Abstract
Objective
This study aimed to determine the effects of the long non-coding (lnc) RNA MT1JP on the apoptosis and migration of hepatocellular carcinoma cells.
Patients and Methods
Patients with liver cancer admitted to the Second People’s Hospital of Liaocheng were included in this study. We transfected hepatocellular carcinoma cells with MT1JP and miR-24-3p and assessed their expression and effects on apoptosis and migration. Correlations were verified using a dual-luciferase reporter and RNA-binding protein coimmunoprecipitation.
Results
The expression of MT1JP was downregulated (P < 0.05), whereas that of miR-24-3p was upregulated in liver cancer. Serum MT1JP levels were correlated with tumor size, alpha-fetoprotein (AFP), TNM stage, differentiation, and lymph node metastasis. Both MT1JP overexpression and miR-24-3p inhibition inhibited cellular proliferation and migration and increased apoptosis rates. They significantly downregulated expression of the cell migration-associated proteins matrix metalloproteinase -2, -9 (MMP-2, MMP-9) (P < 0.05). They upregulated the expression of Bcl-2-related X protein (Bax) and cysteinyl aspartate-specific proteinases (Caspase-3 and -9) proteins that are involved in apoptosis. They decreased expression of B-cell lymphoma/leukemia-2 (Bcl-2; P < 0.05). A target relationship between MT1JP and miR-24-3p was identified using dual-luciferase gene reporter assays and RNA-binding protein coimmunoprecipitations. MT1JP overexpression significantly downregulated miR-24-3p expression (P < 0.05). MT1JP and miR-24-3p expression were negatively correlated in liver cancer tissues (r = −0.561, P < 0.001; Pearson χ2 tests). Rescue experiments showed that upregulating miR-24-3p expression could counteract MT1JP overexpression in hepatocellular carcinoma cells.
Conclusion
MT1JP, even when expressed at low levels, participates in the proliferation, apoptosis, and migration of liver cancer cells by regulating miR-24-3p.
Keywords: lncRNA MT1JP, miR-24-3p, liver cancer, apoptosis, migration
Introduction
Liver cancer is a common malignancy, the sixth most common in the world and second in terms of factors related to tumor-related mortality. Epidemiological studies have shown that the number of new cases and deaths due to liver cancer reached ~800,000 in 2018.1,2 The morbidity of liver cancer is relatively high in China; it has gradually increased along with environmental and lifestyle changes.3 Despite considerable progress in the diagnosis and treatment of liver cancer, the long-term prognosis remains poor. The relative 5-year survival rate is 16.6%; tumors are diagnosed while still localized in 43% of patients. The 5-year relative survival rate is 30.5%.1,4,5 Despite considerable progress in the clinical diagnosis and treatment of cancer, survival rates remain comparatively low.6 Our understanding of liver cancer cells has become progressively detailed with the evolution of molecular biology, but the pathogenesis of liver cancer requires further investigation to determine and refine appropriate treatment strategies.
Long non-coding RNA (lncRNA) is defined as a class of transcripts >200 nt that are involved in the regulation of multiple diseases through involvement in processes including transcriptional regulation, modification, and epigenetic changes.7–9 Accumulating evidence shows that lncRNA play crucial roles in cancer cells;10 MT1JP is an lncRNA that is abnormally expressed in multiple cancers.11,12 The downregulation or upregulation of MT1JP provokes changes in the expression of many cancer-related mRNA molecules,13 suggesting that it is essential to maintain normal cellular activities, playing a crucial role as a tumor inhibitor.14 Overexpression of MT1JP inhibits the proliferation, migration and invasion of gastric cancer cells.15 However, little is known about MT1JP and how it is regulated in liver cancer cells. Therefore, we determined the effects of MT1JP expression on the regulation of their behavior.
Patients and Methods
General Information
The study included 116 patients diagnosed with liver cancer who had been admitted to the Second People’s Hospital of Liaocheng and 112 healthy persons who concurrently underwent physical examinations at the same hospital as controls. Serum, liver cancer and paracancerous tissues were collected from the patients, with only serum collected from the controls. The Ethics Committee at the Second People’s Hospital of Liaocheng approved the experimental protocol and the study was conducted in accordance with the Declaration of Helsinki. All enrolled patients and healthy volunteers provided written informed consent to participate.
The inclusion criteria were: liver cancer diagnosis confirmed by imaging and pathological biopsy, expected survival >3 months, complete clinical data and good compliance, and without targeted tumor therapy (surgery, radiotherapy, chemotherapy) before starting this study.
The exclusion criteria were patients who were not accompanied by their families at the time of admission, autoimmune system defects, infection before admission, and refusal to provide experimental specimens.
Cell Culture and Transfection
Human normal hepatocytes (THLE-3) and the Huh-7, HLE, Hep3B, and MHCC97-H hepatoma cell lines (Cat. Nos. MJ-1908, MJ1403, MJ-1328, CM-1305, CM-1529, respectively; Shanghai Mingjing Biotechnology Co., Ltd., Shanghai, China) were incubated in DMEM containing 10% fetal bovine serum (Cat. No: SH30243.01; HyClone Laboratories Inc., San Angelo, TX, USA) at 37°C in a 5% CO2 atmosphere. When cells reached 85% confluence, 25% pancreatin was added for digestion, then cells were maintained in the medium. The Huh-7 and Hep3B cell lines were transfected with miR-24-3p-inhibitor (inhibitory sequence), miR negative control (miR-NC), blank control (vector), or MT1JP (pcDNA-MT1JP) using Lipofectamine™ 2000 kits (Cat. No: 11668–019; Invitrogen, Carlsbad, CA, USA), as described in the instructions provided with the kit.
Quantitative RT-PCR
The purity and concentration of total RNA extracted using Trizol reagent (Cat. No. 12183555; Invitrogen) were determined using a DR5000 ultraviolet-visible spectrophotometer (Bio-Rad Laboratories Inc., Hercules, CA, USA). The RNA was reverse-transcribed using a kit (Cat. No. AB-4366596; Invitrogen, USA), according to instructions of the manufacturer. The cDNA was amplified and quantified by PCR using custom synthesized primers (Shanghai Sangon Biotech Co., Ltd. Shanghai, China) and SYBR Premix Ex Taq (Cat. No. DRR041A; Takara Bio Inc., Kusatsu, Japan) using an ABI PRISM 7000 (Applied Biosystems, Waltham, MA, USA) instrument. The internal references for MT1JP and miR-214-3p were GAPDH and U6, respectively. The respective forward and reverse primers were: MT1JP, 5′-GCAAAGGGACGTCGGAGA-3′ and 5′-TCCAGGTTGCAGTTGTT-3′; GAPDH, 5′-GCACCGTCAAGGCTGGAAC-3′ and 5′-TGGTGAAGCGCCAGTGGA-3′; miR-214-3p forward, 5′-ACAGCAGGCACAGAGAGGGG-3ʹ and miR-24-3p reverse 5′-CTGGCTCAGTTCAGCAGGAACAG-3′; U6, 5′ -CTCGCTTCGGCAGCACA-3′ and 5′-CTCGCTTCGGCAGCACA-3′. The reaction proceeded by pre-denaturation at 95°C for 30 s, followed by 40 cycles of denaturation at 95°C for 5 s, then annealing and extension at 60°C for 30 s. The results were calculated using 2−ΔΔct.
Western Blotting
Harvested cells were lysed with RIPA buffer (Cat. No. 89901; Thermo Fisher Scientific Co. Ltd., Waltham, MA, USA). Proteins were quantified using BCA kits (Cat. No. P0012S; Beyotime Biotechnology, Shanghai, China), resolved by SDS-PAGE, and transferred to nitrocellulose membranes. Non-specific binding was blocked with 5% skim milk in PBS at room temperature for 1 h. Membranes were immersed in primary antibodies to Caspase 3, Caspase 9, Bax, Bcl-2, MMP-2, MMP-9, and β-actin (Cat. No. ab13847, ab32539, ab32503, ab185002, ab92536, ab76003, ab8226 respectively; Abcam Plc., Cambridge, UK) diluted 1:1000, and left at 4°C overnight. Membranes were washed, incubated with horseradish peroxidase-labeled goat anti-rabbit secondary antibody (Cat. No. ab205718; Abcam Plc. diluted 1:1000) for 1 h at 37°C, then washed with PBS. Proteins were visualized using ECL luminescent reagent (Cat. No. 34096; Thermo Fisher Scientific Co., Ltd.). The internal reference protein was β-actin; the relative expression level of the test protein was calculated as the gray value of the test band/gray value of β-actin.
Detection of Cell Proliferation
Cell proliferation was quantified three times using MTT assay kits (Cat. No. V13154; Thermo Fisher Scientific Co., Ltd.) as follows. Transfected cells (5 × 103/100 μL) were incubated in 96-well plates for 24, 48, 72, and 96 h at 37°C in a 5% CO2 atmosphere. The culture medium on the upper layer was discarded and cells were incubated with MTT solution (20 μL) for 4 h. Thereafter, dimethyl sulfoxide (150 µL) was added. Plates were shaken for 5–10 min to completely dissolve purple formazan crystals. Absorbance was measured at 450 nm using a Multiskan™ GO full wavelength microplate reader (Thermo Fisher Scientific Co., Ltd.).
Apoptosis Assays
Apoptosis was detected using V13241 Annexin V-FITC/PI kits (Thermo Fisher Scientific Co., Ltd.) and digestion was carried out after transfection. Harvested cells were washed twice with PBS and suspended in 100 μL of binding buffer (1 × 106 cells/mL). Annexin V-FITC and PI (5 μL each) were sequentially added and the suspension was incubated for 5 min at room temperature in darkness. Apoptosis was then assessed using a FACScan flow cytometer (Becton Dickinson Becton Dickinson and Co., Franklin Lakes, NJ, USA). Data were analyzed using the Phoenix Flow System (Innovative Cell Technologies Inc., San Diego, CA, USA). Results are presented as averages of three experiments.
Cell Migration
Cell migration was evaluated using scratch-healing tests. Monolayers were scratched using sterilized 100 μL disposable pipette tips perpendicular to the horizontal plane to minimize differences in scratch width. The medium was removed and cells were washed three times with PBS to remove cell debris. Serum-free medium was added and cell migration was assessed by microscopy at 0 and 24 h after cell division.
Detection of Dual Luciferase
Target genes were predicted using starBase v3.0. PrimGLO-MT1JP-wt and primGLO-MT1JP-mut vectors were constructed and co-transfected into cells with a miR-24-3p mimetic or miR-NC, respectively. Cells were cultured for 48 h in 96-well plates. Luciferase was measured using a dual-luciferase reporter gene assay system (Promega Corp., Madison, WI, USA) as described by the manufacturer.
RNA-Binding Protein Immunoprecipitation (RIP)
Binding targets were assessed using RIP kits (Cat. No. 17–701; BIOMARS Technology Development Co., Ltd, Beijing, China) as described by the manufacturer. Cells were suspended in chilled PBS, the supernatant was discarded, and lysates were prepared in RIP lysate solution. Whole-cell protein extracts were washed with RIP wash buffer containing magnetic beads bound to Ago2 antibody or immunoglobulin G (IgG) control. Proteins and immunoprecipitated RNA were digested using protease K. Binding targets in purified RNA were confirmed by qRT-PCR.
Statistical Analyses
Data were statistically analyzed using SPSS 20.0 (IBM Corp., Armonk, NY, USA), and visualized using GraphPad 7 (GraphPad Software, San Diego, CA, USA). Between-group comparisons proceeded using t-tests, and multiple groups were compared using one-way analyses of variance. Post hoc pairwise comparisons were conducted using LSD t-tests. The diagnostic value of MT1JP in liver cancer was determined using receiver operator characteristics (ROC) curves. Correlations between MT1JP and miR-24-3p were analyzed using Pearson χ2 tests. Values of p < 0.05 were considered statistically significant.
Results
lncRNA MT1JP Expression in Liver Cancer
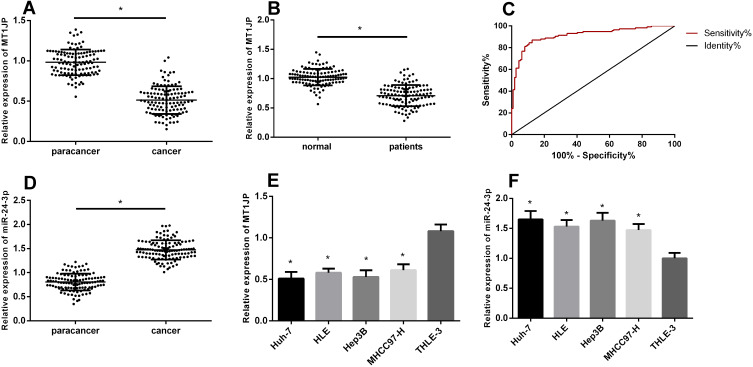
The qRT-PCR results showed significant MT1JP downregulation in cancer tissues and serum samples from patients with liver cancer (P < 0.05; Figure 1), suggesting that serum MT1JP values could be used for diagnosis (AUC, 0.9147; sensitivity, 87.07%; specificity, 87.5%). However, miR-24-3p expression was significantly higher in tumor tissues than paracancerous tissues (P < 0.05). Furthermore, the expression of MT1JP was significantly decreased, whereas that of miR-24-3p was significantly higher in the hepatoma cell lines Huh-7, HLE, Hep3B, and mhc97-h than in the hepatic epithelial cell line THLE-3 (P < 0.05).
Figure 1.
Expression of lncRNA MT1JP in liver cancer. MT1JP expression in hepatocellular carcinoma tissue (A) and in serum of patients with liver cancer (B). Diagnostic value of serum MT1JP for liver cancer (C). Expression of miR-24-3p in hepatocellular carcinoma tissue (D). Comparison of MT1JP (E) and miR-24-3p (F) expression in various hepatocellular carcinoma cell lines (*P < 0.05). lncRNA, long non-coding RNA.
Relationship Between lncRNA MT1JP and Liver Cancer Pathology
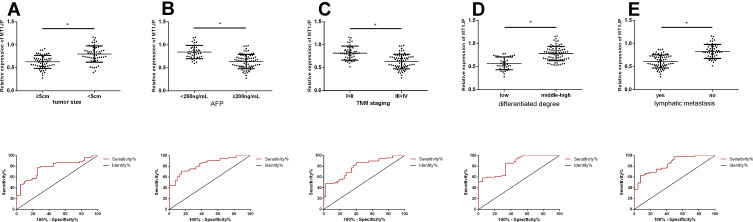
Table 1 shows the relationship between MT1JP expression and patient pathological characteristics. The expression of MT1JP significantly differed according to tumor size, alpha-fetoprotein (AFP), TNM stage, differentiation, lymph node metastasis, and other aspects of liver cancer, but not according to gender, age, and comorbid hepatitis (P > 0.05 for both). ROC curves indicated that MT1JP can distinguish tumor size, AFP, TNM stage, differentiation, and lymph node metastasis (Figure 2), with significant diagnostic value (Table 2).
Table 1.
Relationships Between lncRNA MT1JP and Pathological Characteristics
| n | MT1JP | x2/t | P | |
|---|---|---|---|---|
| Gender | 0.27 | 0.79 | ||
| Male | 70 | 0.71±0.18 | ||
| Female | 46 | 0.70±0.19 | ||
| Age (y) | 1.73 | 0.09 | ||
| <55 | 49 | 0.74±0.18 | ||
| ≥55 | 67 | 0.68±0.18 | ||
| Tumor size (cm) | 5.77 | <0.05 | ||
| ≥5 | 62 | 0.63±0.14 | ||
| <5 | 54 | 0.80±0.18 | ||
| Combined with hepatitis | 1.31 | 0.19 | ||
| Yes | 95 | 0.68±0.19 | ||
| No | 21 | 0.75±0.14 | ||
| AFP (ng/mL) | 6.96 | <0.05 | ||
| <200 | 40 | 0.84±0.14 | ||
| ≥200 | 76 | 0.64±0.16 | ||
| TNM stage | 6.11 | <0.05 | ||
| I+II | 47 | 0.82±0.15 | ||
| III+IV | 69 | 0.63±0.16 | ||
| Differentiation | 7.26 | <0.05 | ||
| Poor | 39 | 0.57±0.14 | ||
| Moderate + high | 77 | 0.78±0.16 | ||
| Lymph node metastasis | 7.85 | <0.05 | ||
| Yes | 65 | 0.61±0.14 | ||
| No | 51 | 0.83±0.15 |
Figure 2.
Relationship between lncRNA MT1JP and pathological characteristics of liver cancer. Expression and diagnostic value of MT1JP in liver tumors with different (A) sizes, (B) AFP levels, (C) TNM stages (D) types of cancer and (E) lymph node metastases (*P < 0.05). lncRNA, long non-coding RNA.
Table 2.
ROC Parameters
| AUC | 95% CI | Sensitivity | Specificity | Cutoff | |
|---|---|---|---|---|---|
| Tumor size | 0.775 | 0.689–0.863 | 77.78% | 74.19% | >0.71 |
| AFP | 0.835 | 0.763–0.907 | 71.05% | 85.00% | <0.71 |
| TNM stage | 0.786 | 0.704–0.868 | 86.96% | 59.58% | <0.80 |
| Differentiation | 0.842 | 0.770–0.915 | 58.44% | 94.87% | >0.75 |
| LN metastasis | 0.840 | 0.770–0.911 | 62.75% | 92.31% | >0.80 |
Abbreviations: AFP, alpha-fetoprotein; AUC, area under the ROC curve; CI, confidence interval; LN, lymph node; ROC, receiver operator characteristics; TNM, tumor, nodes and metastasis.
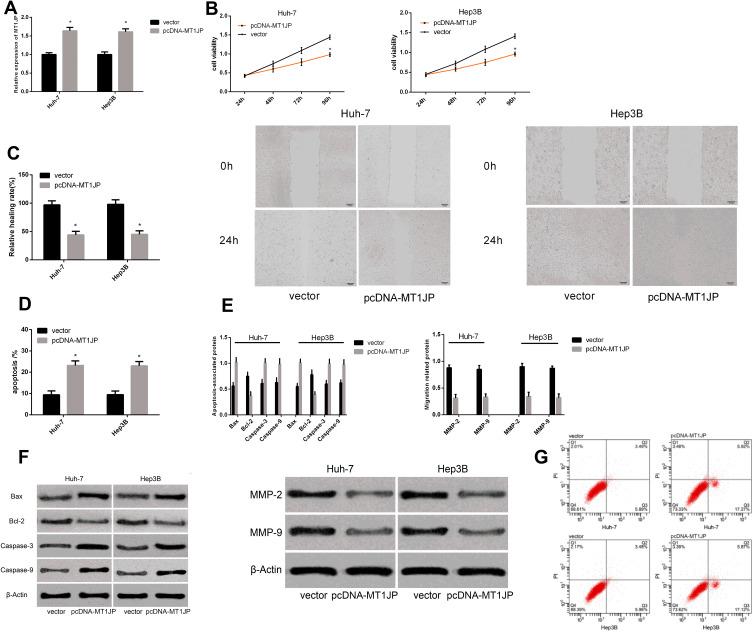
lncRNA MT1JP on Apoptosis and Migration of Hepatocellular Carcinoma Cells
The expression of MT1JP in transfected Huh-7 and Hep3B cell lines was upregulated (Figure 3), whereas cell proliferation and migration were inhibited and apoptosis rates were increased. The expression of the cell migration-associated proteins MMP-2 and MMP-9, as well as that of Bcl-2 was remarkably downregulated (P < 0.05 for all). In contrast, Bax, Caspase-3, and Caspase-9 expression was upregulated (P < 0.05 for all).
Figure 3.
Effect of lncRNA MT1JP on apoptosis and migration of hepatocellular carcinoma cells. Expression of MT1JP after transfection (A). Proliferation (B), migration and (C) apoptosis rates of live cancer cells after transfection with different genes (D). Expression of apoptosis- and migration-associated proteins in liver cancer cells after transfection with different genes (E). Maps of apoptosis- and migration-associated proteins (F). Apoptosis profiles of liver cancer cells after transfection with different genes (G). *P < 0.05 vs vector.
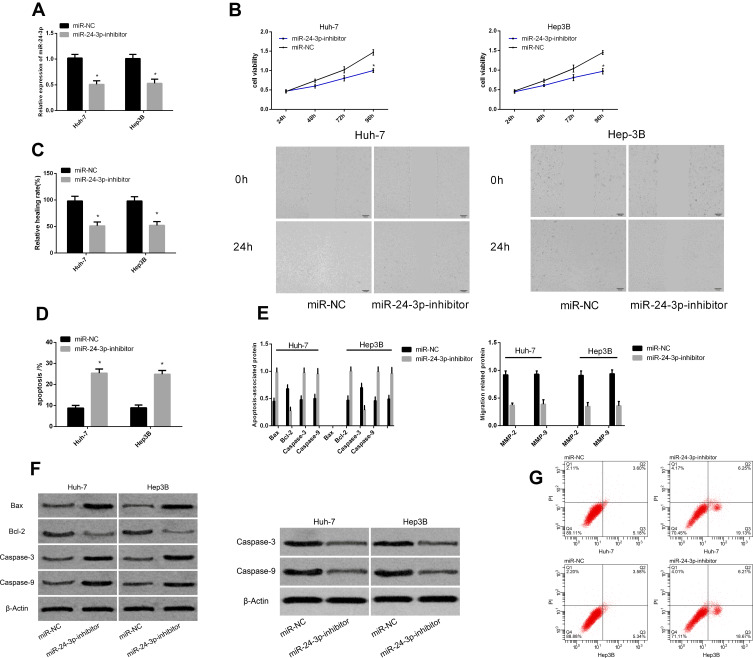
Effects of miR-24-3p on Apoptosis and Migration of Hepatocellular Carcinoma Cells
Figure 4 shows that inhibiting miR-24-3p expression in Huh-7 and Hep3B cells resulted in the downregulation of miR-24-3p, the inhibition of proliferation and migration, and increased apoptosis. In addition, the expression of Bcl-2, MMP-2 and MMP-9 was remarkably downregulated (P < 0.05 for all), whereas that of Bax, Caspase-3, and Caspase-9 was upregulated (P < 0.05 for all).
Figure 4.
Effects of miR-24-3p on apoptosis and migration of hepatocellular carcinoma cells. Expression of miR-24-3p after transfection (A). Proliferation (B), migration and (C) apoptosis rates of cancer cells after transfection with different genes (D). Expression of apoptosis- and migration-associated proteins in liver cancer cells after transfection with different genes (E). Maps of apoptosis- and migration-associated proteins (F). Apoptosis profiles of liver cancer cells after transfection with different genes (G). *P < 0.05 vs miR-NC.
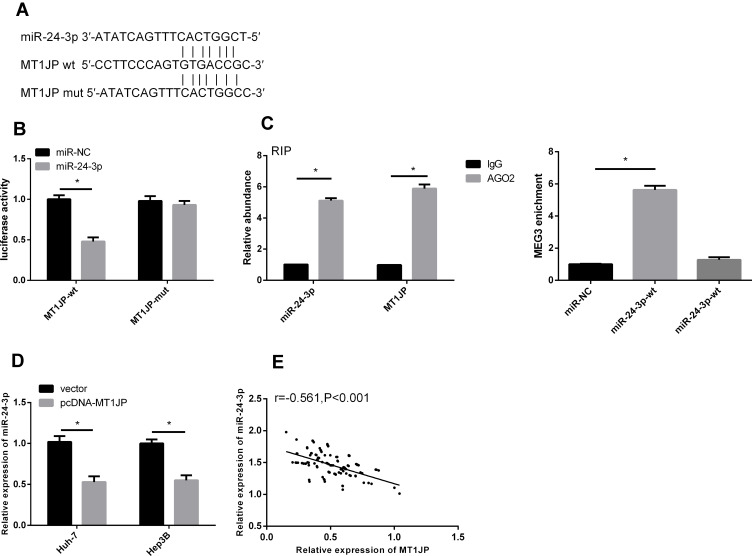
Targeted Relationship Between lncRNA MT1JP and miR-24-3p
We found a targeted binding site between MT1JP and miR-24-3p using starBase v3.0 software (Figure 5). The results of dual-luciferase reporter assays revealed considerably reduced fluorescence activity of MT1JP-Wt. Moreover, RIP assays showed increased MT1JP and miR-24-3p enrichment after immunoprecipitation with anti-Ago2, compared with IgG. The overexpression of MT1JP downregulated miR-24-3p (P < 0.05) MT1JP and miR-24-3p expression were negatively correlated in liver cancer tissues (r = −0.561, P < 0.001; Pearson tests).
Figure 5.
Targeting relationship between lncRNA MT1JP and miR-24-3p. Database prediction that MT1JP and miR-24-3p have binding sites (A). Dual-luciferase reporter gene (B). Relationship between MT1JP and miR-24-3p verified by RIP (C). Change in miR-24-3p level after MT1JP overexpression (D). Correlation between MT1JP and miR-24-3p in liver cancer tissue (E). *P < 0.05; Pearson χ2 tests.
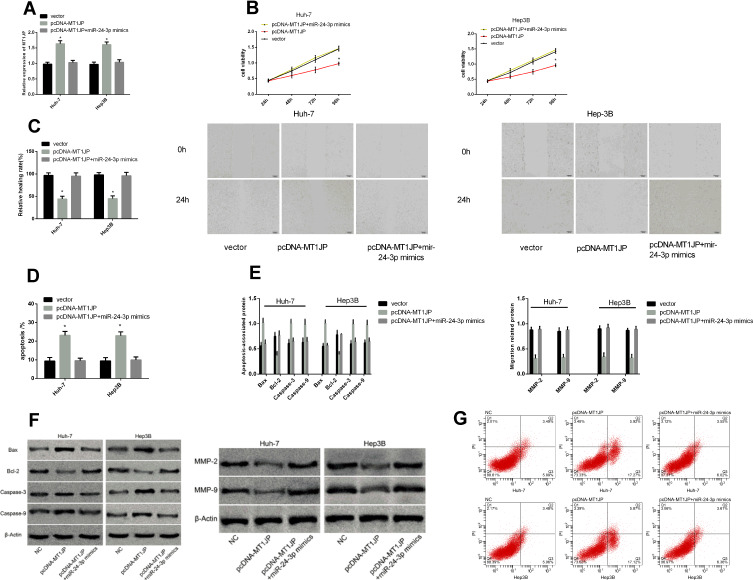
Rescue Experiment
We co-transfected pcDNA-MT1JP + miR-24-3p mimics into Huh-7 and Hep3B cells to further explore the mechanisms underlying the effects of MT1JP on hepatoma cells (Figure 6). The mimics did not alter cell proliferation, migration, and apoptosis compared with the blank vector (P > 0.05), but relieved the inhibition of cell proliferation and migration. They reduced apoptosis compared with pcDNA-MT1JP. Therefore, miR-24-3p upregulation could alleviate the inhibition of proliferation and migration caused by MT1JP overexpression.
Figure 6.
Rescue experiment. MT1JP expression after co-transfection (A). Comparison of hepatoma cell activity (B), migration (C), and apoptosis (D) after co-transfection. Expression of apoptosis- and migration-associated proteins in liver cancer cells after co-transfection (E). Images of apoptosis- and migration-associated proteins (F) and hepatoma cell apoptosis after co-transfection (G). *P < 0.05.
Discussion
Hepatocellular carcinoma and cholangiocarcinoma are common liver cancers worldwide.16 The occurrence of liver cancer varies due to factors including gender and geography. Among these, the primary risk factors include infection (endemic liver flukes, hepatitis B, and C viruses), cigarette smoking, alcohol consumption, obesity, and aflatoxin (AFP),17 which is clinically applied as a liver cancer screen, although it is limited by low sensitivity.18 Accumulated evidence shows that lncRNAs participate in cell differentiation, proliferation, and apoptosis, and can serve as tumor biomarkers.19 They are also involved in the occurrence and development of liver cancer.20 MT1JP is located in a cluster on chromosome 16 with several homologous genes in the metallothionein family.21 As a tumor suppressor, MT1JP affects proliferation by regulating p53 protein expression.21,22
We found that MT1JP was significantly downregulated in liver cancer and that in serum MT1JP could be used to diagnose liver cancer. It is similarly downregulated in breast,21 gastric,23 and bladder24 cancers. Together, these findings suggest that MT1JP plays anti-cancer roles in tumors. The presence of serum markers can distinguish disease states and pathological types in patients.25 Diagnostic ROC curves showed that MT1JP expression has some diagnostic value in terms of differences in tumor size, AFP, TNM stage, differentiation, and lymph node metastasis. Serum MT1JP levels gradually decrease as tumors deteriorate and the degree of tumor differentiation decreases. Therefore, serum MT1JP has potential as a biomarker for diagnosis and treatment. However, the activities of MT1JP in liver cancer have not been reported.
Apoptosis is the orderly and autonomous physiological death of cells under specific physiological or pathological conditions. It is mainly controlled by genes encoding the caspase and Bcl-2 families of proteins.26 Disordered apoptosis causes changes in cell-growth cycles that will eventually lead to the occurrence of pathological states.27 Long non-coding RNA participates in the regulation of apoptosis, affects cell proliferation and differentiation, and thus is relevant to disease processes.28 Migration is a basic cellular function that is involved in physiological and pathological behaviors such as tumor cell proliferation, embryo development, and wound healing.29,30 We showed here that MT1JP overexpression significantly inhibited the proliferation and migration of liver cancer cells, increased apoptosis rates, downregulated the expression of Bcl-2 and migration-related proteins MMP-2 and MMP-9, and upregulated that of the apoptosis-related proteins Bax, Caspase-3, and Caspase-9. These findings suggest that MT1JP could inhibit tumors and therefore serve as a therapeutic target for liver cancer. Nevertheless, further details of the mechanism of MT1JP remain obscure.
An imbalance of miR-24-3p has been shown to contribute to the progression of multiple cancers, including liver cancer, in which it is upregulated.31 Although low levels of miR-24-3p were found in liver cancer tissues and cell lines in the present study, inhibiting such expression still reduced proliferation and migration and significantly increased rates of apoptosis. These findings were similar to those of Fan et al,32 and suggest that miR-24-3p could serve as a target gene for LncRNA CASC2 to treat liver cancer. Notably, lncRNA, acting as a “molecular sponge” by combining with endogenous miRNA, participates in the regulation of tumorigenesis.33 Ma et al33 proposed that lncRNA MT1JP inhibits tumor cell proliferation and increases apoptosis by regulating the miRNA-423-3p/Bim axis in non-small cell lung cancer. MT1JP regulates the progression of gastric cancer through competitively binding to miR-92A-3P.23 We identified a regulatory relationship between MT1JP and miR-24-3p using online tools, then verified it using dual-luciferase reporter assays. Enrichment between the two determined in the RIP experiment further confirmed the ceRNA relationship between MT1JP and miR-24-3p. Pearson χ2 tests negatively correlated MT1JP and miR-24-3p expression in liver cancer tissues. Therefore, MT1JP mediates miR-24-3p expression to regulate the behavior of liver cancer cells. Finally, our rescue experiments showed that upregulating miR-24-3p expression counteracted the inhibition of MT1JP overexpression on proliferation and migration of hepatoma cells, further clarifying the regulatory relationship between MT1JP and miR-24-3p.
The present study has some limitations. The long-term prognosis of patients with liver cancer was not investigated; whether MT1JP is associated with their prognosis awaits further investigation. Growth inhibition effects in tumor-bearing nude mice, the mechanism of MT1JP action, and its relevance to liver cancer should be further explored.
To summarize, MT1JP is expressed at remarkably low levels in liver cancer, but it participates in the proliferation, apoptosis, and migration of liver cancer cells by regulating miR-24-3p.
Acknowledgments
This study was financially supported by the National Natural Science Foundation of China (NSFC) [grant number 21507040], the Natural Science Foundation of Shandong Province [grant number BS2015SW023].
Disclosure
The authors have no conflicts of interest to declare regarding this study.
References
- 1.Ryerson AB,Eheman CR,Altekruse SF,et al. Annual report to the nation on the status of cancer,1975–2012,featuring the increasing incidence of liver cancer. Cancer. 2016;122(9):1312–1337. doi: 10.1002/cncr.29936 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bray F,Ferlay J,Soerjomataram I,Siegel RL,Torre LA,Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2018;68(6):394–424. doi: 10.3322/caac.21492 [DOI] [PubMed] [Google Scholar]
- 3.Wang FS,Fan JG,Zhang Z,Gao B,Wang HY. The global burden of liver disease: the major impact of China. Hepatology 2014;60:2099–2108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ji M, Liu Z, Chang ET, et al. Mass screening for liver cancer: results from a demonstration screening project in Zhongshan City, China. Sci Rep. 2018;8(1):12787. doi: 10.1038/s41598-018-31119-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Endeshaw M, Hallowell BD, Razzaghi H, Senkomago V, McKenna MT, Saraiya M. Trends in liver cancer mortality in the United States: dual burden among foreign- and US-born persons. Cancer. 2019;125(5):726–734. doi: 10.1002/cncr.31869 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Tang B, Qi G, Sun X, et al. HOXA7 plays a critical role in metastasis of liver cancer associated with activation of Snail. Mol Cancer. 2016;15(1):57. doi: 10.1186/s12943-016-0540-4 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 7.Chen X, Sun Y, Cai R, Wang G, Shu X, Pang W. Long noncoding RNA: multiple players in gene expression. BMB Rep. 2018;51(6):280–289. doi: 10.5483/bmbrep.2018.51.6.025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chiu HS, Somvanshi S, Patel E, et al. Pan-cancer analysis of lncRNA regulation supports their targeting of cancer genes in each tumor context. Cell Rep. 2018;23(1):297–312 e212. doi: 10.1016/j.celrep.2018.03.064 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Klingenberg M, Matsuda A, Diederichs S, Patel T. Non-coding RNA in hepatocellular carcinoma: mechanisms, biomarkers and therapeutic targets. J Hepatol. 2017;67(3):603–618. doi: 10.1016/j.jhep.2017.04.009 [DOI] [PubMed] [Google Scholar]
- 10.Mohankumar S, Patel T. Extracellular vesicle long noncoding RNA as potential biomarkers of liver cancer. Brief Funct Genomics. 2016;15(3):249–256. doi: 10.1093/bfgp/elv058 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Al-Sandaqchi AT, Brignell C, Collingwood JF, et al. Metallome of cerebrovascular endothelial cells infected with Toxoplasma gondii using mu-XRF imaging and inductively coupled plasma mass spectrometry. Metallomics. 2018;10(10):1401–1414. doi: 10.1039/c8mt00136g [DOI] [PubMed] [Google Scholar]
- 12.Bi -L-L, Han F, Zhang X-M, Li -Y-Y. LncRNA MT1JP acts as a tumor inhibitor via reciprocally regulating Wnt/β-Catenin pathway in retinoblastoma.. Eur Rev Med Pharmacol Sci. 2018;22(13):4204–4214. doi: 10.26355/eurrev_201807_15414 [DOI] [PubMed] [Google Scholar]
- 13.Lv Z, Zhang Y, Yu X, Y L, Ge Y. The function of long non-coding RNA MT1JP in the development and progression of gastric cancer. Pathol Res Pract. 2018;214(8):1218–1223. doi: 10.1016/j.prp.2018.07.001 [DOI] [PubMed] [Google Scholar]
- 14.Liu L, Yue H, Liu Q, et al. LncRNA MT1JP functions as a tumor suppressor by interacting with TIAR to modulate the p53 pathway. Oncotarget. 2016;7(13):15787–15800. doi: 10.18632/oncotarget.7487 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Xu Y, Zhang G, Zou C, et al. LncRNA MT1JP suppresses gastric cancer cell proliferation and migration through MT1JP/MiR-214-3p/RUNX3 axis. Cell Physiol Biochem. 2018;46(6):2445–2459. doi: 10.1159/000489651 [DOI] [PubMed] [Google Scholar]
- 16.Nayyar M, Imagawa DK, Tirkes T, et al. Composite liver tumors: a radiologic-pathologic correlation. Clin Mol Hepatol. 2014;20(4):406–410. doi: 10.3350/cmh.2014.20.4.406 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Akinyemiju T, Abera S, Ahmed M, et al. The burden of primary liver cancer and underlying etiologies from 1990 to 2015 at the global, regional, and national level: results from the global burden of disease study 2015. JAMA Oncol. 2017;3(12):1683–1691. doi: 10.1001/jamaoncol.2017.3055 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sauzay C, Petit A, Bourgeois AM, et al. Alpha-foetoprotein (AFP): a multi-purpose marker in hepatocellular carcinoma. Clin Chim Acta. 2016;463:39–44. doi: 10.1016/j.cca.2016.10.006 [DOI] [PubMed] [Google Scholar]
- 19.Martens-Uzunova ES, Bottcher R, Croce CM, Jenster G, Visakorpi T, Calin GA. Long noncoding RNA in prostate, bladder, and kidney cancer. Eur Urol. 2014;65(6):1140–1151. doi: 10.1016/j.eururo.2013.12.003 [DOI] [PubMed] [Google Scholar]
- 20.Smekalova EM, Kotelevtsev YV, Leboeuf D, et al. lncRNA in the liver: prospects for fundamental research and therapy by RNA interference. Biochimie. 2016;131:159–172. doi: 10.1016/j.biochi.2016.06.007 [DOI] [PubMed] [Google Scholar]
- 21.Zhu D, Zhang X, Lin Y, Liang S, Song Z, Dong C. MT1JP inhibits tumorigenesis and enhances cisplatin sensitivity of breast cancer cells through competitively binding to miR-24-3p. Am J Transl Res. 2019;11(1):245–256. [PMC free article] [PubMed] [Google Scholar]
- 22.Chaudhary R, Lal A. Long noncoding RNAs in the p53 network. Wiley Interdiscip Rev RNA. 2017;8(3):e1410. doi: 10.1002/wrna.1410 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhang G, Li S, Lu J, et al. LncRNA MT1JP functions as a ceRNA in regulating FBXW7 through competitively binding to miR-92a-3p in gastric cancer. Mol Cancer. 2018;17(1):87. doi: 10.1186/s12943-018-0829-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yu H, Wang S, Zhu H, Rao D. LncRNA MT1JP functions as a tumor suppressor via regulating miR-214-3p expression in bladder cancer. J Cell Physiol. 2019;234(9):16160–16167. doi: 10.1002/jcp.28274 [DOI] [PubMed] [Google Scholar]
- 25.Steinacker P, Semler E, Anderl-Straub S, et al. Group FTS. Neurofilament as a blood marker for diagnosis and monitoring of primary progressive aphasias. Neurology. 2017;88(10):961–969. doi: 10.1212/WNL.0000000000003688 [DOI] [PubMed] [Google Scholar]
- 26.Edlich F. BCL-2 proteins and apoptosis: recent insights and unknowns. Biochem Biophys Res Commun. 2018;500(1):26–34. doi: 10.1016/j.bbrc.2017.06.190 [DOI] [PubMed] [Google Scholar]
- 27.Hassani S, Ghaffari P, Chahardouli B, et al. Disulfiram/copper causes ROS levels alteration, cell cycle inhibition, and apoptosis in acute myeloid leukaemia cell lines with modulation in the expression of related genes. Biomed Pharmacother. 2018;99:561–569. doi: 10.1016/j.biopha.2018.01.109 [DOI] [PubMed] [Google Scholar]
- 28.Qi Y, Wang Z, Wu F, et al. Long noncoding RNA HOXD-AS2 regulates cell cycle to promote glioma progression. J Cell Biochem. 2018. doi: 10.1002/jcb.28117 [DOI] [PubMed] [Google Scholar]
- 29.Butler AE, Kirakossian D, Gurlo T, Gao F, Coppola G, Butler PC. In the setting of beta-cell stress, the pancreatic duct gland transcriptome shows characteristics of an activated regenerative response. Am J Physiol Gastrointest Liver Physiol. 2018;315(5):G848–G854. doi: 10.1152/ajpgi.00177.2018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Williams SP, Gould CM, Nowell CJ, et al. Systematic high-content genome-wide RNAi screens of endothelial cell migration and morphology. Sci Data. 2017;4(1):170009. doi: 10.1038/sdata.2017.9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Dong X, Ding W, Ye J, et al. MiR-24-3p enhances cell growth in hepatocellular carcinoma by targeting metallothionein 1M. Cell Biochem Funct. 2016;34(7):491–496. doi: 10.1002/cbf.3213 [DOI] [PubMed] [Google Scholar]
- 32.Fan JC, Zeng F, Le YG, Xin L. LncRNA CASC2 inhibited the viability and induced the apoptosis of hepatocellular carcinoma cells through regulating miR-24-3p. J Cell Biochem. 2018;119(8):6391–6397. doi: 10.1002/jcb.26479 [DOI] [PubMed] [Google Scholar]
- 33.Ma J, Yan H, Zhang J, Tan Y, Gu W. Long-chain non-coding RNA (lncRNA) MT1JP suppresses biological activities of lung cancer by regulating miRNA-423-3p/Bim axis. Med Sci Monit. 2019;25:5114–5126. doi: 10.12659/MSM.914387 [DOI] [PMC free article] [PubMed] [Google Scholar]