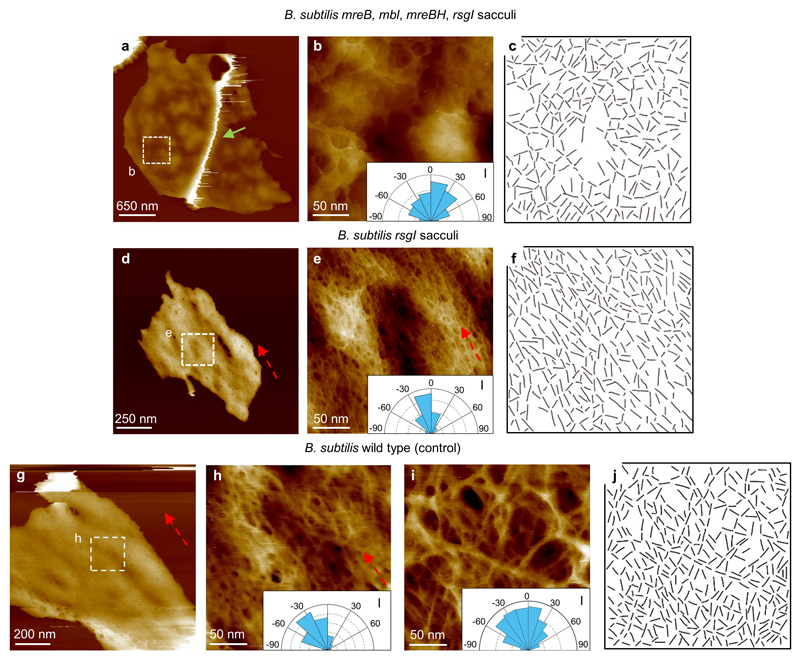
Extended Data Figure 8. B. subtilis strand orientation analysis and mechanistic insights.
a, B. subtilis mreB mbl mreBH, rsgI mutant sacculus image of the internal structure of cell wall, with a defined ridge (green arrow). This mutant lacks the MreB family of proteins, which leads to a spheroidal shape. DS= 411 nm. b, Higher resolution image from a showing the fine structure of the internal surface of a B. subtilis mreB mbl mreBH, rsgI mutant sacculus, with a disordered structure (see Inset, I, indicating a broad distribution of strand orientations) similar to the internal structure of S. aureus, suggesting that when Gram positive cells cannot maintain a rod shape, no cylindrical orientation is required. DS = 52 nm. c, Data used to obtain the orientation distributions for the glycan strands. Lines were hand drawn over the images along the clearly visible strands, using short straight strokes. A custom MATLAB routine (SI2) was then used to obtain the orientation distribution of these lines. The routine successfully recognised ~ 70% of the lines and determined their orientation. d, The mutations of the MreB family, shown in a-c, was created in a B. subtilis rsgI background strain. We prepared a purified sample from this B. subtilis rsgI mutant as a control. This image corresponds to the internal CW structure. DS = 76 nm. e, Higher resolution image from within d showing glycan strand orientation along the short axis of the rod morphology of the bacteria (dashed red arrow and inset, I). This sample had the same characteristics and structures as the WT strain. DS = 23 nm. f, Data used to plot the orientation distribution rose from inset in e. g, As an additional control, a sacculi sample of B. subtilis WT was grown as the mutants. This image shows another internal CW structure from a broken sacculus. DS = 125 nm. h, Higher resolution image from within g showing glycan strand orientation cylindrical along the short axis of the rod morphology of the bacteria (see dashed red arrow and inset, I). DS = 16 nm. i, The same strand orientation analysis as shown in h was applied to the external surface of B.subtilis cells (Fig. 3c in the main text), the angle distribution is broad (inset, I), indicating the glycan strands on the external surface have no predominant orientation in contrast with the internal surface. j, Data used to plot the orientation distribution rose from the inset in i, using the same procedure as c and f. This experiment provides mechanistic insight into the cylindrical orientation architecture in the inside of the rod-shaped of Gram-positive species B. subtilis. The protein complex of MreB, Mbl and MBH is vital for the synthesis of the peptidoglycan in this cylindrical architecture22–24.