ji M, Guo H, Shi H. Metformin combined with p38 MAPK inhibitor improves cisplatin sensitivity in cisplatin‑resistant ovarian cancer. Mol Med Rep. 2014;10(5):2346–2350. doi: 10.3892/mmr.2014.2490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Choi YJ, et al. Identifying an ovarian cancer cell hierarchy regulated by bone morphogenetic protein 2. Proc Natl Acad Sci U S A. 2015;112(50):E6882–E6888. doi: 10.1073/pnas.1507899112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Silva IA, et al. Aldehyde dehydrogenase in combination with CD133 defines angiogenic ovarian cancer stem cells that portend poor patient survival. Cancer Res. 2011;71(11):3991–4001. doi: 10.1158/0008-5472.CAN-10-3175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Shank JJ, et al. Metformin targets ovarian cancer stem cells in vitro and in vivo. Gynecol Oncol. 2012;127(2):390–397. doi: 10.1016/j.ygyno.2012.07.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chae YK, et al. Repurposing metformin for cancer treatment: current clinical studies. Oncotarget. 2016;7(26):40767–40780. doi: 10.18632/oncotarget.8194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Elbere I, et al. Significantly altered peripheral blood cell DNA methylation profile as a result of immediate effect of metformin use in healthy individuals. Clin Epigenetics. 2018;10(1):156. doi: 10.1186/s13148-018-0593-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tang G, et al. Metformin inhibits ovarian cancer via decreasing H3K27 trimethylation. Int J Oncol. 2018;52(6):1899–1911. doi: 10.3892/ijo.2018.4343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sabit H, Abdel-Ghany SE, M Said OA, Mostafa MA, El-Zawahry M. Metformin reshapes the methylation profile in breast and colorectal cancer cells. Asian Pac J Cancer Prev. 2018;19(10):2991–2999. doi: 10.22034/APJCP.2018.19.10.2991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yu X, et al. Anti-tumor activity of metformin: from metabolic and epigenetic perspectives. Oncotarget. 2017;8(3):5619–5628. doi: 10.18632/oncotarget.13639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Coffman LG, Choi YJ, McLean K, Allen BL, di Magliano MP, Buckanovich RJ. Human carcinoma-associated mesenchymal stem cells promote ovarian cancer chemotherapy resistance via a BMP4/HH signaling loop. Oncotarget. 2016;7(6):6916–6932. doi: 10.18632/oncotarget.6870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.McLean K, et al. Human ovarian carcinoma–associated mesenchymal stem cells regulate cancer stem cells and tumorigenesis via altered BMP production. J Clin Invest. 2011;121(8):3206–3219. doi: 10.1172/JCI45273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Coffman LG, et al. Ovarian carcinoma-associated mesenchymal stem cells arise from tissue-specific normal stroma. Stem Cells. 2019;37(2):257–269. doi: 10.1002/stem.2932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Parmar MK, et al. Paclitaxel plus platinum-based chemotherapy versus conventional platinum-based chemotherapy in women with relapsed ovarian cancer: the ICON4/AGO-OVAR-2.2 trial. Lancet. 2003;361(9375):2099–2106. doi: 10.1016/S0140-6736(03)13718-X. [DOI] [PubMed] [Google Scholar]
- 41.Wagner U, et al. Final overall survival results of phase III GCIG CALYPSO trial of pegylated liposomal doxorubicin and carboplatin vs paclitaxel and carboplatin in platinum-sensitive ovarian cancer patients. Br J Cancer. 2012;107(4):588–591. doi: 10.1038/bjc.2012.307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Chan JK, et al. Weekly vs. every-3-week paclitaxel and carboplatin for ovarian cancer. N Engl J Med. 2016;374(8):738–748. doi: 10.1056/NEJMoa1505067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Katsumata N, et al. Long-term results of dose-dense paclitaxel and carboplatin versus conventional paclitaxel and carboplatin for treatment of advanced epithelial ovarian, fallopian tube, or primary peritoneal cancer (JGOG 3016): a randomised, controlled, open-label trial. Lancet Oncol. 2013;14(10):1020–1026. doi: 10.1016/S1470-2045(13)70363-2. [DOI] [PubMed] [Google Scholar]
- 44.Perren TJ, et al. A phase 3 trial of bevacizumab in ovarian cancer. N Engl J Med. 2011;365(26):2484–2496. doi: 10.1056/NEJMoa1103799. [DOI] [PubMed] [Google Scholar]
- 45.Banerjee S, et al. A multicenter, randomized trial of flat dosing versus intrapatient dose escalation of single-agent carboplatin as first-line chemotherapy for advanced ovarian cancer: an SGCTG (SCOTROC 4) and ANZGOG study on behalf of GCIG. Ann Oncol. 2013;24(3):679–687. doi: 10.1093/annonc/mds494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lindemann K, et al. First-line treatment of advanced ovarian cancer with paclitaxel/carboplatin with or without epirubicin (TEC versus TC)--a gynecologic cancer intergroup study of the NSGO, EORTC GCG and NCIC CTG. Ann Oncol. 2012;23(10):2613–2619. doi: 10.1093/annonc/mds060. [DOI] [PubMed] [Google Scholar]
- 47.Pignata S, et al. Carboplatin plus paclitaxel once a week versus every 3 weeks in patients with advanced ovarian cancer (MITO-7): a randomised, multicentre, open-label, phase 3 trial. Lancet Oncol. 2014;15(4):396–405. doi: 10.1016/S1470-2045(14)70049-X. [DOI] [PubMed] [Google Scholar]
- 48.Qin M, Jin Y, Ma L, Zhang YY, Pan LY. The role of neoadjuvant chemotherapy followed by interval debulking surgery in advanced ovarian cancer: a systematic review and meta-analysis of randomized controlled trials and observational studies. Oncotarget. 2018;9(9):8614–8628. doi: 10.18632/oncotarget.23808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Shackelford DB, et al. LKB1 inactivation dictates therapeutic response of non-small cell lung cancer to the metabolism drug phenformin. Cancer Cell. 2013;23(2):143–158. doi: 10.1016/j.ccr.2012.12.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Pierotti MA, et al. Targeting metabolism for cancer treatment and prevention: metformin, an old drug with multi-faceted effects. Oncogene. 2013;32(12):1475–1487. doi: 10.1038/onc.2012.181. [DOI] [PubMed] [Google Scholar]
- 51.Eikawa S, Nishida M, Mizukami S, Yamazaki C, Nakayama E, Udono H. Immune-mediated antitumor effect by type 2 diabetes drug, metformin. Proc Natl Acad Sci U S A. 2015;112(6):1809–1814. doi: 10.1073/pnas.1417636112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Liu S, et al. Breast cancer stem cells are regulated by mesenchymal stem cells through cytokine networks. Cancer Res. 2011;71(2):614–624. doi: 10.1158/0008-5472.CAN-10-0538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.McLean K, et al. Leukemia inhibitory factor functions in parallel with interleukin-6 to promote ovarian cancer growth. Oncogene. 2019;38(9):1576–1584. doi: 10.1038/s41388-018-0523-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Scharping NE, Menk AV, Whetstone RD, Zeng X, Delgoffe GM. Efficacy of PD-1 blockade is potentiated by metformin-induced reduction of tumor hypoxia. Cancer Immunol Res. 2017;5(1):9–16. doi: 10.1158/2326-6066.CIR-16-0103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Kordes S, et al. Metformin in patients with advanced pancreatic cancer: a double-blind, randomised, placebo-controlled phase 2 trial. Lancet Oncol. 2015;16(7):839–847. doi: 10.1016/S1470-2045(15)00027-3. [DOI] [PubMed] [Google Scholar]
- 56.Eisenhauer EA, et al. New response evaluation criteria in solid tumours: revised RECIST guideline (version 1.1) Eur J Cancer. 2009;45(2):228–247. doi: 10.1016/j.ejca.2008.10.026. [DOI] [PubMed] [Google Scholar]
- 57.Mutch DG, Prat J. 2014 FIGO staging for ovarian, fallopian tube and peritoneal cancer. Gynecol Oncol. 2014;133(3):401–404. doi: 10.1016/j.ygyno.2014.04.013. [DOI] [PubMed] [Google Scholar]
- 58. National Cancer Institute. Common terminology criteria for adverse events (CTCAE). National Cancer Institute website. https://ctep.cancer.gov/protocolDevelopment/electronic_applications/ctc.htm Updated March 27, 2020. Accessed May 4, 2020.
- 59.Pulaski HL, et al. Identifying alemtuzumab as an anti-myeloid cell antiangiogenic therapy for the treatment of ovarian cancer. J Transl Med. 2009;7:49. doi: 10.1186/1479-5876-7-49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Raghavan S, et al. Formation of stable small cell number three-dimensional ovarian cancer spheroids using hanging drop arrays for preclinical drug sensitivity assays. Gynecol Oncol. 2015;138(1):181–189. doi: 10.1016/j.ygyno.2015.04.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Raghavan S, et al. Personalized medicine-based approach to model patterns of chemoresistance and tumor recurrence using ovarian cancer stem cell spheroids. Clin Cancer Res. 2017;23(22):6934–6945. doi: 10.1158/1078-0432.CCR-17-0133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Raghavan S, Mehta P, Horst EN, Ward MR, Rowley KR, Mehta G. Comparative analysis of tumor spheroid generation techniques for differential in vitro drug toxicity. Oncotarget. 2016;7(13):16948–16961. doi: 10.18632/oncotarget.7659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Mehta P, Novak C, Raghavan S, Ward M, Mehta G. Self-renewal and CSCs in vitro enrichment: growth as floating spheres. In: Papaccio G, Desiderio V eds. Cancer Stem Cells: Methods and Protocols. New York, New York, USA: Springer New York; 2018:61–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Zhou W, Triche TJ, Laird PW, Shen H. SeSAMe: reducing artifactual detection of DNA methylation by Infinium BeadChips in genomic deletions. Nucleic Acids Res. 2018;46(20):e123. doi: 10.1093/nar/gky691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Triche TJ, Weisenberger DJ, Van Den Berg D, Laird PW, Siegmund KD. Low-level processing of Illumina Infinium DNA Methylation BeadArrays. Nucleic Acids Res. 2013;41(7):e90. doi: 10.1093/nar/gkt090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Zhou W, Laird PW, Shen H. Comprehensive characterization, annotation and innovative use of Infinium DNA methylation BeadChip probes. Nucleic Acids Res. 2017;45(4):e22. doi: 10.1093/nar/gkw967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Wang W, Shen H, Xie J, Zhou Q, Chen Y, Lu H. Bioinformatics analysis of time-series genes profiling to explore key genes affected by age in fracture healing. Mol Biol Rep. 2014;41(6):3881–3889. doi: 10.1007/s11033-014-3255-x. [DOI] [PubMed] [Google Scholar]
- 68.Peters TJ, et al. De novo identification of differentially methylated regions in the human genome. Epigenetics Chromatin. 2015;8:6. doi: 10.1186/1756-8935-8-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.