Abstract
New chemical probes have been designed to facilitate the identification of adenosine-to-inosine (A-to-I) edited RNAs. These reagents combine a conjugate acceptor for selective inosine covalent modification with functional groups for bioorthogonal biotinylation. The resulting biotinylated RNA was enriched and verified with RT-qPCR. This powerful chemical approach provides new opportunities to identify and quantify A-to-I editing sites.
Graphical Abstract

RNA molecules perform a wide variety of functions, from regulating gene expression to scaffolding protein complexes to controlling enzymatic reactions. Beyond the canonical chemical structure of RNA nucleosides, it is now appreciated that RNA can be modified and that such modifications can alter RNA localization, decay, and translatability.
One such RNA modification that can significantly alter RNA structure and function is adenosine-to-inosine (A-to-I) RNA editing (Scheme 1a). A-to-I editing has been widely identified in a range of species, including mammals.1 This editing process is primarily catalyzed by three different adenosine deaminases acting on RNA (ADARs), including two isoforms of ADAR1 and one of ADAR2; no deaminase activity has been detected for ADAR3.2, 3 These enzymes are conserved for A-to-I RNA editing throughout the animal kingdom and have been suggested to have different subcellular localization in a cell-type/tissue-type dependent manner.4–6
Scheme 1.

Comparison of the previous and current work on adenosine-to-inosine identification. a. Conversion of adenosine to inosine. b. Known selective alkylation of inosine by conjugate addition to acrylonitrile. c. Proposed reagent for selective inosine modification and identification.
A-to-I RNA editing plays essential roles in the control of cellular functions. The editing event changes the hydrogen bonding patterns of involved nucleobase and the resulting inosines readily base pair with cytidine. As a result, the editing site will be recorded as guanosine instead of originally adenosine.7 If the editing site happens in the protein-coding region, a different amino acid sequence will be generated, which results in protein diversity. For example, the protein recoding capacity has been demonstrated to be crucial for cell survival when it comes to the editing of Gria2 at the Q/R site to achieve functional GluA2 protein expression.8
If the editing site occurs in the non-coding region, the biogenesis and target specificity of the corresponding small-interfering RNAs (siRNAs) and microRNAs (miRNAs) will be significantly affected, thus impacting gene transcription and protein translation.9 Dysregulation of A-to-I RNA editing could potentially lead to neurodegenerative diseases, autoimmune disorders and even cancers.10, 11
Despite the high importance of A-to-I RNA editing it remains challenging to identify RNA editing sites with single nucleotide resolution. Chemical approaches have been developed to understand the chemical raeactivty of ADAR enzymes and substrate specificity.12–14 Recent methods to screen the editing sites have mainly relied on the development of deep sequencing and the advancements in bioinformatics.15, 16 Although the approach did allow transcriptome-wide analysis of editing sites, the process is intrinsically prone to false positive results owing to the abundant and unexhausted lists of single nucleotide polymorphisms (SNPs) as well as sequencing errors. Suzuki and coworkers reported a chemical approach to facilitate the identification of editing sites with confidence.17, 18 This new method coupled the use of a traditional inosine alkylator, acrylonitrile, with reverse transcription that stops at the modification site (Scheme 1b). Despite the initial success in global and unbiased screening of new A-to-I editing sites, this biochemical identification method could be considerably enhanced with the integration of a protocol for enrichment of edited sites for downstream sequencing analysis (Scheme 1c).
A-to-I editing sites are sparse in cellular RNA when compared with the whole RNA populations, although it is one of the most abundant RNA modifications. No A-to-I modifications have been found in rRNA, which accounts for 90% of cellular RNAs. Many key edited sites are present only on rare transcripts.16 To reveal A-to-I editing sites with confidence, large quantities of material and cost-prohibitive RNA sequencing depth are required. In addition to the relatively sparse signals, A-to-I editing sites are dynamic, which not only depends heavily on cell and tissue types but also varies at different cell states throughout developmental processes or disease progression.6, 19, 20 Enrichment of edited RNA would allow targeted RNA sequencing, thus eliminating unnecessary RNA sequencing reads; while some progress has been made for designing alkylating reagents that could enable enrichment,21 there still is a critical need to develop bi-functional agents that could be used for analyzing inosine in RNA.
We designed the new A-to-I RNA editing probes based on the structure of acrylonitrile. (Scheme 2). We envisioned the 2-(N-alkyl-N-methylamino)acrylonitrile scaffold as an attractive target for probe development, because the tertiary amine linker should ensure solubility in biological media and the alkyl residue on the amine can be freely chosen. We elected to incorporate an azidoethyl group (1; AtoI_N3) and a propargyl group (2; AtoI_yne), each of which offers the capability of attaching a biotin tag via different “click” reaction modalities. The 2-(N,N-dialkylamino)acrylonitrile scaffold could be easily accessed via a Mannich condensation, decarboxylation, aldol condensation cascade of cyanoacetic acid, formalin, and the corresponding amine22 or ammonium salt in a one-pot reaction. The triethylammonium acetate buffer (pH = 4.5) ensures fast carbonyl condensation and decarboxylation at ambient temperatures in aqueous media and makes it especially useful for thermally labile substrates. Applying this strategy, the AtoI_N3 probe could be prepared in 71% yield and the more volatile AtoI_yne was accessed in 54% yield. Furthermore, the more complex, biotinylated probe 3 was prepared by a CuAAC “click” reaction of 2 with commercially available N-(prop-2-ynyl)biotinamide (31% yield).
Scheme 2.

Synthesis of acrylonitrile probes for inosine alkylation. a. A three-component coupling from cyanoacetic acid, formaldehyde, and a secondary amine directly provides alkyne- and azide-bearing probe molecules. b. A Cu-catalyzed azide/alkyne dipolar cycloaddition generates the biotinylated reagent.
Probes 1 and 2 each demonstrated reasonable aqueous solubility, whereas solutions of 3 were observed to be quite turbid, suggesting limited solubility. This observation, coupled with the fact that RNA degradation can result from CuAAC “click” reactions23, prompted us to focus on utilizing 1 for all subsequent characterizations. We therefore could rely on strain-promoted azide-alkyne cycloadditions (SPAAC), which do not require catalysis by copper complexes.
We next tested alkylation selectivity by incubating 1 with inosine and the canonical nucleosides (adenosine, cytidine, uridine, guanosine) and by analyzing relative reactivities. Mixtures of the probe and the corresponding nucleoside were heated at 70 °C for either 15 or 30 minutes, followed by HPLC analysis. This systematic study revealed that AtoI_N3 is significantly more selective for inosine over the other nucleosides (Figure S1).
To further test the selectivity, we utilized synthetic oligos, one with an inosine and one without. (Figure 1, a). First, we demonstrated selective inosine alkylation by reverse transcription (RT; Figure 1, b, c). As shown, we observed a very strong RT stop at the inosine site that was absent in control oligos. We also demonstrated the selectivity of modification through dot blot analysis, whereby each of the oligos was alkylated, subjected to SPAAC with dibenzocyclooctyne-biotin (DIBO-biotin). Biotinylation was evaluated using dot blot (Figure 1, d–f). More detailed labeling condition optimizations could be found in Figure S2.
Figure 1.

Characterizing acrylonitrile-based probe alkylation of RNA. a. Oligonucleotide sequences used in this study. b. Schematic of experiment to characterize alkylation by reverse transcription. c. Denaturing gel electrophoresis demonstrates selective RT-stop at inosine site. d. Schematic of experiment to characterize alkylation by dot-blot. e. Dot blot demonstrating selective alkylation of inosine-containing oligo. f. Integration of signal in panel e.
Having demonstrated that our novel reagents can selectively alkylate inosine in synthetic oligos, we aimed to test their utility in a complex pool of RNAs. To test the specificity of enrichment by 1, a known A-to-I transcript was evaluated using RT-qPCR. Human brain total RNA was modified with 1 and later biotinylated by SPAAC, followed by enrichment with streptavidin (Figure 2a). We then compared the RT-qPCR profiles for a known A-to-I transcript, dihydrofolate reductase (DHFR)24, and a negative control, branched chain ketoacid dehydrogenase kinase (BCKDK)16. As predicted, DHFR, which was highly edited in the 3’ UTR, was significantly enriched (Figure 2b). These results clearly demonstrate that RNA incubation with AtoI_N3 results in transcript-selective alkylation and subsequent enrichment.
Figure 2.
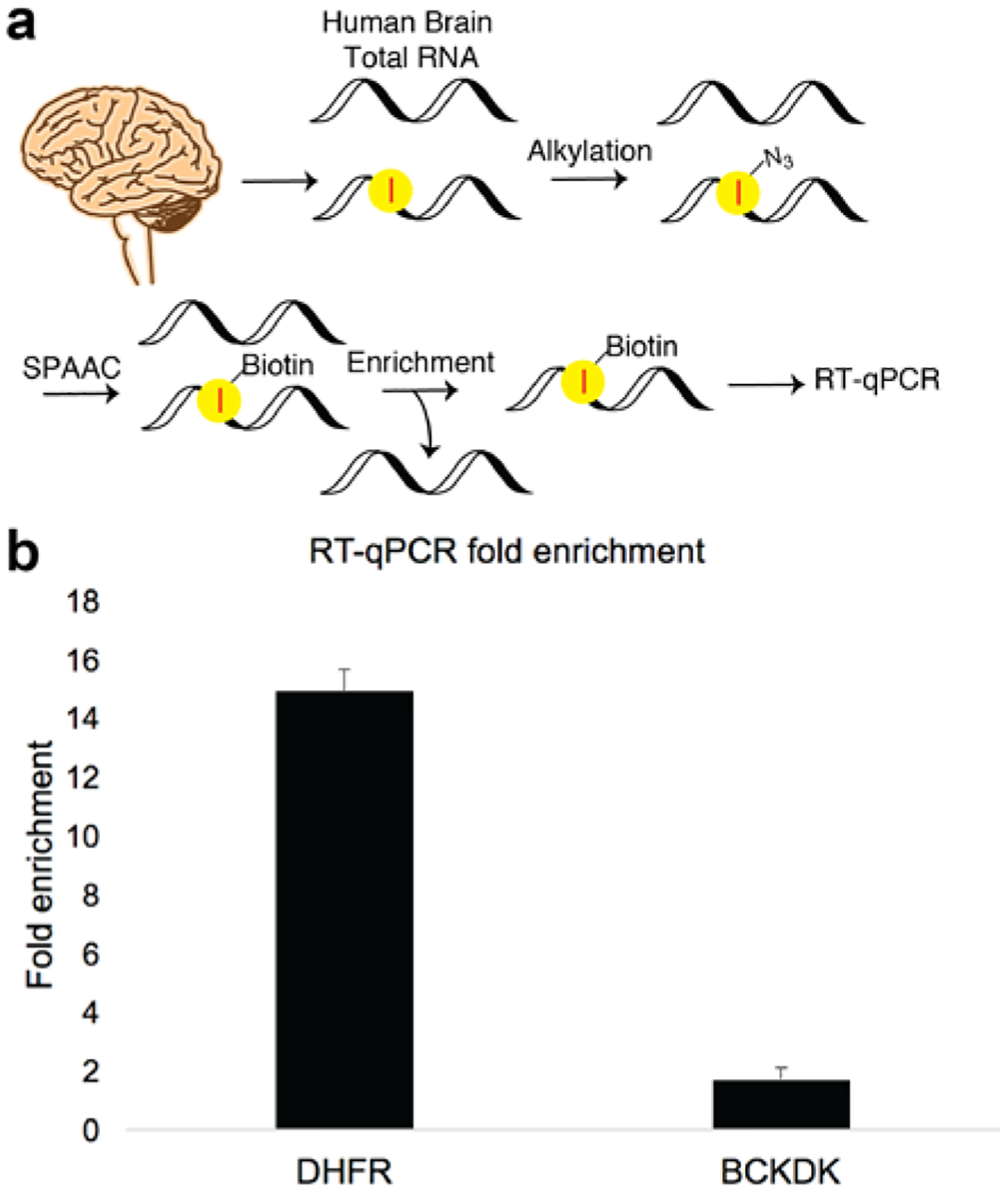
a. RT-qPCR of AtoI edited transcript compared with transcript without modification. b. Enrichment was calculated against a negative control with the ΔΔCt method (SI), in biological duplicates.
In conclusion, we have successfully designed a new acrylonitrile-based chemical probe for inosine-selective alkylation. In addition, we have demonstrated through a battery of tests that AtoI_N3 can modify inosine in the context of intact RNA species to enable inosine site identification by reverse transcription and dot-blot analyses. Lastly, we have utilized AtoI_N3 to demonstrate the stringency of inosine modification through transcript-specific enrichment to enable profiling by RT-qPCR. We envision that AtoI_N3 will be invaluable to the discovery of novel RNA editing sites, in particular on lowly expressed RNAs or highly complex mixtures of RNAs such as those derived from patient samples or complex tissues such as the brain. Such analyses are currently underway in our lab and will be reported in due course.
Supplementary Material
ACKNOWLEDGMENT
The research was funded by NIH (1DP2GM119164-01 to RCS). RCS is a Pew Biomedical Scholar. MG was supported by a postdoctoral fellowship from the Deutsche Forschungsgemeinschaft.
Footnotes
Supporting Information
The Supporting Information is available free of charge on the ACS Publications website at DOI: Experimental procedures, characterization data of synthetic compounds, NMR spectra and biology experimental details (PDF)
The authors declare no conflict of interest.
REFERENCES
- 1.Wagner RW; Smith JE; Cooperman BS; Nishikura K, A double-stranded RNA unwinding activity introduces structural alterations by means of adenosine to inosine conversions in mammalian cells and Xenopus eggs. Proceedings of the National Academy of Sciences 1989, 86, 2647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chen CX; Cho DS; Wang Q; Lai F; Carter KC; Nishikura K, A third member of the RNA-specific adenosine deaminase gene family, ADAR3, contains both single- and double-stranded RNA binding domains. RNA (New York, N.Y.) 2000, 6, 755–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Oakes E; Anderson A; Cohen-Gadol A; Hundley HA, Adenosine Deaminase That Acts on RNA 3 (ADAR3) Binding to Glutamate Receptor Subunit B Pre-mRNA Inhibits RNA Editing in Glioblastoma. The Journal of biological chemistry 2017, 292, 4326–4335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Patterson JB; Samuel CE, Expression and regulation by interferon of a double-stranded-RNA-specific adenosine deaminase from human cells: evidence for two forms of the deaminase. Molecular and cellular biology 1995, 15, 5376–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Snyder EM; Licht K; Braun RE, Testicular adenosine to inosine RNA editing in the mouse is mediated by ADARB1. Biology of reproduction 2017, 96, 244–253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Tan MH; Li Q; Shanmugam R; Piskol R; Kohler J; Young AN; Liu KI; Zhang R; Ramaswami G; Ariyoshi K; Gupte A; Keegan LP; George CX; Ramu A; Huang N; Pollina EA; Leeman DS; Rustighi A; Goh YPS; Chawla A; Del Sal G; Peltz G; Brunet A; Conrad DF; Samuel CE; O’Connell MA; Walkley CR; Nishikura K; Li JB, Dynamic landscape and regulation of RNA editing in mammals. Nature 2017, 550, 249–254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bass BL, RNA editing by adenosine deaminases that act on RNA. Annual review of biochemistry 2002, 71, 817–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sommer B; Kohler M; Sprengel R; Seeburg PH, RNA editing in brain controls a determinant of ion flow in glutamate-gated channels. Cell 1991, 67, 11–9. [DOI] [PubMed] [Google Scholar]
- 9.Bass BL, Double-stranded RNA as a template for gene silencing. Cell 2000, 101, 235–8. [DOI] [PubMed] [Google Scholar]
- 10.Han L; Diao L; Yu S; Xu X; Li J; Zhang R; Yang Y; Werner HMJ; Eterovic AK; Yuan Y; Li J; Nair N; Minelli R; Tsang YH; Cheung LWT; Jeong KJ; Roszik J; Ju Z; Woodman SE; Lu Y; Scott KL; Li JB; Mills GB; Liang H, The Genomic Landscape and Clinical Relevance of A-to-I RNA Editing in Human Cancers. Cancer cell 2015, 28, 515–528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Slotkin W; Nishikura K, Adenosine-to-inosine RNA editing and human disease. Genome medicine 2013, 5, 105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Monteleone LR; Matthews MM; Palumbo CM; Thomas JM; Zheng Y; Chiang Y; Fisher AJ; Beal PA, A Bump-Hole Approach for Directed RNA Editing. Cell Chem Biol 2019, 26, 269–277 e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wang Y; Park S; Beal PA, Selective Recognition of RNA Substrates by ADAR Deaminase Domains. Biochemistry 2018, 57, 1640–1651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mizrahi RA; Shin D; Sinkeldam RW; Phelps KJ; Fin A; Tantillo DJ; Tor Y; Beal PA, A Fluorescent Adenosine Analogue as a Substrate for an A-to-I RNA Editing Enzyme. Angew Chem Int Ed Engl 2015, 54, 8713–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Oakes E; Vadlamani P; Hundley HA, Methods for the Detection of Adenosine-to-Inosine Editing Events in Cellular RNA. Methods in molecular biology (Clifton, N.J.) 2017, 1648, 103–127. [DOI] [PubMed] [Google Scholar]
- 16.Park E; Williams B; Wold BJ; Mortazavi A, RNA editing in the human ENCODE RNA-seq data. Genome Res 2012, 22, 1626–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sakurai M; Yano T; Kawabata H; Ueda H; Suzuki T, Inosine cyanoethylation identifies A-to-I RNA editing sites in the human transcriptome. Nature chemical biology 2010, 6, 733–40. [DOI] [PubMed] [Google Scholar]
- 18.Suzuki T; Ueda H; Okada S; Sakurai M, Transcriptome-wide identification of adenosine-to-inosine editing using the ICE-seq method. Nature protocols 2015, 10, 715–32. [DOI] [PubMed] [Google Scholar]
- 19.Paul MS; Bass BL, Inosine exists in mRNA at tissue-specific levels and is most abundant in brain mRNA. The EMBO journal 1998, 17, 1120–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yang JH; Luo X; Nie Y; Su Y; Zhao Q; Kabir K; Zhang D; Rabinovici R, Widespread inosine-containing mRNA in lymphocytes regulated by ADAR1 in response to inflammation. Immunology 2003, 109, 15–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Knutson SD; Ayele TM; Heemstra JM, Chemical Labeling and Affinity Capture of Inosine-Containing RNAs Using Acrylamidofluorescein. Bioconjug Chem 2018, 29, 2899–2903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Krawczyk H, A New Access to 2-(Chloromethyl)Acrylonitryle. Synthetic Commun 1995, 25, 3357–3362. [Google Scholar]
- 23.Mack S; Fouz MF; Dey SK; Das SR, Pseudo-Ligandless Click Chemistry for Oligonucleotide Conjugation. Curr Protoc Chem Biol 2016, 8, 83–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Nakano M; Fukami T; Gotoh S; Nakajima M, A-to-I RNA Editing Up-regulates Human Dihydrofolate Reductase in Breast Cancer. J Biol Chem 2017, 292, 4873–4884. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


