Abstract
In severe cases of coronavirus disease 2019 (COVID-19), viral pneumonia progresses to respiratory failure. Neutrophil extracellular traps (NETs) are extracellular webs of chromatin, microbicidal proteins, and oxidant enzymes that are released by neutrophils to contain infections. However, when not properly regulated, NETs have the potential to propagate inflammation and microvascular thrombosis — including in the lungs of patients with acute respiratory distress syndrome. We now report that sera from patients with COVID-19 have elevated levels of cell-free DNA, myeloperoxidase-DNA (MPO-DNA), and citrullinated histone H3 (Cit-H3); the latter 2 are specific markers of NETs. Highlighting the potential clinical relevance of these findings, cell-free DNA strongly correlated with acute-phase reactants, including C-reactive protein, D-dimer, and lactate dehydrogenase, as well as absolute neutrophil count. MPO-DNA associated with both cell-free DNA and absolute neutrophil count, while Cit-H3 correlated with platelet levels. Importantly, both cell-free DNA and MPO-DNA were higher in hospitalized patients receiving mechanical ventilation as compared with hospitalized patients breathing room air. Finally, sera from individuals with COVID-19 triggered NET release from control neutrophils in vitro. Future studies should investigate the predictive power of circulating NETs in longitudinal cohorts and determine the extent to which NETs may be novel therapeutic targets in severe COVID-19.
Keywords: Infectious disease, Inflammation
Keywords: Neutrophils
Serum levels of neutrophil extracellular traps identify COVID-19 patients with more severe respiratory disease.
Introduction
As of late May 2020, the coronavirus disease 2019 (COVID-19) pandemic has affected more than 5 million individuals from over 180 countries and has resulted in unprecedented health, social, and economic crises (1). The disease is caused by severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), manifesting with flu-like symptoms and a viral pneumonia that progresses to acute respiratory distress syndrome (ARDS) and even multiorgan failure in some individuals (2).
Elevated levels of blood neutrophils are an early indicator of SARS-CoV-2 infection, predicting severe respiratory disease and worse outcomes (3, 4). Over the past decade, our group and many others have revealed a pathogenic role for neutrophil-derived neutrophil extracellular traps (NETs) in various thrombo-inflammatory states, including sepsis (5, 6), thrombosis (7–9), and respiratory failure (10, 11). NETs are extracellular webs of DNA, histones, microbicidal proteins, and oxidant enzymes that are released by neutrophils to corral infections; however, when not properly regulated, NETs have the potential to initiate and propagate inflammation and thrombosis (12, 13). Indeed, inhibition of neutrophils and NETs is protective in various models of influenza-associated ARDS (14–17). Although it has yet to be assessed whether NETs contribute to the inflammatory storm that leads to respiratory failure in many patients with COVID-19, there is emerging evidence to implicate inflammatory cytokines, such as IL-1β and IL-6, in the COVID-19 milieu (18). Not surprisingly, NETs are intimately intertwined with both cytokines, and especially IL-1β, in many pulmonary and cardiovascular diseases (9, 19–24).
Other pandemic viruses, including influenza H1N1, SARS-CoV, and Middle East respiratory syndrome coronavirus, are associated with neutrophilic infiltration at sites of infection and development of ARDS (25, 26). The acute, exudative phase of ARDS is characterized by an exuberant immune response producing proinflammatory cytokines and chemokines, increased neutrophil infiltration and accumulation in the alveoli, and disruption of the alveolar epithelial-capillary barrier (27). Culturing neutrophils in vitro with influenza-infected lung epithelial cells triggers NETosis and augments endothelial damage by culture supernatants (14). Neutrophil-depleted mice demonstrate milder lung pathology in response to influenza infection, including lower levels of thrombomodulin, matrix metalloproteinases, and myeloperoxidase (MPO) in bronchoalveolar lavage fluid (14). At the same time, influenza-infected mice are protected by strategies that prevent NETosis including inhibition of superoxide dismutase (14) and MPO (14, 28). In patients with influenza A infection, high levels of NETs predict a poor prognosis (29).
Work to date exploring the pathophysiology of COVID-19 has focused especially on macrophages and epithelial cells, with little attention paid to neutrophils and their catalysts, checkpoints, and effector mechanisms — all of which could add actionable context to our understanding of the COVID-19 inflammatory storm. Here, as a first step toward assessing the role of NETs in COVID-19, we sought to measure various markers of NETs in sera of hospitalized patients and to determine their relationship to severity of illness.
Results
Detection of NETs in sera of COVID-19 patients.
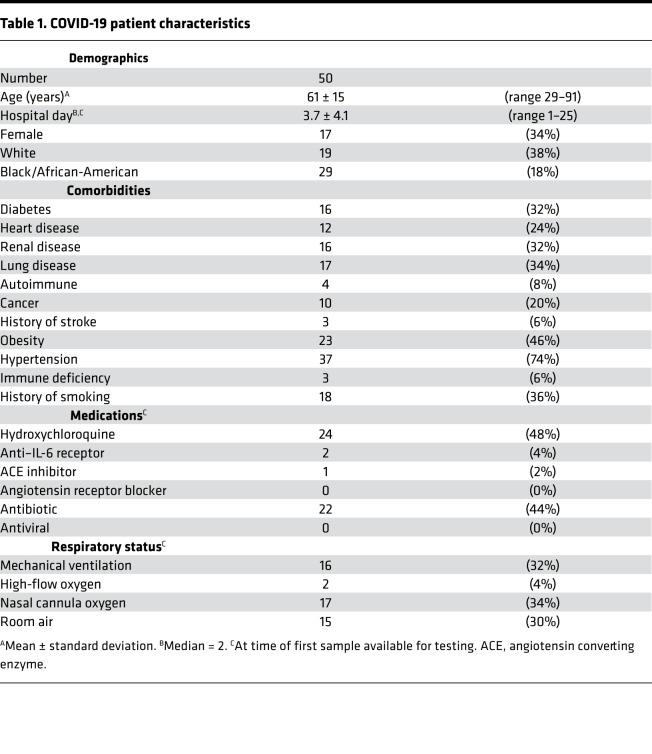
Serum samples were obtained from 50 patients hospitalized with COVID-19 at a large academic hospital (Table 1). As compared with serum samples from 30 healthy controls, the COVID-19 samples showed higher levels of cell-free DNA (Figure 1A), MPO-DNA complexes (Figure 1B), and citrullinated histone H3 (Cit-H3, Figure 1C). The latter 2 markers are generally regarded as specific for NET remnants. While cell-free DNA and MPO-DNA demonstrated a significant correlative relationship (Figure 1D), the association between cell-free DNA and Cit-H3 was not significant (Figure 1E). For a subset of the patients (n = 22), longitudinal serum samples were available. Three of those patients showed worsening oxygenation during the period of collection (room air to nasal cannula oxygen, nasal cannula oxygen to high-flow oxygen, and room air to mechanical ventilation, respectively). For all 3, markers of NETs trended up as oxygenation worsened (Supplemental Figure 1; supplemental material available online with this article; https://doi.org/10.1172/jci.insight.138999DS1). All available samples (n = 84) were included in the subsequent correlation analyses. In summary, 3 markers indicative of NET remnants were elevated in sera of patients with COVID-19 as compared with controls.
Table 1. COVID-19 patient characteristics.
Figure 1. Detection of NETs in sera of COVID-19 patients.
Sera from COVID-19 patients (n = 50) and healthy controls (n = 30) were assessed for cell-free DNA (A), myeloperoxidase-DNA (MPO-DNA) complexes (B), or citrullinated histone H3 (Cit-H3) (C). COVID-19 samples were compared with controls by Mann-Whitney U test; ***P < 0.001, ****P < 0.0001. For the COVID-19 samples, correlation of cell-free DNA with MPO-DNA (D) and Cit-H3 (E) was assessed. Spearman’s correlation coefficients were calculated and are shown in the panels.
Association between NETs and clinical biomarkers.
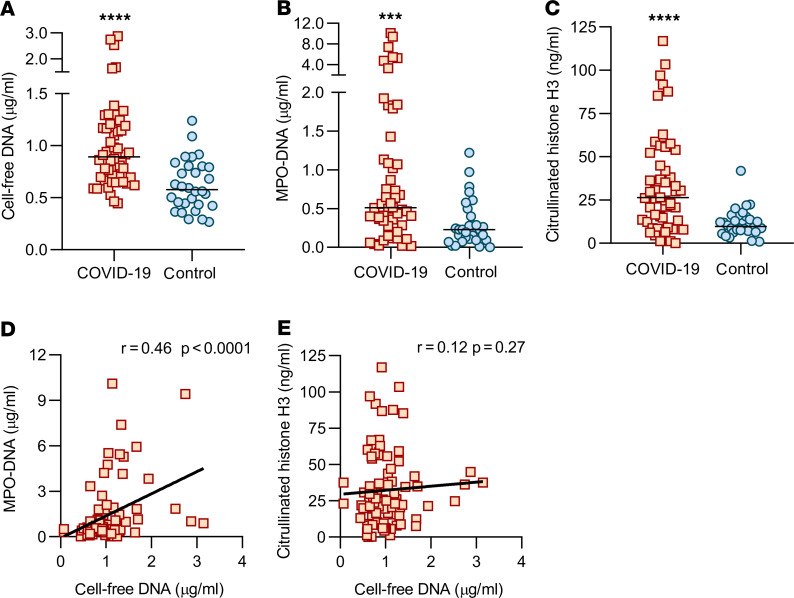
Given that circulating NETs may be drivers of, or form in response to, other blood products, we next asked how the aforementioned NET markers compared with several commonly available clinical tests. Specifically, we assessed potential correlations with C-reactive protein, D-dimer, lactate dehydrogenase, absolute neutrophil count, and platelet count. To draw integral comparisons, we limited the analysis of clinical laboratory measurements to those performed on the same day as serum used for NET assays. Cell-free DNA demonstrated a strong positive correlation with all the clinical tests other than platelet count (Figure 2, A–D). When we compared the clinical labs with MPO-DNA, we detected a strong positive correlation with absolute neutrophil count (Figure 2E), while C-reactive protein (r = 0.10, P = 0.44), D-dimer (r = 0.05, P = 0.72), and lactate dehydrogenase (r = 0.17, P = 0.22) demonstrated positive slopes that were not statistically significant. Interestingly, Cit-H3 levels were positively correlated with platelet counts (Figure 2F) but not the other clinical laboratory measurements. All available samples (n = 84) were included in the correlation analyses of Figure 2. When including only the first available sample from each patient (n = 50), correlations were similar (Supplemental Figure 2). Approximately half of the patients were treated with hydroxychloroquine (Table 1). We did not find a difference in NET markers from samples collected on days of hydroxychloroquine administration versus days without (Supplemental Figure 3). In summary, cell-free DNA and to a lesser extent MPO-DNA and Cit-H3 demonstrated significant correlations with clinical studies routinely used in the care of patients with COVID-19.
Figure 2. Association between NETs and clinical biomarkers in all available serum samples.
Cell-free DNA was compared with clinical laboratory results (when available on the same day), and correlation coefficients were calculated for C-reactive protein (A, n = 64), D-dimer (B, n = 56), lactate dehydrogenase (C, n = 55), and absolute neutrophil count (D, n = 69). In E (n = 69), MPO-DNA was compared with absolute neutrophils count, and in F (n = 81), Cit-H3 was compared with platelet count. The results of other relevant comparisons are discussed in the text. Pearson’s correlation coefficients were calculated and are shown in the panels.
NETs associate with severe disease including mechanical ventilation.
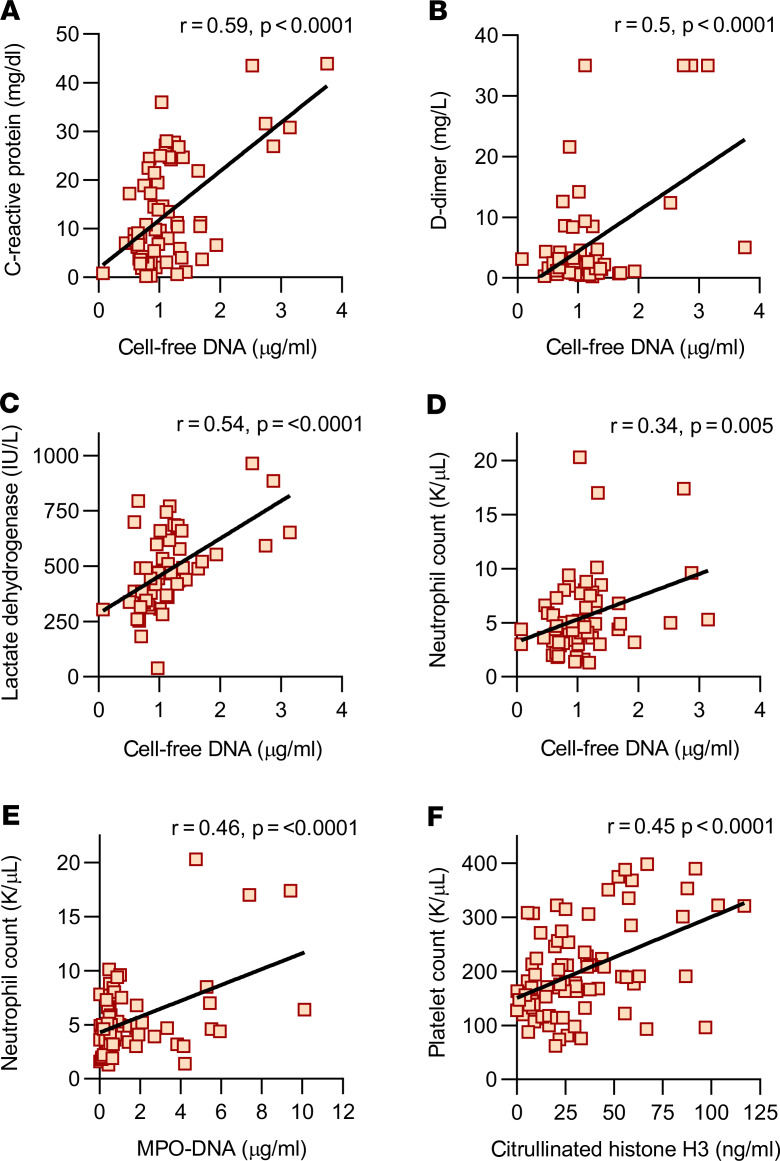
We next determined the clinical status associated with each available serum sample. We compared samples from patients with severe COVID-19 (those requiring mechanical ventilation, n = 27 samples) with patients with milder COVID-19 (oxygen saturation > 94% on ambient air, n = 24 samples). As compared with patients breathing room air, patients requiring mechanical ventilation had significantly higher levels of cell-free DNA (Figure 3A) and MPO-DNA (Figure 3B) but not Cit-H3 (Figure 3C). Absolute neutrophil counts were not significantly higher in the ventilated patients (Figure 3D). Taken together, these data suggest a possible relationship between level of serum NETs and severity of COVID-19.
Figure 3. Levels of NETs associate with mechanical ventilation in all available serum samples.
Serum samples were grouped by clinical status (room air versus mechanical ventilation) and analyzed for cell-free DNA (A, n = 51), MPO-DNA complexes (B, n = 51), Cit-H3 (C, n = 51), and absolute neutrophil count (D, n = 42). Groups were compared by Mann-Whitney U test; ****P < 0.0001, *P < 0.05. For D, the P value was 0.08.
COVID-19 sera trigger control neutrophils to release NETs.
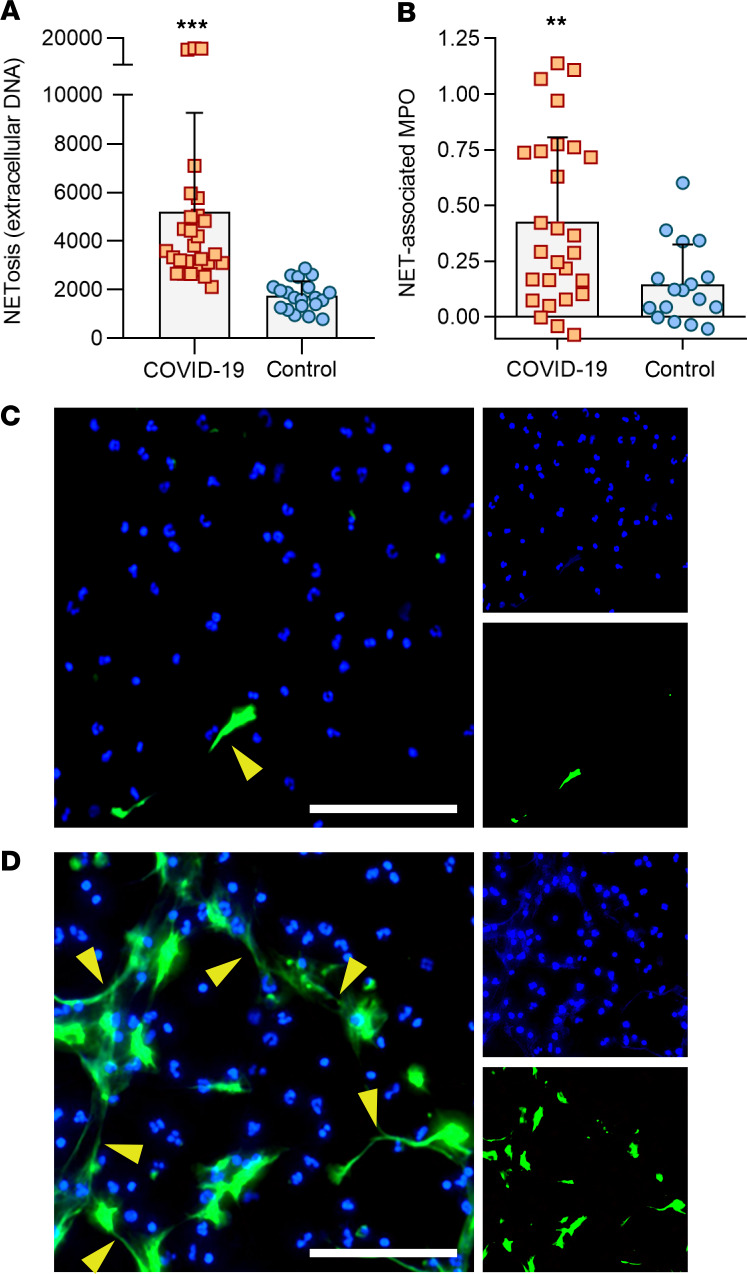
If COVID-19 presents a milieu favoring NETosis, we reasoned that direct exposure of control neutrophils to patient sera (without any additional agonist) would trigger these healthy neutrophils to undergo NETosis. As compared with heterologous control sera, the patient sera robustly promoted NETosis whether measured by externalization of DNA (Figure 4A) or release of DNA-bound MPO enzyme (Figure 4B). Immunofluorescence microscopy demonstrated extracellular chromatin structures decorated with neutrophil elastase characteristic of NETs (Figure 4C). In summary, serum samples from patients with COVID-19 robustly triggered healthy neutrophils to undergo NETosis.
Figure 4. COVID-19 sera trigger control neutrophils to release NETs.
COVID-19 samples (for which sufficient sera were available) were tested for their ability to trigger neutrophils isolated from healthy controls to undergo NETosis. (A) NETosis was quantified using the cell-impermeant dye SYTOX Green as described in Methods (n = 27 COVID-19 samples, and n = 20 controls). Fluorescence intensity (excitation/emission 504 nm/523 nm) is shown on the y axis. Bars demonstrate mean and standard deviation while each data point represents a unique patient/control; ***P < 0.001 by t test. (B) In an independent set of experiments, NETosis was quantified as nuclease-liberated MPO activity (n = 27 COVID-19 samples, and n = 17 controls). Absorbance at 450 nm is shown on the y axis after subtracting background from untreated cells. Bars demonstrate mean and standard deviation while each data point represents a unique patient/control; **P < 0.01 by t test. (C) Representative image of control neutrophils cultured with 10% heterologous control serum (upper) or COVID-19 serum (lower). Neutrophil elastase is stained green and DNA is stained blue. Scale bar: 100 μm. The yellow arrows highlight some examples of NET strands.
Discussion
Here, we report for the first time to our knowledge elevated levels of serum NETs in many hospitalized patients with COVID-19. We measured 3 markers commonly used to detect NET remnants in blood (cell-free DNA, MPO-DNA, and Cit-H3) and found significant elevations in all 3. We also found that COVID-19 sera are potent stimulators of NETosis when added to control neutrophils. Taken together, these data provide evidence that COVID-19, at least in hospitalized patients, is a pro-NETotic state. The triggers of NETosis in COVID-19 are potentially myriad and will require further investigation. Possibilities include virus-damaged epithelial cells (14, 30), activated platelets (31, 32), activated endothelial cells (33), and inflammatory cytokines, such as IL-1β (20, 34), IL-8 (35, 36), granulocyte colony-stimulating factor (37, 38), and likely others (18).
Of the markers we tested, cell-free DNA was most closely aligned with traditional inflammatory markers used to track COVID-19, including C-reactive protein, D-dimer, and lactate dehydrogenase. Notably, although cell-free DNA is not a highly specific marker for NETs, it was strongly correlated with absolute neutrophil count, as was the more specific marker of NETs, MPO-DNA. Somewhat unexpectedly, Cit-H3 did not correlate well with the other 2 markers but did associate strongly with platelet levels. It is believed that the predominant driver of histone citrullination (i.e., production of Cit-H3) in NETs is the enzyme peptidylarginine deiminase 4 (PAD4) (39). However, neutrophils can be triggered to undergo NETosis by a variety of stimuli, and in vitro studies demonstrate that not all pathways to NETosis are equally reliant on PAD4 activity (40); for example, stimuli that lead to robust reactive oxygen species production may be relatively PAD4 independent (41). The dichotomy between MPO-DNA and Cit-H3 levels in the COVID-19 sera tested here potentially suggests that 2 or more pathways to NETosis are active in patients with COVID-19, with the pathway leading to Cit-H3 perhaps having some relationship to platelets (42). It should also be noted that neutrophils are relatively short-lived cells that may experience cell death through many pathways, including apoptosis, necrosis, pyroptosis, NETosis, and others. Markers such as lactate dehydrogenase, cell-free DNA, and Cit-H3 may therefore also be produced by neutrophil cell death that is independent of NETosis (43, 44). The activation of other cell death programs, and their relationship to the inflammatory storm, certainly warrant further investigation in COVID-19.
NETs were first described in 2004 as a novel pathogen eradication strategy that could function as an alternative to phagocytosis (36), but it is now recognized that NETs have double-edged-sword properties and likely exacerbate (and in some cases even initiate) autoimmune and vascular diseases (45). NETs present and stabilize a variety of oxidant enzymes in the extracellular space, including MPO, NADPH oxidase, and nitric oxide synthase (46), while serving as a source of extracellular histones that carry significant cytotoxic potential (47, 48). In light of these toxic cargo, it is not surprising that NETs play a role in a variety of lung diseases, including cystic fibrosis (where they occlude larger airways) (49),smoking-related lung disease (50), and, with particular relevance here, pathogen-induced acute lung injury and ARDS (14, 51, 53). NETs have also been very well studied in the setting of cardiovascular disease, where they infiltrate and propagate inflammation in the vessel wall (53) and, when formed intravascularly, occlude arteries (54), veins (55), and microscopic vessels (56). Early studies of COVID-19 suggest a high risk of morbid arterial events (57), and one can speculate that the risk of venous thrombosis will increasingly reveal itself as more data become available (58).
Severe COVID-19 appears to be defined by neutrophilia, as well as elevations in IL-1β, IL-6, and D-dimer (18), the latter suggesting hyperactivity of the coagulation system. All these findings have significant potential for crosstalk with NETs. NETs are linked to IL-1β (both upstream and downstream) in cardiovascular and pulmonary diseases (19–22), including as described by our group for venous thrombosis (9). The same is true for IL-6, either directly (23), or perhaps with IL-1β as an intermediary (24). Of course, as discussed above, examples of NETs as drivers of thrombosis are myriad because intravascular NETosis is responsible for initiation and accretion of thrombotic events in arteries, veins, and — particularly pertinent to COVID-19 — the microvasculature, where thrombotic disease can drive end organ damage in lungs, heart, kidneys, and other organs (59, 60). Mechanistically, NETs, via electrostatic interactions, activate the contact pathway of coagulation (61), while presenting tissue factor to activate the intrinsic pathway (62). Simultaneously, serine proteases in NETs dismantle natural brakes on coagulation, such as tissue factor pathway inhibitor and antithrombin (63). Bidirectional interplay between NETs and platelets may also be critical for COVID-19–associated microvascular thrombosis, as has been characterized in a variety of disease models (60, 61).
Of interest, a recent small study performed in China suggested potential efficacy of the adenosine receptor agonist, dipyridamole, in severe cases of COVID-19 (64). Dipyridamole is an FDA-approved drug that our group recently discovered to inhibit NET formation by activation of adenosine A2A receptors (7). In the aforementioned trial, patients with COVID-19–associated bilateral pneumonia were treated with oral dipyridamole for 7 days, in addition to treatment with antiviral agents (64). As compared with controls, dipyridamole-treated patients demonstrated improvements in platelet counts and D-dimer levels (64). Given the urgent need for effective treatments for COVID-19, a randomized study to characterize the impact of dipyridamole on COVID-19–related NETosis, thrombo-inflammatory storm, and, of course, outcomes may be warranted. Other approaches to combating NETs have been reviewed (65, 66) and include the dismantling of already-formed NETs (deoxyribonucleases) and strategies that might prevent initiation of NET release, including inhibitors of neutrophil elastase and PAD4.
This study is not without limitations, including the use of serum samples retrieved from the clinical laboratory, rather than samples drawn specifically for research purposes. Here, it is certainly possible that NETs were partially degraded over time, thereby lowering our measurements. It should also be emphasized that it is not clear whether the NET remnants described here are drivers of disease severity or a mere consequence of acute inflammation in patients. Indeed, the definitive accounting of COVID-19 pathophysiology and answering questions of causality will likely await the development of model systems. Our hope, though, is that these findings will ignite further research into the role of neutrophil effector functions in the complications of COVID-19 (67). As a first step, future studies should investigate the predictive power of circulating NETs in well-phenotyped longitudinal cohorts. Furthermore, given the dichotomy we found here between MPO-DNA and Cit-H3, investigators should be encouraged to continue to include diverse markers of NETosis in future studies. As we await definitive antiviral and immunological solutions to the current pandemic, we posit that antineutrophil therapies may be part of a personalized strategy for some individuals affected by COVID-19 who are at risk for progression to respiratory failure.
Methods
Human samples.
Serum samples from 50 hospitalized COVID-19 patients (84 total samples) were used in this study. Blood was collected into serum separator tubes containing clot activator and serum separator gel by a trained hospital phlebotomist. After completion of biochemical testing ordered by the clinician, the remaining serum was stored at 4°C for up to 48 hours before it was deemed “discarded” and released to the research laboratory. Serum samples were immediately divided into small aliquots and stored at –80°C until the time of testing. All 50 patients had a confirmed COVID-19 diagnosis based on FDA-approved RNA testing. This study complied with all relevant ethical regulations and was approved by the University of Michigan Institutional Review Board (HUM00179409), which waived the requirement for informed consent given the discarded nature of the samples. Healthy volunteers were recruited through a posted flyer; exclusion criteria for these controls included history of a systemic autoimmune disease, active infection, and pregnancy. For preparation of control serum, blood was collected into serum separator tubes containing clot activator and serum separator gel by a trained hospital phlebotomist. Samples were centrifuged at approximately 2000 g, similar to the clinical samples. These serum samples were divided into small aliquots and stored at –80°C until the time of testing.
Quantification of cell-free DNA.
Cell-free DNA was quantified in sera using the Quant-iT PicoGreen dsDNA Assay Kit (Invitrogen, Thermo Fisher Scientific, P11496) according to the manufacturer’s instructions.
Quantification of Cit-H3.
Cit-H3 was quantified in sera using the Citrullinated Histone H3 (clone 11D3) ELISA Kit (Cayman, 501620) according to the manufacturer’s instructions.
Quantification of MPO-DNA complexes.
MPO-DNA complexes were quantified similarly to what has been previously described (68). This protocol used several reagents from the Cell Death Detection ELISA kit (Roche). First, a high-binding EIA/RIA 96-well plate (Costar) was coated overnight at 4°C with anti-human MPO antibody (Bio-Rad 0400-0002), diluted to a concentration of 1 μg/mL in coating buffer (Cell Death kit). The plate was washed 2 times with wash buffer (0.05% Tween-20 in PBS), then blocked with 4% bovine serum albumin (MilliporeSigma) in PBS (supplemented with 0.05% Tween-20) for 2 hours at room temperature. The plate was again washed 5 times, before incubating for 90 minutes at room temperature with 10% serum or plasma in the aforementioned blocking buffer (without Tween-20). The plate was washed 5 times, then incubated for 90 minutes at room temperature with 10× anti-DNA antibody (HRP conjugated; from the Cell Death kit) diluted 1:100 in blocking buffer. After 5 more washes, the plate was developed with 3,3′,5,5′-Tetramethylbenzidine (TMB) substrate (Invitrogen, Thermo Fisher Scientific) followed by a 2N sulfuric acid stop solution. Absorbance was measured at a wavelength of 450 nm using a Cytation 5 Cell Imaging Multi-Mode Reader (BioTek). Data were normalized to in vitro–prepared NET standards included on every plate, which were quantified based on their DNA content.
Human neutrophil purification.
For neutrophil preparation, blood from healthy volunteers was collected into sodium citrate tubes by standard phlebotomy techniques. The anticoagulated blood was then fractionated by density-gradient centrifugation using Ficoll-Paque Plus (GE Healthcare). Neutrophils were further purified by dextran sedimentation of the red blood cell layer, before lysing residual red blood cells with 0.2% sodium chloride. Neutrophil preparations were at least 95% pure as confirmed by nuclear morphology.
NETosis assay (SYTOX Green).
A cell-impermeant dye, SYTOX Green (Thermo Fisher Scientific), was used to measure NETosis. Briefly, purified neutrophils were resuspended in 1× PBS (Gibco, Thermo Fisher Scientific). One hundred thousand neutrophils were seeded into each well of a 0.001% poly-l-lysine–coated (MilliporeSigma), 96-well, black, clear-bottom non-tissue culture plate (Costar) and were allowed to adhere for 20 minutes at 37°C and 5% CO2. PBS was gently removed and control/patient serum (diluted to 10% in RPMI culture medium from Thermo Fisher Scientific, supplemented with l-glutamine) was carefully added without disrupting adherent cells. SYTOX Green was added at the same time to a final concentration of 500 nM. All treatments were done in triplicate. Cells were allowed to undergo NETosis for 4 hours. Culture medium was then gently removed, and fresh 1× PBS was added to each well. Fluorescence was quantified at excitation and emission wavelengths of 504 nm and 523 nm, respectively, using a Cytation 5 Cell Imaging Multi-Mode Reader (BioTek). Data were collected using the area-scan setting of the plate reader.
NETosis assay (NET-associated MPO).
Purified neutrophils were resuspended in RPMI medium (Gibco, Thermo Fisher Scientific) supplemented with 0.5% bovine serum albumin (MilliporeSigma), 0.5% heat-inactivated fetal bovine serum (FBS, Gibco, Thermo Fisher Scientific), and l-glutamine. Neutrophils (1 × 105/well) were cultured in a 96-well tissue culture plate (Costar) in the presence of either patient or control serum, diluted to a final concentration of 10%. Plates were incubated for 3 hours at 37°C and 5% CO2. To collect NET-associated MPO, the culture medium was discarded (to remove any soluble MPO) and replaced with 100 μL of PBS supplemented with 5 U/mL Micrococcal Nuclease (Thermo Fisher Scientific). After 10 minutes at 37°C, digestion of NETs was stopped with 10 mM EDTA. Supernatants were transferred to a V-shaped, 96-well plate, and centrifuged at 400 g for 5 minutes to remove debris. Supernatants were then transferred into a new flat-bottom, 96-well plate. To quantify MPO activity, an equal volume of TMB substrate (Thermo Fisher Scientific) was added to each well. After 10 minutes of incubation in the dark, the reaction was stopped by the 2N sulfuric acid solution. Absorbance was measured at 450 nm using a Cytation 5 Cell Imaging Multi-Mode Reader.
NETosis assay (microscopy).
For immunofluorescence microscopy, 1 × 105 neutrophils were seeded onto coverslips coated with 0.001% poly-l-lysine (MilliporeSigma) and cultured as for the above assays. Samples were then fixed with 4% paraformaldehyde for 10 minutes at room temperature, followed by blocking with 10% FBS in PBS. The primary antibody was against neutrophil elastase (Abcam 21595, diluted 1:100), and the FITC-conjugated secondary antibody was from SouthernBiotech (4052-02, diluted 1:250). DNA was stained with Hoechst 33342 (Invitrogen, Thermo Fisher Scientific). Images were collected with a Cytation 5 Cell Imaging Multi-Mode Reader.
Statistics.
Normally distributed data were analyzed by 2-sided t test, and skewed data were analyzed by Mann-Whitney U test. Data analysis was with GraphPad Prism software version 8. Correlations were tested by Pearson’s or Spearman’s correlation coefficient as indicated. Statistical significance was defined as P < 0.05 unless stated otherwise.
Study approval.
The study was approved by the University of Michigan Institutional Review Board (HUM00179409), which waived the requirement for informed consent given the discarded nature of the patient samples. Healthy volunteers were recruited under University of Michigan protocol HUM00044257 and provided informed consent.
Author contributions
YZ, SY, HS, KG, JAM, MZ, and CB conducted experiments and analyzed data. YZ, AW, BJB, ME, RJW, YK, and JSK conceived the study and analyzed data. All authors participated in writing the manuscript and gave approval before submission.
Supplementary Material
Acknowledgments
The work was supported by a COVID-19 Cardiovascular Impact Research Ignitor Grant from the Michigan Medicine Frankel Cardiovascular Center as well as by the A. Alfred Taubman Medical Research Institute. YZ was supported by career development grants from the Rheumatology Research Foundation and APS ACTION. JAM was partially supported by the VA Healthcare System. YK was supported by the NIH (K08HL131993, R01HL150392), the Falk Medical Research Trust Catalyst Award, and the JOBST-American Venous Forum Award. JSK was supported by grants from the NIH (R01HL115138), Lupus Research Alliance, and Burroughs Wellcome Fund. The team also thanks all members of the “NETwork to target neutrophils in COVID-19 collaborative working group” for their helpful advice and encouragement.
Version 1. 04/24/2020
In-Press Preview
Version 2. 06/04/2020
Electronic publication
Footnotes
Conflict of interest: ME reports receipt of lonodelestat from Santhera for preclinical studies.
Copyright: © 2020, American Society for Clinical Investigation.
Reference information: JCI Insight. 2020;5(11):e138999.https://doi.org/10.1172/jci.insight.138999.
Contributor Information
Yu Zuo, Email: yzu@med.umich.edu.
Srilakshmi Yalavarthi, Email: syalavar@med.umich.edu.
Hui Shi, Email: hushi@med.umich.edu.
Kelsey Gockman, Email: kego@med.umich.edu.
Melanie Zuo, Email: zuom@med.umich.edu.
Jacqueline A. Madison, Email: madisonj@med.umich.edu.
Andrew Weber, Email: AWeber4@northwell.edu.
Betsy J. Barnes, Email: bbarnes1@northwell.edu.
Mikala Egeblad, Email: egeblad@cshl.edu.
Robert J. Woods, Email: robertwo@med.umich.edu.
Yogendra Kanthi, Email: ykanthi@med.umich.edu.
Jason S. Knight, Email: jsknight@umich.edu.
References
- 1. Center for Systems Science and Engineering. Coronavirus COVID-19 Global Cases by the Center for Systems Science and Engineering (CSSE) at Johns Hopkins University (JHU). https://coronavirus.jhu.edu/map.html Updated May 21, 2020. Accessed May 21, 2020.
- 2.Pedersen SF, Ho YC. SARS-CoV-2: a storm is raging. J Clin Invest. 2020;130(5):2202–2205. doi: 10.1172/JCI137647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. doi: 10.1101/2020.03.12.20035048. Zhang B, et al. Immune phenotyping based on neutrophil-to-lymphocyte ratio and IgG predicts disease severity and outcome for patients with COVID-19 [preprint]. Posted on medrxiv March 16, 2020. Accessed May 14, 2020. [DOI] [PMC free article] [PubMed]
- 4. doi: 10.1101/2020.03.05.20031906. Song CY, Xu J, He JQ, Lu YQ. COVID-19 early warning score: a multi-parameter screening tool to identify highly suspected patients [preprint]. Posted on medrxiv March 8, 2020. Accessed May 14, 2020. [DOI]
- 5.Iba T, Levy JH, Raj A, Warkentin TE. Advance in the management of sepsis-induced coagulopathy and disseminated intravascular coagulation. J Clin Med. 2019;8(5):E728. doi: 10.3390/jcm8050728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ward PA, Fattahi F. New strategies for treatment of infectious sepsis. J Leukoc Biol. 2019;106(1):187–192. doi: 10.1002/JLB.4MIR1118-425R. [DOI] [PubMed] [Google Scholar]
- 7.Ali RA, et al. Adenosine receptor agonism protects against NETosis and thrombosis in antiphospholipid syndrome. Nat Commun. 2019;10(1):1916. doi: 10.1038/s41467-019-09801-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Meng H, et al. In vivo role of neutrophil extracellular traps in antiphospholipid antibody-mediated venous thrombosis. Arthritis Rheumatol. 2017;69(3):655–667. doi: 10.1002/art.39938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yadav V, et al. Ectonucleotidase tri(di)phosphohydrolase-1 (ENTPD-1) disrupts inflammasome/interleukin 1β-driven venous thrombosis. J Clin Invest. 2019;129(7):2872–2877. doi: 10.1172/JCI124804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Potey PM, Rossi AG, Lucas CD, Dorward DA. Neutrophils in the initiation and resolution of acute pulmonary inflammation: understanding biological function and therapeutic potential. J Pathol. 2019;247(5):672–685. doi: 10.1002/path.5221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Frantzeskaki F, Armaganidis A, Orfanos SE. Immunothrombosis in acute respiratory distress syndrome: cross talks between inflammation and coagulation. Respiration. 2017;93(3):212–225. doi: 10.1159/000453002. [DOI] [PubMed] [Google Scholar]
- 12.Twaddell SH, Baines KJ, Grainge C, Gibson PG. The emerging role of neutrophil extracellular traps in respiratory disease. Chest. 2019;156(4):774–782. doi: 10.1016/j.chest.2019.06.012. [DOI] [PubMed] [Google Scholar]
- 13.Porto BN, Stein RT. Neutrophil extracellular traps in pulmonary diseases: too much of a good thing? Front Immunol. 2016;7:311. doi: 10.3389/fimmu.2016.00311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Narasaraju T, et al. Excessive neutrophils and neutrophil extracellular traps contribute to acute lung injury of influenza pneumonitis. Am J Pathol. 2011;179(1):199–210. doi: 10.1016/j.ajpath.2011.03.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Moorthy AN, Tan KB, Wang S, Narasaraju T, Chow VT. Effect of high-fat diet on the formation of pulmonary neutrophil extracellular traps during influenza pneumonia in BALB/c mice. Front Immunol. 2016;7:289. doi: 10.3389/fimmu.2016.00289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Rudd JM, et al. Neutrophils induce a novel chemokine receptors repertoire during influenza pneumonia. Front Cell Infect Microbiol. 2019;9:108. doi: 10.3389/fcimb.2019.00108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ashar HK, et al. The role of extracellular histones in influenza virus pathogenesis. Am J Pathol. 2018;188(1):135–148. doi: 10.1016/j.ajpath.2017.09.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mehta P, et al. COVID-19: consider cytokine storm syndromes and immunosuppression. Lancet. 2020;395(10229):1033–1034. doi: 10.1016/S0140-6736(20)30628-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Liberale L, et al. Interleukin-1β mediates arterial thrombus formation via NET-associated tissue factor. J Clin Med. 2019;8(12):E2072. doi: 10.3390/jcm8122072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Meher AK, et al. Novel role of IL (interleukin)-1β in neutrophil extracellular trap formation and abdominal aortic aneurysms. Arterioscler Thromb Vasc Biol. 2018;38(4):843–853. doi: 10.1161/ATVBAHA.117.309897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Josefs T, et al. Neutrophil extracellular traps promote macrophage inflammation and impair atherosclerosis resolution in diabetic mice. JCI Insight. 2020;5(7):134796. doi: 10.1172/jci.insight.134796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lachowicz-Scroggins ME, et al. Extracellular DNA, neutrophil extracellular traps, and inflammasome activation in severe asthma. Am J Respir Crit Care Med. 2019;199(9):1076–1085. doi: 10.1164/rccm.201810-1869OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Merza M, et al. Neutrophil extracellular traps induce trypsin activation, inflammation, and tissue damage in mice with severe acute pancreatitis. Gastroenterology. 2015;149(7):1920–1931.e8. doi: 10.1053/j.gastro.2015.08.026. [DOI] [PubMed] [Google Scholar]
- 24.Ridker PM. From C-reactive protein to interleukin-6 to interleukin-1: moving upstream to identify novel targets for atheroprotection. Circ Res. 2016;118(1):145–156. doi: 10.1161/CIRCRESAHA.115.306656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Perlman S, Dandekar AA. Immunopathogenesis of coronavirus infections: implications for SARS. Nat Rev Immunol. 2005;5(12):917–927. doi: 10.1038/nri1732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Blondonnet R, Constantin JM, Sapin V, Jabaudon M. A Pathophysiologic approach to biomarkers in acute respiratory distress syndrome. Dis Markers. 2016;2016:3501373. doi: 10.1155/2016/3501373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Matthay MA, et al. Acute respiratory distress syndrome. Nat Rev Dis Primers. 2019;5(1):18. doi: 10.1038/s41572-019-0069-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sugamata R, et al. Contribution of neutrophil-derived myeloperoxidase in the early phase of fulminant acute respiratory distress syndrome induced by influenza virus infection. Microbiol Immunol. 2012;56(3):171–182. doi: 10.1111/j.1348-0421.2011.00424.x. [DOI] [PubMed] [Google Scholar]
- 29.Zhu L, et al. High level of neutrophil extracellular traps correlates with poor prognosis of severe influenza A infection. J Infect Dis. 2018;217(3):428–437. doi: 10.1093/infdis/jix475. [DOI] [PubMed] [Google Scholar]
- 30.Cortjens B, et al. Neutrophil extracellular traps cause airway obstruction during respiratory syncytial virus disease. J Pathol. 2016;238(3):401–411. doi: 10.1002/path.4660. [DOI] [PubMed] [Google Scholar]
- 31.Clark SR, et al. Platelet TLR4 activates neutrophil extracellular traps to ensnare bacteria in septic blood. Nat Med. 2007;13(4):463–469. doi: 10.1038/nm1565. [DOI] [PubMed] [Google Scholar]
- 32.McDonald B, et al. Platelets and neutrophil extracellular traps collaborate to promote intravascular coagulation during sepsis in mice. Blood. 2017;129(10):1357–1367. doi: 10.1182/blood-2016-09-741298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gupta AK, et al. Activated endothelial cells induce neutrophil extracellular traps and are susceptible to NETosis-mediated cell death. FEBS Lett. 2010;584(14):3193–3197. doi: 10.1016/j.febslet.2010.06.006. [DOI] [PubMed] [Google Scholar]
- 34.Sil P, Wicklum H, Surell C, Rada B. Macrophage-derived IL-1β enhances monosodium urate crystal-triggered NET formation. Inflamm Res. 2017;66(3):227–237. doi: 10.1007/s00011-016-1008-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gupta AK, Hasler P, Holzgreve W, Gebhardt S, Hahn S. Induction of neutrophil extracellular DNA lattices by placental microparticles and IL-8 and their presence in preeclampsia. Hum Immunol. 2005;66(11):1146–1154. doi: 10.1016/j.humimm.2005.11.003. [DOI] [PubMed] [Google Scholar]
- 36.Brinkmann V, et al. Neutrophil extracellular traps kill bacteria. Science. 2004;303(5663):1532–1535. doi: 10.1126/science.1092385. [DOI] [PubMed] [Google Scholar]
- 37.Demers M, et al. Cancers predispose neutrophils to release extracellular DNA traps that contribute to cancer-associated thrombosis. Proc Natl Acad Sci U S A. 2012;109(32):13076–13081. doi: 10.1073/pnas.1200419109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Cedervall J, et al. Neutrophil extracellular traps accumulate in peripheral blood vessels and compromise organ function in tumor-bearing animals. Cancer Res. 2015;75(13):2653–2662. doi: 10.1158/0008-5472.CAN-14-3299. [DOI] [PubMed] [Google Scholar]
- 39.Wong SL, Wagner DD. Peptidylarginine deiminase 4: a nuclear button triggering neutrophil extracellular traps in inflammatory diseases and aging. FASEB J. 2018;32(12):6358–6370. doi: 10.1096/fj.201800691R. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Tatsiy O, McDonald PP. Physiological stimuli induce PAD4-dependent, ROS-independent NETosis, with early and late events controlled by discrete signaling pathways. Front Immunol. 2018;9:2036. doi: 10.3389/fimmu.2018.02036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kenny EF, et al. Diverse stimuli engage different neutrophil extracellular trap pathways. Elife. 2017;6:e24437. doi: 10.7554/eLife.24437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zucoloto AZ, Jenne CN. Platelet-neutrophil interplay: insights into neutrophil extracellular trap (NET)-driven coagulation in infection. Front Cardiovasc Med. 2019;6:85. doi: 10.3389/fcvm.2019.00085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Romero V, et al. Immune-mediated pore-forming pathways induce cellular hypercitrullination and generate citrullinated autoantigens in rheumatoid arthritis. Sci Transl Med. 2013;5(209):209ra150. doi: 10.1126/scitranslmed.3006869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Elkon KB. Review: cell death, nucleic acids, and immunity: inflammation beyond the grave. Arthritis Rheumatol. 2018;70(6):805–816. doi: 10.1002/art.40452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wigerblad G, Kaplan MJ. NETs spread ever wider in rheumatic diseases. Nat Rev Rheumatol. 2020;16(2):73–74. doi: 10.1038/s41584-019-0352-1. [DOI] [PubMed] [Google Scholar]
- 46.Smith CK, et al. Neutrophil extracellular trap-derived enzymes oxidize high-density lipoprotein: an additional proatherogenic mechanism in systemic lupus erythematosus. Arthritis Rheumatol. 2014;66(9):2532–2544. doi: 10.1002/art.38703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Silvestre-Roig C, et al. Externalized histone H4 orchestrates chronic inflammation by inducing lytic cell death. Nature. 2019;569(7755):236–240. doi: 10.1038/s41586-019-1167-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Saffarzadeh M, et al. Neutrophil extracellular traps directly induce epithelial and endothelial cell death: a predominant role of histones. PLoS One. 2012;7(2):e32366. doi: 10.1371/journal.pone.0032366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Dwyer M, et al. Cystic fibrosis sputum DNA has NETosis characteristics and neutrophil extracellular trap release is regulated by macrophage migration-inhibitory factor. J Innate Immun. 2014;6(6):765–779. doi: 10.1159/000363242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Dicker AJ, et al. Neutrophil extracellular traps are associated with disease severity and microbiota diversity in patients with chronic obstructive pulmonary disease. J Allergy Clin Immunol. 2018;141(1):117–127. doi: 10.1016/j.jaci.2017.04.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Lefrançais E, Mallavia B, Zhuo H, Calfee CS, Looney MR. Maladaptive role of neutrophil extracellular traps in pathogen-induced lung injury. JCI Insight. 2018;3(3):98178. doi: 10.1172/jci.insight.98178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Liu S, et al. Neutrophil extracellular traps are indirectly triggered by lipopolysaccharide and contribute to acute lung injury. Sci Rep. 2016;6:37252. doi: 10.1038/srep37252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Warnatsch A, Ioannou M, Wang Q, Papayannopoulos V. Inflammation. Neutrophil extracellular traps license macrophages for cytokine production in atherosclerosis. Science. 2015;349(6245):316–320. doi: 10.1126/science.aaa8064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Döring Y, Soehnlein O, Weber C. Neutrophil extracellular traps in atherosclerosis and atherothrombosis. Circ Res. 2017;120(4):736–743. doi: 10.1161/CIRCRESAHA.116.309692. [DOI] [PubMed] [Google Scholar]
- 55.Brill A, et al. Neutrophil extracellular traps promote deep vein thrombosis in mice. J Thromb Haemost. 2012;10(1):136–144. doi: 10.1111/j.1538-7836.2011.04544.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Tanaka K, et al. In vivo characterization of neutrophil extracellular traps in various organs of a murine sepsis model. PLoS One. 2014;9(11):e111888. doi: 10.1371/journal.pone.0111888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Zhou F, et al. Clinical course and risk factors for mortality of adult inpatients with COVID-19 in Wuhan, China: a retrospective cohort study. Lancet. 2020;395(10229):1054–1062. doi: 10.1016/S0140-6736(20)30566-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Bunce PE, High SM, Nadjafi M, Stanley K, Liles WC, Christian MD. Pandemic H1N1 influenza infection and vascular thrombosis. Clin Infect Dis. 2011;52(2):e14–e17. doi: 10.1093/cid/ciq125. [DOI] [PubMed] [Google Scholar]
- 59.Pfeiler S, Massberg S, Engelmann B. Biological basis and pathological relevance of microvascular thrombosis. Thromb Res. 2014;133(suppl 1):S35–S37. doi: 10.1016/j.thromres.2014.03.016. [DOI] [PubMed] [Google Scholar]
- 60.Ma AC, Kubes P. Platelets, neutrophils, and neutrophil extracellular traps (NETs) in sepsis. J Thromb Haemost. 2008;6(3):415–420. doi: 10.1111/j.1538-7836.2007.02865.x. [DOI] [PubMed] [Google Scholar]
- 61.Gould TJ, et al. Neutrophil extracellular traps promote thrombin generation through platelet-dependent and platelet-independent mechanisms. Arterioscler Thromb Vasc Biol. 2014;34(9):1977–1984. doi: 10.1161/ATVBAHA.114.304114. [DOI] [PubMed] [Google Scholar]
- 62.Kambas K, et al. Tissue factor expression in neutrophil extracellular traps and neutrophil derived microparticles in antineutrophil cytoplasmic antibody associated vasculitis may promote thromboinflammation and the thrombophilic state associated with the disease. Ann Rheum Dis. 2014;73(10):1854–1863. doi: 10.1136/annrheumdis-2013-203430. [DOI] [PubMed] [Google Scholar]
- 63.Massberg S, et al. Reciprocal coupling of coagulation and innate immunity via neutrophil serine proteases. Nat Med. 2010;16(8):887–896. doi: 10.1038/nm.2184. [DOI] [PubMed] [Google Scholar]
- 64. doi: 10.1016/j.apsb.2020.04.008. Liu X, et al. Potential therapeutic effects of dipyridamole in the severely ill patients with COVID-19 [published online April 20, 2020]. Acta Pharm Sin B. https://dx.doi.org/10.1016%2Fj.apsb.2020.04.008. [DOI] [PMC free article] [PubMed]
- 65.Thålin C, Hisada Y, Lundström S, Mackman N, Wallén H. Neutrophil extracellular traps: villains and targets in arterial, venous, and cancer-associated thrombosis. Arterioscler Thromb Vasc Biol. 2019;39(9):1724–1738. doi: 10.1161/ATVBAHA.119.312463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Barnado A, Crofford LJ, Oates JC. At the bedside: neutrophil extracellular traps (NETs) as targets for biomarkers and therapies in autoimmune diseases. J Leukoc Biol. 2016;99(2):265–278. doi: 10.1189/jlb.5BT0615-234R. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Barnes BJ, et al. Targeting potential drivers of COVID-19: neutrophil extracellular traps. J Exp Med. 2020;217(6):e20200652. doi: 10.1084/jem.20200652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Kessenbrock K, et al. Netting neutrophils in autoimmune small-vessel vasculitis. Nat Med. 2009;15(6):623–625. doi: 10.1038/nm.1959. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.