Abstract
Background
Melanoma is aggressive and lethal melanocytic neoplasm, and its incidence has increased worldwide in recent decades. Accumulating evidence has showed that various long noncoding RNAs (lncRNAs) participated in occurrence of malignant tumors, including melanoma. The present study was designed to investigate function of lncRNA colon cancer-associated transcript-1 (CCAT1) in melanoma.
Methods
The expression levels of CCAT1, miR-296-3p and Integrin alpha9 (ITGA9) in melanoma tissues or cells were measured using real-time quantitative polymerase chain reaction (RT-qPCR). The concentrations of glucose and lactate were measured for assessing glycolysis of melanoma cells. 3-(4,5-dimethyl-2-thiazolyl)-2,5-diphenyl-2H-tetrazol-3-ium bromide (MTT), flow cytometry, and transwell assays were conducted to assess proliferation, apoptosis, and migration of melanoma cells. Western blot assay was performed to measure the protein expression of ITGA9, hexokinase 2 (HK2), and epithelial–mesenchymal transition (EMT)-related proteins in melanoma tissues or cells. The relationship among CCAT1, miR-296-3p, and ITGA9 was predicted and confirmed by bioinformatics analysis, dual-luciferase reporter, and RNA immunoprecipitation (RIP) assay, respectively. A xenograft experiment was established to assess the effect of CCAT1 knockdown in vivo.
Results
CCAT1 was effectively increased in melanoma tissues and cells compared with matched controls, and deficiency of CCAT1 impeded cell glycolysis, proliferation, migration while induced apoptosis, which were abrogated by knockdown of miR-296-3p in melanoma cells. In addition, our findings revealed that ITGA9 overexpression abolished miR-296-3p overexpression-induced effects on melanoma cells. Importantly, CCAT1 regulated ITGA9 expression by sponging miR-296-3p. The results of xenograft experiment suggested that CCAT1 silencing inhibited melanoma cell growth in vivo.
Conclusion
LncRNA CCAT1 promoted ITGA9 expression by sponging miR-296-3p in melanoma.
Keywords: lncRNA CCAT1, miR-296-3p, ITGA9, melanoma
Introduction
Melanoma is a common diagnosed cancer that responsible for 80% of deaths from all skin cancer, with high aggressive and metastasis.1,2 Although technology had made progress, melanoma patients still carries a poor prognosis with high incidence rates and high mortality.3 In the recent years, early diagnosis, control metabolism, and surgical/drug intervention were the good treatment ways for melanoma patients,4–6 nevertheless, pathogenesis of metastatic melanoma at the molecular level remained largely unknown, which needed to be further investigated.7,8 Thus, it was urgently to screen effective and specific biomarkers for early detection and diagnosis of melanoma.
Long noncoding RNA (lncRNA) composed of more than 200 nucleotides in length and were commonly regarded as untranslational transcripts without protein-coding ability.9 However, lncRNA could limit regulatory effect of miRNA on mRNA expression by acting as miRNA sponge.10 In addition, similar to protein-coding genes, lncRNA has important roles in diverse cellular processes by silencing miRNA, which was a canonical competing endogenous RNA (ceRNA) regulating pattern; moreover, frequent dysregulation of lncRNA already associated with many vital cellular processes and pathogenesis of tumors.11,12 A study has shown that abnormal expression of lncRNA participated in initiation, progression, metastasis, and drug resistance of human malignancies.13 For instance, colon cancer-associated transcript-1 (CCAT1) has been defined as an oncogenic lncRNA in glioma and acute myeloid leukemia.14,15 In addition, Chen et al identified that noncoding RNA (MHENCR) was increased in melanoma tissues, and mechanism experiment revealed that MHENCR as a competitively endogenous RNA enhanced insulin-like growth factor 1 (IGF1) expression by specifically sponging miR-425 and miR-489, and further activated protein kinase B pathway in vitro.16 In addition, Tang et al found HOTAIR was overexpressed in metastatic tissue, which involved in the cell motility, invasion, and metastasis of melanoma.17 Chen et al affirmed the therapeutic potential of lncRNA in the future cancer treatment.13 In conclusion, various lncRNAs have been reported to be associated with the oncogenesis of melanoma.18 Mechanistically, aberrant expression of lncRNA was involved in cancer cell dysfunction and many molecules processes by modulating transcriptional regulation, dysregulation of multiple targets and pathways, so as to cause the occurrence pathogenesis of tumors. Recently, CCAT1 has been proposed to be an oncogenic lncRNA that was associated with initiation and progression of multiple cancers.19,20 Importantly, Lv et al reported that the silencing of CCAT1 remarkably curbed proliferation, migration, and invasion of melanoma cells.21
MicroRNAs (miRNAs), noncoding RNAs with approximate 22 nucleotides in length, play a key role during the development of malignant tumor due to their regulatory effects on gene expression by functioning as repressor of translation and gene expression.22 For instance, Muhammad et al reported that miR-203 targeted cytokine signaling 3 (SOCS3) to regulate proliferation and mammospheres formation of breast cancer cells.23 In addition, the amount of study results have shown that many miRNAs were dysregulated in melanoma,24 indicating the miRNAs might function as prognostic markers for this disease.
Therefore, further investigation on the mechanism by which CCAT1 inducing might provide the novel therapies for melanoma. In this study, we will probe the underlying mechanisms by which lncRNA CCAT1 mediating effects in melanoma.
Materials and Methods
Clinical Samples
The study was approved by the Ethics Committee of Xi’an Central Hospital Affiliated to Xi’an Jiaotong University and performed according to the Declaration of Helsinki. Human melanoma tissues, including 40 primary melanomas and 40 adjacent normal tissues (away from the primary tumors at least 3 cm), were collected from all of participants prior to surgery at Xi’an Central Hospital Affiliated to Xi’an Jiaotong University, promptly frozen in liquid nitrogen and then immediately stored at −80°C until RNAs or proteins were extracted. All tumor tissue samples were confirmed as melanoma tissues by detailed histological diagnosis. The written informed consent was provided for all patients or their guardians.
Cells Culture
Melanoma cell lines (A375, SK-MEL-28, and A875) and normal human epidermal melanocytes (NHEM) were obtained from China center for type culture collection (Wuhan, China). Cells were cultured in IMDM medium (GIBCO BRL, Grand Island, NY, USA) supplemented with 10% fetal bovine serum (GIBCO BRL), 100 unit/L of penicillin (GIBCO BRL), and 100 μg/L of streptomycin (GIBCO BRL) in a humidified incubator containing 5% CO2 at 37°C.
Plasmid Construction, RNA Interference and Cell Transfection
Specific small interfering RNA (siRNA) against CCAT1 (si-CCAT1) and its negative control (si-NC), miR-296-3p inhibitor (anti-miR-296-3p) and its negative control (anti-miR-NC), miR-296-3p mimic (miR-296-3p) and its negative control (miR-NC), overexpressed vector of ITGA9 (ITGA9) and its negative control (vector) and specific short hairpin RNA (shRNA) objecting to CCAT1 (sh-CCAT1) and shRNA scrambled control (sh-NC) were purchased from RiboBio (Guangzhou, China). The above oligonucleotides or plasmids were transfected into melanoma cells utilizing Lipofectamine 2000 reagent (Thermo Fisher Scientific, Waltham, MA, USA) referring to manufacturer’s instructions. After transfection 48 h, A375 and A875 cells were collected for subsequence assays.
RNA Extraction and Real-Time Quantitative Polymerase Chain Reaction (RT-qPCR)
Total RNA was isolated from tissues or cells using TRIzol reagent (Invitrogen, Carlsbad, CA, USA) according to the instructions of manufacturer. RNA was quantified on NanoDrop ND-1000 (Applied Biosystems, Foster City, CA, USA) and then synthesized complementary DNA using Prime Script RT Reagent kit (Takara, Dalian, China) or microRNA Reverse Transcription Kit (Thermo Fisher Scientific). RT-qPCR was carried out using SYBR-Green PCR kit (Takara) on ABI 7500 HT system (Applied Biosystems). The relative expression levels of genes were calculated by 2−ΔΔCt method. Relative expression was normalized to glyceraldehyde-3-phosphate dehydrogenase (GAPDH) for CCAT1 and Integrin alpha9 (ITGA9) or endogenous small nuclear RNA U6 for miR-296-3p.
The sequences of the specific primers were listed:
CCAT1 (Forward, 5ʹ-GCCGTGTTAAGCATTGCGAA-3ʹ; Reverse, 5ʹ- TCATGTCTCGGCACCTTTCC-3ʹ);
miR-296-3p (Forward, 5ʹ-TGCCTAATTCAGAGGGTTGG-3ʹ; Reverse, 5ʹ-CTCCACTCCTGGCACACAG-3ʹ);
ITGA9 (Forward, 5ʹ-TCGTTCTTCGGCTACGCAG-3ʹ; Reverse, 5ʹ- CCCCATCCACTCATCATCGC −3ʹ);
GAPDH (Forward, 5ʹ-TCCCATCACCATCTTCCAGG-3ʹ; Reverse, 5ʹ-GATGACCCTTTTGGCTCCC-3ʹ);
U6 (Forward, 5ʹ-CTCGCTTCGGCAGCACA-3ʹ; Reverse, 5ʹ-AACGCTTCACGAATTTGCGT-3ʹ).
Glucose Consumption and Lactate Production Assay
After 48 hours transfection, supernatant of A375 and A875 cells was collected to for measurement of glucose concentration with Glucose Assay Kit (Nanjing Jiancheng Bioengineering Institute, Nanjing, China) in line with the manufacturer’s manuals, Relative glucose consumption was quantified by subtracting the amount of glucose present in cell culture medium without any cells. Analogously, concentration of lactate production was measured with lactate assay kit (Solarbio, Beijing, China) as instructed by the manufacturer. Moreover, the standard curve was established to calculate the test samples.
3-(4,5-Dimethyl-2-Thiazolyl)-2,5-Diphenyl-2H-Tetrazol-3-Ium Bromide (MTT) Assay
Exponentially growing cells were seeded onto 96-well plates (3000 cells/well) with 200 μL of medium. A375 and A875 cells were cultured in an incubator at 37°C for indicated time. Subsequently, 20 μL of MTT (Thermo Fisher Scientific; 5 mg/mL) was added to each well and cultured for an additional 4 h. Dimethyl sulfoxide (DMSO) was used to dissolve the generated formazan crystals in 96-well plates after discarding supernatant. In general, cell viability was determined by measuring absorbance at 490 nm wavelength on a microplate reader (Applied Biosystems) at 0 h, 24 h, 48 h and 72 h post-transfection, respectively.
Flow Cytometry
Briefly, transfected A375 and A875 cells were washed with cold phosphate buffer saline and suspended in binding buffer. Apoptotic cells were stained with Annexin V-fluorescein isothiocyanate (FITC)/Propidium Iodide (PI) (Thermo Fisher Scientific) for 30 min as per the manufacturer’s instructions. Finally, apoptotic rate was calculated using a FACScan flow cytometer (Becton Dickinson, San Jose, CA, USA). Furthermore, cell cycle was determined with cell cycle detection CycleTEST PLUS DNA Reagent Kit (BD Biosciences) as described by Liu et al.25
Cell Migration Assay
Melanoma cells (2×105) were seeded into the transwell upper chambers wells of 24-well with 8-μm poly (ethylene terephthalate) membranes (Millipore Corp, Billerica, MA, USA) with 150 μL of serum-free medium, while lower wells were added 600 μL of the complete medium with 10% fetal bovine serum. After incubation for 48 h, cotton swabs were used to remove non-migrating cells. The migrated cells were fixed with 20% methanol and stained with 0.1% crystal violet for 15 min. Cell migration assay was performed by counting cells in five random fields from each of the migration chambers under a microscope (Applied Biosystems). Cell migration rate was analyzed with Image J software (National Institutes of Health, Bethesda, MD, USA).
Western Blot Assay
A375 and A875 cells were harvested with RIPA buffer (Thermo Fisher Scientific) in compliance with the manufacturer’s instructions. Then, the protein concentration was examined using BCA Protein Assay Kit (Thermo Fisher Scientific). Equal amount protein samples were loaded on sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and transferred onto polyvinylidene fluoride (PVDF) membranes. Subsequently, membranes were blocked with 1% bovine serum albumin for 1 h at room temperature and probed with primary antibodies at 4°C overnight, and GAPDH (1:2000 dilution; Abcam, Cambridge, MA, USA) was used for normalization. After extensive washing, the membranes were incubated with a goat anti-rabbit horseradish peroxidase-conjugated secondary antibody (1:2000 dilution; Abcam) for 1 h. Finally, protein bands were visualized and analyzed with enhanced chemiluminescent method and Image J software (National Institutes of Health), respectively. The primary antibodies were listed as follow: hexokinase 2 (HK2; 1:1000 dilution; Abcam), E-cadherin (1:1000 dilution; Abcam), vimentin (1:1000 dilution; Abcam), N-cadherin (1:1000 dilution; Abcam), ITGA9 (1:1000 dilution; Abcam). ProliferatingCellNuclearAntigen (PCNA; 1:1000 dilution; Abcam), cyclin D1 (1:1000 dilution; Abcam), B-cell lymphoma-2 (Bcl-2; 1:1000 dilution; Abcam).
Dual-Luciferase Reporter Assay
Online database Starbase (http://starbase.sysu.edu.cn/) and TargetScan (http://www.targetscan.org/vert_72/) were utilized to predict the potential target gene of CCAT1 and miR-296-3p, respectively. In addition, to construct the luciferase report vector, the sequences containing the presumed putative binding sites on miR-296-3p were amplified by PCR and then cloned into the luciferase report vectors (Ambion, Foster City, CA, USA) to generate the wild-type reporter or mutant-type reporter. A375 and A875 cells were co-transfected with miR-296-3p mimic or miR-NC and 500 nM of vector using Lipofectamine 2000 (Thermo Fisher Scientific) following the standard protocol. Luciferase activities were detected at 24 h after transfection using Dual-Luciferase Reporter Assay System (Thermo Fisher Scientific). The results were expressed as relative luciferase activity, and luciferase of Renilla was used as an internal control.
RNA Immunoprecipitation (RIP) Assay
RIP assay was implemented using a Magna RIP RNA-Binding Protein Immunoprecipitation kit (Millipore) in line with the manufacturer’s manuals. In short, A375 and A875 cells were harvested and lysed in lysis buffer, then 200 μL of lysates were incubated with magnetic beads conjugated with anti-Ago2 antibody (Abcam), with IgG as a reference control. After washing off unbound material, RNA was purified with proteinase K and extracted for RT-qPCR assay as above description.
Tumor Xenografts Growth in Nude Mice
All animal studies were performed according to instruction of the American Veterinary Medical Association guidelines, which was permitted by the Institutional Animal Care and Use Committee of Xi’an Central Hospital Affiliated to Xi’an Jiaotong University. A total of 12 BALB/c nude mice (Shanghai Experimental Animal Center, Shanghai, China) were housed in specific pathogen-free conditions. A375 (1×105 cells) transfected with sh-CCAT1 or sh-NC were suspended in 200 μL of free culture medium and then subcutaneously injected into right flank of nude mice. The tumor sizes were recorded at the indicated times based on formula: V = 1/2× ab2 method (maximum (a) and minimum (b) length of the tumor). After 35 days the mice were euthanized, and the tumors were removed for the weight detection and other experiments.
Statistical Analysis
All quantitative data were expressed as mean ± standard deviation. GraphPad Prism 7 (GraphPad Inc, La Jolla, CA, USA) was performed to assess P value between 2 groups or more groups with Student’s t-test or one-way analysis of variance, respectively. Statistically significant was defined at P value less than 0.05.
Results
CCAT1 Was Upregulated in Human Melanoma Tissues and Cells
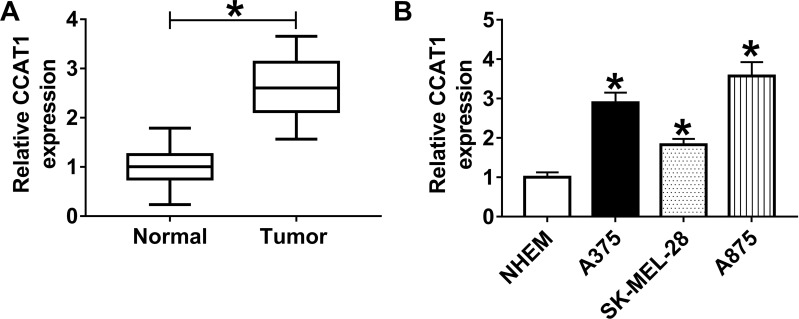
To confirm the previous observations, we first examined the expression level of CCAT1 in 40 melanoma tissue samples and corresponding non-tumor tissues by RT-qPCR. The data showed that CCAT1 level was remarkably elevated in melanoma tissues in comparison with matched normal tissues (Figure 1A). Analogously, CCAT1 was remarkably increased in melanoma cell lines compared with NHEM cells (Figure 1B). These results indicated that CCAT1 was upregulated in melanoma tissues and cells.
Figure 1.
The expression level of CCAT1 in melanoma tissues and cells. (A and B) RT-qPCR assay was performed to examine the expression level of CCAT1 in 40 pairs of melanoma tissues and adjacent normal tissues, as well as in melanoma cell lines (A375, SK-MEL-28, and A875) and normal human epidermal melanocytes (NHEM cells). *P < 0.05.
CCAT1 Knockdown Suppressed Glycolysis, Proliferation and Migration While Accelerated the Apoptosis of Melanoma Cells
As CCAT1 level was increased in human melanoma tissues and cells, we knocked down CCAT1 expression in A375 and A875 cells. As shown in Figure 2A and B, the loss-of-function experiments showed that CCAT1 was remarkably decreased in A375 and A875 cells treatment with si-CCAT1 compared with si-NC group. In addition, glucose consumption and lactate production were declined in A375 and A875 cells transfected with si-CCAT1 compared with si-NC group, suggesting glucose metabolism was repressed by CCAT1 knockdown (Figure 2C–F). Consistently, CCAT1 knockdown triggered a decrease of HK2 expression in A375 and A875 cells (Figure 2G and H). MTT assay indicated that CCAT1 knockdown inhibited proliferation of A375 and A875 cells (Figure 2I and J). Additionally, enhancement of apoptosis in A375 and A875 cells was observed in si-CCAT1 group when compared with si-NC group (Figure 2K). G0/G1 phase-cells were increased and S phase-cells were decreased in si-CCAT1 group when compared with si-NC group (Figure 2L and M). In addition, PCNA, cyclin D1, and Bcl-2 were declined in melanoma cells after silencing of CCAT1 (Figure 2N and O). Cell migration capacity was significantly decreased in A375 and A875 cells transfected with interference sequence of CCAT1 (Figure 2P and Q). Also, we estimated the expression levels of the epithelial marker E-cadherin and the mesenchymal markers vimentin and N-cadherin expression in A375 and A875 cells. The results revealed that E-cadherin was highly expressed, in contrast, the expression levels of N-cadherin and vimentin were markedly downregulated in A375 and A875 cells after silencing of CCAT1 (Figure 2R and S). Above results showed that the absence of CCAT1 might play an important suppressor role during the development of melanoma.
Figure 2.
CCAT1 knockdown suppressed proliferation, glycolysis, and migration, while induced apoptosis and cell cycle arrest in melanoma cells. (A–S) A375 and A875 cells were transfected with si-NC or si-CCAT1. (A and B) The expression level of CCAT1 was analyzed by RT-qPCR assay in transfected A375 and A875. (C–F) The glucose consumption and lactate production level were displayed in A375 and A875 at post-transfection. (G and H) Western blot analysis was used for analysis of HK2 level in A375 and A875. (I and J) MTT assay was utilized to estimate the cell viability of A375 and A875 cells at the indicated time points. (K–M) Apoptosis rate and cell cycle of melanoma cell lines (A375 and A875) were detected using the flow cytometry assay. (N and O) The expression levels of PCNA, cyclin D1, and Bcl-2 were assessed by Western blot analysis. (P and Q) Cells capacity of migration was evaluated by transwell assay in melanoma cells transfected with si-NC or si-CCAT1. (R and S) The EMT markers (E-cadherin, vimentin and N-cadherin) in melanoma cells were assessed by Western blot analysis. *P < 0.05.
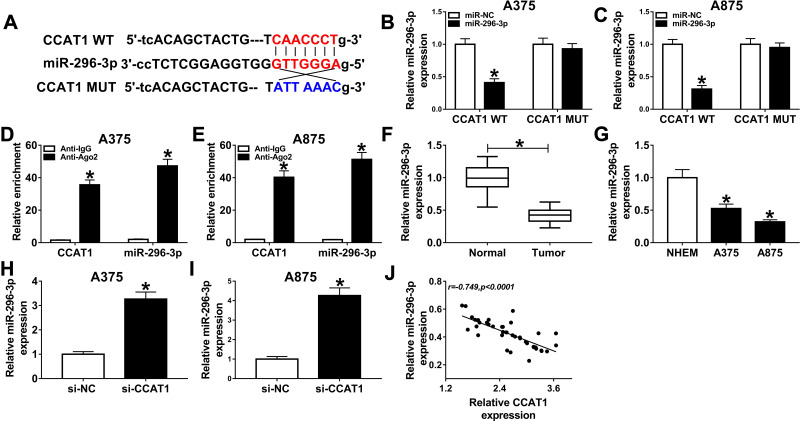
MiR-296-3p was a potential direct target of CCAT1 in melanoma cells. To identify potential target genes of CCAT1, StarBase online database was used in the current study. As presented in Figure 3A, CCAT1 had the potential binding sites on miR-296-3p. The luciferase reporter was established to confirm whether miR-296-3p was a true molecular target of CCAT1. Luciferase activity results revealed miR-296-3p mimic effectively decreased the luciferase activity of the CCAT1 WT report vector in melanoma cells, while luciferase activity had no significant change in CCAT1 MUT (Figure 3B and C). RIP results suggested that CCAT1 and miR-296-3p were enriched in anti-Ago2 group compared with control group, confirming the binding relationship between CCAT1 and miR-296-3p (Figure 3D and E). Next, we investigated the expression level of miR-296-3p in melanoma tissues and cells. MiR-296-3p was declined in melanoma tissues and cells in comparison with matched controls (Figure 3F and G). As shown in Figure 3H and I, knockdown of CCAT1 dramatically enhanced expression of miR-296-3p expression in A375 and A875 cells. Interestingly, miR-296-3p was negatively correlated with CCAT1 in melanoma tissues (Figure 3J). Collectively, the above data illustrated that miR-296-3p was negatively regulated by CCAT1 in melanoma cells.
Figure 3.
CCAT1 negatively regulated miR-296-3p expression in melanoma cells. (A) Putative complementary sites of miR-296-3p on CCAT1, and the matched mutant sites were shown. (B–E) The binding association between miR-296-3p and CCAT1 was confirmed with dual-luciferase reporter and RIP assays. (F and G) RT-qPCR assay was applied to measure the expression level of miR-296-3p in melanoma tissues and cells together with matched control groups. (H and I) The expression level of miR-296-3p was measured by RT-qPCR assay in melanoma cells (A375 and A875) transfected with si-NC, or si-CCAT1. (J) The relationship between miR-296-3p and CCAT1 was analyzed in melanoma tissues. *P < 0.05.
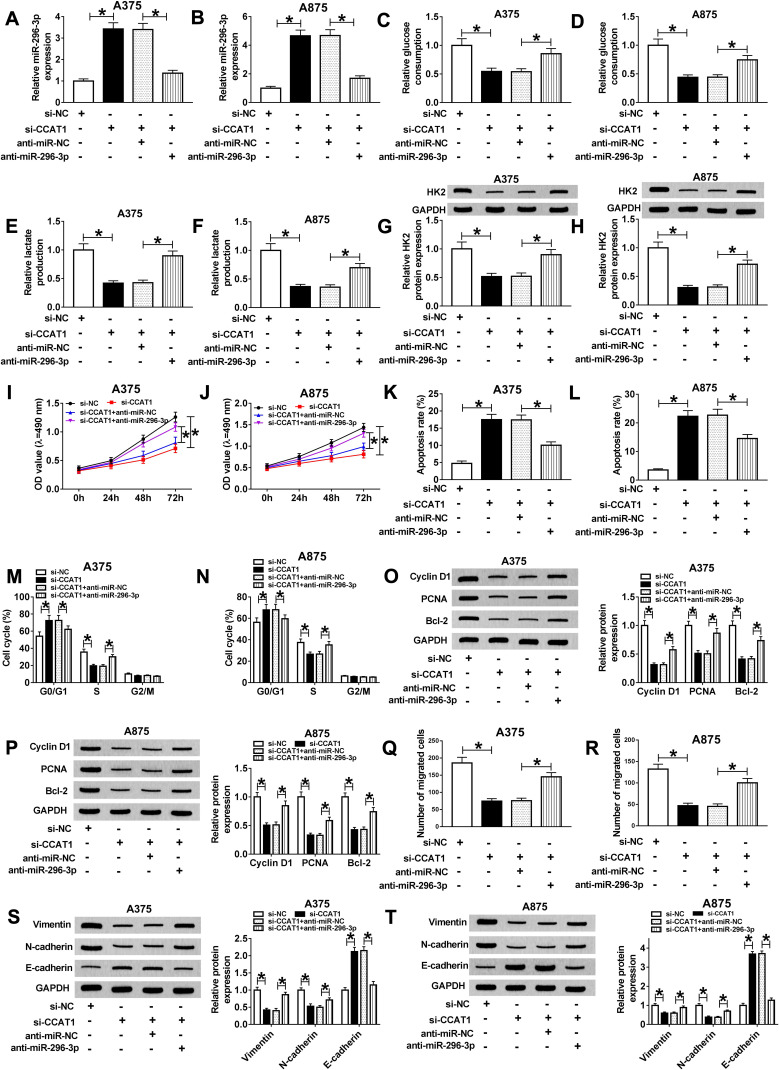
MiR-296-3p Knockdown Recovered the CCAT1 Silencing-Mediated Effects on Glycolysis, Proliferation, Migration, and Apoptosis of Melanoma Cells
To observe the regulatory mechanism between CCAT1 and miR-296-3p in melanoma cells, functional experiments were performed to investigate the effects of CCAT1 and miR-296-3p on glycolysis, proliferation, migration, and apoptosis of melanoma cells. As presented in Figure 4A and B, knockdown of CCAT1 dramatically increased expression of miR-296-3p, while transfection of miR-296-3p inhibitor evidently abolished this reaction. The glucose consumption and lactate production were increased in A375 and A875 cells co-transfected with si-CCAT1 and anti-miR-296-3p compared with those only transfected with si-CCAT1, revealing that miR-296-3p inhibitor could partially recuperate the inhibitory effect of si-CCAT1 on glucose metabolism (Figure 4C–F). Moreover, the downregulation of miR-296-3p abolished the repression effect on HK2 expression in A375 and A875 cells caused by CCAT1 knockdown (Figure 4G and H). MTT analysis revealed that proliferation was remarkably repressed in A375 and A875 cells transfected with si-CCAT1, which was abrogated after co-transfection with si-CCAT1 and miR-296-3p inhibitor (Figure 4I and J). In addition, flow cytometry analysis showed that high apoptosis rates of both A375 and A875 cells were triggered by CCAT1 knockdown, analogously, miR-296-3p knockdown weakened enhancement of apoptosis in A375 and A875 cells induced by si-CCAT1 (Figure 4K and L). The cell cycle arrest in A375 and A875 cells induced by si-CCAT1 was abolished by anti-miR-296-3p (Figure 4M and N). The downregulation of PCNA, cyclin D1, and Bcl-2 in si-CCAT1 induced A375 and A875 cells were wreaked by silencing of miR-296-3p (Figure 4O and P). Furthermore, CCAT1 knockdown weakened the migration ability of A375 and A875 cells, which was eliminated by knockdown of miR-296-3p (Figure 4Q and R). Additionally, knockdown of CCAT1 drastically hindered the expression of Vimentin and N-cadherin while reinforced E-cadherin expression, which were inverted by miR-296-3p knockdown (Figure 4S and T). These data revealed that CCAT1 knockdown-mediated effect on glycolysis, proliferation, migration, and apoptosis in melanoma cells by targeting miR-296-3p in vitro.
Figure 4.
Interference of miR-296-3p restored the CCAT1 knockdown-mediated effects on glycolysis, proliferation, migration, apoptosis, and cell cycle of melanoma cells. (A–T) A375 and A875 cells were transfected with si-NC, si-CCAT1, si-CCAT1+anti-miR-NC or si-CCAT1+anti-miR-296-3p. (A and B) The expression level of miR-296-3p in transfected A375 and A875 cells was detected by RT-qPCR. (C–F) Glucose consumption and lactate production levels in A375 and A875 were displayed. (G and H) The protein expression level of HK2 was detected with Western blot assay in A375 and A875. (I and J) MTT assay was conducted to evaluate the viability of transfected A375 and A875 cells. (K–N) Apoptosis and cell cycle of A375 and A875 cells were detected by flow cytometry assay. (O and P) The expression levels of PCNA, cyclin D1, and Bcl-2 were assessed by Western blot analysis. (Q and R) The migration ability of treated melanoma cells was measured by transwell assay. (S and T) Western blot was used to estimate the expression levels of EMT-related proteins in A375 and A875 cells at post-transcription. *P < 0.05.
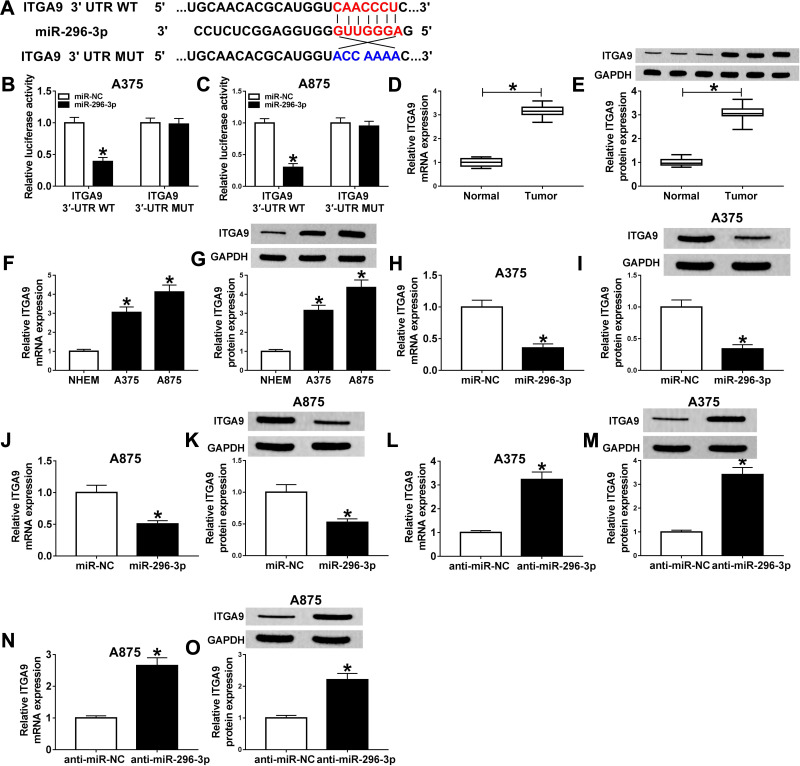
ITGA9 Was a Target of MiR-296-3p in Melanoma Cells
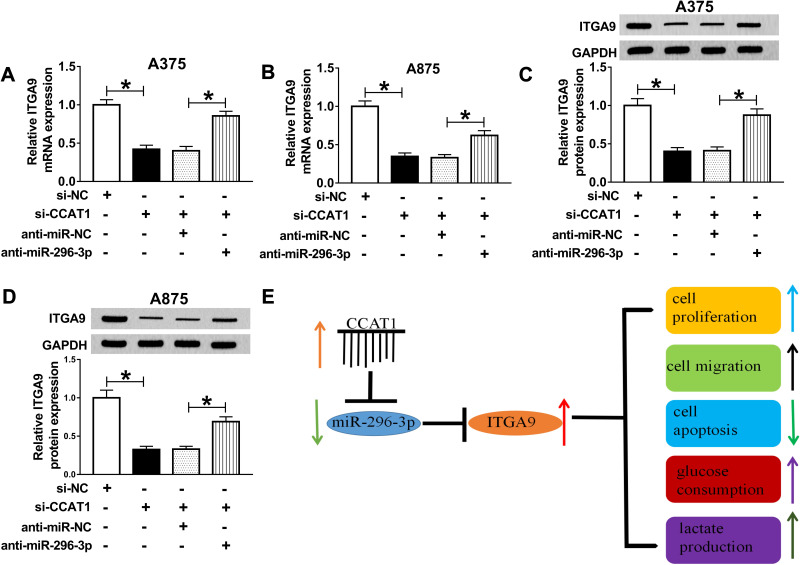
The binding sequences of miR-296-3p on 3ʹ UTR of ITGA9 and its mutant were shown in Figure 5A. To investigate whether ITGA9 mRNA was a target of miR-296-3p in melanoma cells, dual-luciferase report assay was conducted. The results showed that enforced expression of miR-296-3p inhibited the luciferase activity of the ITGA9 3ʹUTR WT report vector compared to negative control, while similar result was not observed in ITGA9 3ʹUTR MUT report vector (Figure 5B and C). Subsequently, we investigated the mRNA and protein expression of ITGA9 in melanoma tissues and cells, and ITGA9 was overexpressed in melanoma tissues and cells relative to corresponding control, no matter mRNA or protein (Figure 5D–G). In addition, the expression level of ITGA9 was effectively downregulated in A375 and A875 cells transfected with miR-296-3p compared with control (Figure 5L–O). Moreover, knockdown of miR-296-3p inhibited ITGA9 expression in melanoma cells (Figure 5H–K). From these data, it was speculated that ITGA9 was a direct target of miR-296-3p in melanoma cells.
Figure 5.
ITGA9 was a direct target of miR-296-3p in melanoma cells. (A) The predicted binding sequences between miR-296-3p on 3ʹ UTR of ITGA9, and its mutant were shown. (B and C) Dual-luciferase reporter assay was carried out to assess luciferase activity in A375 and A875 cells following co-transfection with wild or mutant reporter vector and miR-296-3p or miR-NC, respectively. (D–G) The mRNA and protein levels of ITGA9 were measured by RT-qPCR and Western blot assays in melanoma tissues and cells, respectively. (H–O) The expression levels of ITGA9 were evaluated with RT-qPCR and Western blot assays in A375 and A875 cells transfected with miR-NC, or miR-296-3p, as well as in A375 and A875 cells transfected with anti-miR-NC, or anti-miR-296-3p. *P < 0.05.
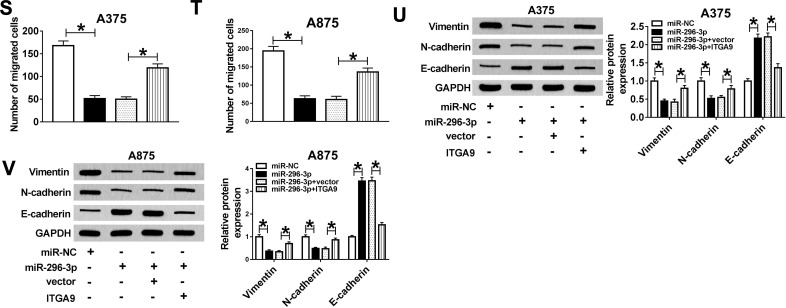
Overexpression of MiR-296-3p Impeded Glycolysis, Proliferation, Migration While Induced Apoptosis in Melanoma Cells Through Targeting ITGA9
According to above results, overexpression of miR-296-3p inhibited ITGA9 expression in melanoma cells. A375 and A875 cells were transfected with miR-NC, miR-296-3p, miR-296-3p+vector, or miR-296-3p+ITGA9. As shown in Figure 6A–D, elevated expression of miR-296-3p obviously inhibited ITGA9 expression, while gain of ITGA9 overturned these effects. In addition, as indicated in Figure 6E–H, miR-296-3p treatment evidently decreased glucose consumption and lactate production in A375 and A875 cells, which were rescued through upregulation of ITGA9. We also analyzed the level of HK2 by Western blot assay. The results revealed that upregulation of ITGA9 apparently reversed the inhibition effect on HK2 expression A375 and A875 cells caused by miR-296-3p mimic (Figure 6I and J). MTT assay suggested that the upregulation of ITGA9 abolished the inhibition impact on cell proliferation caused by miR-296-3p (Figure 6K and L). The high apoptosis rate of melanoma cells induced by upregulation of miR-296-3p was abrogated by treatment with overexpressed vector of ITGA9 (Figure 6M and N). As shown in Figure 6O and P, miR-296-3p induced-effects on cell cycle were abolished by overexpression of ITGA9. Furthermore, overexpression of ITGA9 counteracted the suppressive effects on PCNA, cyclin D1, and Bcl-2 expression in melanoma cells (Figure 6Q and R). Analogously, introduction with ITGA9 also restored loss of migration capability in melanoma cells induced by miR-296-3p mimic (Figure 6S and T). Finally, biomarkers of epithelial–mesenchymal transition (EMT) process were measured in melanoma cells after transfection by Western blot assay, and the data showed that overexpression of miR-296-3p increased E-cadherin and decreased Vimentin and N-cadherin in melanoma cells, which were overturned by ITGA9 overexpression (Figure 6U and V). Collectively, miR-296-3p overexpression inhibited glucose metabolism, proliferation and migration, as well as induced apoptosis of melanoma cells by targeting ITGA9 expression.
Figure 6.
Continued.
Figure 6.
MiR-296-3p overexpression induced-effects on glycolysis, proliferation, migration, apoptosis, and cell cycle were abolished by overexpression of ITGA9 in melanoma cells. (A–V) A375 and A875 cells were transfected with miR-NC, miR-296-3p, miR-296-3p+vector or miR-296-3p+ITGA9. (A–D) RT-qPCR and Western blot assay were used to assess the expression levels of ITGA9 in transfected A375 and A875 cells. (E–H) The consumption of glucose and lactate production was measured. (I and J) The protein expression level of HK2 was examined with Western blot assay. (K and L) MTT assay was performed for examining the cell viability of A375 and A875 cells. (M–P) The apoptotic rate and cell cycle were quantified in transfected A375 and A875 cells by flow cytometry analysis. (Q and R) The Western blot assay was used to assess the expression levels of PCNA, cyclin D1, and Bcl-2 in A375 and A875 cells. (S and T) The migration ability of melanoma cells was evaluated with transwell assay. (U and V) Biomarkers of EMT process were measured by Western blot assay in melanoma cells after transfection. *P < 0.05.
CCAT1 Knockdown Inhibited ITGA9 Expression by Sponging MiR-296-3p
To determine the relationship between CCAT1 and ITGA9 in melanoma, we analyzed the expression levels of ITGA9 in transfected cells. RT-qPCR and Western blot assays indicated that mRNA and protein expression levels of ITGA1 were obviously downregulated in cells with CCAT1 knockdown, while introduction of miR-296-3p inhibitor effectively recovered effects of CCAT1 silencing on ITGA9 expression (Figure 7A–D). Furthermore, a schematic diagram of our findings is shown in Figure 7E. These results demonstrated that CCAT1 knockdown suppressed ITGA9 expression in melanoma cells by affecting miR-296-3p.
Figure 7.
CCAT1 knockdown inhibited ITGA9 expression by sponging miR-296-3p in vitro. (A–D) The mRNA and protein levels of ITGA9 were measured in A375 and A875 cells transfected with si-NC, si-CCAT1, si-CCAT1+anti-miR-NC or si-CCAT1+anti-miR-296-3p by RT-qPCR and Western blot assays, respectively. (E) The schematic diagram of our findings was shown. *P < 0.05.
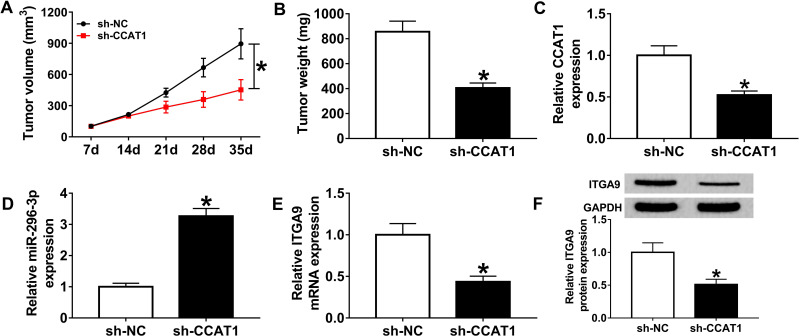
CCAT1 Silencing Impeded Tumor Growth in a Xenograft Model
As it was reported that knockdown of CCAT1 regulated proliferation of melanoma cells in vitro, we measured whether CCAT1 has a suppressive effect on tumor growth. The sh-CCAT1 group showed the smaller tumor volume and heavier tumor weight compared with sh-NC group (Figure 8A and B). Additionally, we also found that CCAT1 was declined in sh-CCAT1 group compared with sh-NC group (Figure 8C). Furthermore, the results of RT-qPCR and Western blot assay revealed that inhibition of CCAT1 inhibited ITGA9 expression while aggrandized miR-296-3p expression in removed tumor tissues (Figure 8D–F). In summary, all results confirmed that knockdown of CCAT1 inhibited tumor growth in vivo.
Figure 8.
CCAT1 knockdown impeded tumor growth in vivo. (A and B) The volume and weight of tumor were presented. (C and D) RT-qPCR assay was implemented to determine the expression levels of CCAT1 and miR-296-3p in removed tumor tissues. (E and F) The mRNA and protein levels of ITGA9 were measured in removed tumor tissues by RT-qPCR and Western blot assay, respectively. *P < 0.05.
Discussion
Recent study has discussed and reported that the ceRNAs interaction network and lncRNA-directed therapeutics were widespreadly existed in the pathogenesis of cancer.26 LncRNA functioned as key regulator in malignant tumors by binding miRNA to modulate mRNA expression, which was a common regulation mode.27,28 The review conducted by Aftab et al implied that miRNAs and lncRNAs were implicated during the development of melanoma, which might be useful biomarkers for future therapeutic.29 Not surprisingly, CCAT1 could act as biomarker in the prognosis of breast cancer patients, and its expression level was closely correlated with differentiation grades and lymph node metastases.30 Guo and Hua also reported that CCAT1 acted as diagnostic and prognostic biomarker in human cancers.31 Collectively, CCAT1 had the huge application value for diagnostic of malignant tumors.
The results from Prensner and Chinnaiyan have shown that lncRNA contributed to cancer treatment by exerting anti-metastatic function.32 Consistent with previous conclusions,21 our results suggested that the silencing of CCAT1 remarkably curbed proliferation, migration, and invasion of melanoma cells. Additionally, knockdown of CCAT1 impeded glycolysis and decreased vimentin and N-cadherin expression, while increased the expression E-cadherin in melanoma cells. As we all known, cell migration, invasion and cell substrate adhesion were regulated by epithelial–mesenchymal transition (EMT) process by altering the epithelial marker E-cadherin and mesenchymal markers Snail, vimentin, and N-cadherin,33 which was involved in metastasis and the development of tumor.34 Our study revealed the CCAT1 was highly expressed in melanoma tumor tissues compared with adjacent normal tissues, besides, we concluded that low-expression of CCAT1 increased the expression of E-cadherin and decreased expression of vimentin and N-cadherin in melanoma cells, which was an unfavorable factors for migration of melanoma cells. Consistently, Zhuang et al reported that CCAT1 was overexpressed in laryngeal squamous cell carcinoma that indicated poor prognosis and advanced clinical stage, and ectopic expression of CCAT1 suppressed E-cadherin expression while enhanced the expression of N-cadherin and Vimentin in tumor cells by targeting the let-7/HMGA2/Myc pathway.35 Additionally, mechanical analysis demonstrated that CCAT1 promoted glioma cell proliferation via inhibiting miR-410,14 as well as functioned as let-7 sponge in hepatocellular carcinoma.36 In this paper, miR-296-3p was a target genes of CCAT1 with dual-luciferase luciferase reporter and RIP assays. Additionally, miR-296-3p inhibitor recovered the CCAT1 siRNA-mediated effects on glycolysis, proliferation, migration, apoptosis, and EMT process in melanoma cells.
As for miR-296, it has been confirmed as a potential tumor suppressor in lung cancer,37 pancreatic cancer,38 and colorectal cancer.39 A previous study reported that miR-296-3p directly targeted the oncogenic protein mitogen-activated protein kinase-activated protein kinase-2 to weaken chemoresistance of nasopharyngeal carcinoma by inhibiting EMT.40 In this study, miR-296-3p was decreased in melanoma tissues and cells, and it was negatively regulated by CCAT1.
Additionally, ITGA9 has been predicted as a target gene of miR-296-3p by bioinformatics tools. ITGA9, an extracellular matrix component, exerted crucial function to regulate multi-step processes, including cell adhesion, migration and tissue invasion.41 Schreiber et al confirmed that ITGA9 played key roles in angiogenesis, proliferation, and migration in various cancers.42 Importantly, ITGA9 has been reported to be implicated in the EMT and contribute to cell adhesion in the extracellular matrix.43 Analogously, a previous research identified that miR-125b suppressed the EMT and cell invasion by targeting ITGA9 in melanoma.44 Therefore, we aimed to further insight into the role of CCAT1 in melanoma and its relationship to ITGA9. Currently, ITGA9 was upregulated in melanoma tissues and cells relative to corresponding controls. In addition, miR-296-3p overexpression-induced effects on melanoma cells could be restored by ITGA9 overexpression in vitro. Importantly, CCAT1 silencing increased ITGA9 expression by sponging miR-296-3p by mechanism experiments. Nevertheless, certain limitations in this should be listed: Our conclusion was acquired with a limited sample size, only 40 samples were collected; Considering CCAT1 acted as an oncogene in high aggressive melanoma cells, the overexpression of CCAT1 in less aggressive cells might be a meaningful research. Although our data suggested that CCAT1/miR-296-3p/ITGA9 axis plays vital function in primary function of human melanoma cells such as proliferation, invasion, and migration, and the detailed mechanism that CCAT1/miR-296-3p/ITGA9 axis controlled biological behavior need be explored in the next work.
We demonstrated that CCAT1 affected glycolysis, proliferation, migration, apoptosis and EMT in melanoma cells through regulation of miR-296-3p/ITGA9 axis. Mechanistically, our studies identified that CCAT1 was involved in melanoma process by acting as a competing endogenous RNA to promote ITGA9 by sponging miR-296-3p in melanoma.
Conclusion
In this study, we confirmed that CCAT1 exerted oncogene role by increasing expression of ITGA9 via targeting miR-296-3p, indicating that CCAT1, miR-296-3p and ITGA9 might be useful biomarkers or future treatment targets for melanoma patients. Mechanistically, CCAT1 functioned as competing endogenous RNA to sponge miR-296-3p, providing novel insight into the treatment of tumorigenesis melanoma in the future.
Highlights
CCAT1 was upregulated in melanoma cancer tissues and cells.
Silencing of CCAT1 repressed glycolysis, proliferation, and migration, while accelerated the apoptosis in melanoma cells.
CCAT1 regulated ITGA9 by sponging miR-296-3p was firstly reported.
Funding Statement
This work was hosted by the Xi’an Central Hospital Affiliated to Xi’an Jiaotong University.
Abbreviations
lncRNAs, long noncoding RNAs; CCAT1, colon cancer-associated transcript-1; ITGA9, Integrin alpha9; RT-qPCR, real-time quantitative polymerase chain reaction; HK2, hexokinase 2; EMT, epithelial–mesenchymal transition; RIP, RNA immunoprecipitation; IGF1, growth factor 1; NHEM, normal human epidermal melanocytes; shRNA, short hairpin RNA; GAPDH, glyceraldehyde-3-phosphate dehydrogenase; ITGA9, Integrin alpha9; FITC, fluorescein isothiocyanate; PI, propidium Iodide; SDS-PAGE, sodium dodecyl sulfate-polyacrylamide gel electrophoresis; PVDF, polyvinylidene fluoride.
Data Sharing Statement
The analyzed data sets generated during the present study are available from the corresponding author on reasonable request.
Ethics Approval and Consent to Participate
The present study was approved by the ethical review committee of Xi’an Central Hospital Affiliated to Xi’an Jiaotong University. Written informed consent was obtained from all enrolled patients.
Author Contributions
All authors made substantial contributions to conception and design, acquisition of data, or analysis and interpretation of data; took part in drafting the article or revising it critically for important intellectual content; gave final approval of the version to be published; and agree to be accountable for all aspects of the work.
Disclosure
The authors declare that they have no competing interests.
References
- 1.Miller AJ, Mihm MC. Melanoma. N Engl J Med. 2006;355(1):51–65. doi: 10.1056/NEJMra052166 [DOI] [PubMed] [Google Scholar]
- 2.McNally RJQ, Basta NO, Errington S, James PW, Norman PD, Craft AW. Socioeconomic patterning in the incidence and survival of children and young people diagnosed with malignant Melanoma in Northern England. J Investig Dermatol. 2014;134(11):2703–2708. doi: 10.1038/jid.2014.246 [DOI] [PubMed] [Google Scholar]
- 3.Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2018;68(6):394–424. doi: 10.3322/caac.21492 [DOI] [PubMed] [Google Scholar]
- 4.Malvi P, Chaube B, Singh SV, et al. Elevated circulatory levels of leptin and resistin impair therapeutic efficacy of dacarbazine in melanoma under obese state. Cancer Metab. 2018;6:2. doi: 10.1186/s40170-018-0176-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Malvi P, Chaube B, Singh SV, et al. Weight control interventions improve therapeutic efficacy of dacarbazine in melanoma by reversing obesity-induced drug resistance. Cancer Metab. 2016;4:21. doi: 10.1186/s40170-016-0162-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mohammad N, Malvi P, Meena AS, et al. Cholesterol depletion by methyl-β-cyclodextrin augments tamoxifen induced cell death by enhancing its uptake in melanoma. Mol Cancer. 2014;13:204. doi: 10.1186/1476-4598-13-204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Abildgaard C, Guldberg P. Molecular drivers of cellular metabolic reprogramming in melanoma. Trends Mol Med. 2015;21(3):164–171. doi: 10.1016/j.molmed.2014.12.007 [DOI] [PubMed] [Google Scholar]
- 8.Spencer KR, Mehnert JM. Mucosal melanoma: epidemiology, biology and treatment. Cancer Treat Res. 2016;167:295–320. doi: 10.1007/978-3-319-22539-5_13 [DOI] [PubMed] [Google Scholar]
- 9.Diederichs S. The four dimensions of noncoding RNA conservation. Trends Genet. 2014;30(4):121–123. doi: 10.1016/j.tig.2014.01.004 [DOI] [PubMed] [Google Scholar]
- 10.Paraskevopoulou MD, Hatzigeorgiou AG. Analyzing MiRNA-LncRNA Interactions. Methods Mol Biol. 2016;1402:271–286. doi: 10.1007/978-1-4939-3378-5_21 [DOI] [PubMed] [Google Scholar]
- 11.Wan J, Deng D, Wang X, Wang X, Jiang S, Cui R. LINC00491 as a new molecular marker can promote the proliferation, migration and invasion of colon adenocarcinoma cells. Onco Targets Ther. 2019;12:6471–6480. doi: 10.2147/OTT.S201233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sun Q, Li J, Li F, et al. LncRNA LOXL1-AS1 facilitates the tumorigenesis and stemness of gastric carcinoma via regulation of miR-708-5p/USF1 pathway. Cell Prolif. 2019:e12687. doi: 10.1111/cpr.12687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chen Q-N, Wei -C-C, Wang Z-X, Sun M. Long non-coding RNAs in anti-cancer drug resistance. Oncotarget. 2016;8(1):1925–1936. doi: 10.18632/oncotarget.12461 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wang Z-H, Guo X-Q, Zhang Q-S, et al. Long non-coding RNA CCAT1 promotes glioma cell proliferation via inhibiting microRNA-410. Biochem Biophys Res Commun. 2016;480(4):715–720. doi: 10.1016/j.bbrc.2016.10.047 [DOI] [PubMed] [Google Scholar]
- 15.Chen L, Wang W, Cao L, et al. Long non-coding RNA CCAT1 Acts as a competing endogenous RNA to regulate cell growth and differentiation in acute myeloid leukemia. Mol Cells. 2016;39(4):330–336. doi: 10.14348/molcells.2016.2308 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chen X, Dong H, Liu S, et al. Long noncoding RNA MHENCR promotes melanoma progression via regulating miR-425/489-mediated PI3K-Akt pathway. Am J Transl Res. 2017;9(1):90–102. [PMC free article] [PubMed] [Google Scholar]
- 17.Tang L, Zhang W, Su B, Yu B. Long noncoding RNA HOTAIR is associated with motility, invasion, and metastatic potential of metastatic melanoma. Biomed Res Int. 2013;2013:1–7. doi: 10.1155/2013/251098 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Guo L, Yao L, Jiang Y. A novel integrative approach to identify lncRNAs associated with the survival of melanoma patients. Gene. 2016;585(2):216–220. doi: 10.1016/j.gene.2016.03.036 [DOI] [PubMed] [Google Scholar]
- 19.Nissan A, Stojadinovic A, Mitrani-Rosenbaum S, et al. Colon cancer associated transcript-1: a novel RNA expressed in malignant and pre-malignant human tissues. Int J Cancer. 2012;130(7):1598–1606. doi: 10.1002/ijc.26170 [DOI] [PubMed] [Google Scholar]
- 20.Yang F, Xue X, Bi J, et al. Long noncoding RNA CCAT1, which could be activated by c-Myc, promotes the progression of gastric carcinoma. J Cancer Res Clin Oncol. 2013;139(3):437–445. doi: 10.1007/s00432-012-1324-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lv L, Jia JQ, Chen J. The lncRNA CCAT1 upregulates proliferation and invasion in melanoma cells via suppressing miR-33a. Oncol Res. 2018;26(2):201–208. doi: 10.3727/096504017X14920318811749 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hayes J, Peruzzi PP, Lawler S. MicroRNAs in cancer: biomarkers, functions and therapy. Trends Mol Med. 2014;20(8):460–469. doi: 10.1016/j.molmed.2014.06.005 [DOI] [PubMed] [Google Scholar]
- 23.Muhammad N, Bhattacharya S, Steele R, Ray RB. Anti-miR-203 suppresses ER-positive breast cancer growth and stemness by targeting SOCS3. Oncotarget. 2016;7(36):58595–58605. doi: 10.18632/oncotarget.11193 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wozniak M, Mielczarek A, Czyz M. miRNAs in melanoma: tumor suppressors and oncogenes with prognostic potential. Curr Med Chem. 2016;23(28):3136–3153. doi: 10.2174/1389557516666160831164544 [DOI] [PubMed] [Google Scholar]
- 25.Liu W, Liu X, Pan Z, et al. Ailanthone induces cell cycle arrest and apoptosis in melanoma B16 and A375 cells [retracted in: biomolecules. 2020 Apr 17;10(4):]. Biomolecules. 2019;9(7):275. doi: 10.3390/biom9070275 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 26.Salmena L, Poliseno L, Tay Y, Kats L, Pandolfi Pier P. A ceRNA hypothesis: the rosetta stone of a hidden RNA language? Cell. 2011;146(3):353–358. doi: 10.1016/j.cell.2011.07.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Qi X, Zhang D-H, Wu N, Xiao J-H, Wang X, Ma W. ceRNA in cancer: possible functions and clinical implications. J Med Genet. 2015;52(10):710. doi: 10.1136/jmedgenet-2015-103334 [DOI] [PubMed] [Google Scholar]
- 28.Jalali S, Bhartiya D, Lalwani MK, Sivasubbu S, Scaria V. Systematic transcriptome wide analysis of lncRNA-miRNA Interactions. PLoS One. 2013;8(2):e53823. doi: 10.1371/journal.pone.0053823 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Aftab MN, Dinger ME, Perera RJ. The role of microRNAs and long non-coding RNAs in the pathology, diagnosis, and management of melanoma. Arch Biochem Biophys. 2014;563:60–70. doi: 10.1016/j.abb.2014.07.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhang X-F, Liu T, Li Y, Li S. Overexpression of long non-coding RNA CCAT1 is a novel biomarker of poor prognosis in patients with breast cancer. Int J Clin Exp Pathol. 2015;8(8):9440–9445. [PMC free article] [PubMed] [Google Scholar]
- 31.Guo X, Hua Y. CCAT1: an oncogenic long noncoding RNA in human cancers. J Cancer Res Clin Oncol. 2017;143(4):555–562. doi: 10.1007/s00432-016-2268-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Prensner JR, Chinnaiyan AM. The Emergence of lncRNAs in Cancer Biology. Cancer Discov. 2011;1(5):391. doi: 10.1158/2159-8290.CD-11-0209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Thiery JP, Sleeman JP. Complex networks orchestrate epithelial–mesenchymal transitions. Nat Rev Mol Cell Biol. 2006;7(2):131–142. doi: 10.1038/nrm1835 [DOI] [PubMed] [Google Scholar]
- 34.Voulgari A, Pintzas A. Epithelial–mesenchymal transition in cancer metastasis: mechanisms, markers and strategies to overcome drug resistance in the clinic. Biochim Biophys Acta Rev Cancer. 2009;1796(2):75–90. doi: 10.1016/j.bbcan.2009.03.002 [DOI] [PubMed] [Google Scholar]
- 35.Zhuang K, Wu Q, Jiang S, Yuan H, Huang S, Li H. CCAT1 promotes laryngeal squamous cell carcinoma cell proliferation and invasion. Am J Transl Res. 2016;8(10):4338–4345. [PMC free article] [PubMed] [Google Scholar]
- 36.Deng L, Yang S-B, Xu -F-F, Zhang J-H. Long noncoding RNA CCAT1 promotes hepatocellular carcinoma progression by functioning as let-7 sponge. J Exp Clin Cancer Res. 2015;34(1):18. doi: 10.1186/s13046-015-0136-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ge T, Wu HC, Zhou YY, Shen SM, Zhu LG, You GX. MiR-296-3p may affect the proliferation and migration of non-small cell lung cancer cells via regulating RABL3. Eur Rev Med Pharmacol Sci. 2019;23(13):5823–5830. doi: 10.26355/eurrev_201907_18321 [DOI] [PubMed] [Google Scholar]
- 38.Li H, Li J, Shi B, Chen F. MicroRNA296 targets AKT2 in pancreatic cancer and functions as a potential tumor suppressor. Mol Med Rep. 2017;16(1):466–472. doi: 10.3892/mmr.2017.6602 [DOI] [PubMed] [Google Scholar]
- 39.He Z, Yu L, Luo S, et al. miR-296 inhibits the metastasis and epithelial-mesenchymal transition of colorectal cancer by targeting S100A4. BMC Cancer. 2017;17(1):140. doi: 10.1186/s12885-017-3121-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Deng X, Liu Z, Liu X, et al. miR-296-3p negatively regulated by nicotine stimulates cytoplasmic translocation of c-Myc via MK2 to suppress chemotherapy resistance. Mol Ther. 2018;26(4):1066–1081. doi: 10.1016/j.ymthe.2018.01.023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Gupta SK, Oommen S, Aubry MC, Williams BP, Vlahakis NE. Integrin α9β1 promotes malignant tumor growth and metastasis by potentiating epithelial-mesenchymal transition. Oncogene. 2012;32:141. doi: 10.1038/onc.2012.41 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Schreiber TD, Steinl C, Essl M, et al. The integrin α9β1 on hematopoietic stem and progenitor cells: involvement in cell adhesion, proliferation and differentiation. Haematologica. 2009;94(11):1493. doi: 10.3324/haematol.2009.006072 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Mostovich LA, Prudnikova TY, Kondratov AG, et al. Integrin alpha9 (ITGA9) expression and epigenetic silencing in human breast tumors. Cell Adh Migr. 2011;5(5):395–401. doi: 10.4161/cam.5.5.17949 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zhang J, Na S, Liu C, Pan S, Cai J, Qiu J. MicroRNA-125b suppresses the epithelial–mesenchymal transition and cell invasion by targeting ITGA9 in melanoma. Tumor Biol. 2016;37(5):5941–5949. doi: 10.1007/s13277-015-4409-8 [DOI] [PubMed] [Google Scholar]