Abstract
The gut microbiome and circadian rhythms both exhibit unique influence on mammalian hosts and have been implicated in the context of many diseases, particularly metabolic disorders. It has become increasingly apparent that these systems also interact closely to alter host physiology and metabolism. However, the mechanisms that underlie these observations remain largely unknown. Recent findings have implicated microbially-derived mediators as potential signals between the gut microbiome and host circadian clocks; two specific mediators are discussed in this review: short-chain fatty acids and bile acids. Key gaps in knowledge and major challenges that remain in the circadian and microbiome fields are also discussed, including animal vs human models and the need for precise timed sample collection.
Keywords: Microbiome, circadian rhythms, metabolism, short-chain fatty acids, bile acids
Linking Gut Microbes and Circadian Rhythms
The gut microbiome has been largely recognized for its importance in regulating several physiological systems, including the immune, metabolic, and nervous systems. Like any other organ of the body, the gut microbial “organ” is wholly interconnected with other vital life systems, and its composition and functions are very responsive to environmental and dietary stimuli. There is growing evidence that the gut microbiome plays an essential role in regulating host metabolism (see Box 1), particularly in its action on and reaction to host circadian rhythms (CRs). CRs are governed by a feedback loop of transcription factors that regulate gene networks that constitute our biological clock and are responsible for temporal organization of processes that oppose one another, thereby supporting energy efficiency (see Box 2). Similar to the microbiome, the clock is key to maintaining normal functions in countless physiological processes, from temperature regulation and hormone secretion to digestion, metabolism, and behavior. The functional link between gut microbiota and CRs ultimately impacts host metabolism. There are potentially hundreds of other bioactive molecules, from either the host or microbiota, that contribute to this complex, bi-directional system, and researchers are only just beginning to identify and delineate their mechanisms of action. Some host-derived mediators that have been shown to influence the gut microbiome in a circadian context include anti-microbial peptides[1], glucocorticoid hormones[2], and intestinal mucus secretion[3]. Microbially-derived mediators have also been identified as circadian modulators, such as hydrogen sulfide, certain vitamins, and tryptophan derivatives[4]. Although these mediators are likely key components of the circadian-microbiome network and must be explored further, little is currently known concerning their mechanisms of action. This review describes the current state of knowledge of two microbially-derived mediators that have been studied and characterized at length, short-chain fatty acids (SCFAs) and modified bile acids (Figure 1). Remaining challenges that must be overcome in order to advance discovery in such a nascent field are also discussed.
Box 1: Gut microbes regulate metabolism.
Comprised of trillions of microorganisms that reside in various niches throughout the gastrointestinal tract, gut microbiota are involved in the development, maintenance, and deterioration of many host systems and processes. A broad range of hosts, from plants to animals, tolerate and nurture these commensal microbes because of the vast number of functions they perform; To name a few, gut microbes aid in digestion of specific dietary nutrients, produce vitamins absorbed by the host, influence development of specific organ systems, and are essential for immune system programming. Imbalance of microbes are also associated with the development of several diseases[60,61]. Emerging research on host-microbe interactions reveals that gut microbes are capable of programming host energy balance, such as lipid absorption and storage, and have a major influence on metabolic health[31,62]. Studies that utilize germ-free (GF) mice, raised in complete absence of microbes, describe a unique metabolic profile compared to conventionally-raised, Specific Pathogen Free (SPF) mice, including reduced adiposity, improved glucose tolerance, and insulin sensitivity[63]. GF mice are resistant to diet-induced obesity, and microbiota transplantation quickly results in increased adiposity, implying that gut microbes can alter host energy balance and metabolic programming [5,63–65]. Conventionalization of GF mice induces changes in intestinal and peripheral tissue transcription patterns of key metabolic genes relating to nutrient absorption and metabolism, demonstrating that gut microbes directly influence host metabolic programming[66,67].
Gut microbiota are exquisitely sensitive to environmental changes, including dietary and physiological, and exhibit rapid responses in both community membership and functions – effects that can feedback onto host health outcomes[40,68,69]. Humans with obesity exhibit similar dysbiosis in comparison to lean individuals, while leptin-deficient obese mice display a similar phenomenon[70,71]. This has also been demonstrated by GF mouse conventionalization; mice that receive obese SPF donor sample display increased adiposity compared to lean SPF donor, despite no differences in food consumption between groups[31]. However, human microbiome studies are difficult to replicate and sufficiently control all variables, and community composition often differs between cohorts and studies. Altogether, these observations suggest that diet can functionally alter host physiology via gut microbes, ultimately inducing metabolic disease, but more must be done to understand why and how gut microbes induce such a profound and unique metabolic phenotype. This topic has been covered extensively by this source[72].
Box 2: Circadian rhythms regulate metabolism.
CRs are essential to coordinate behaviors to the rising and setting of the sun, but also to compartmentalize key physiological processes. CRs are observable in almost all animal species regularly exposed to light, including prokaryotes and cyanobacteria[73]. Defined by their self-sustaining and cell-autonomous nature, CRs are perfectly positioned to synchronize the myriad of events necessary to adjust metabolism to the active and inactive phases of life[74]. The molecular components of the circadian clock are expressed in nearly every cell and tissue-type, and form an elegant transcriptional-translational feedback loop that controls gene expression patterns. In short, Transcription factors BMAL1 and CLOCK dimerize and activate transcription of many genes, including Cryptochrome (Cry) 1-2 and Period (Per) 1–3 which serve as the negative arm to repress Bmal1 and Clock transcription. The expression of Cry1-2 and Per1–3 are subsequently reduced, releasing the negative feedback arm and allowing the positive arm to resume expression. Additional components also regulate the components of this loop, including Retinoic Receptor-related Orphan Receptor (Ror) and Rev-erbs. When translated, these two components act in opposition; ROR promotes Bmal1 transcription, and REV-ERBs inhibit it[75].
Host CRs are coordinated by a central, master clock located in the suprachiasmatic nuclei (SCN) of the hypothalamus. The central clock receives light and dark cues from the environment and transmits this information to peripheral tissues to keep the body functioning along the same rhythm. Although the genes involved in maintaining the circadian network drive many biological pathways, they are especially relevant in metabolism[74,76]. Clock components, particularly Bmal1, exhibit promoter-binding patterns that are enriched for key metabolic genes, which likely contribute to the vast number of metabolic genes that display rhythmic transcription and translation[77–79]. Transcriptome expression analysis of several mouse organs indicate that almost half of all protein-coding genes exhibit 24-hour oscillation in at least one tissue, with the highest percentage of 16% in the liver[80]. Following circadian disruption, metabolism and energy networks become imbalanced, leading to disorders such as obesity and diabetes[81,82]. This topic has been covered extensively by this source[83].
Figure 1: Microbe-derived mediators that influence host metabolic outcomes.
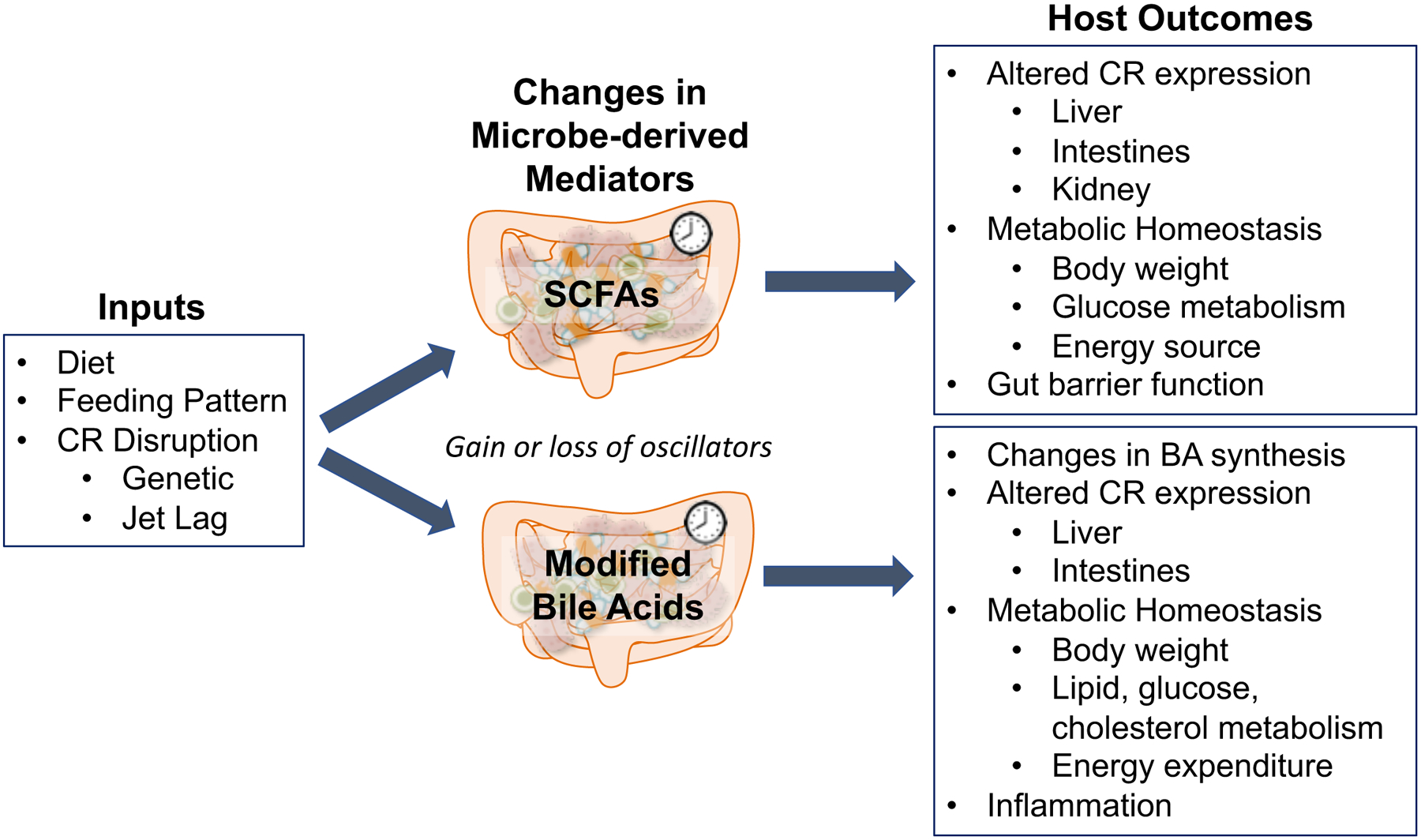
Inputs to the host that can include altered diet, feeding schedule (such as time-restricted feeding), or circadian rhythm disruption (by genetic deletion of core clock genes or jet lag protocol in animal models), which can significantly affect the synthesis, metabolism, and dynamics of gut microbe-derived metabolites. SCFAs and modified secondary bile acids are two such groups of metabolites that also exhibit oscillations and influence host CRs, both of which are subject to changes of the preceding inputs. Gain or loss of gut mediators as oscillators can alter host metabolism, although specific dynamics concerning how this occurs has not been thoroughly described. As illustrated by the studies discussed in this review, both of these metabolite groups exhibit unique influence on host outcomes, particularly relating to metabolism and circadian rhythm expression.
Gut microbes and circadian rhythms are closely linked and drive metabolic regulation
Recent studies have revealed elegant associations between host CRs and gut microbiota in mice. Although gut microbes are not exposed to light, diurnal host signals induce oscillations in both abundance and function of gut bacteria[5–7]. Specific microbial taxa, as well as key microbially-derived products, have been demonstrated to exhibit diurnal oscillations in relative abundance over a 24-hour period[3,5,8]. Despite potential confounders in starting microbiomes across mouse cohorts, differences between animal facilities, and unique study designs to manipulate the circadian system, microbial oscillations persist across studies[9,10]. Total biomass of gut bacteria also fluctuates over 24-hours, as does ~20% of KEGG functional pathways[8,10].
A handful of studies have outlined molecular mechanisms of complex microbial-circadian dynamics (of note the squid-vibrio model, see Box 3). However, the basic mechanisms that underlie how microbial communities within a multicellular host maintain oscillations remains a mystery. It is clear that the host circadian clock status and integrity, particularly that of peripheral clocks (see Box 4), has great influence on the dynamics of intestinal microbes. Mice with genetic mutations in Bmal1 and Per1/2 exhibit significantly altered microbial community profiles and loss of rhythms in specific microbial taxa[6,8]. Gut microbes have also been linked as drivers of host molecular rhythms; without gut microbes, germ-free (GF) mice exhibit entirely different suprachiasmatic nuclei (SCN) and hepatic transcriptional patterns, particularly in core circadian and metabolic pathways[5]. A similar effect has also been observed in intestinal epithelial cells (IECs) of antibiotic-treated or GF mice, in which clock and metabolic gene expression is reduced, and diurnal histone signaling is significantly dampened, via lack of HDAC3[11,12]. This begs the question of whether the absence of gut microbes or core clock components, resulting in loss of homeostatic rhythmicity, allows for gain of secondary oscillations that can exert entrainment of the host, which could explain the emergence of unique rhythmic patterns. The reprogramming of these host rhythms may contribute to the development of certain disease states that are known to be associated with disturbed microbiome and circadian disorganization.
Box 3: The squid-vibrio model.
The squid-vibrio model of symbiosis involves a specific a microbial species that interacts with the host rhythmically. Consisting of a relationship between the bobtail squid Euprymna scolopes and the gram-negative bacterium Vibrio fischeri, the squid swim in shallow oceanic waters to feed at night, at which point V. fisceri enter the squid’s light organ and luminesce to camouflage the squid[84]. These two organisms also exhibit a circadian-relationship; V. fischeri colonize the light organ each evening and a significant portion of community is expelled in the morning, exhibiting a rhythm[85]. This pattern is based on the host provision of specific amino acids as metabolic substrates for the microbes, as well as a structural change within the light organ crypts where the microbes reside, that induces a metabolic shift within the microbes towards anaerobic glycerol respiration. In turn, the microbial luminescence induces diurnal host cryptochrome expression which may induce the critical changes in host transcriptional metabolic and signaling pathways for the night feeding period. The reduced complexity of this system with a single microbial species has enabled researchers to manipulate specific mechanisms, such as the luminescence activity of V. fischeri, and delineate the direct effects of that signal on specific host physiology[86].
Box 4: Peripheral circadian clocks uniquely influence metabolism.
Peripheral clocks can display local control and receive external signals independent of the SCN[87]. For example, intestinal epithelial cells (IECs) exhibit unique transcription oscillations of the core clock genes in comparison to the master clock, which allow for local control of gut functions such as nutrient absorption and intestinal motility[88–90]. Time-restricted feeding (TRF) experiments with mice show that liver transcription patterns can be entrained independently of the SCN[91,92] and can restore hepatic transcription rhythms in circadian-disrupted or high-fat fed[93] mice, even while SCN expression remains arrhythmic[94]. Peripheral clocks can also influence the brain, such as the peripheral adipocyte clock and modulation of feeding behavior[95].
Numerous mouse models using genetic tissue-specific deficiency of a functional clock display metabolic phenotypes that vary from both global knockout and wild-type counterparts[76,96]. This is especially evident with Bmal1; mice lacking Bmal1 only in specific tissues exhibit a variety of phenotypes completely distinct from the global knockout. For example, the liver-specific Bmal1 knockout mouse does not display the arrhythmia, accelerated aging, or increased adiposity relative the global KO counterpart, but instead has a unique glucose tolerance profile with more efficient glucose clearance, as well as reduced mitochondrial respiration[97–99]. Muscle-specific loss of Bmal1 in mice results in glucose intolerance and fasting hyperglycemia[100], beta-cell-specific loss results in a dampened insulin response to available nutrients[101], and adipocyte-specific loss results in larger adipocyte size, reduced energy expenditure, and altered feeding behavior during the rest phase[95]. Tissue-specific transgenics have shown that peripheral circadian clocks have more nuanced effects on host physiology than previously considered.
Perturbations to metabolic homeostasis also influence both host CRs and gut microbiome oscillations. When mice are fed a high-fat diet, not only is circadian clock gene expression significantly modulated in both the SCN and liver, but many of the microbial oscillations in relative abundance observed in lean mice are also disturbed or dampened[5,7,13]. Another study showed that exposure to high-fat liquid diet subjected mice to more significant changes in microbial community profile following weekly light-dark phase reversals, while mice given regular chow maintained a more stable community[14]. Functional output of microbes is also affected by high-fat feeding, observed by the overall reduction and absence of diurnal abundance of SCFAs, which are known to heavily influence host metabolism[5]. Additional dietary regimens, including ketogenic and high-protein low-carbohydrate, have begun to be investigated in this context and revealed changes in both circadian organization and microbial community dynamics that is worth further study[15–18]. These misalignments persist in consistent light entrainment, and appear to contribute to the negative metabolic consequences, suggested by time-restricted feeding (TRF) in comparison to ad libitum feeding. Not only does TRF of high-fat fed mice during the animal’s active phase prevent the negative metabolic effects of the diet, including abnormal fat deposition and glucose tolerance, but it also partially restores the microbial oscillations that are lost due to dietary intervention[7]. Thus, the environmental cue of consolidated feeding may prevent metabolic disturbance via the maintenance of microbial oscillations.
Several pieces of data support the notion that diurnal dynamics of gut microbes are also important in humans. Fecal microbes originating from jet-lagged humans induce obesity and glucose intolerance in GF mice receiving microbial transplantation, as compared to GF mice receiving non-jet-lagged stool[8]. More recently, a study revealed that time of meal intake in humans corresponded with diurnal dynamics of the oral microbiome[19]. It is clear that CRs, especially peripheral clocks, and gut microbes respond to similar signals, such as diet and behavioral feeding patterns. The entrainment of these systems, both to environmental cues and each other, has a great influence on metabolic health and status, which thus far has been difficult to target clinically. Investigating the specific mechanisms that these two “organs” utilize to regulate metabolism will allow researchers to adapt that information and eventually manipulate these and other systems for medical intervention of metabolic diseases.
Elucidating drivers of the gut microbiome-peripheral clock axis
Thus far, we have illustrated a clear association between circadian regulation and gut microbes that has functional metabolic significance, and some distinct mechanisms have been described to explain these observations. One example is a study by Wang et al., who showed that the expression of transcription factor NFIL3 within IECs in mice is regulated by gut microbial signals via the circadian clock factor REV-ERBα, and disruption to this signaling relay directly worsens metabolic outcomes[20]. By serial deletion of each signaling component and following the downstream result, this study outlined a precise signaling pathway for how gut microbes can specifically impact host CRs and metabolism. This reductive approach allowed for manipulation of each component and observation of the result, which would not have been possible if the method was to examine all the aspects and interactions simultaneously. Herein lies the difficulty in delineating the complex networks of gut microbial and circadian regulation on host metabolism. To move beyond association-based studies, research groups may require hypotheses and experimental designs focused on specific components, such as peripheral clocks and certain metabolites, as opposed to the whole system. Currently, a major gap in knowledge in this field is the identification and characterization of specific microbially-derived mediators that are directly responsible for inducing changes in host circadian dynamics and metabolism. Next, we will describe two potential mediators and the current evidence implicating their involvement in the host-microbe circadian-metabolic dynamic.
Short-Chain Fatty Acids
A major functional by-product of gut microbes is the class of metabolites called short-chain fatty acids (SCFAs). Fermentation of specific fibers by gut microbes results in the production of the metabolic products known as SCFAs, the majority of which are butyrate, propionate, and acetate[21]. SCFA production in the intestine is largely attributed to fermentation by microbes belonging to the phylum Firmicutes[22,23]. While these products can be used as an energy source for certain host cells (colonocytes, hepatocytes, adipocytes)[21], some signaling mechanisms of SCFA action have also been identified. For example, it has been shown that SCFAs are sufficient to induce specific chromatin states via modulation of histone acetylation and methylation in GF mice[24]. SCFAs also stimulate certain G-protein coupled receptors, such as FFA receptors, which influence gut hormone secretion[25,26]. Although they have been observed to influence many aspects of host metabolism, glucose metabolism is directly regulated by SCFAs via hormone signaling, modulation of gut barrier function, and more[27]. Additionally, propionate can be directly taken up by the liver and intestines to serve as a precursor for gluconeogenesis.
SCFAs are generally associated with improved metabolic outcomes. For example, colonic delivery of propionate to humans for 6 months drastically reduced body weight and adipose tissue in overweight humans[28]. Similar studies have also been performed with rodents in which administration of butyrate, propionate, acetate, or a mixture of all three significantly reduced body weight gain due to high-fat diet feeding[26,29]. However, conflicting evidence has also emerged suggesting that SCFAs may be associated with obesity and other negative metabolic outcomes. For example, obese human subjects have increased levels of SCFAs as compared to lean subjects[30], and an “obese” gut microbial community in leptin-deficient rodents has also been associated with increased SCFA levels[31]. Additionally, GF mice are resistant to diet-induced obesity and exhibit very low levels of SCFAs, apart from acetate which can also be produced by the host. Whether or not this obesity-resistance is directly caused by reduced SCFA levels remains to be determined. Taken together, SCFAs clearly play a role in and are associated with mammalian metabolic disorders, but the details have yet to be clearly described. It is possible that differential timing of SCFA production and delivery can have specific effects on the host; if the diurnal pattern of delivery is altered or disrupted, perhaps due to circadian disruption, it could lead to negative metabolic consequences. Further exploration is required to determine causality of this hypothesis.
As briefly mentioned above, SCFAs display an additional layer of regulation and interaction with the host circadian system. Leone et al. measured SCFA levels in cecal and fecal contents from C57Bl/6 WT mice and found that both butyrate and propionate exhibit diurnal oscillations in abundance, which are lost under high fat feeding[5]. This was validated by quantitative measurement of key genes involved in butyrate production, which displayed significant oscillation in regular chow but not high-fat dietary conditions. To explore potentially direct effects on hepatic regulation, Leone et al. also treated murine, hepatic-derived organoids with individual SCFAs. Quantitative PCR revealed that both butyrate and acetate significantly shifted the period of expression of major circadian genes. A similar experimental approach validated these findings in vivo; treatment of C57Bl/6 WT mice with SCFAs caused significant changes in hepatic Bmal1 and Per2 gene expression patterns. This study provided support that microbially-derived SCFAs can directly modulate in hepatic circadian gene expression, possibly serving as a mechanism by which gut microbes and peripheral circadian clocks regulate host metabolism.
Further work supports the notion that SCFAs and CRs are linked. Mice with global Bmal1 knockout lack any rhythmicity in fecal SCFA levels[32]. Treatment of murine intestinal organoids with individual SCFAs (acetate, butyrate, or propionate) directly caused the circadian clock gene expression to advance, indicated by PER2::LUC expression, while formate, another SCFA, did not induce these changes[33]. Tahara et al. showed that direct administration of SCFAs induced circadian entrainment of certain tissues in mice[34]. Using antibiotic-treated PER2::LUC mice, they found that oral administration of mixed SCFAs induced significant phase change in luciferase bioluminescence in peripheral tissues (kidney, liver, and submandibular gland), but only when administered at specific times of day. However, when treated with a single SCFA species, few significant phase shifts were observed, suggesting that a combination of mixed SCFAs is required for adequate circadian signaling. Additionally, jet-lagged mice given SCFA treatment entrained faster to a new light-dark cycle, implicating SCFAs in potentially resetting the central clock. They also showed that exposure of GF mice to specific SCFAs directly modulates hepatic circadian gene expression. However, application of SCFAs directly to hepatic tissue ex vivo did not induce any changes in PER2 expression, implying that the complexity of an intact in vivo system is required for the signal mediation involved in SCFA-peripheral circadian regulation.
Bile Acids
Another major group of metabolites involved in gut microbial and circadian regulation are bile acids (BAs). Derived from cholesterol, primary BAs are synthesized in the liver and circulated to and from the intestinal tract to facilitate nutrient digestion and absorption[35]. BAs are major regulators of metabolic pathways via activation of receptors, notably nuclear receptor farnesoid X receptor (FXR) and membrane receptor TGR5 [36]. Of particular interest is the role that gut microbes play in BA metabolism and regulation. First, they have been shown to regulate hepatic expression of key rate-limiting enzymes involved in BA synthesis, including CYP7a1, CYP7b1, and CYP27a1, by ileal signaling[37]. Second, they are responsible for the deconjugation and dehydroxylation of primary BAs that feed back onto the host via hepatic circulation, which occurs primarily by taxa in the phylum Firmicutes via bile salt hydrolase (BSH) activity[38]. Although most primary BAs, conjugated or otherwise, are resorbed back into circulation, some escape absorption and are metabolized by colonic gut bacteria to be transformed from primary into secondary BAs. When absorbed systemically, they can act as signaling molecules in the host, such as via FXR activation in response to deconjugated BAs, and via TGR5 which is activated by secondary BAs and influences host energy homeostasis[39]. The bi-directional relationship between gut microbes and BA populations is particularly complex because they respond to and influence each other, as well as respond to additional cues such as diet[40]. These interactions can activate signaling pathways via the mentioned receptors and impact a wide array of host metabolic pathways, including cholesterol, glucose, and lipid homeostasis[35]. Inflammation, energy expenditure, atherosclerosis, and thermogenesis are also known to be under similar BA signaling regulation[41,42].
Microbial modifications of host-derived BAs have also been shown to influence circadian gene expression[43], providing additional evidence of a direct physical link between gut microbiota and CRs on host metabolism. Several key enzymes in BA synthesis exhibit diurnal expression patterns following regulation by core circadian clock genes[44,45]. In mice, both primary and secondary BAs exhibit significant oscillations in serum abundance, with peak levels occurring near the end of the dark phase, whereas unconjugated BA levels peak during the light phase[46]. This indicates that altered microbial regulation of BA metabolism occurs at different times in the circadian cycle of mice. Similar patterns were also observed of liver BA levels, implying that this particular microbial influence reaches such a key metabolic organ and peripheral clock, and potential perturbations to either BA synthesis or gut microbes could significantly impact the host.
Apart from homeostatic BA regulation, circadian disruption in mice by sleep disruption, time-restricted feeding, or global knockout of Rev-erbα, Per1 and Per2, significantly alters expression of key genes involved in BA regulation[44,47,48]. Increased expression of BSH activity among gut microbiota induces significant changes in lipid metabolism and CR gene expression in both ileum and liver tissue [49]. Increased BSH activity also reduced body weight, serum cholesterol, and liver triglycerides, supporting the notion that deconjugated BAs derived from gut microbes directly influence host metabolic physiology. However, this study was conducted on conventionally-raised SPF mice that were treated with antibiotics, which could have contributed to some of the effects observed. It has also been shown that GF mice monocolonized with nonfunctional-BSH strains, as compared to BSH-functional strains, revealed different changes in metabolism and both hepatic and intestinal gene expression, notably CR genes[50]. Another study measured circadian gene expression in vitro (Caco-2 cells) and in vivo (C57Bl/6 mice) settings following treatment with conjugated versus deconjugated BAs[43]. Unconjugated BAs uniquely induced significant oscillation in circadian gene expression in vitro. Similar treatment in vivo also altered ileal, colonic, and hepatic gene expression, although the presence of significant oscillation patterns could not be determined because only one timepoint for data collection was performed. This work showed that direct exposure to microbially-altered BAs alters peripheral circadian clock gene expression, implying the potential circadian influence of certain BAs on a complex in vivo host, but requires further investigation to validate these findings.
Gaps in Knowledge: Challenges in the Field
Although significant progress has been made to elucidate the mechanistic interactions between CRs and the microbiome, there remain many gaps in knowledge. Not only are each incredibly complex in themselves, but their interactions are essentially a set of positive feedback loops; one system signals to the other, causing the other to respond and signal back to the first one. This is demonstrated by evidence that when either system is impaired, the other is subsequently affected. When key circadian genes are knocked out in mice, shifts in gut microbiota ensue[8]; similarly, GF mice exhibit impaired circadian gene expression patterns[5].
This brings about the question of how the hierarchy of these systems are built and interact with one another. If the ultimate goal is to manipulate one or both of these systems to improve or reset metabolic health, hierarchical drivers and downstream processes must first be thoroughly defined in the proper context and relative to time. Where and when do you intervene in such a dynamic and complex circuit? What are the central drivers that can be manipulated with accuracy and precision? In the absence of central drivers, how does the emergence of secondary drivers contribute to reprogramming of the host in the context of disease states associated with microbiome or circadian disorganization? Similar to obesity, diseases such as aging[51], type 2 diabetes[8], colorectal cancer[52], and inflammatory bowel disease[53] exhibit unique microbiome and circadian profiles, but therapies that target or attempt to reset these altered profiles will not be possible until the key drivers, either primary or secondary, are identified and characterized. In addition to gastrointestinal and metabolic diseases, neurological disorders are also of interest in this context by influence of the gut-brain axis, the microbiome serving as a major signaling component of the gut and the central circadian clock housed in the brain. For example, hepatic encephalopathy is associated with both severe changes in gut microbiome function[54] and central circadian disorganization[55], so it follows that functional alterations of either or both would influence each other and the host in turn. However, it remains unknown how the circadian-microbiome network influences manifestation of the disease, or how commonly prescribed antibiotic treatments affect these interactions. These questions remain elusive and further progress in the circadian-microbiome research field are hindered by challenges presented by time and space.
To understand temporal relationships between host-microbe events, frequent and accurate time sequence samples must be obtained from specific locations. Reliance on stool for human microbiome sampling is limiting in this regard because subjects are rarely able to provide multiple samples throughout the day and night. Transit time through the colon can vary considerably among individuals and is usually on the order of hours, confounding any attempt to correlate microbiome changes with blood and biological assessments made elsewhere in real time. It is also technically infeasible to sample microbiomes of other intestinal regions that exhibit vastly different community profiles and functions. Finally, the inter-individual variations of the gut microbiome are enormous, making any type of cross-sectional analysis of human populations difficult to power and perform. For host circadian-microbiome studies, strong consideration should be given to longitudinal study designs where time-sequence samples of individual subjects are obtained and where they serve as their own controls. As mentioned above, sequential stool samples from human study subjects are very difficult to obtain. An alternative is to sample from the other end of the GI tract – specifically the salivary microbiome where time sequence samples can be readily obtained. These results can also be correlated with the timing of food intake which likely affects host and microbiome rhythms[19].
Many of the challenges associated with human studies can be circumvented by using animal models where time-relevant events from host tissues and regional gut microbiomes are more readily assessed. However, these studies often require large numbers of animals to be sacrificed a specific time points within a 24–48 hour period under highly controlled environmental and feeding conditions. Inbred mice commonly used in most circadian studies are also nocturnal, awake and eating during the dark cycle and sleeping and inactive during the light cycle – species variations from humans that could result in significantly different temporal relationships between microbes and host. On the other hand, time-sequence stool collections can be performed on individually-housed animals, providing opportunity to assess changes in colonic microbiota over time.
Yet another complex layer of these interactions is the sexual dimorphism of both the microbiome and host circadian expression patterns. Microbiome profiles in mice differ significantly by sex, and gonadectomy of both males and females depend upon dietary content[56]. Sexual dimorphism is also observed in reference to CRs; gonadectomy of male mice drastically reduces locomotor activity in comparison to females, and testosterone treatment eliminates that difference[57]. Mice also exhibit sexual dimorphism in hepatic gene expression of key metabolic genes, while those differences are lost in Cry1/2 knockout mice[58]. The CR-microbiome dynamic also presents sexual dimorphism. Liang et al. showed that female mice exhibit more significant microbial oscillations in stool than male counterparts; this difference is lost in Bmal1 global knockout mice[6]. More recently, Weger et al. showed that many of the differences in hepatic diurnal gene expression patterns between the sexes in mice are lost in GF conditions[59]. Understanding how sex and sex hormones affect diurnal microbiome patterns may be crucial to elucidating how these systems communicate and develop relationships. Integration of large-scale sequencing and metabolomics studies may inform on how sex hormones and sexual development play such a major role in circadian-microbiome interactions and influence on host metabolism.
Concluding Remarks and Future Perspectives
Taken together, there is clear evidence that gut microbes and CRs interact to influence host metabolism and energy balance. SCFAs and microbially-modified BAs are potential mediators of this network, and many others likely exist that have yet to be identified in this context. Further validation and mechanistic description of how these mediators accomplish signaling is required. With this knowledge, we may be able to leverage their connection to the microbiome-circadian network to develop treatment for metabolic disorders. This could include direct manipulation of peripheral clocks or specific functions of gut microbes, or these systems could be bypassed with timed delivery of mediators or compounds known to induce relevant signaling cascades. To implement timed drug delivery for optimal efficacy, particularly for potential gut microbiome modulation, we must first understand how essential mediators interact within the circadian-microbiome network and the downstream pathways that will be impacted.
However, many experimental hurdles in the field remain that must be overcome to achieve these goals and develop potential chronotherapies. Reliable methods of human microbiome sampling over a precise timescale are necessary to push the field forward and away from animal models that exhibit vastly different microbiome and circadian patterns. Differences in both environment and dietary composition with human studies remain difficult and expensive to control. The importance of observed sexual dimorphisms must also be addressed in future studies to fully grasp how these systems communicate and how sex development and hormones influence the effects of the microbiome-circadian network. In addition, we are just at the beginning at identifying the myriad of bioactive molecules made by the gut microbiome that are continuously taken up by the host. In turn, we recognize the essential role of host circadian networks on diurnal patterns of the gut microbiome. The integration of largescale sequencing and metabolomics studies may inform on exactly how these signals interact with host circadian networks to have such a major impact on energy balance and metabolic diseases. While this area of research is becoming recognized as a key feature of host physiology, there remains much to be discovered before the potential of clinical application becomes a reality (see Outstanding Questions).
Outstanding Questions:
How do gut microbial signals influence the complex hierarchy of peripheral and central circadian clocks?
How do sex hormones and the gut microbial programming of sex development interact with the microbiome-circadian network?
How can we overcome the challenges of human subject research to better understand the relationships between gut microbiomes and host circadian networks?
Can clinical interventions targeting host circadian networks and the gut microbiome be developed to effectively reset metabolic health or status long-term? How can these findings be applied to the future development of chronotherapy, i.e. timed drug delivery?
Trends:
Gut microbiota contribute significantly to host metabolism, including nutrient digestion, vitamin synthesis, and immune programming. Disrupted balance of gut microbiota can lead to the development of several metabolic disorders.
Gut microbes and circadian rhythms are intertwined via metabolic regulation, but the mechanisms that underlie their interactions are still not understood.
Diurnal variations in regional gut microbiota affect host circadian rhythms to regulate many core processes, particularly metabolism.
Microbe-derived metabolites such as short-chain fatty acids and bile acids are likely mediators for gut microbiota- host circadian communication.
Further progress in the field requires more precise timed sampling, less reliance on stool, and consideration of inter-individual human microbiome differences and sex hormones.
References
- 1.Bellet MM et al. (2013) Circadian clock regulates the host response to Salmonella. Proc. Natl. Acad. Sci 110, 9897–9902 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wu T et al. (2018) Chronic glucocorticoid treatment induced circadian clock disorder leads to lipid metabolism and gut microbiota alterations in rats. Life Sci. 192, 173–182 [DOI] [PubMed] [Google Scholar]
- 3.Thaiss CA et al. (2016) Microbiota Diurnal Rhythmicity Programs Host Transcriptome Oscillations. Cell 167, 1495–1510.e12 [DOI] [PubMed] [Google Scholar]
- 4.Parkar SG et al. (2019) Potential Role for the Gut Microbiota in Modulating Host Circadian Rhythms and Metabolic Health. Microorganisms 7, [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Leone V et al. (2015) Effects of diurnal variation of gut microbes and high fat feeding on host circadian clock function and metabolism. Cell Host Microbe 17, 681–689 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Liang X et al. (2015) Rhythmicity of the intestinal microbiota is regulated by gender and the host circadian clock. Proc. Natl. Acad. Sci 112, 10479–10484 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zarrinpar A et al. (2014) Diet and Feeding Pattern Affect the Diurnal Dynamics of the Gut Microbiome. Cell Metab. 20, 1006–1017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Thaiss CA et al. (2014) Transkingdom Control of Microbiota Diurnal Oscillations Promotes Metabolic Homeostasis. Cell 159, 514–529 [DOI] [PubMed] [Google Scholar]
- 9.Deloris Alexander A et al. (2006) Quantitative PCR assays for mouse enteric flora reveal strain-dependent differences in composition that are influenced by the microenvironment. Mamm. Genome 17, 1093–1104 [DOI] [PubMed] [Google Scholar]
- 10.Rausch P et al. (2016) Analysis of factors contributing to variation in the C57BL/6J fecal microbiota across German animal facilities. Int. J. Med. Microbiol 306, 343–355 [DOI] [PubMed] [Google Scholar]
- 11.Mukherji A et al. (2013) Homeostasis in intestinal epithelium is orchestrated by the circadian clock and microbiota cues transduced by TLRs. Cell 153, 812–827 [DOI] [PubMed] [Google Scholar]
- 12.Kuang Z et al. (2019) The intestinal microbiota programs diurnal rhythms in host metabolism through histone deacetylase 3. bioRxiv DOI: 10.1101/580613 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Eckel-Mahan KL et al. (2013) Reprogramming of the Circadian Clock by Nutritional Challenge. Cell 155, 1464–1478 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Voigt RM et al. (2014) Circadian disorganization alters intestinal microbiota. PloS One 9, e97500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tognini P et al. (2017) Distinct Circadian Signatures in Liver and Gut Clocks Revealed by Ketogenic Diet. Cell Metab. 26, 523–538.e5 [DOI] [PubMed] [Google Scholar]
- 16.Lindefeldt M et al. (2019) The ketogenic diet influences taxonomic and functional composition of the gut microbiota in children with severe epilepsy. Npj Biofilms Microbiomes 5, 1–13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Oishi K et al. (2012) Low-carbohydrate, high-protein diet affects rhythmic expression of gluconeogenic regulatory and circadian clock genes in mouse peripheral tissues. Chronobiol. Int 29, 799–809 [DOI] [PubMed] [Google Scholar]
- 18.Mu C et al. (2016) The Colonic Microbiome and Epithelial Transcriptome Are Altered in Rats Fed a High-Protein Diet Compared with a Normal-Protein Diet. J. Nutr 146, 474–483 [DOI] [PubMed] [Google Scholar]
- 19.Collado MC et al. (2018) Timing of food intake impacts daily rhythms of human salivary microbiota: a randomized, crossover study. FASEB J. Off. Publ. Fed. Am. Soc. Exp. Biol 32, 2060–2072 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wang Y et al. (2017) The intestinal microbiota regulates body composition through NFIL3 and the circadian clock. Science 357, 912–916 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.den Besten G et al. (2013) The role of short-chain fatty acids in the interplay between diet, gut microbiota, and host energy metabolism. J. Lipid Res 54, 2325–2340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Louis P and Flint HJ (2009) Diversity, metabolism and microbial ecology of butyrate-producing bacteria from the human large intestine. FEMS Microbiol. Lett 294, 1–8 [DOI] [PubMed] [Google Scholar]
- 23.Vital M et al. (2014) Revealing the Bacterial Butyrate Synthesis Pathways by Analyzing (Meta)genomic Data. mBio 5, e00889–14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Krautkramer KA et al. (2016) Diet-Microbiota Interactions Mediate Global Epigenetic Programming in Multiple Host Tissues. Mol. Cell 64, 982–992 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tolhurst G et al. (2012) Short-chain fatty acids stimulate glucagon-like peptide-1 secretion via the G-protein-coupled receptor FFAR2. Diabetes 61, 364–371 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lu Y et al. (2016) Short Chain Fatty Acids Prevent High-fat-diet-induced Obesity in Mice by Regulating G Protein-coupled Receptors and Gut Microbiota. Sci. Rep 6, 37589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chambers ES et al. (2018) Role of Gut Microbiota-Generated Short-Chain Fatty Acids in Metabolic and Cardiovascular Health. Curr. Nutr. Rep 7, 198–206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chambers ES et al. (2015) Effects of targeted delivery of propionate to the human colon on appetite regulation, body weight maintenance and adiposity in overweight adults. Gut 64, 1744–1754 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Vinolo MAR et al. (2012) Tributyrin attenuates obesity-associated inflammation and insulin resistance in high-fat-fed mice. Am. J. Physiol. Endocrinol. Metab 303, E272–282 [DOI] [PubMed] [Google Scholar]
- 30.Schwiertz A et al. (2010) Microbiota and SCFA in lean and overweight healthy subjects. Obes. Silver Spring Md 18, 190–195 [DOI] [PubMed] [Google Scholar]
- 31.Turnbaugh PJ et al. (2006) An obesity-associated gut microbiome with increased capacity for energy harvest. Nature 444, 1027–1031 [DOI] [PubMed] [Google Scholar]
- 32.Segers A et al. (2018) The circadian clock regulates the diurnal levels of microbial short-chain fatty acids and their rhythmic effects on colon contractility in mice. Acta Physiol. Oxf. Engl DOI: 10.1111/apha.13193 [DOI] [PubMed] [Google Scholar]
- 33.Luzader D et al. (2018) Gut Microbial Metabolites Modulate the Amplitude and Phase of PER2 and BMAL1 Circadian Rhythms in Intestinal Epithelial Cells and Organoids. Gastroenterology 154, S–67 [Google Scholar]
- 34.Tahara Y et al. (2018) Gut Microbiota-Derived Short Chain Fatty Acids Induce Circadian Clock Entrainment in Mouse Peripheral Tissue. Sci. Rep 8, [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.de Aguiar Vallim TQ et al. (2013) Pleiotropic Roles of Bile Acids in Metabolism. Cell Metab. 17, 657–669 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hylemon PB et al. (2009) Bile acids as regulatory molecules. J. Lipid Res 50, 1509–1520 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sayin SI et al. (2013) Gut Microbiota Regulates Bile Acid Metabolism by Reducing the Levels of Tauro-beta-muricholic Acid, a Naturally Occurring FXR Antagonist. Cell Metab. 17, 225–235 [DOI] [PubMed] [Google Scholar]
- 38.Devlin AS and Fischbach MA (2015) A biosynthetic pathway for a prominent class of microbiota-derived bile acids. Nat. Chem. Biol 11, 685–690 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wahlström A et al. (2016) Intestinal Crosstalk between Bile Acids and Microbiota and Its Impact on Host Metabolism. Cell Metab. 24, 41–50 [DOI] [PubMed] [Google Scholar]
- 40.Devkota S et al. (2012) Dietary-fat-induced taurocholic acid promotes pathobiont expansion and colitis in Il10−/− mice. Nature 487, 104–108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Pols TWH (2014) TGR5 in inflammation and cardiovascular disease. Biochem. Soc. Trans 42, 244–249 [DOI] [PubMed] [Google Scholar]
- 42.Ding L et al. (2018) Trimethylamine-N-oxide (TMAO)-induced atherosclerosis is associated with bile acid metabolism. Lipids Health Dis. 17, 286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Govindarajan K et al. (2016) Unconjugated Bile Acids Influence Expression of Circadian Genes: A Potential Mechanism for Microbe-Host Crosstalk. PloS One 11, e0167319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Duez H et al. (2008) Regulation of Bile Acid Synthesis by the Nuclear Receptor Rev-erbα. Gastroenterology 135, 689–698.e5 [DOI] [PubMed] [Google Scholar]
- 45.Lavery DJ and Schibler U (1993) Circadian transcription of the cholesterol 7 alpha hydroxylase gene may involve the liver-enriched bZIP protein DBP. Genes Dev. 7, 1871–1884 [DOI] [PubMed] [Google Scholar]
- 46.Zhang Y-KJ et al. (2011) Diurnal Variations of Mouse Plasma and Hepatic Bile Acid Concentrations as well as Expression of Biosynthetic Enzymes and Transporters. PLOS ONE 6, e16683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ferrell JM and Chiang JYL (2015) Short-Term Circadian Disruption Impairs Bile Acid and Lipid Homeostasis in Mice. Cell. Mol. Gastroenterol. Hepatol 1, 664–677 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ma K et al. (2009) Circadian Dysregulation Disrupts Bile Acid Homeostasis. PLOS ONE 4, e6843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Joyce SA et al. (2014) Regulation of host weight gain and lipid metabolism by bacterial bile acid modification in the gut. Proc. Natl. Acad. Sci 111, 7421–7426 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Yao L et al. (2018) A selective gut bacterial bile salt hydrolase alters host metabolism. eLife 7, [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Paschos GK and FitzGerald GA (2017) Circadian Clocks and Metabolism: Implications for Microbiome and Aging. Trends Genet. 33, 760–769 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Bishehsari F et al. (2016) Light/Dark Shifting Promotes Alcohol-Induced Colon Carcinogenesis: Possible Role of Intestinal Inflammatory Milieu and Microbiota. Int. J. Mol. Sci 17, [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Gombert M et al. (2019) The connection of circadian rhythm to inflammatory bowel disease. Transl. Res 206, 107–118 [DOI] [PubMed] [Google Scholar]
- 54.Rai R et al. (2015) Gut Microbiota: Its Role in Hepatic Encephalopathy. J. Clin. Exp. Hepatol 5, S29–S36 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Bruyneel M and Sersté T (2018) Sleep disturbances in patients with liver cirrhosis: prevalence, impact, and management challenges. Nat. Sci. Sleep 10, 369–375 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Org E et al. (2016) Sex differences and hormonal effects on gut microbiota composition in mice. Gut Microbes 7, 313–322 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Iwahana E et al. (2008) Gonadectomy reveals sex differences in circadian rhythms and suprachiasmatic nucleus androgen receptors in mice. Horm. Behav 53, 422–430 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Bur IM et al. (2009) The Circadian Clock Components CRY1 and CRY2 Are Necessary to Sustain Sex Dimorphism in Mouse Liver Metabolism. J. Biol. Chem 284, 9066–9073 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Weger BD et al. (2019) The Mouse Microbiome Is Required for Sex-Specific Diurnal Rhythms of Gene Expression and Metabolism. Cell Metab. 29, 362–382.e8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Brestoff JR and Artis D (2013) Commensal bacteria at the interface of host metabolism and the immune system. Nat. Immunol 14, 676–684 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Holmes E et al. (2011) Understanding the role of gut microbiome–host metabolic signal disruption in health and disease. Trends Microbiol. 19, 349–359 [DOI] [PubMed] [Google Scholar]
- 62.Martinez-Guryn K et al. (2018) Small Intestine Microbiota Regulate Host Digestive and Absorptive Adaptive Responses to Dietary Lipids. Cell Host Microbe 23, 458–469.e5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Bäckhed F et al. (2004) The gut microbiota as an environmental factor that regulates fat storage. Proc. Natl. Acad. Sci 101, 15718–15723 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Bäckhed F et al. (2007) Mechanisms underlying the resistance to diet-induced obesity in germ-free mice. Proc. Natl. Acad. Sci 104, 979–984 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.El Aidy S et al. (2013) The gut microbiota elicits a profound metabolic reorientation in the mouse jejunal mucosa during conventionalisation. Gut 62, 1306–1314 [DOI] [PubMed] [Google Scholar]
- 66.Larsson E et al. (2012) Analysis of gut microbial regulation of host gene expression along the length of the gut and regulation of gut microbial ecology through MyD88. Gut 61, 1124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.El Aidy S et al. (2013) The gut microbiota elicits a profound metabolic reorientation in the mouse jejunal mucosa during conventionalisation. Gut 62, 1306–1314 [DOI] [PubMed] [Google Scholar]
- 68.Huang E et al. (2013) Composition of Dietary Fat Source Shapes Gut Microbiota Architecture and Alters Host Inflammatory Mediators in Mouse Adipose Tissue. JPEN J. Parenter. Enteral Nutr 37, [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.David LA et al. (2014) Diet rapidly and reproducibly alters the human gut microbiome. Nature 505, 559–563 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Turnbaugh PJ et al. (2009) A core gut microbiome in obese and lean twins. Nature 457, 480–484 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Ley RE et al. (2005) Obesity alters gut microbial ecology. Proc. Natl. Acad. Sci. U. S. A 102, 11070–11075 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Sonnenburg JL and Bäckhed F (2016) Diet–microbiota interactions as moderators of human metabolism. Nature 535, 56–64 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Hut RA and Beersma DGM (2011) Evolution of time-keeping mechanisms: early emergence and adaptation to photoperiod. Philos. Trans. R. Soc. Lond. B. Biol. Sci 366, 2141–2154 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Sancar G and Brunner M (2014) Circadian clocks and energy metabolism. Cell. Mol. Life Sci. CMLS 71, 2667–2680 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Buhr ED and Takahashi JS (2013) Molecular components of the Mammalian circadian clock. Handb. Exp. Pharmacol DOI: 10.1007/978-3-642-25950-0_1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Mayeuf-Louchart A et al. (2017) Circadian control of metabolism and pathological consequences of clock perturbations. Biochimie 143, 42–50 [DOI] [PubMed] [Google Scholar]
- 77.Koike N et al. (2012) Transcriptional Architecture and Chromatin Landscape of the Core Circadian Clock in Mammals. Science 338, 349–354 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Zhang Y et al. (2017) The hepatic circadian clock fine-tunes the lipogenic response to feeding through RORα/γ. Genes Dev. 31, 1202–1211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Rey G et al. (2011) Genome-Wide and Phase-Specific DNA-Binding Rhythms of BMAL1 Control Circadian Output Functions in Mouse Liver. PLOS Biol. 9, e1000595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Zhang R et al. (2014) A circadian gene expression atlas in mammals: Implications for biology and medicine. Proc. Natl. Acad. Sci. U. S. A 111, 16219–16224 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Evans JA and Davidson AJ (2013) Health Consequences of Circadian Disruption in Humans and Animal Models In Progress in Molecular Biology and Translational Science 119 (Gillette MU, ed), pp. 283–323, Academic Press; [DOI] [PubMed] [Google Scholar]
- 82.Rudic RD et al. (2004) BMAL1 and CLOCK, Two Essential Components of the Circadian Clock, Are Involved in Glucose Homeostasis. PLoS Biol. 2, [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Reinke H and Asher G (2019) Crosstalk between metabolism and circadian clocks. Nat. Rev. Mol. Cell Biol 20, 227–241 [DOI] [PubMed] [Google Scholar]
- 84.McFall-Ngai M (2014) Divining the Essence of Symbiosis: Insights from the Squid-Vibrio Model. PLOS Biol. 12, e1001783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Boettcher KJ et al. (1996) Bioluminescence in the symbiotic squid Euprymna scolopes is controlled by a daily biological rhythm. J. Comp. Physiol. A 179, 65–73 [Google Scholar]
- 86.Heath-Heckman EAC et al. (2013) Bacterial Bioluminescence Regulates Expression of a Host Cryptochrome Gene in the Squid-Vibrio Symbiosis. mBio 4, e00167–13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Dibner C et al. (2010) The Mammalian Circadian Timing System: Organization and Coordination of Central and Peripheral Clocks. Annu. Rev. Physiol 72, 517–549 [DOI] [PubMed] [Google Scholar]
- 88.Pardini L et al. (2005) Human intestinal circadian clock: expression of clock genes in colonocytes lining the crypt. Chronobiol. Int 22, 951–961 [DOI] [PubMed] [Google Scholar]
- 89.Hoogerwerf WA et al. (2007) Clock gene expression in the murine gastrointestinal tract: endogenous rhythmicity and effects of a feeding regimen. Gastroenterology 133, 1250–1260 [DOI] [PubMed] [Google Scholar]
- 90.Gachon F et al. (2006) The circadian PAR-domain basic leucine zipper transcription factors DBP, TEF, and HLF modulate basal and inducible xenobiotic detoxification. Cell Metab. 4, 25–36 [DOI] [PubMed] [Google Scholar]
- 91.Stokkan KA et al. (2001) Entrainment of the circadian clock in the liver by feeding. Science 291, 490–493 [DOI] [PubMed] [Google Scholar]
- 92.Damiola F et al. (2000) Restricted feeding uncouples circadian oscillators in peripheral tissues from the central pacemaker in the suprachiasmatic nucleus. Genes Dev. 14, 2950–2961 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Chaix A et al. (2014) Time-restricted feeding is a preventative and therapeutic intervention against diverse nutritional challenges. Cell Metab. 20, 991–1005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Vollmers C et al. (2009) Time of feeding and the intrinsic circadian clock drive rhythms in hepatic gene expression. Proc. Natl. Acad. Sci. U. S. A 106, 21453–21458 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Paschos GK et al. (2012) Obesity in mice with adipocyte-specific deletion of clock component Arntl. Nat. Med 18, 1768–1777 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Meyer-Kovac J et al. (2017) Hepatic gene therapy rescues high-fat diet responses in circadian Clock mutant mice. Mol. Metab 6, 512–523 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.McDearmon EL et al. (2006) Dissecting the Functions of the Mammalian Clock Protein BMAL1 by Tissue-Specific Rescue in Mice. Science 314, 1304–1308 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Lamia KA et al. (2008) Physiological significance of a peripheral tissue circadian clock. Proc. Natl. Acad. Sci. U. S. A 105, 15172–15177 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Jacobi D et al. (2015) Hepatic Bmal1 Regulates Rhythmic Mitochondrial Dynamics and Promotes Metabolic Fitness. Cell Metab. 22, 709–720 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Harfmann BD et al. (2016) Muscle-specific loss of Bmal1 leads to disrupted tissue glucose metabolism and systemic glucose homeostasis. Skelet. Muscle 6, 12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Perelis M et al. (2015) Pancreatic β cell enhancers regulate rhythmic transcription of genes controlling insulin secretion. Science 350, aac4250. [DOI] [PMC free article] [PubMed] [Google Scholar]


