Abstract
Electronic health records (EHRs) have been widely used to help physicians to make decisions by predicting medical events such as diseases, prescriptions, outcomes, and so on. How to represent patient longitudinal medical data is the key to making these predictions. Recurrent neural network (RNN) is a popular model for patient longitudinal medical data representation from the view of patient status sequences, but it cannot represent complex interactions among different types of medical information, i.e., temporal medical event graphs, which can be represented by graph neural network (GNN). In this paper, we propose a hybrid method of RNN and GNN, called RGNN, for next-period prescription prediction from two views, where RNN is used to represent patient status sequences, and GNN is used to represent temporal medical event graphs. Experiments conducted on the public MIMIC-III ICU data show that the proposed method is effective for next-period prescription prediction, and RNN and GNN are mutually complementary.
Keywords: Medical prediction, Recurrent neural network, Graph neural network, Next-period prescription prediction
Background
In the last decade, with the rapid growth of electronic health records (EHRs), data-driven studies, such as drug repurposing and medical event prediction, have become more and more popular in the medical domain. An EHR is composed of a sequence of a patient’s visits in chronological order, each of which contains various medical information, such as demographics, vital signs, diagnoses, medications, procedures, laboratory test results, etc. A medical event prediction task is to predict some types of medical events, including diseases, prescriptions, outcomes, etc., using other types of medical information or medical history. For example, next-period prescription prediction is to predict all medications of a patient in the next time using his/her medical history. The critical challenge of medical event prediction driven by EHRs is how to represent patient longitudinal medical data accurately, also known as patient representation.
A large number of methods have been proposed to predict medical events, As medical data of each patient is time series data, the typical time series analysis methods such as machine learning methods based on manually-crafted features [1] and Autoregressive Integrated Moving Average (ARIMA) models applied for medical event prediction [2]. The manually-crafted features used in traditional machine learning methods are usually task-specific, and do not cover complex relations related to different types of medical information and time. In recent years, with the development of deep learning methods, a number of them have been gradually used for medical event prediction and have achieved good results.
The basic deep learning methods such as RNN [3] and Convolution Neural Networks (CNN) [4] have shown great potential for patient representation. However they still have limitations in heterogeneous information fusion and temporal information utilization.
To deal with these limitations mentioned above, researchers have attempted to extend the basic deep learning methods. For example, Jin et al. proposed a new Long Short-Term Memory (LSTM, a variant of RNN) learning framework to model inter-correlations of different types of medical information [5]. They developed three variants of LSTM based on demographics, laboratory test results and history prescriptions to predict next-period prescriptions. Among these variants, the decomposed LSTM that used LSTM to model history prescription sequence and integrated laboratory test results into each cell achieved the best results. An et al. proposed an attention-based LSTM for high-risk prediction [6]. The model first adopted attention-based LSTM to individually represent each type of medical information, including diagnosis sequence, laboratory sequence, and their combination sequence, and then concatenate these three representations together for prediction. All these methods aim to represent patient status sequences, but without considering complex interactions among different types of medical information, that is, temporal medical event graphs. GNN is a potential method to represent temporal medical event graphs.
In this study, we focus on next-period prescription prediction, and propose a novel hybrid method of RNN and GNN (called RGNN) for this task. In RGNN, RNN and GNN are first individually used to represent patient longitudinal medical data from two views, and then they are combined organically. The decomposed LSTM, a state-of-the-art method for next-period prescription prediction, is used to represent patient status sequences, and GNN is used to represent temporal event graphs.
In the case of GNN, we present two methods to represent medical event graphs. Experiments conducted on the public MIMIC-III ICU data show that the proposed method is effective for next-period prescription prediction and RNN and GNN are mutually complementary.
In summary, our main contributions are:
We propose a novel hybrid method of RNN and GNN, called RGNN, to represent patient longitudinal medical data from two views and apply it to next-period prescription prediction. In RGNN, RNN is used for patient status sequence representation, and GNN for temporal medical event graph representation.
We present two methods to build a temporal medical event graphs and represent them by GNN. We first build the graphs according to co-occurrences of different types of medical events at two neighbor time points, where the nodes are medical events and the edges are co-occurrences relations, and then further add time intervals between medical events as edge weights.
We evaluate the performance of RGNN on the public MIMIC-III ICU data. Experiment results demonstrate that RGNN can achieve promising performance on next-period prescription prediction, and RNN and GNN are mutually complementary.
Related work
Medical event prediction
In the medical domain, medical event prediction is a promising research topic. The main task of medical event prediction is to predict future medical events including risk of diseases [6], prescriptions [5], mortality rate [7], hospital readmission [8], length of stay in hospital [9], postoperative complications [10], survival time [11] and so on. In this paper, we mainly focus on next-period prescription prediction.
As patient medical data is typical time series data, almost all time series data analysis methods can be deployed for medical event prediction. They may fall into the following three categories: (1) statistic analysis methods such as Cox proportional hazards model [12] and hierarchical Association Rule Model (HARM) [13]; (2) statistic machine learning methods based on manually-crafted features such as artificial neural network, decision tree, logistic regression, Support Vector Machines (SVM) [14, 15]; (3) deep learning methods such as auto-encode model [16], CNN [11], RNN [17], LSTM and Bi-LSTM [18]. For example, in the early studies, McCormick et al.’s proposed a Hierarchical Association Rule Model (HARM) to predict disease risk from medical data using association analysis and Bayesian estimation [13]. Deken et al. are investigated ANN, decision tree, and logistic regression for breast cancer survival time prediction [19]. Choi et al. are presented an RNN-based system, i.e., doctor AI, to predict multiple medical events in the next time [20]. Baytas et al. proposed a time-aware LSTM network, called T-LSTM, for patient subtyping [21]. Among the three categories, deep learning methods attract more and more attention from researchers in recent years due to their excellent ability to model complex non-linear relationships within data and their remarkable results [22]. In the case of next-period prescription prediction, one of the latest studies is Jin et al.’s work [5]. In this work, they proposed a variant LSTM, called decomposed LSTM, that can fuse different types of medical information effectively.
Graph neural network
Graph neural network (GNN) is a kind of deep neural network powerful for complex graphs [23]. Several methods are recently proposed to compute representations of nodes, edges, and graphs [24–26]. Among them, Graph Convolutional Network (GCN) that computes the representation of a node recursively from its neighbors is the most common one [27] and is widely applied to many domains such as natural language processing and knowledge graph representation. In the medical domain, GCN starts to be applied to many tasks. For example, Choi et al.’s deployed GCN to learn medical concept representations from the graph of medical ontology knowledge [28]. Ma et al.’s recognized the drug–drug interaction (DDI) problem as a graph classification problem and solve it by GCN [29]. Besides GCN, some other GNNs also have been proposed recently such as GAMENet [30] and Decagon [31]. GAMENet is a Graph Augmented Memory Network designed to integrate the DDI knowledge graph for the personalized recommendation of medication combination. Decagon is a multi-modal GNN for drug side effect prediction. Decagon is a multi-modal GNN for drug side effect prediction.
Method
Task introduction
Next-period prescription prediction can be recognized as a multi-label classification problem as follows: given a set of patients P = { } with their medical histories X = { , , …, } and demographics D = { , , …, }, we need to predict their prescriptions in the next time. For patient pk, is a visit sequence that comprises sequences of different types of medical events such as diagnosis sequence, laboratory test sequence, prescription sequence, etc. In this study, following Jin et al.’s work, we only consider laboratory test sequence and prescription sequence, denoted by and respectively, where and are medications and laboratory tests at the time i, represented by a one-hot vector of medication vocabulary (denoted as ∑) and a one-hot vector of laboratory test item vocabulary (denoted as L). We adopt to denote the status of pk at the time t. In the case of M, . Figure 1 gives an example of medication vocabulary, where the numbers in parentheses are indices of items in the vocabularies.
Fig. 1.
Example of the medical history of a patient (pk)
Model description
The structure of our proposed hybrid method of decomposed LSTM and GNN (RGNN) is shown in Fig. 2. The medical history of patient, is represented by decomposed LSTM and GNN separately, and then the representations from the two views are combined in different ways. Subsequently, the final representation of (denoted by ) and the demographics of patient pk are concatenated, transformed by a Rectified Linear Unit (ReLU) activation function, and fed into a Sigmod function to predict medications in the next time.
Fig. 2.
Structure of the hybrid method of decomposed LSTM and GNN (RGNN)
Following the Jin et al.’s work [5], we use the following decomposed LSTM (as shown in Fig. 3) to represent the status sequence of patient :
| 1 |
| 2 |
| 3 |
| 4 |
| 5 |
| 6 |
where , and denote the logistic function, hyperbolic tangent function, and element-wise multiplication, respectively; denotes one of three gates (forget f, input i and output o gates); Ws are weight matrices and bs are bias vectors.
Fig. 3.
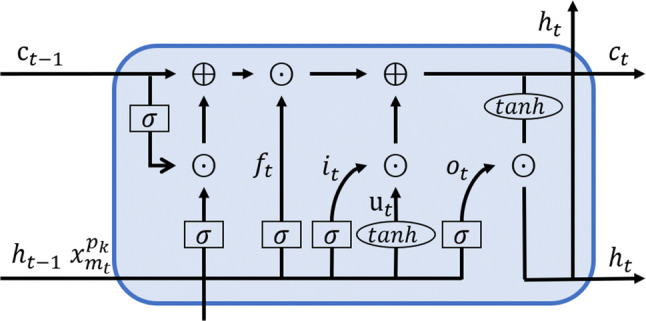
Structure of decomposed LSTM
We propose two methods to build temporal medical event graph: (1) co-occurrence graph and (2) time-aware graph. In the co-occurrence graph, medical events at the time i () are linked to medical events at time i + 1 (). The graph can be denoted by (T = {1, 2, …, t}) and is the adjacency matrix regarding and . After obtaining co-occurrence graph A, we deploy GCN, a kind of GNN, to represent A as follows:
| 7 |
| 8 |
where . is the degree matrix of A, I is the identity matrix, Ws are weight matrices, and MEAN{.} is the mean function.
Considering different time intervals between two neighbor time points, we build a time-aware graph by replacing by , where is the time interval between the time i and time i + 1.
We combine the representations from decomposed LSTM and GNN in the following three ways:
Concatenation mechanism:
-
Attention mechanism:
9 Gate mechanism:
The final medical history representation of (i.e.,) and the demographics of patient are concatenated and transformed into by a ReLU function defined as follows:
| 10 |
where is the weight matrix and is the bias vector.
At the end of the hybrid method, the medications in the next time are predicted by a Sigmod function as follows:
| 11 |
where is the weight matrix and is the bias vector.
To obtain model parameters, we employ the cross-entropy as the loss function as follows:
| 12 |
Experiments
Dataset
In this paper, we select the records of a group of patients that have at least two visits from MIMIC-III, a publicly available dataset that contains realistic medical data of 53,423 adults (age ≥ 16 years) in the ICU of Beth Israel medical center, Massachusetts Institute of technology from 2001 to 2012, as well as clinical data of 7787 newborns from 2001 to 2008. To avoid sparse data problem, we follow previous studies [5, 30] to remove laboratory test items appearing less than 50 times and keep the most frequent 200 medications. Finally, as shown in Table 1, we obtain a dataset of 7121 patients, where there are 11,269,796 laboratory test items of 462 types, 1,030,083 medications of 200 types. On average, a patient has 2.68 visits, each visit has 590 laboratory test items (lab test) and 54 prescriptions, each prescription has 6.26 medications, the time interval between two visit is 2.26 years, and the time interval between two prescriptions is 1 day. We randomly split the selected dataset into a training set, a development set and a test set in patients with a ratio of 80:10:10. The statistics of the dataset are listed in Table 2, where #* is the number of *.
Table 1.
Statistics of patient data
| Items | Value |
|---|---|
| Patients | 7121 |
| Laboratory test | 11,269,796 |
| Laboratory test code | 462 |
| Medications | 1,030,083 |
| Medications code | 200 |
| Avg visits per patient | 2.68 |
| Avg laboratory test per visit | 590 |
| Avg prescriptions per visit | 54 |
| Avg medications per prescription | 6.26 |
| Avg time interval between two neighbor prescription | 1 day |
| Avg time interval between two neighbor visits | 2.26 years |
Table 2.
Statistics of the dataset used in this study
| Dataset | #Patient | #Visit | #Medication | #Labtest |
|---|---|---|---|---|
| Training | 4557 | 13,966 | 752,960 | 8,338,978 |
| Development | 1139 | 2278 | 121,985 | 1,293,216 |
| Test | 1425 | 2875 | 155,138 | 1,637,602 |
| Total | 7121 | 19,094 | 1,030,083 | 11,269,796 |
Evaluation metrics
As next-period medication prediction is a class imbalance problem, we utilize the Area Under ROC (Receiver Operating Characteristic) curve (AUC) and Area Under Precision-Recall Curve (AUPR) to evaluate model performance. The AUC is the primary metric.
Experimental settings
We start with the basic LSTM considering different types of medical events separately, doctor AI [20], T-LSTM [21], decomposed LSTM [5] and GNN, and then compare different RGNN variants, denoted by RGNN-G-C, where G∈{“co-occurrence graph (CG)”, “time-aware graph (TG)”} represents graph construction method and C∈{“concatenation mechanism (CAT)”, “attention mechanism (ATT)”, “gate mechanism” (GAT)} represents the way to combine the representations from decomposed LSTM and GNN. As the same as RGNN, GNN also contains two variants according to the graph construction methods, which are denoted by GNN-CG and GNN-TG. All methods are implemented in TensorFlow and all models are trained three times with the following hyperparameters:
Results
A comparison of our method with other methods is shown in Table 3. Among the methods used for comparison, decomposed LSTM and GNN designed to fuse different types of medical events outperform the other LSTM variants that considers different types of medical events separately by more about 2% in AUC. Compared to decomposed LSTM and GNN, RGNN that combines them obtains an AUC improvement of 0.9% to 2.9%. In the case of RGNN, RGNN using TG is a little better than RGNN using CG, RGNN using ATT or GAT achieves much better performance than RGNN using CAT. The AUC difference between RGNN using ATT or GAT and RGNN using CAT is around 0.8%. RGNN-TG-ATT achieves the highest AUC of 0.8387 and the highest AUPR of 0.2848.
Table 3.
Comparison of our method with other methods
| Method | AUC | AUPR |
|---|---|---|
| LSTM | 0.7884 ± 0.0155 | 0.2450 ± 0.0070 |
| GNN-CG | 0.8091 ± 0.0003 | 0.1929 ± 0.0021 |
| GNN-TG | 0.8106 ± 0.0007 | 0.1950 ± 0.0023 |
| Doctor AI [20] | 0.7555 ± 0.0008 | 0.1662 ± 0.0048 |
| T-LSTM [21] | 0.7898 ± 0.0049 | 0.1289 ± 0.0048 |
| Decompos LSTM [5] | 0.8194 ± 0.0075 | 0.2669 ± 0.0097 |
| RGNN-CG-CAT | 0.8284 ± 0.0038 | 0.2704 ± 0.0083 |
| RGNN-CG-ATT | 0.8364 ± 0.0044 | 0.2741 ± 0.0007 |
| RGNN-CG-GAT | 0.8365 ± 0.0033 | 0.2634 ± 0.0043 |
| RGNN-TG-CAT | 0.8316 ± 0.0025 | 0.2766 ± 0.0052 |
| RGNN-TG-ATT | 0.8387 ± 0.0049 | 0.2848 ± 0.0011 |
| RGNN-TG-GAT | 0.8381 ± 0.0038 | 0.2643 ± 0.0038 |
The highest values are highlighted in bold
Moreover, we investigate the performance of RGNN-TG-ATT on the top 10 diagnoses (as shown in Table 4). The AUCs of RGNN-TG-ATT on all the top 10 diagnoses are higher than 0.81. The highest AUC is 0.8739 on chest pain, and the lowest AUC is 0.8130 on altered mental status. RGNN-TG-ATT achieves the highest AUPR of 0.3471 on coronary artery disease and the lowest AUPR of 0.2427 on upper gastrointestinal bleeding.
Table 4.
Performance of RGNN-TG-ATT on the top 10 diagnoses
| Diagnosis | AUC | AUPR |
|---|---|---|
| Pneumonia | 0.8293 ± 0.0084 | 0.2850 ± 0.0117 |
| Sepsis | 0.8225 ± 0.0115 | 0.3107 ± 0.0226 |
| Congestive heart failure (CHF) | 0.8552 ± 0.0061 | 0.3322 ± 0.0161 |
| Coronary artery disease (CAD) | 0.8582 ± 0.0129 | 0.3471 ± 0.0235 |
| Chest pain (CP) | 0.8739 ± 0.0091 | 0.2855 ± 0.0137 |
| Intracranial hemorrhage (ICH) | 0.8136 ± 0.0092 | 0.2453 ± 0.0156 |
| Altered mental status (AMS) | 0.8130 ± 0.0111 | 0.2481 ± 0.0080 |
| Gastrointestinal bleeding (GIB) | 0.8370 ± 0.0081 | 0.2542 ± 0.0127 |
| Upper GI | 0.8514 ± 0.0056 | 0.2427 ± 0.0135 |
| Abdominal pain | 0.8455 ± 0.0105 | 0.3318 ± 0.0306 |
The highest values are highlighted in bold
Discussion
In this study, we investigate two methods for patient longitudinal and heterogeneous medical data representation, that is, decomposed LSTM and GNN. A common characteristic of them is that the two methods are specially designed to fuse different types of medical information that are dependent on each other. As we mentioned in the method section, decomposed LSTM is used to represent a patient status sequence, while GNN is used to represent the temporal medical event graph. The former mainly focuses on patient status at each time, and the latter one mainly focuses on the whole medical events. This may be the main reason that both decomposed LSTM and GNN outperform the basic LSTM that considers different types of medical information separately as shown in Table 3. Because patient medical data is time-sequential data, RGNN using TG is a little better than RGNN using CG. To assess the effect of time on GNN, we further compare RGNN-TG-ATT with RGNN-CG-ATT on the top 10 diagnoses as shown in Table 5, where time volatility is the standard deviation of time intervals divided by the mean of time intervals. In most cases, the larger the time volatility, the effect the time on AUC. For example, as the largest time volatility of 2.8871 brings the biggest AUC difference of 0.0161, and the smallest time volatility of 2.3731 brings the smallest AUC difference of − 0.0003. From Table 4, we find that the effect of time on GNN is not very remarkable. The reason may be that the times in the data used in this study are relatively concentrated as the data comes from the ICU of Beth Israel medical center, Massachusetts Institute of technology. In the case of the combination of the two representations from RNN and GNN, the reason why both the attention mechanism and the gate mechanism show better performance than the concatenation mechanism is that the two representations share some information and are complementary to each other.
Table 5.
Effect of time on GNN
| Diagnosis | AUC | Time volatility | ||
|---|---|---|---|---|
| RGNN-CG-ATT | RGNN-TG-ATT | ∆AUC | ||
| Pneumonia | 0.8230 ± 0.0046 | 0.8293 ± 0.0084 | 0.0063 | 2.7016 |
| Sepsis | 0.8135 ± 0.0088 | 0.8225 ± 0.0115 | 0.0090 | 2.8092 |
| CHF | 0.8513 ± 0.0102 | 0.8552 ± 0.0061 | 0.0039 | 2.7968 |
| CAD | 0.8575 ± 0.0071 | 0.8582 ± 0.0129 | 0.0007 | 2.5006 |
| CP | 0.8578 ± 0.1057 | 0.8739 ± 0.0091 | 0.0161 | 2.8871 |
| ICH | 0.8139 ± 0.0105 | 0.8136 ± 0.0092 | − 0.0003 | 2.3731 |
| AMS | 0.8103 ± 0.0076 | 0.8130 ± 0.0111 | 0.0027 | 2.5725 |
| GIB | 0.8320 ± 0.0022 | 0.8370 ± 0.0081 | 0.0050 | 2.5993 |
| Upper GI | 0.8491 ± 0.0016 | 0.8514 ± 0.0056 | 0.0023 | 2.5530 |
| Abdominal pain | 0.8440 ± 0.0068 | 0.8455 ± 0.0105 | 0.0015 | 2.5325 |
| All | 0.8364 ± 0.0044 | 0.8387 ± 0.0049 | 0.0023 | 2.6739 |
The highest values are highlighted in bold
Although the proposed method shows promising performance, there may be three directions for further improvement: (1) considering more types of medical information such as diagnosis sequence beside laboratory test sequence and prescription sequence; (2) introducing values into the one-hot vectors of medical history at each time; (3) integrating medical knowledge.
Conclusions
In this study, a novel hybrid method of RNN and GNN, called RGNN, is proposed for next-period prescription prediction. RGNN represents patient longitudinal medical data from two views, that is, RNN for patient status sequence representation and GNN for temporal medical event graph representation. Experiments on a benchmark dataset prove the effectiveness of RGNN on next-period prescription prediction. RGNN can also be applied to other medical event prediction tasks.
Acknowledgements
This paper is supported in part by grants: National Natural Science Foundations of China (U1813215, 61876052 and 61573118), Special Foundation for Technology Research Program of Guangdong Province (2015B010131010), National Natural Science Foundations of Guangdong, China (2019A1515011158), Guangdong Province Covid-19 Pandemic Control Research Fund (2020KZDZX1222), Strategic Emerging Industry Development Special Funds of Shenzhen (JCYJ20180306172232154 and JCYJ20170307150528934) and Innovation Fund of Harbin Institute of Technology (HIT.NSRIF.2017052). We also thank Jin’s team for sharing their source code of decomposed LSTM.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Sicen Liu, Email: liusicen@stu.hit.edu.cn.
Tao Li, Email: litao@stu.hit.edu.cn.
Haoyang Ding, Email: Hongyang.DING@Yiducloud.cn.
Buzhou Tang, Email: tangbuzhou@gmail.com.
Xiaolong Wang, Email: wangxl@insun.hit.edu.cn.
Qingcai Chen, Email: qingcai.chen@gmail.com.
Jun Yan, Email: Jun.YAN@Yiducloud.cn.
Yi Zhou, Email: zhouyi@sysu.edu.cn.
References
- 1.Fu L, Li F, Zhou J, Wen X, Yao J, Shepherd M. Event prediction in healthcare analytics: beyond prediction accuracy. In: Cao H, Li L, Wang R, editors. Trends and applications in knowledge discovery and data mining. Cham: Springer International Publishing; 2016. pp. 181–189. [Google Scholar]
- 2.Bui C, Pham N, Vo A, Tran A, Nguyen A, Le T (2018) Time series forecasting for healthcare diagnosis and prognostics with the focus on cardiovascular diseases. In: Van Vo T, Le Nguyen TA, Nguyen Duc T (eds) 6th international conference on the development of biomedical engineering in Vietnam (BME6). Springer, Singapore, pp 809–818. 10.1007/978-981-10-4361-1_138
- 3.Mikolov T, Karafiat M, Burget L, Cernocky J, Khudanpur S (2010) Recurrent neural network based language model. In: Conference of the international speech communication association, pp 1045–1048
- 4.Suresh H, Hunt N, Johnson A, Celi LA, Szolovits P, Ghassemi M (2017) Clinical intervention prediction and understanding with deep neural networks. In: Proceedings of the 2nd machine learning for healthcare conference, PMLR 68, pp 322–337
- 5.Jin B, Yang H, Sun L, Liu C, Qu Y, Tong J (2018) A treatment engine by predicting next-period prescriptions. In: KDD’18: proceedings of the 24th ACM SIGKDD international conference on knowledge discovery & data mining. ACM Press, London, pp 1608–1616. 10.1145/3219819.3220095
- 6.An Y, Huang N, Chen X, Wu F, Wang J. High-risk prediction of cardiovascular diseases via attention-based deep neural networks. IEEE/ACM Trans Comput Biol Bioinformat. 2019 doi: 10.1109/TCBB.2019.2935059. [DOI] [PubMed] [Google Scholar]
- 7.Muisyo T, Bernardo EO, Camazine M, Colvin R, Thomas KA, Borgman MA, Spinella PC. Mortality prediction in pediatric trauma. J Pediatr Surg. 2019;54:1613–1616. doi: 10.1016/j.jpedsurg.2018.08.045. [DOI] [PubMed] [Google Scholar]
- 8.Rajkomar A, Oren E, Chen K, Dai AM, Hajaj N, Hardt M, Liu PJ, Liu X, Marcus J, Sun M, Sundberg P, Yee H, Zhang K, Zhang Y, Flores G, Duggan GE, Irvine J, Le Q, Litsch K, Mossin A, Tansuwan J, Wang D, Wexler J, Wilson J, Ludwig D, Volchenboum SL, Chou K, Pearson M, Madabushi S, Shah NH, Butte AJ, Howell MD, Cui C, Corrado GS, Dean J. Scalable and accurate deep learning with electronic health records. Npj Digital Med. 2018;1:18. doi: 10.1038/s41746-018-0029-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Meadows K, Gibbens R, Gerrard C, Vuylsteke A. Prediction of patient length of stay on the intensive care unit following cardiac surgery: a logistic regression analysis based on the cardiac operative mortality risk calculator. EuroSCORE, J Cardiothorac Vasc Anesth. 2018;32:2676–2682. doi: 10.1053/j.jvca.2018.03.007. [DOI] [PubMed] [Google Scholar]
- 10.Okada S, Shimada J, Teramukai S, Kato D, Tsunezuka H, Miyata N, Ishihara S, Furuya T, Nakazono C, Ishikawa N, Inoue M. Risk stratification according to the prognostic nutritional index for predicting postoperative complications after lung cancer surgery. Ann Surg Oncol. 2018;25:1254–1261. doi: 10.1245/s10434-018-6368-y. [DOI] [PubMed] [Google Scholar]
- 11.Ourselin S, Joskowicz L, Sabuncu MR, Unal G, Wells W (eds) (2016) Medical image computing and computer-assisted intervention—MICCAI 2016. In: 19th international conference, Athens, Greece, October 17–21, Part 2. Springer, Greece [DOI] [PubMed]
- 12.Wu X, Spitz MR, Lee JJ, Lippman SM, Ye Y, Yang H, Khuri FR, Kim E, Gu J, Lotan R, Hong WK. Novel susceptibility loci for second primary tumors/recurrence in head and neck cancer patients: large-scale evaluation of genetic variants. Cancer Prevent Res. 2009;2:617–624. doi: 10.1158/1940-6207.CAPR-09-0025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.McCormick T, Rudin C, Madigan D. A hierarchical model for association rule mining of sequential events: an approach to automated medical symptom prediction. SSRN J. 2011 doi: 10.2139/ssrn.1736062. [DOI] [Google Scholar]
- 14.Murphy KP. Machine learning: a probabilistic perspective. Cambridge: MIT Press; 2012. [Google Scholar]
- 15.Yao HR, Chang DC, Frieder O, Huang W, Lee TS (2019) Graph Kernel prediction of drug prescription. In: 2019 IEEE EMBS international conference on biomedical & health informatics (BHI). IEEE, Chicago, IL, pp 1–4. 10.1109/BHI.2019.8834676
- 16.Xie R, Wen J, Quitadamo A, Cheng J, Shi X. A deep auto-encoder model for gene expression prediction. BMC Genomics. 2017;18:11. doi: 10.1186/s12864-017-4226-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jagannatha AN, Yu H. Structured prediction models for RNN based sequence labeling in clinical text. Proc Conf Empir Methods Nat Lang Process. 2016;2016:856–865. doi: 10.18653/v1/d16-1082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jagannatha AN, Yu H. Bidirectional RNN for medical event detection in electronic health records. Proc Conf. 2016;2016:473–482. doi: 10.18653/v1/n16-1056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Delen D, Walker G, Kadam A. Predicting breast cancer survivability: a comparison of three data mining methods. Artif Intell Med. 2005;34:113–127. doi: 10.1016/j.artmed.2004.07.002. [DOI] [PubMed] [Google Scholar]
- 20.Choi E, Bahadori MT, Schuetz A, Stewart WF, Sun J. Doctor AI: predicting clinical events via recurrent neural networks. JMLR Workshop Conf Proceed. 2016;56:301–318. [PMC free article] [PubMed] [Google Scholar]
- 21.Baytas IM, Xiao C, Zhang X, Wang F, Jain AK, Zhou J (2017) Patient subtyping via time-aware LSTM networks. In: Proceedings of the 23rd ACM SIGKDD international conference on knowledge discovery and data mining. ACM, Halifax NS, pp 65–74. 10.1145/3097983.3097997
- 22.Esteva A, Robicquet A, Ramsundar B, Kuleshov V, DePristo M, Chou K, Cui C, Corrado G, Thrun S, Dean J. A guide to deep learning in healthcare. Nat Med. 2019;25:24–29. doi: 10.1038/s41591-018-0316-z. [DOI] [PubMed] [Google Scholar]
- 23.Zhou J, Cui G, Zhang Z, Yang C, Liu Z, Wang L, Li C, Sun M (2018) Graph neural networks: a review of methods and applications. ArXiv:1812.08434 [Cs, Stat] http://arxiv.org/abs/1812.08434. Accessed 4 Sept 2019
- 24.Scarselli F, Gori M, Tsoi AC, Hagenbuchner M, Monfardini G. The graph neural network model. IEEE Trans Neural Netw. 2009;20:61–80. doi: 10.1109/TNN.2008.2005605. [DOI] [PubMed] [Google Scholar]
- 25.Uddin S, Khan A, Piraveenan M (2015) Administrative claim data to learn about effective healthcare collaboration and coordination through social network. In: 2015 48th Hawaii international conference on system sciences, pp 3105–3114. 10.1109/HICSS.2015.375
- 26.Beck D, Haffari G, Cohn T (2018) Graph-to-sequence learning using gated graph neural networks. In: Proceedings of the 56th annual meeting of the association for computational linguistics (ACL), vol 1, pp 273–283
- 27.Kipf TN, Welling M (2017) Semi-supervised classification with graph convolutional networks. In: International conference on learning representations (ICLR)
- 28.Choi E, Bahadori MT, Song L, Stewart WF, Sun J (2016) GRAM: graph-based attention model for healthcare representation learning. ArXiv:1611.07012 [Cs, Stat]. http://arxiv.org/abs/1611.07012. Accessed 4 Sept 2019 [DOI] [PMC free article] [PubMed]
- 29.Ma T, Xiao C, Zhou J, Wang F (2018) Drug similarity integration through attentive multi-view graph auto-encoders. ArXiv:1804.10850 [Cs, Stat]. http://arxiv.org/abs/1804.10850. Accessed 1 Nov 2019
- 30.Shang J, Xiao C, Ma T, Li H, Sun J (2019) GAMENet: graph augmented memory networks for recommending medication combination. In: Thirty-third AAAI conference on artificial intelligence/thirty-first innovative applications of artificial intelligence conference/ninth AAAI symposium on educational advances in artificial intelligence, pp 1126–1133
- 31.Zitnik M, Agrawal M, Leskovec J. Modeling polypharmacy side effects with graph convolutional networks. Bioinformatics. 2018;34:i457–i466. doi: 10.1093/bioinformatics/bty294. [DOI] [PMC free article] [PubMed] [Google Scholar]




