Abstract
N-alpha-acetyltransferase 80 (NAA80) was recently demonstrated to acetylate the N-terminus of actin, with NAA80 knockout cells showing actin cytoskeleton-related phenotypes, such as increased formation of membrane protrusions and accelerated migration. Here we report that NAA80 knockout cells additionally display fragmentation of the Golgi apparatus. We further employed rescue assays to demonstrate that this phenotype is connected to the ability of NAA80 to modify actin. Thus, re-expression of NAA80, which leads to re-establishment of actin`s N-terminal acetyl group, rescued the Golgi fragmentation, whereas a catalytic dead NAA80 mutant could neither restore actin Nt-acetylation nor Golgi structure. The Golgi phenotype of NAA80 KO cells was shared by both migrating and non-migrating cells and live-cell imaging indicated increased Golgi dynamics in migrating NAA80 KO cells. Finally, we detected a drastic increase in the amount of F-actin in cells lacking NAA80, suggesting a causal relationship between this effect and the observed re-organization of Golgi structure. The findings further underscore the importance of actin Nt-acetylation and provide novel insight into its cellular roles, suggesting a mechanistic link between actin modification state and Golgi organization.
Keywords: Actin cytoskeleton, cell migration, Golgi structure, N-terminal acetylation, N-alpha-acetyltransferase 80 (NAA80), posttranslational modification
Introduction
Recently, an actin modifier, the N-alpha-acetyltransferase 80 (NAA80/NatH), was identified and shown to perform the final step in actin’s unique N-terminal maturation process [1–3]. NAA80 is a member of the N-terminal acetyltransferase (NAT) family of enzymes, which are characterized by their ability to catalyse the transfer of an acetyl group from Acetyl-CoA to the N-terminal alpha-amino group of a protein [4]. N-terminal (Nt)-acetylation is probably the most abundant modification of human proteins, typically occurring co-translationally [5, 6]. Importantly, various links between this modification and essential biological processes were unravelled [reviewed in 4, 7], extending from folding, degradation, and molecular interactions at the protein level [8–11], to severe malfunctions like cancer [12] and inheritable developmental syndromes [13–16]. When it comes to actin Nt-acetylation, knockout of NAA80 in human cells leads to an increased ratio of filamentous to globular actin and a higher number of filopodia and lamellipodia, as well as accelerated migration [1, 17]. Several factors control the actin motor underlying cell migration [18], thus it is likely that actin Nt-acetylation by NAA80 is crucial for normal cytoskeletal regulation. The considerably altered cellular phenotype of NAA80 KO cells, combined with the known key roles of the actin cytoskeleton in several cellular processes, makes it likely that the absence of actin Nt-acetylation affects the functional organization of cellular organelles; however, this has not been previously explored.
Indeed, the actin cytoskeleton plays an important role in the organization of the Golgi apparatus [19–21], although the molecular mechanisms of this connection remain incompletely understood. The Golgi is composed of stacks of flat disc-like membranes termed cisternae, and acts as a key organelle of the secretory pathway and central station in intracellular trafficking [22–24]. The polarized Golgi stacks are divided into cis, medial and trans compartments, which accommodate distinct sets of Golgi enzymes that successively process secretory cargo proteins passing through this organelle on their way from the endoplasmic reticulum (ER) to the plasma membrane (PM) [22]. Mammalian cells typically contain multiple cisternal stacks, which are laterally linked to form the Golgi ribbon, a continuous structure, which is positioned in the perinuclear region due to its close association with the centrosome [25, 26].
Several findings support the notion of an intimate link between the actin machinery and the structural organization of the Golgi apparatus. Depletion or overexpression of different actin binding proteins (ABPs) has been reported to affect Golgi structure. For example, Golgi dispersion has been shown to result from depletion or overexpression of the actin nucleation promoting factor WHAMM (WASP homolog associated with actin, membranes and microtubules) [27], or constitutive activation of Diaph1 [28]. It also takes place in response to overexpression of INF1 [29], depletion of certain formins like FMNL1γ [30], FMNL2 and -3 [31], or a splice variant of INF2 [32], as well as depletion of certain myosins, such as myosin 1C [33]. On the other hand, Golgi compaction was observed upon depletion of cortactin [34], or the unconventional myosin 18A [35]. Furthermore, a Golgi-specific tropomyosin isoform was identified which might stabilise actin filaments at lateral rims of Golgi cisternae for vesicle formation [36].
Moreover, Golgi structure is affected by drugs targeting actin polymerization. Stabilising drugs like Jasplakinolide were reported to cause Golgi fragmentation, whereas actin depolymerising drugs like Latrunculin A were associated with Golgi compaction [37, 38]. Interestingly, Nt-acetylation of actin was reported to affect the balance between G- and F-actin in the favour of F-actin [1], as well as actin elongation and depolymerization properties, with the proposed net result being the stabilisation of filaments [1, 17]. However, neither the absolute F-actin levels of NAA80 KO cells, nor their Golgi morphology, have been previously addressed.
Results and Discussion
Absence of NAA80 and actin Nt-acetylation cause Golgi fragmentation
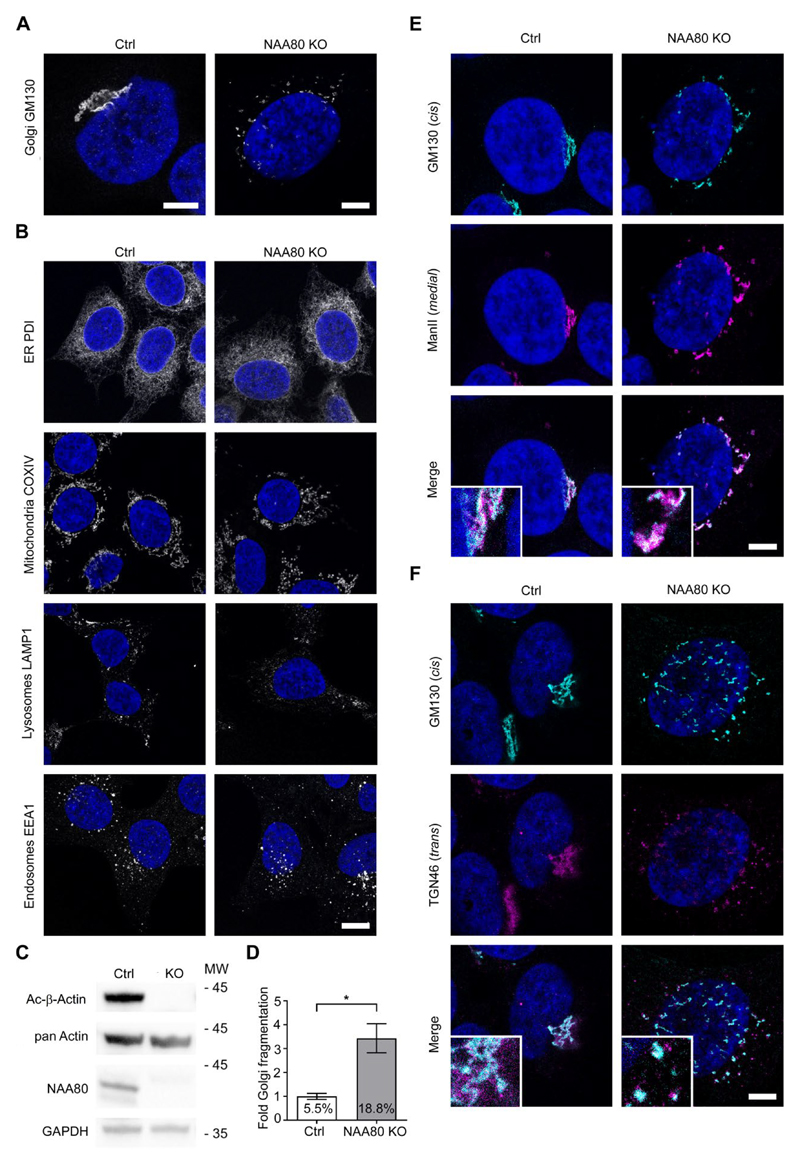
Initially, we subjected NAA80 KO cells to a small-scale organelle morphology screen using specific markers in immunofluorescence staining (Fig. 1A and B). These cells were validated to lack both the NAA80 protein and actin Nt-acetylation (Fig. 1C). We found that the NAA80 KO cultures harbour an increased proportion of cells with a fragmented Golgi, as initially detected by antibodies towards the cis-Golgi protein GM130 (Fig. 1A). Further quantification validated this observation (Fig. 1D). Use of different Golgi markers revealed that Golgi fragmentation can also be detected with antibodies against Golgi residents, which preferentially localize to medial (Fig. 1E) or trans compartments (Fig. 1F). The observed colocalisation of GM130 with the medial- and trans-Golgi markers in the KO cells indicated that while the Golgi ribbon becomes fragmented, the individual stacks remain relatively intact. Corresponding results for an additional knockout clone are shown in Supplemental Fig. S1. This analysis did not reveal any other organelles affected by the lack of NAA80 among those investigated (ER, mitochondria, lysosomes and endosomes) (Fig. 1B). However, it should be noted that this observation-based cell stain analysis does not completely exclude possible effects of NAA80 KO on other organelles potentially detectable by for example electron microscopy.
Fig. 1. NAA80 knockout results in Golgi fragmentation.
A Confocal STED images showing the Golgi morphology in fixed HAP1 Ctrl and NAA80 KO cells labelled with GM130, a cis-Golgi protein. Nuclei were stained with DAPI. Representative images from three independent experiments, scale bars = 5 μm. B. Fixed HAP1 Ctrl and NAA80 KO cells were IF labelled with different subcellular markers as indicated in addition to DAPI nuclear stain. Representative confocal STED images from two independent experiments, scale bar = 10 μm. C. Western blot analysis of HAP1 Ctrl and NAA80 KO whole cell lysates showing the Nt-acetylation status of actin (Ac-β-actin) and detection of pan actin and NAA80. GAPDH was used as a loading control. One of three independent experiments is shown. D. Quantification of Golgi fragmentation in HAP1 NAA80 KO cells, expressed relative to Ctrl and with absolute % of fragmented Golgi complexes in the culture indicated. The mean ± sd from three independent experiments (each with n ≥ 500 cells) is shown. *p ≤ 0.05, unpaired t-test with Welch’s correction. E. HAP1 Ctrl and NAA80 KO cells co-stained for GM130 (cis-Golgi) and Mannosidase II (medial-Golgi). Confocal STED images out of two independent experiments are shown. Zoomed-in inset images are in 3x magnification, scale bar = 5 μm. F. As in E, but showing co-staining of GM130 (cis-Golgi) and TGN46 (trans-Golgi).
Golgi fragmentation in NAA80 KO cells is a specific effect of missing actin Nt-acetylation
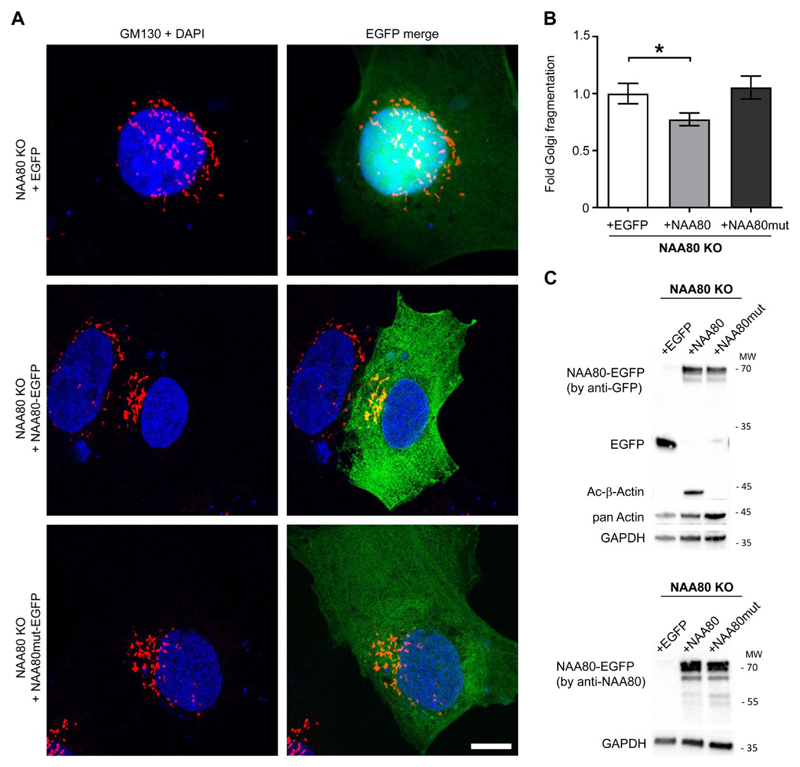
We next tested whether there exists a direct link between the Golgi phenotype of the NAA80 KO cells and the Nt-acetylation status of actin. Therefore, NAA80 KO cells were transiently transfected with EGFP constructs containing either the functional NAA80 enzyme or a catalytically non-functional NAA80 mutant, in addition to an empty EGFP control (Fig. 2A). Subsequently, cells were stained for GM130 and the degree of Golgi fragmentation was evaluated in transfected cells (Fig. 2 B). Only transfection with the functional NAA80 enzyme could significantly decrease Golgi fragmentation (Fig. 2B). A modest rescue effect is likely due to limited time of NAA80 expression in this transient transfection assay. Unspecific effects of the NAA80 knockout is unlikely since rescue assay with the other NAA80 knockout clone showed the same trend (Fig. S1 G and F). From previous work, we know that NAA80 is highly likely to act very specifically on actin [1, 2]. Assessing the actin Nt-acetylation status by western blot (Fig. 2C), demonstrated that the actin modification was restored to a high degree by this transient transfection. Following reacetylation of actin in this rescue assay, it is likely that reversing the Golgi fragmentation will take additional time. Combined, the data in Figure 2 show that Nt-acetylation of actin corresponds to a more compact Golgi phenotype, whereas lacking Nt-acetylation of actin is associated with conditions of increased Golgi fragmentation. Thus, NAA80 enzymatic activity, i.e. actin Nt-acetylation, is necessary to obtain normal Golgi structure.
Fig. 2. Restoring actin Nt-acetylation decreases Golgi fragmentation in NAA80 KO cells.
A. HAP1 NAA80 KO cells were transfected with either EGFP-control vector, NAA80-EGFP or catalytically dead NAA80mut-EGFP, fixed 24 h post transfection and IF stained for GM130. Representative confocal STED images from three independent experiments are shown; scale bar = 20 μm. B. Quantification of Golgi fragmentation in the transfected NAA80 KO cells shown in A. The mean ± sd of three independent experiments (each with n ≥ 200 cells) is shown; *p ≤ 0.05, one-way ANOVA with Tukey’s multiple comparison test. C. Western blot analysis of EGFP, actin Nt-acetylation (Ac-β-actin), pan actin and NAA80 in whole cell lysates of transfected NAA80 KO cells. Cells were transfected as in A and harvested 24 h post transfection. Representative blots from two independent experiments are shown.
Golgi fragmentation and hypermotility phenotypes of NAA80 KO are distinct
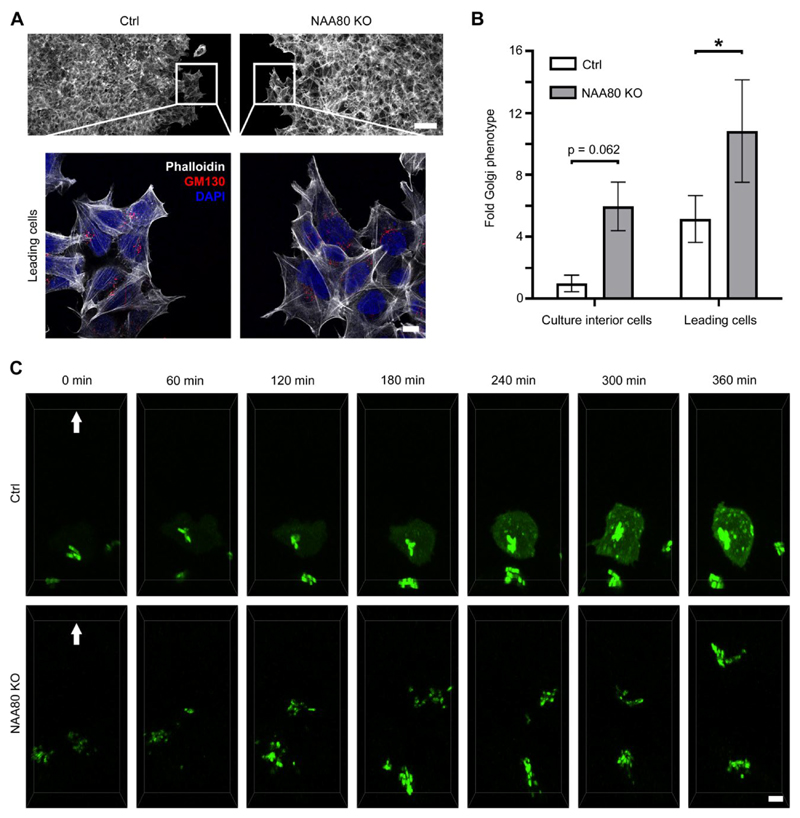
We previously showed that HAP1 NAA80 KO cells display a hypermotile phenotype, as shown by an increase in both random single cell movements and directed migration in a wound healing assay. In the latter case the NAA80 KO cells were able to fill the experimental wound significantly faster than control cells [1]. Since re-positioning of the Golgi apparatus is a pre-requisite for cell polarization and motility [26, 39], we tested whether the observed Golgi fragmentation of NAA80 KO cells could be connected to their hypermotile phenotype. Accordingly, Golgi morphology of HAP1 Ctrl and NAA80 KO cells was investigated during a wound healing assay. After 18 h of cell migration, the wounded cultures were fixed and co-stained for GM130 and phalloidin to visualise cis-Golgi and the actin filament networks, respectively (Fig. 3A). The Golgi fragmentation phenotypes of cells at the wound edge, or residing within the confluent inner cell mass, at least 180 μm from the wound edge, were compared (Fig. 3A). In general, we observed an increased tendency for Golgi fragmentation in the migrating (wound edge) vs. the non-migrating (inner mass) cells. For example, the actively migrating HAP1 Ctrl cells showed an about five-fold increase of Golgi fragmentation, as compared to the non-motile cell population. Nevertheless, in comparison to the control cells, the actively migrating NAA80 KO cells at the wound edge still showed about two-times higher degree of Golgi fragmentation. Comparing the non-migrating cells in the inner confluent areas, an approx. six-fold increase in Golgi fragmentation was observed in NAA80 KO vs. Ctrl cells. In summary, a fragmented Golgi can typically be observed more frequently in migrating wild-type cells as compared to non-migrating wild-type cells [26]. Importantly, however we disproved that the Golgi fragmentation observed in the NAA80 KO could simply be a secondary outcome of the previously established hypermotility phenotype of NAA80 KO cells [1, 17], but rather is specifically connected to the Nt-acetylation status of actin.
Fig. 3. Increased Golgi fragmentation in NAA80 KO cells is independent of cell motility and involves enhanced Golgi dynamics.
A. HAP1 Ctrl and NAA80 KO cells in a wound-healing assay, fixed 18 h post wounding, labelled with GM130 antibodies and stained with phalloidin. Confocal STED images show an overview of the wound edge (top, only phalloidin, 25x objective) and zoomed-in images showing the Golgi morphology in cells at the leading edge (100x objective). Representative images from three independent experiments are shown; scale bars correspond to 50 or 10 μm (overview or zoomed-in images, respectively). B. Quantification of Golgi fragmentation in the motile (leading edge) and relatively stationary (culture interior) cells from A. The mean ± sd from three independent experiments is shown, including n ≥ 200 cells per experiment. * p ≤ 0.05, one-way ANOVA with Tukey’s multiple comparison test. C. Selected images from live-cell recordings of HAP1 Ctrl or NAA80 KO cells transiently transfected with mannosidase II–mNeonGreen and migrating into the wound area during the total 6 h period of imaging. Cells were seeded 24 h and transfected 22 h prior to wound infliction and subsequent initiation of imaging. Shown are maximum projections of selected z-stacks (5 optical sections) acquired with spinning disk microscopy. Arrows point in the direction of cell movement. Representative cells at the very wound edge were chosen from a minimum of three independent experiments. Scale bar = 5 μm.
In addition to the fixed cells of the wound-healing assays, we investigated Golgi morphology in living migrating cells using an mNeonGreen-conjugated mannosidase II construct (Fig. 3C and Supplementary movie 1 and 2). In HAP1 Ctrl cells, the Golgi maintained its compact morphology and static localisation for almost the entire 6 h recording period. Only during the last two hours, noticeable repositioning of the Golgi towards the wound was seen. By contrast, in NAA80 KO cells the Golgi seemed to be much more dynamic, switching several times between compact and more fragmented morphologies, without assuming a preferred positioning in front of the nucleus facing the leading edge. In agreement with their hypermotile character, the NAA80 KO cells containing the Golgi signal advanced well into the open wound during the 6 h of live imaging. Supplementary movies related to this article can be found at https://doi.org/10.1016/j.yexcr.2020.111961
In summary, imaging of living cells suggested that increased Golgi dynamics and continuous remodelling of Golgi membranes might be the underlying reason for the Golgi fragmentation encountered in fixed cells. Furthermore, these results raise the possibility that the actin cytoskeleton could also increase its dynamics in the absence of actin Nt-acetylation; however, this point is not supported by current in vitro data [1], and remains to be investigated in intact cells. Additionally, based on the present live-imaging data, it can be speculated whether the lack of Nt-acetylation of actin renders the cell incapable of proper re-orientation of Golgi towards the leading edge of migrating cells. However, even if this were the case, it does not impair the speed of migration of the NAA80 KO cells, which remains constant in the course of wound healing [1]. It is possible that the total lack of Nt-acetylated actin in these cells represents a highly artificial condition, which is unrelated to normal cell physiology. Under these conditions it is possible that several actin-cytoskeletal functions are affected simultaneously, creating an imbalance in these tightly regulated processes and making the connection between migration and Golgi phenotypes difficult to interpret.
Increased F-actin content of NAA80 KO cells may contribute to Golgi fragmentation
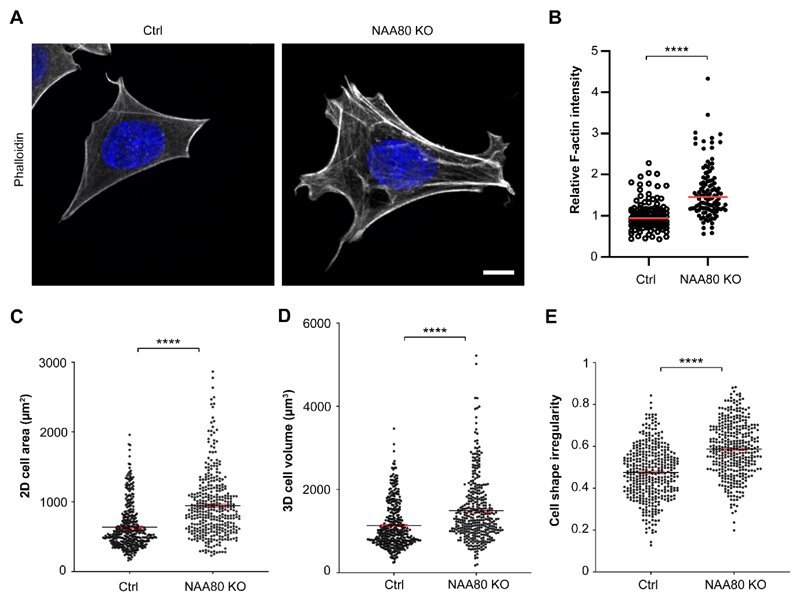
Since Golgi structure is affected by altered F-actin content [37, 38], and Nt-acetylation of actin was reported to decrease the G/F-actin ratio [1], we wanted to directly assess the F-actin content of NAA80 KO cells. We therefore optimized an F-actin measurement procedure based on intra-well simultaneous phalloidin staining of NAA80 KO and Ctrl cells (see Methods). Notably, the intensity of the F-actin signal was clearly higher in the NAA80 KO cells (Fig. 4A). Quantification of the F-actin signal relative to cell size revealed that NAA80 KO cells display a considerably (approx. 1.6-fold) more intensive F-actin signal, as compared to Ctrl cells (Fig. 1B). In order to quantitatively assess the cell size for this purpose, we subjected cells to live-cell holographic image analysis. Here we detected an increased cell size in both 2D cell area (Fig. 4C) and 3D cell volume (Fig 4D), of which the latter has previously been linked to F-actin content [40, 41]. In addition, a parameter called cell shape irregularity was also noticeably increased for NAA80 KO cells in this analysis (Fig. 4E), possibly reflecting the increased tendency of NAA80 KO cells to form the cytoskeleton-supported membrane protrusions reported previously [1]. The holographic time lapse movies S3 and S4 display differences in morphological parameters detected in the quantitative analysis.
Fig. 4. Increased level of F-actin and morphological differences in NAA80 KO cells.
A. HAP1 NAA80 KO and Ctrl cells were subjected to intra-well simultaneous fluorescent phalloidin staining and imaged with equal settings using a high-speed confocal spinning disk. Shown are representative cells for Ctrl and NAA80 KO imaged as z-stacks. DAPI was used to stain nuclei. Scale bar = 10 µm. B. The sum of F-actin intensity (based on the phalloidin staining) was measured in single HAP1 Ctrl and NAA80 KO cells from A. Shown are single values for the relative F-actin intensity normalised to the Ctrl. The mean is marked as a red line. A total of n ≥ 100 cells pooled out of three independent experiments are shown. **** p ≤ 0.0001, unpaired t-test with Welch’s correction. C-E. Morphological analysis measuring 2D cell area (C, 49 % increase), 3D cell volume (D, 32 % increase) and cell shape irregularity (E, 24 % increase) by live-cell holographic imaging using the HoloMonitor M4 system (see Methods). Shown is the mean (black line) with ± SEM (red lines) of n = 364 (NAA80 KO) and n = 383 (Ctrl) cells pooled together out of three independent experiments. **** p ≤ 0,0001, unpaired t-test with Welch’s correction.
Intriguingly, the simultaneous occurrence of increased F-actin levels and a fragmented Golgi has already been observed in other contexts, for example, when employing F-actin stabilizing or destabilizing drugs or up/down regulation of certain ABPs, as mentioned earlier. Certain ABPs could conceivably be dependent on the Nt-acetylated state of actin, in line with known implications of Nt-acetylation for other proteins [4, 9, 42, 43]. This could potentially either affect the Golgi structure directly or contribute to increased F-actin content. But, it remains currently unknown whether the binding behaviour of ABPs are sensitive towards this N-terminal modification. Another proposed general mechanism of Nt-acetylation is its impact on protein turnover [44]. Thus, one may consider whether actin itself might accumulate over time due to an increased half-life in the absence of the Nt-acetylation. Absolute levels of total actin in NAA80 KO cells relative to Ctrl have however not been precisely determined yet.
It has already been shown that the proliferation of HAP1 NAA80 KO cells is somewhat slowed down [1]. Hence, it is possible that the increased fragmentation of the Golgi in NAA80 KO cells is partly caused by delay(s) in cell cycle progression, since Golgi morphology is closely regulated and undergoes dramatic changes during the cell cycle. At late G2 phase, as cells prepare for mitosis, the Golgi ribbon is unlinked, as the individual cisternal stacks lose their lateral connections and become separated. This step in particular has been suggested to be regulated also by the actin cytoskeleton [45] and Golgi inheritance phenotypes connected to this has been reported [46]. Conversely, Golgi fragmentation might in turn affect cell cycle progression. Note however, that visually detectable mitotic cells were excluded from the Golgi phenotype analysis and that the most dramatic fold increase in Golgi fragmentation was obtained from cells in the confluent inner mass in the wound assay, where one can expect a limited amount of cell division events.
In conclusion, we here showed that Nt-acetylation of actin by NAA80 is essential for maintaining the structural integrity of the Golgi and point to a balance in F-actin amount as a possible important mechanistic link. Hence, we reveal new insight into the cellular function of this newly identified actin modifying enzyme. It remains a topic for future studies to determine whether actin Nt-acetylation dynamics might have biological regulatory roles to increase migration speed and/or Golgi fragmentation under certain conditions and whether Nt-acetylation-dependent ABP interactions might be involved.
Methods
Plasmids and Antibodies
NAA80-EGFP and NAA80mut-EGFP were described previously [1]. mNeonGreen-ManII-N-10 was obtained from Allele Biotechnology, San Diego, CA, USA.
Primary antibodies used were mouse anti-Nt-acetylated β-Actin (Abcam, ab6276, Nt-Ac specificity validated in [1]), mouse anti-Actin-pan (Abcam, ab8224), rabbit anti-COXIV (Cell signalling Technology, 4850), rabbit anti-EEA1 (Santa Cruz Biotechnology, sc-33585), rabbit anti-GAPDH (Santa Cruz Biotechnology, sc-25778), rabbit anti-GM130 (Abcam, ab52649), mouse anti-GM130 (BD Biosciences, 610822), mouse anti-LAMP1 (Santa Cruz Biotechnology, sc-18821), rabbit anti-Mannosidase II (kind gift from Kelley Moremen University of Georgia, USA), rabbit anti-NAA80 (custom made, Biogenes GmbH, Berlin, Germany), mouse-PDI (Thermo Fisher Scientific Inc., MA3-018), sheep anti-TGN46 (Bio-Rad Laboratories Inc., AHP5006).
Cell culture
HAP1 cells were obtained from Horizon Genomics and cultured as recommended. NAA80 KO1 (HZGHC003171c003) and KO2 (HZGHC003171c012) and wild-type HAP1 cells (C631) were grown in Iscove’s Modified Dulbecco’s Medium with 10% FBS and 1% penicillin/streptomycin. In the present study, NAA80 KO2 (HZGHC003171c012, harbouring a 404 bp insertion in exon 2) was used as the primary clone and central results were validated with NAA80 KO1 (HZGHC003171c003, harbouring a 17 bp deletion in exon 2) shown in the Supplementary data. The original KO numbering previously introduced [1] was kept. Since these near-haploid cells typically diploidize spontaneously over time, all HAP1 cell lines were passaged until diploid status was confirmed by an Accuri BD C6 flow cytometer using propidium iodide, as described previously [1] and detailed in (Beigl, Kjosås, Seljeseth, Glomnes and Aksnes, unpublished).
All cells were kept at 37 °C and 5% CO2. In transfection experiments, cells were seeded on plates or coverslips and transfected with X-tremeGENE 9 (Roche) as recommended and imaged ~24 h posttransfection.
Immunofluorescence staining of cells
Cells grown on glass coverslips (1.5 H, Assistant) were fixed with 3% (wt/vol) paraformaldehyde (PFA) in 0.1 M phosphate buffer for 30 min, permeabilized with 0.1% Triton X-100 for 10 min, washed in PBS, blocked with 8% BSA/2% goat serum in PBS for 1h, incubated with primary antibody diluted in 2% BSA/2% goat serum for 1-4 hours, washed in PBS, incubated with secondary antibody for 45-60 min, washed three times in PBS for a total of at least 1 h, and finally washed in water before mounting in Prolong Diamond with DAPI. F-actin was detected by incubation with fluorescent conjugated phalloidin (diluted 1:50 in PBS) along with the secondary antibodies.
F-actin intensity measurement
For the quantification of F-actin by phalloidin-fluorescence intensity, additional steps were performed to secure comparable intensity quantification. Knockout and control cells were seeded in each side of an ibidi two chamber silicone inlet placed on a 12 mm coverslip. The following day, prior to fixation, the silicone inlet was removed to allow the simultaneous intra-well processing of the two different cell lines. Cells were immediately fixed as described above, washed three times with PBS and subsequently permeabilised like IF samples (0.1% Triton X-100, 10 min, RT). Phalloidin-Atto 647N was diluted 1:50 in PBS and incubated for 20 min at RT in the well. For the imaging, randomly chosen single cells without any directly adjacent cells were imaged as z-stacks including the whole F-actin cytoskeleton. High-speed confocal imaging was done with the Andor Dragonfly system as further described below. The total intensity of the F-actin signal in z-stacks was quantified using the Imaris software.
Imaging
Confocal (STED) images were obtained using a Leica TCS SP8 STED 3x confocal laser microscope equipped with an HC PL APO STED 100x 1.4 NA oil objective, 1 Airy unit pinhole aperture and the appropriate filter combinations. The used lasers were a blue-diode (405 nm, 50 mW), white-light (470-670 nm lambda range, ~ 1.5 mW per line, pulsed supercontinuum), and a depletion laser (STED, 775 nm). Images were acquired with the Leica Application Suite X software, exported and processed in ImageJ.
High-speed confocal imaging of fixed samples was performed using an Andor Dragonfly 500 spinning disk confocal system mounted on a Nikon Eclipse Ti2 microscope with a CFI SR HP Apo TIRF 100x 1.49 NA oil objective, iXion camera and appropriate filter combinations. The system was coupled to an Andor Integrated Laser Engine multi-line laser source in combination with the Andor Borealis Beam Conditioning Unit. Images were acquired with the Fusion software and analysed using the Imaris software.
Golgi phenotype quantification was done using a Leica DMI6000 B wide field fluorescence microscope in combination with an HCX PL APO 100x 1.4 NA oil objective (+1.5x magnification lens). In randomly chosen fields of view cell number was counted using the DAPI stained nuclei and number of cells with compact and fragmented Golgi morphology was determined using the Golgi marker GM130, similar as previously [47]. Mitotic cells recognisable by the DAPI staining were excluded. In quantification approaches with transient transfected cells, only visibly transfected (EGFP expressing) cells were included. Cells only partly visible in the field of view were excluded from the quantification.
For the live-cell fluorescent imaging, HAP1 NAA80 KO and control cells were seeded simultaneously in one 35 mm μ-dish (ibidi) on each side of a silicone insert (ibidi). Cell attachment was facilitated 20 min at RT followed by incubation at 37°C, 5% CO2 for 2 hours before transfection with the ManII-mNeonGreen construct. After approx. 24 h, the silicone inlet was removed and the dish gently rinsed with PBS. Using a thin sterile needle, micro scratch wounds were inflicted in both Ctrl and KO cell patches. Dishes were filled with IMDM w/o phenol red and immediately imaged using the Andor Dragonfly spinning disk system in combination with a 60x CFI Plan Apochromat Lambda oil objective. Small z-stacks with 5 planes were taken every 15 min at a maximum of four positions in one session using the Fusion software. Images were subsequently exported to the Imaris software for further processing.
For holographic imaging and analysis of live cells a HoloMonitor M4 (PHI AB, Sweden) was used. Data in form of holographic 3D time-lapse images were further processed in the HStudio software. 50,000 cells were seeded in 35 mm μ-dishes (ibidi) in 3 ml cell culture medium. This resulted in an initial cell confluency of 2-5%. Cells were imaged every 5 min for 13 hours with at least 3 different fields of view per dish (20x magnification). Morphological analysis of cell area, volume and cell shape irregularity was done for images taken 13 hours post seeding. Cells were identified and segmented by defining an object size threshold combined with background threshold Auto-Otsu, pre-smoothing “off”.
SDS PAGE and Western blot
SDS PAGE and Western blot was performed as described previously [48].
Supplementary Material
Summary statement.
The authors report on a newly identified posttranslational modification of actin and show here how its absence fragments Golgi structure, possibly through elevated levels of F-actin.
Acknowledgements
The confocal and spinning-disk imaging was performed at the Molecular Imaging Center (MIC), Department of Biomedicine, University of Bergen. We thank Hege Avsnes Dale in particular for help with Imaris image processing.
Funding
The work was funded by the Research Council of Norway (Project 249843), the Norwegian Health Authorities of Western Norway (Project 912176), the Norwegian Cancer Society, and the European Research Council (ERC) under the European Union Horizon 2020 Research and Innovation Program under grant agreement 772039.
Footnotes
Competing interests
The authors declare no competing or financial interests.
CRediT authorship contribution statement
Tobias B. Beigl: Conceptualization, Writing - original draft, Writing - review & editing, Data curation, Formal analysis, Investigation, Methodology, Visualization. Monica Hellesvik: Writing - review & editing, Data curation, Formal analysis, Investigation, Methodology, Visualization. Jaakko Saraste: Writing - review & editing. Thomas Arnesen: Writing - review & editing, Supervision, Funding acquisition. Henriette Aksnes: Conceptualization, Writing - original draft, Writing - review & editing, Supervision, Project administration.
References
- 1.Drazic A, et al. NAA80 is actin's N-terminal acetyltransferase and regulates cytoskeleton assembly and cell motility. Proc Natl Acad Sci U S A. 2018;115(17):4399–4404. doi: 10.1073/pnas.1718336115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Goris M, et al. Structural determinants and cellular environment define processed actin as the sole substrate of the N-terminal acetyltransferase NAA80. Proc Natl Acad Sci U S A. 2018;115(17):4405–4410. doi: 10.1073/pnas.1719251115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wiame E, et al. NAT6 acetylates the N-terminus of different forms of actin. FEBS J. 2018;285(17):3299–3316. doi: 10.1111/febs.14605. [DOI] [PubMed] [Google Scholar]
- 4.Aksnes H, Ree R, Arnesen T. Co-translational, Post-translational, and Non-catalytic Roles of N-Terminal Acetyltransferases. Mol Cell. 2019;73(6):1097–1114. doi: 10.1016/j.molcel.2019.02.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Aksnes H, et al. First Things First: Vital Protein Marks by N-Terminal Acetyltransferases. Trends Biochem Sci. 2016;41(9):746–760. doi: 10.1016/j.tibs.2016.07.005. [DOI] [PubMed] [Google Scholar]
- 6.Arnesen T, et al. Proteomics analyses reveal the evolutionary conservation and divergence of N-terminal acetyltransferases from yeast and humans. Proc Natl Acad Sci U S A. 2009;106(20):8157–62. doi: 10.1073/pnas.0901931106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Millar AH, et al. The Scope, Functions, and Dynamics of Posttranslational Protein Modifications. Annu Rev Plant Biol. 2019;70:119–151. doi: 10.1146/annurev-arplant-050718-100211. [DOI] [PubMed] [Google Scholar]
- 8.Holmes WM, et al. Loss of amino-terminal acetylation suppresses a prion phenotype by modulating global protein folding. Nat Commun. 2014;5 doi: 10.1038/ncomms5383. 4383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Scott DC, et al. N-terminal acetylation acts as an avidity enhancer within an interconnected multiprotein complex. Science. 2011;334(6056):674–8. doi: 10.1126/science.1209307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Shemorry A, Hwang CS, Varshavsky A. Control of protein quality and stoichiometries by N-terminal acetylation and the N-end rule pathway. Mol Cell. 2013;50(4):540–51. doi: 10.1016/j.molcel.2013.03.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Elia F, et al. The N-terminus of Sec61p plays key roles in ER protein import and ERAD. PLoS One. 2019;14(4):e0215950. doi: 10.1371/journal.pone.0215950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kalvik TV, Arnesen T. Protein N-terminal acetyltransferases in cancer. Oncogene. 2013;32(3):269–76. doi: 10.1038/onc.2012.82. [DOI] [PubMed] [Google Scholar]
- 13.Cheng H, et al. Truncating Variants in NAA15 Are Associated with Variable Levels of Intellectual Disability, Autism Spectrum Disorder, and Congenital Anomalies. Am J Hum Genet. 2018;102(5):985–994. doi: 10.1016/j.ajhg.2018.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cheng H, et al. Phenotypic and biochemical analysis of an international cohort of individuals with variants in NAA10 and NAA15. Hum Mol Genet. 2019;28(17):2900–2919. doi: 10.1093/hmg/ddz111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Myklebust LM, et al. Biochemical and cellular analysis of Ogden syndrome reveals downstream Nt-acetylation defects. Hum Mol Genet. 2015;24(7):1956–76. doi: 10.1093/hmg/ddu611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Rope AF, et al. Using VAAST to identify an X-linked disorder resulting in lethality in male infants due to N-terminal acetyltransferase deficiency. Am J Hum Genet. 2011;89(1):28–43. doi: 10.1016/j.ajhg.2011.05.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Aksnes H, et al. Actin polymerization and cell motility are affected by NAA80-mediated posttranslational N-terminal acetylation of actin. Commun Integr Biol. 2018;11(4):e1526572. doi: 10.1080/19420889.2018.1526572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Grantham J, Lassing I, Karlsson R. Controlling the cortical actin motor. Protoplasma. 2012;249(4):1001–15. doi: 10.1007/s00709-012-0403-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Egea G, et al. Actin acting at the Golgi. Histochem Cell Biol. 2013;140(3):347–60. doi: 10.1007/s00418-013-1115-8. [DOI] [PubMed] [Google Scholar]
- 20.Gurel PS, Hatch AL, Higgs HN. Connecting the cytoskeleton to the endoplasmic reticulum and Golgi. Curr Biol. 2014;24(14):R660–R672. doi: 10.1016/j.cub.2014.05.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ravichandran Y, Goud B, Manneville JB. The Golgi apparatus and cell polarity: Roles of the cytoskeleton, the Golgi matrix, and Golgi membranes. Curr Opin Cell Biol. 2019;62:104–113. doi: 10.1016/j.ceb.2019.10.003. [DOI] [PubMed] [Google Scholar]
- 22.Klumperman J. Architecture of the mammalian Golgi. Cold Spring Harb Perspect Biol. 2011;3(7) doi: 10.1101/cshperspect.a005181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mironov AA, Beznoussenko GV. Molecular mechanisms responsible for formation of Golgi ribbon. Histol Histopathol. 2011;26(1):117–33. doi: 10.14670/HH-26.117. [DOI] [PubMed] [Google Scholar]
- 24.Yadav S, Linstedt AD. Golgi positioning. Cold Spring Harb Perspect Biol. 2011;3(5) doi: 10.1101/cshperspect.a005322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gosavi P, Gleeson PA. The Function of the Golgi Ribbon Structure - An Enduring Mystery Unfolds! Bioessays. 2017;39(11) doi: 10.1002/bies.201700063. [DOI] [PubMed] [Google Scholar]
- 26.Saraste J, Prydz K. A New Look at the Functional Organization of the Golgi Ribbon. Front Cell Dev Biol. 2019;7:171. doi: 10.3389/fcell.2019.00171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Campellone KG, et al. WHAMM is an Arp2/3 complex activator that binds microtubules and functions in ER to Golgi transport. Cell. 2008;134(1):148–61. doi: 10.1016/j.cell.2008.05.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zilberman Y, et al. Involvement of the Rho-mDia1 pathway in the regulation of Golgi complex architecture and dynamics. Mol Biol Cell. 2011;22(16):2900–11. doi: 10.1091/mbc.E11-01-0007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Copeland SJ, Thurston SF, Copeland JW. Actin- and microtubule-dependent regulation of Golgi morphology by FHDC1. Mol Biol Cell. 2016;27(2):260–76. doi: 10.1091/mbc.E15-02-0070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Colon-Franco JM, Gomez TS, Billadeau DD. Dynamic remodeling of the actin cytoskeleton by FMNL1gamma is required for structural maintenance of the Golgi complex. J Cell Sci. 2011;124(Pt 18):3118–26. doi: 10.1242/jcs.083725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kage F, et al. FMNL2 and -3 regulate Golgi architecture and anterograde transport downstream of Cdc42. Sci Rep. 2017;7(1) doi: 10.1038/s41598-017-09952-1. 9791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ramabhadran V, et al. Splice variant-specific cellular function of the formin INF2 in maintenance of Golgi architecture. Mol Biol Cell. 2011;22(24):4822–33. doi: 10.1091/mbc.E11-05-0457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Capmany A, et al. MYO1C stabilizes actin and facilitates the arrival of transport carriers at the Golgi complex. J Cell Sci. 2019;132(8) doi: 10.1242/jcs.225029. [DOI] [PubMed] [Google Scholar]
- 34.Kirkbride KC, et al. Regulation of late endosomal/lysosomal maturation and trafficking by cortactin affects Golgi morphology. Cytoskeleton (Hoboken) 2012;69(9):625–43. doi: 10.1002/cm.21051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Dippold HC, et al. GOLPH3 bridges phosphatidylinositol-4- phosphate and actomyosin to stretch and shape the Golgi to promote budding. Cell. 2009;139(2):337–51. doi: 10.1016/j.cell.2009.07.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Percival JM, et al. Targeting of a tropomyosin isoform to short microfilaments associated with the Golgi complex. Mol Biol Cell. 2004;15(1):268–80. doi: 10.1091/mbc.E03-03-0176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.di Campli A, et al. Morphological changes in the Golgi complex correlate with actin cytoskeleton rearrangements. Cell Motil Cytoskeleton. 1999;43(4):334–48. doi: 10.1002/(SICI)1097-0169(1999)43:4<334::AID-CM6>3.0.CO;2-3. [DOI] [PubMed] [Google Scholar]
- 38.Lazaro-Dieguez F, et al. Actin filaments are involved in the maintenance of Golgi cisternae morphology and intra-Golgi pH. Cell Motil Cytoskeleton. 2006;63(12):778–91. doi: 10.1002/cm.20161. [DOI] [PubMed] [Google Scholar]
- 39.Kupfer A, Louvard D, Singer SJ. Polarization of the Golgi apparatus and the microtubule-organizing center in cultured fibroblasts at the edge of an experimental wound. Proc Natl Acad Sci U S A. 1982;79(8):2603–7. doi: 10.1073/pnas.79.8.2603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Henson JH. Relationships between the actin cytoskeleton and cell volume regulation. Microsc Res Tech. 1999;47(2):155–62. doi: 10.1002/(SICI)1097-0029(19991015)47:2<155::AID-JEMT7>3.0.CO;2-T. [DOI] [PubMed] [Google Scholar]
- 41.Hoffmann EK, Lambert IH, Pedersen SF. Physiology of cell volume regulation in vertebrates. Physiol Rev. 2009;89(1):193–277. doi: 10.1152/physrev.00037.2007. [DOI] [PubMed] [Google Scholar]
- 42.Arnaudo N, et al. The N-terminal acetylation of Sir3 stabilizes its binding to the nucleosome core particle. Nat Struct Mol Biol. 2013;20(9):1119–21. doi: 10.1038/nsmb.2641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Yang D, et al. Nalpha-acetylated Sir3 stabilizes the conformation of a nucleosome-binding loop in the BAH domain. Nat Struct Mol Biol. 2013;20(9):1116–8. doi: 10.1038/nsmb.2637. [DOI] [PubMed] [Google Scholar]
- 44.Varshavsky A. N-degron and C-degron pathways of protein degradation. Proc Natl Acad Sci U S A. 2019;116(2):358–366. doi: 10.1073/pnas.1816596116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kondylis V, et al. The golgi comprises a paired stack that is separated at G2 by modulation of the actin cytoskeleton through Abi and Scar/WAVE. Dev Cell. 2007;12(6):901–15. doi: 10.1016/j.devcel.2007.03.008. [DOI] [PubMed] [Google Scholar]
- 46.Magliozzi R, et al. Inheritance of the Golgi Apparatus and Cytokinesis Are Controlled by Degradation of GBF1. Cell Rep. 2018;23(11):3381–3391 e4. doi: 10.1016/j.celrep.2018.05.031. [DOI] [PubMed] [Google Scholar]
- 47.Aksnes H, et al. An organellar Nα-acetyltransferase, Naa60, acetylates cytosolic N termini of transmembrane proteins and maintains Golgi integrity. Cell Rep. 2015;10(8):1362–74. doi: 10.1016/j.celrep.2015.01.053. [DOI] [PubMed] [Google Scholar]
- 48.Aksnes H, et al. Molecular determinants of the N-terminal acetyltransferase Naa60 anchoring to the Golgi membrane. J Biol Chem. 2017;292(16):6821–6837. doi: 10.1074/jbc.M116.770362. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.