Abstract
Plants uniquely have a family of proteins called extra-large G proteins (XLG) that share homology in their C-terminal half with the canonical Gα subunits; we carefully detail here that Arabidopsis XLG2 lacks critical residues requisite for nucleotide binding and hydrolysis which is consistent with our quantitative analyses. Based on microscale thermophoresis, Arabidopsis XLG2 binds GTPγS with an affinity 100 times lower than that to canonical Gα subunits. This means that given the concentration range of guanine nucleotide in plant cells, XLG2 is not likely bound by GTP in vivo. Homology modeling and molecular dynamics simulations provide a plausible mechanism for the poor nucleotide binding affinity of XLG2. Simulations indicate substantially stronger salt bridge networks formed by several key amino-acid residues of AtGPA1 which are either misplaced or missing in XLG2. These residues in AtGPA1 not only maintain the overall shape and integrity of the apoprotein cavity but also increase the frequency of favorable nucleotide-protein interactions in the nucleotide-bound state. Despite this loss of nucleotide dependency, XLG2 binds the RGS domain of AtRGS1 with an affinity similar to the Arabidopsis AtGPA1 in its apo-state and about 2 times lower than AtGPA1 in its transition state. In addition, XLG2 binds the Gβγ dimer with an affinity similar to that of AtGPA1 XLG2 likely acts as a dominant negative Gα protein to block G protein signaling. We propose that XLG2, independent of guanine nucleotide binding, regulates the active state of the canonical G protein pathway directly by sequestering Gβγ and indirectly by promoting heterodimer formation.
Keywords: Arabidopsis, AGB1, Gβγ, AtGPA1, AtRGS1, Extra-large G protein, XLG2, dominant negative G alpha
INTRODUCTION
The canonical heterotrimeric guanosine nucleotide-binding protein complex, consisting of Gα, Gβ and Gγ subunits, serves as a molecular on-off switch in the cell. The inactive or “off–state” form consists of the guanosine diphosphate (GDP) bound to the Gα subunit in complex with the Gβγ dimer. For the active or “on–state”, exchange of GDP for GTP in Gα, either spontaneously or catalyzed by a guanine nucleotide exchange factor, changes the Gα conformation leading to dissociation, partly or entirely (Lambert, 2008), from the Gβγ dimer and thus enabling both Gα and Gβγ to propagate signaling to downstream components (Sprang, 1997; Urano, Chen, Botella, & Jones, 2013; Urano & Jones, 2014). Signaling is terminated when the Gα subunit hydrolyzes GTP thus returning to the inactive GDP-bound state. The rate of GTP hydrolysis is an intrinsic property of each Gα subunit but it can be accelerated by Regulator of G protein Signaling (RGS) proteins (Kleuss, Raw, Lee, Sprang, & Gilman, 1994; Sprang, 2016). The Gα structure required for nucleotide binding and hydrolysis and for interaction with Gα-Gβγ and Gα-RGS interactions are well understood (Gilman, 1987; S. R. Sprang, 1997).
In humans, there are multiple genes encoding G protein subunits resulting in 23 Gα, 5 Gβ and 12 Gγ subunits. The Gα subunits are divided into four subclasses (Gs, Gi, Gq and G12/13) based on function and sequence similarity. However, in Arabidopsis, there is only one canonical Gα (AtGPA1) which approximates the sequence of the ancestral Gα subunit that evolved into these four animal Gα subclasses (Temple, Jones, & Jones, 2010). AtGPA1 has a near identical structure to that of human Giα1 (Jones et al., 2011). In addition to the canonical Gα subunit AtGPA1, the Arabidopsis genome encodes three atypical Extra-large G proteins (XLG1, XLG2, and XLG3) (Chakravorty, Gookin, Milner, Yu, & Assmann, 2015). The other components of the Arabidopsis G protein core are a Gβ subunit (AGB1) (Ullah, Chen, Temple, Boyes, & Alonso, 2003), one of three Gγ subunits (AGG1, AGG2, and AGG3) (Thung, Trusov, Chakravorty, & Botella, 2012) , and one receptor-like RGS protein (AtRGS1) (Christopher A. Johnston et al., 2007).
The presence of these atypical G proteins makes G protein signaling in plants unique and paradoxical (Chakravorty, et al., 2015; Maruta, Trusov, Brenya, Parekh, & Botella, 2015; Urano et al., 2016). Specifically, the N-terminal half of XLG proteins lacks homology to any characterized domain but contains a putative nuclear localization signal and a cysteine-rich region while the C-terminal half of XLG proteins shares homology (i.e. evolutionary history, (Urano, et al., 2016)) with canonical Gα subunits (~30% identity). However, there is controversy to what extent that these atypical Gα homologs bind and hydrolyze nucleotides and interact with AtRGS1 and AGB1 (Liang et al., 2018; Urano et al., 2016).
For canonical Gα subunits, there are three major conformational changes between the GDP and GTP-bound states of the protein located in what are called Switch I, II and III. Switch I and Switch II directly contact the bound guanine nucleotide and include residues critical for catalyzing GTP hydrolysis, while Switch III contacts Switch II when in the activated conformation (Coleman et al., 1994). These switches are linked between the nucleotide-binding domain and the RGS binding domain and are represented by five conserved sequence motifs named G1 to G5 (Berman, Wilkie, & Gilman, 1996). The G1–G3 boxes provide critical contacts for the β and γ phosphates of the guanine nucleotide and are essential for the coordination of Mg2+. The G4 and G5 loops are involved primarily in binding the guanine ring. The G2 and G3 boxes overlap with Switches I and II that are also the key Gβγ binding sites. The RGS domain directly binds to the three switch regions and stabilizes them in a transition state conformation.
It is paramount to resolve unequivocally if XLG proteins bind guanine nucleotide and relevant signaling elements such as RGS and Gβγ to elucidate its atypical mechanism. Here, we combine structure-based and physicochemical experimental methods along with molecular simulations to analyze the binding of XLG2 with both the nucleotide and with a candidate binding partner, AtRGS1/ Gβγ dimer. We describe for the first time in great detail the structural issues that should raise concern among those who claim that XLG proteins are nucleotide-dependent switches. In fact, we show that XLG2 binds nucleotide so poorly that it is essentially nucleotide free in the cell, yet despite its nucleotide-free, “empty” state, XLG2 interacts with its partners AtRGS1 and AGB1 with an affinity similar to AtGPA1 in its transition state. We used molecular dynamic simulations to explain how this binding is disrupted and how these protein-protein interactions are maintained.
RESULTS AND DISCUSSION
XLG proteins lack critical residues for coordination of the γ and β phosphates on the guanine nucleotide
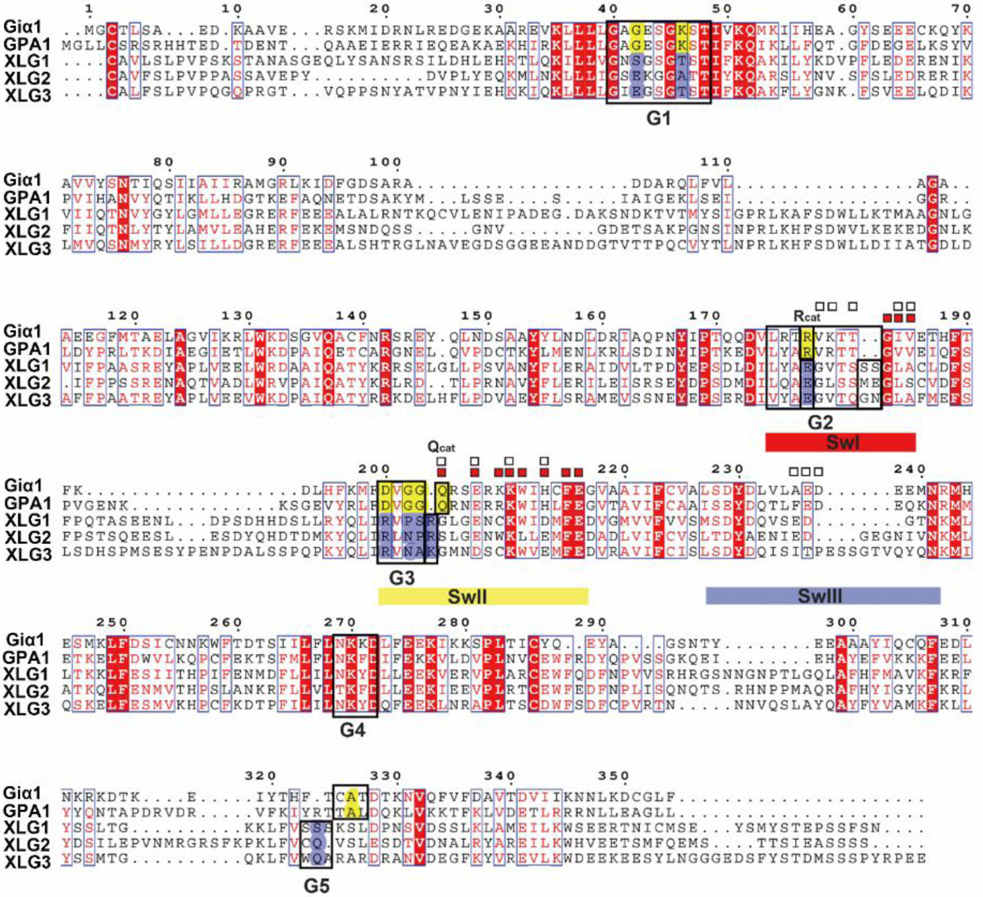
A multiple sequence alignment of XLGs Gα domain, AtGPA1, and human Giα1 is shown in Fig. 1 with the G1-G5 motifs and switches I-III regions highlighted (noted as SwI-III). To compare the protein structures between XLGs and canonical Gα subunits, we created high-quality models of the Gα homology domains of XLG2 using MODELLER and the aligned sequences shown in Fig. 1. The human RGS4 and Giα1 transition state (Ligand: AlF4 and GDP) complex (PDB [1AGR]) was used as template to generate the models of XLG2. Models were created using the Automodel script based on the template of human Giα1 in complex with AlF4 and GDP (PDB [1AGR]). For evaluation and selection of the "best" model, we calculated the objective function (molpdf) DOPE score, GA341 assessment score between the model and the template (Fig. S1). The final model (XLG2-1) was selected given the lowest average value of the molpdf and the DOPE assessment scores.
Figure 1. Alignment between human Giα1 Arabidopsis AtGPA1 and the C terminal G alpha domain of the three XLGs.
The G1-G5 motifs are shown in black boxes. The switches I-III regions (SwI-III) are highlighted (SwI in red, SwII in yellow and SwIII in blue). A percentage of equivalent residues is calculated per columns, considering physico-chemical properties. Blue boxes highlight residues with the same physico-chemical properties and red solid highlighting means the same residues. The contact residues to RGS protein are labeled with white boxes □ and the contact residues with Gβ are labeled with red boxes ■. The residues which are conserved in human Giα1 and AtGPA1 for GTP/GDP binding and hydrolysis but are missing in XLGs are highlighted with yellow and blue respectively. The residues essential for the catalysis of the nucleotide are highlighted as Rcat and Qcat. (The C domain of XLGs start with first C residues in the paper, C436 in XLG1, C435 in XLG2 and C396 in XLG3).
As shown in Fig. S2, human Giα1 and Arabidopsis GPA1 have two domains: a Ras-like domain and an all-helical domain. Animal Gα subunits and AtGPA1 are extremely similar in structure (RMSD= 1.8 Ȧ (J. C. Jones, J. W. Duffy, et al., 2011)). The Ras-like domain is essential for the nucleotide and RGS proteins binding which contains the five guanine nucleotide binding motifs (G1-G5) and three flexible switch regions (SwI-III) (Fig. S2A). The all-helical domain is important for the intrinsic nucleotide exchange rate (Jones, Jones, Temple, & Dohlman, 2012; J. C. Jones, B. R. Temple, A. M. Jones, & H. G. Dohlman, 2011). XLG2-1 shares a similar overall 3D structure with human Giα1 and AtGPA1 even though the sequence identity is ~ 30%. XLG2-1 contains a globally similar Ras-like domain and α helix domain. The three switch regions and the G1-G5 boxes are highlighted (Fig. S2B). The RMSD between Giα1 and XLG2-1 is 0.67 Ȧ. However, despite similar global structure between XLG2-1 and human Giα1, many of the conserved motifs which are essential for nucleotide binding and hydrolysis are missing, including key residues within the G1, G3 and G5 motifs for nucleotide binding and some dominant residues in the P loop, Switch I and Switch II for coordinating water and Mg2+ to catalyze GTP hydrolysis (Fig. 1 and Fig. 2B). These critical differences between the canonical Gα and the XLG Gα domain are described in detail next.
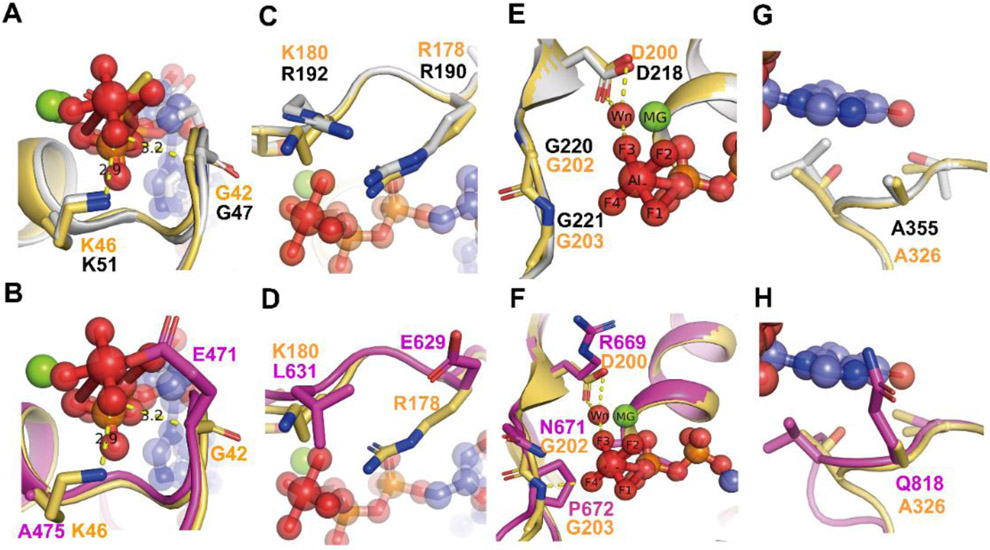
Figure 2. Comparison of the G motifs of AtGPA1 PDB [2XTZ] and the XLG2 G alpha domain model with human Giα1 (PDB [1AGR]).
Grey: AtGPA1, Magenta: XLG2-1 model, Light orange: Giα1. The substrate GDP and AlF4 are shown as sticks and spheres. Mg+2 is shown as green sphere. Wn: nucleophilic water. The main different residues in G1 motif of XLG2 compared with Giα1 and AtGPA1 are shown as sticks (A, B). Both AtGPA1 and Giα1 have the same G and K residues in G1 motif (A). the G42 and K46 in Giα1 were replaced by E471 and Al475 in the counterpart position of XLG2 (B). The main different residues in G2 motif of XLG2 compared with Giα1 and AtGPA1 are shown as sticks (C, D). Both AtGPA1 and Giα1 have the same R residues (known as arginine finger) and similar charged K180 and R192 in G2 motif (C). But in XLG2 no arginine finger exists rather a Glu is at this position. Also, the charged K or R was replaced by a L (D). The main different residues in G3 motif of XLG2 compared with Giα1 and AtGPA1 are shown as sticks E, F. Both AtGPA1 and Giα1 have the same DVGG residues in G3 motif (E). But in XLG2 the DVGG was replaced by R669/N671/P672 relatively (F).The main different residues in G5 motif of XLG2 compared with Giα1 and AtGPA1 are shown as sticks in G and H. Both AtGPA1 and Giα1 have the same A residues in G5 motif (G). While in XLG2 the conserved A was replaced by Q818 (H).
The highly conserved G1 motif is a phosphate-binding region containing a flexible structure designated “P-loop” (Saraste, Sibbald, & Wittinghofer, 1990). The G1 motif has a consensus sequence of GXXXXGKS/T for the heterotrimeric Gα subunits (Sprang, 1997). The P-loop envelopes the phosphates allowing the main chain and side-chain nitrogen atoms to interact tightly with the negatively-charged phosphates (Fig 2 A, B). In animal Gα subunits as well as in AtGPA1, the sequence in the P loop and G1 motif are mostly conserved as“GAGESGKS” (Fig 1, see G1 box). However, in XLG2-1, the G42 residue of Giα1 in the P loop is replaced by E471 and the K46 residue of Giα1 is replaced by A475, respectively (Fig 1 and Fig 2B). The G42 residue of Giα1 or G47 residue of AtGPA1 in the P loop play a dominant role in binding the substrate with the main chain forms hydrogen bond with the γ phosphate oxygen atom (Coleman et al., 1994; Jones et al., 2011). This G residue is shown in Fig 2A and B. More importantly, only a G residue side chain is small enough to avoid steric clash with the nucleotide and mutation of the corresponding P-loop residue in Giα1, G42 to V, also drastically reduces its GTP hydrolysis activity (Bosch et al., 2012; Raw, Coleman, Gilman, & Sprang, 1997; Seeburg, Colby, Capon, Goeddel, & Levinson, 1984). Structural studies of G42V mutant in Giα1 suggest that the introduced valine side chain sterically prevents appropriate positioning of Q204 which coordinates a nucleophilic water molecule during GTP hydrolysis and steric pressure will induce the reconfiguration of switch II (Bosch, et al., 2012; Raw, et al., 1997; Sprang, 2016). Thus, we assume that the substitution of the large side chain of E471 in XLG2-1 reduces GTPase activity (Fig 2B).
Additional differences were found with the P loop of the XLG proteins. The lysine (K46 of Giα1 and K51 of AtGPA1 ) residue in the G1 motif directly interacts with the β- and γ-phosphate oxygens of the GTP and thus is crucial for the required free energy change (Sprang, 2016) (Fig. 2A). Given that there are two dominant residues mutations in the nucleotide pocket of XLG2 (G42 in Giα1 to E471 and K46 to A475 ) (Fig. 3B), we hypothesize that XLG2 binds the nucleotide with a reduced affinity in vitro and that XLG2 is nucleotide free in vivo.
Figure 3. Interactions between the catalysis center of the Ras domain and critical residues of RGS proteins.
(A) The critical contact residues between Gαi1 and RGS4 (PDB 1AGR) are shown in light orange. Arg 178 (Rcat) is within hydrogen bonding distance of the leaving group β-γ bridge oxygen and Q204 (Qcat) is a hydrogen bond donor to a fluorine (or O–) Al substituent and accepts a hydrogen bond from the presumptive water nucleophile (Wn). The hydrogen bond network (yellow dashed lines) involving N128 (Asn thumb of RGS4), Qcat, G42 and the the γ phosphate (modeled by AlF4) orient Wn for nucleophilic attack and stabilize developing charge at the β-γ bridge leaving group oxygen. RGS4 residues Asn 128 constrain the conformation of Gαi1 Q204 (Qcat) to the pre-transition state conformation. AtGPA1 contains the same catalysis network (A) however the catalysis network was disrupted in XLG2 with the loss of the Glncat and Arg finger and replaced by R673 and E629 respectively (B). Grey: AtGPA1, Magenta: XLG2, Light orange: Giα1. The substrate GDP and AlF4 are shown as sticks and spheres. Main catalysis residues between Giα1, AtGPA1, XLG2 and RGS4 are highlighted as sticks. Wn: nucleophilic water.
The G3 box contains the signature sequence DXGG conserved throughout the heterotrimeric G-protein superfamily. Similar to the P loop, residues with the G3 motif interact with the γ-phosphate of GTP but also orients the Mg2+ ion that is critical for coordination of the guanine nucleotide. In AtGPA1 and Giα1, the G3 box is invariant “DVGG” (Fig. 2E), however in the XLG2 Gα domain, the residues are replaced by “RLNP” (Fig. 2F). The conserved Asp residue of canonical Gα subunits provides the water-molecule-mediated coordination of Mg2+ and therefore, the substitution of Asp for this critical Arg disrupts the ability to bind Mg2+ (Noel, Hamm, & Sigler, 1993; Sprang, 1997; Sprang, 2016). Moreover, the main chain amide of the signature Gly residue is essential for nucleotide binding through hydrogen bonding to the γ-phosphate oxygen of the GTP (Noel, et al., 1993). The main chain amide of this Gly is hydrogen bonded to the γ-phosphate and mutation of the two Gly residues in the G3 box confer dominant negative phenotypes (Barren & Artemyev, 2007; Noel, et al., 1993; Sprang, 1997). Gilman’s group showed that GDP-bound Gαs G226A mutant (the second Gly in the G3 DVGG motif) has a higher affinity for Gβγ than the wild-type subunit and is incapable of undergoing a GTP-induced conformational change (Lee, Taussig, & Gilman, 1992). Taken together, stark differences in the three dominant residues in the G3 motif in the XLG2 protein nucleotide binding pocket which is conserved among all XLG proteins (Fig. 1) suggest that XLGs exist in the empty nucleotide state.
The G5 motif consensus is S/C/T-A-K/L/T. In Giα1, the G5 motif sequence is C-A-T (A = residue 326) and in AtGPA1, it is T-A-L (A = residue 355, Fig. 2G and H). In either form, the main chain of A326 in Giα1 is essential for the binding of GTP/GDP specifically forming a hydrogen bond with the oxygen of the guanidine nucleotide and S substitution at this site weakens the affinity for GTPγS through steric crowding (Posner, Mixon, Wall, Sprang, & Gilman, 1998). Also, the equivalent A366S mutation in the G5 motif of Gαs decreases Gαs’s affinity for GDP and GTPγS by steric crowding and shifting Gα towards the empty nucleotide pocket state (Iiri, Herzmark, Nakamoto, van Dop, & Bourne, 1994; Posner, et al., 1998). However, in XLG2, the equivalent residues are C-Q-V (Q = residue 818, Fig. 2H). Thus, this substitution of A326 with Q818 in XLG2-1 is predicted to create a steric clash for nucleotide binding providing further inference that XLG is nucleotide-free.
XLG proteins lack key residues to catalyze GTP hydrolysis
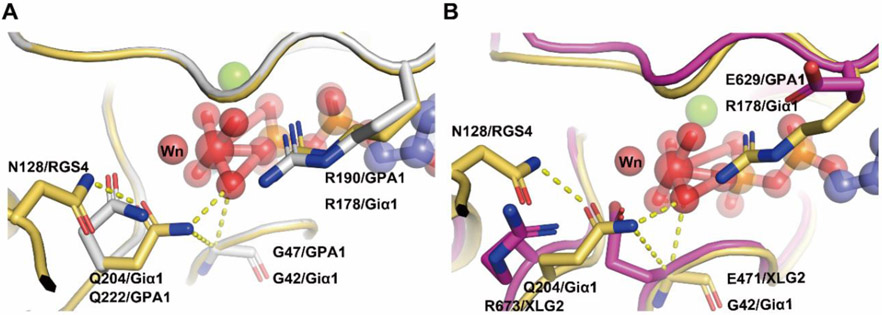
Two amino acids, one from the Gα subunit (the conserved catalytic glutamine residue in Switch II region which is named “Qcat” ) and one from the RGS protein (the so-called “Asn thumb”), together with nucleophilic water and a Mg2+ in the catalytic center are essential elements for the catalytic reaction (Sprang, 2016) (Fig. 3). In Giα1, the Qcat in Switch II is Q204 is essential for catalytic activity in the Gα subunit. A conserved Arg residue in Switch I region designated “Rcat” here, is also a major determinant of the catalytic activity. A water molecule designated “Wn” occupies the position for the nucleophile engaged in an in-line attack on the phosphate. The Asn thumb (N128 in RGS4) in the RGS domain reorients the Qcat allowing the carboximido moiety to form hydrogen bonds with AlF4 mimicking a γ phosphate oxygen atom and Wn. Rcat forms electrostatic interactions with the β phosphate oxygen and with one of the fluoride substituents of AlF4 (Fig.3). Mutations in these residues of Switch I and Switch II are known to drastically alter GTPase activity (Sprang, 2016).
All XLG proteins lack both essential Rcat and Qcat for the catalysis (Fig. 3B). In XLG proteins, the Rcat residue in Switch I is E, creating a charge reversal that disrupts electrostatic interactions with the β and γ phosphates of the guanine nucleotide. The equivalent mutation in Gαi1 exist as a stable protein in a nucleotide-free state and lacks the capacity to form the active conformation (D. E. Coleman, et al., 1994). In all XLG proteins, the Qcat residue of Switch II is R/K which is unable to coordinate with either the Asn thumb of the RGS protein or the nucleophilic water to hydrolyze GTP. Both Q204R and R178C mutations abrogate nucleotide hydrolysis (Coleman, et al., 1994). The structural characteristic of the XLG proteins catalysis center suggests that XLGs lack the ability both to coordinate with RGS to hydrolyze GTP and the intrinsic GTPase activity of Gα subunits.
XLG2 has a much lower binding affinity towards nucleotide than canonical G subunits yet interacts with similar affinities towards Gβγ and AtRGS1
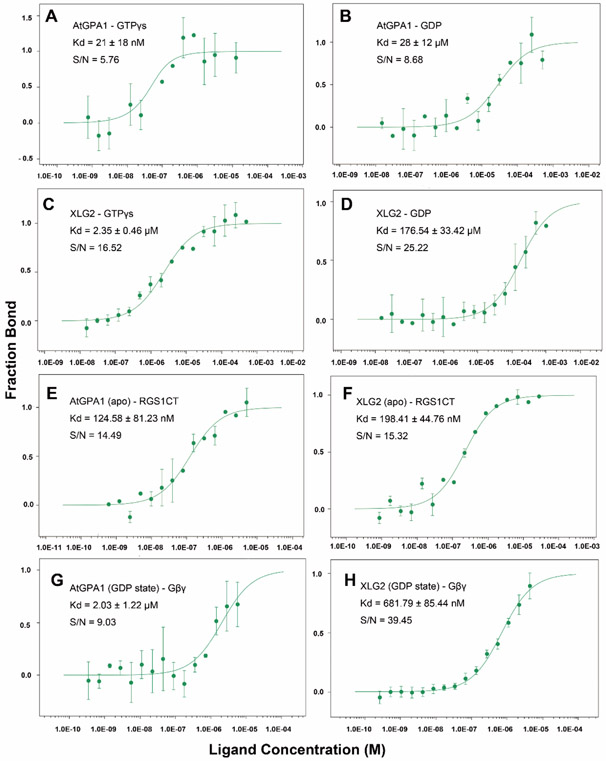
Assessments of nucleotide binding to XLG proteins to date lack quantitation for binding constants (Heo, Sung, & Assmann, 2012). Similarly, XLG protein interaction with AtRGS1 and Gβγ have been indirect measurements (Chakravorty, et al., 2015; Liang, et al., 2018; Urano, et al., 2016). To correct this deficit, we used microscale thermophoresis (MST) to measure the binding affinity of XLG2 and AtGPA1 with guanine nucleotides (GDP and GTPγS) and with binding partners AtRGS1 and Gβγ. The advantages of this new technique are the capability of obtaining accurate affinities in the low affinity (μM-mM Kd) range with small amounts of protein. Note that, unlike MST, traditional radioisotope binding assays are not accurate for low affinity interactions. Raw data with the quality control parameters provided are in Fig 4 and S3 and are summarized in Table 1. The observed Kd of AtGPA1 binding GTPγS was ~ 21 nM. This is within the range of Kds reported for animal G subunits (10-100 nM, (Malinski, Zera, Angleson, & Wensel, 1996)). XLG2 bound GTPγS with a Kd of ~ 2 μM which is 100 times lower affinity than GTPγS binds AtGPA1 when tested under the same conditions. Note that this difference is a conservative estimate because the Kd for GTPγS binding to AtGPA1 using a radioactive ligand was shown to be ~ 2nM (Johnston, Willard, Kimple, Siderovski, & Willard, 2008). The affinity of XLG2 to GDP is ~6 fold lower (177 μM) than of AtGPA1 binding to GDP . However both XLG2 and AtGPA1 showed much lower binding affinity towards GDP compared with GTPγS. Quantitative analyses clearly show that XLG2 is severely impaired in guanine nucleotide binding (Table 1).
Figure 4. Binding isotherms for nucleotide, RGS1-C domain and Gβγ to AtGPA1 and XLG2.
Microscale Thermophoresis was used. (A) Binding isotherm and Kd value of AtGPA1 binding GTPγS and (B) GDP. (C) Binding isotherm and Kd value of XLG2 binding GTPγS and (D) GDP. (E) Binding isotherm and Kd value of RGS1 C terminal domain to AtGPA1 apo state and (F) XLG2 apo state. (G) Gβγ binding to AtGPA1 and (H) XLG2 Gα domain. S/N: signal to noise ratio. Each experiment was repeated at least once. Binding curve and Kd were fitted as described in Methods. Error bars represent StdDEV. Each experiment was repeated at least once.
Table 1. Summary of the binding affinity (Kd) among AtGPA1 and XLG2 with nucleotide, RGS1-C domain and Gβγ.
Top row in the inset indicates the tested interactors. The values were determined from the binding isotherms shown in Figure 4. The values are averages and StdDev for all the experimental replicates. Each experiment was replicated at least once.
| GTPγS | GDP | RGS1-C domain Gα in apo state |
RGS1-C domain Gα in transition state |
Gβγ Gα in GDP state |
|
|---|---|---|---|---|---|
| AtGPA1 | 21 ± 18 nM | 28 ± 12 μM | 125 ±81 nM | 67 ± 18 nM | 2 ± 1.22 μM |
| XLG2 | 2350 ± 460 nM | 177 ±33 μM | 198 ±45 nM | NA | 0.7 ±0.09 μM |
With this poor affinity toward guanine nucleotides, the concentration of GTP in plant cells would need to be 100 times greater than in animal cells for XLG2 to be GTP bound, however, for several reasons, this explanation of a mechanism to compensate the weak GTP affinity by XLG proteins is not reasonable. First, such a saturating concentration of GTP would eliminate the switch-like behavior of the canonical plant Gα subunit. Second, protein translation uses the same machinery in both plant and animal cells and the GTP hydrolyzed for its proof reading and is sensitive to its cytoplasmic concentration. Similarly, plant and animal microtubules requires GTP binding and hydrolysis. Both translation and cytoskeletal dynamics would cease at this high concentration of GTP. Third, nucleotide synthesis uses product inhibition to control the levels accordingly. A 100-fold higher concentration of GTP would be incompatible with enzymes involved in nucleotide synthesis. Fourth, the highest known concentration of GTP in plant cells is equivalent to only one Kd for GTP binding to XLG2 (Ashihara & Nygaard, 1989; Meyer & Wagner, 1985; Yin, Katahira, & Ashihara, 2014). As such, the concentration of GTP in plant cells, especially non-dividing cells, may be rate-limiting for full occupancy of XLG2 by GTP. For these reasons, we conclude that XLG2 is not likely bound by GTP in vivo.
Assmann’s group reported the unusual finding that the three XLG proteins bind and hydrolyze GTP using Ca2+ instead of Mg2+ as a coordinating factor (Heo, et al., 2012). To test this, we performed MST experiments to measure the binding affinity of XLG2 with nucleotide in the presence of Ca2+. The results showed lower binding affinity towards GTPγS (~186 μM) with Ca2+vs. Mg2+(~2 μM) (Fig S3).This indicates that Ca2+ may not act as the cofactor for XLGs binding GTPγS. The Kd for XLG2 binding to GDP in the presence of Ca2+ is ~28 μM vs. 177 μM in the presence of Mg2+ (Fig S3). However it is still too low affinity to consider Ca2+ as a cofactor for XLGs. Interestingly, despite XLG2 having much lower binding affinity towards GTPγS and GDP compared with AtGPA1, it had a similar binding affinity to the C-terminal RGS domain of AtRGS1 and to the Arabidopsis Gβγ dimer (AGB1/AGG1). AtGPA1 bound AtRGS1 with a Kd ~125 nM with in AtGPA1 in its apo state and ~ 67 nM in its transition state (Fig.S3). XLG2 has a similar Kd of ~198 nM towards AtRGS1 when in its apo state (Table 1 and Fig. 4). The Kd for a transition state XLG2 was not determined because this state is not relevant due to its nucleotide independence. Moreover, XLG2 showed a ~0.7 μM binding affinity towards Gβγ similar to that of AtGPA1 which is ~2 μM (Table 1 and Fig. 4). This suggests that XLG2 exists as a nucleotide-independent inhibitor of G signaling through its ability to sequester Gβγ directly or indirectly by binding to AtRGS1 thus enabling freed AtGPA1 to sequester Gβγ.
A mechanistic explanation: Relative instability of XLG2 confers the reduced nucleotide interaction
We applied several computational modeling and simulation approaches to understand the underlying molecular mechanisms differentiating AtGPA1 and XLG2 proteins. We sought to provide structural and molecular dynamics rationales for the experimentally observed differences in nucleotide binding preferences by the two proteins. To this point, we performed microseconds of molecular dynamics (MD) simulations of four molecular complexes, involving GDP and GTP nucleotides, each in complex with both AtGPA1 and the homology-modeled XLG2-1 Gα domain, followed by comparative analyses of the respective MD trajectories. The main finding of our simulations is that the molecular dynamic behaviors of XLG2-1 differs from that of AtGPA1. We observed that overall XLG2-1 was more mobile in comparison with AtGPA1, which generally retained its original crystallographic structure over the course of simulations. Furthermore, to distinguish the two proteins with respect to their nucleotide binding capabilities, we focused on analyzing the behavior of the ligand binding site both in the context of the intra-protein and ligand-protein interactions in order to more clearly understand the key factors contributing to the experimental findings of the lower nucleotide binding affinity in XLG2.
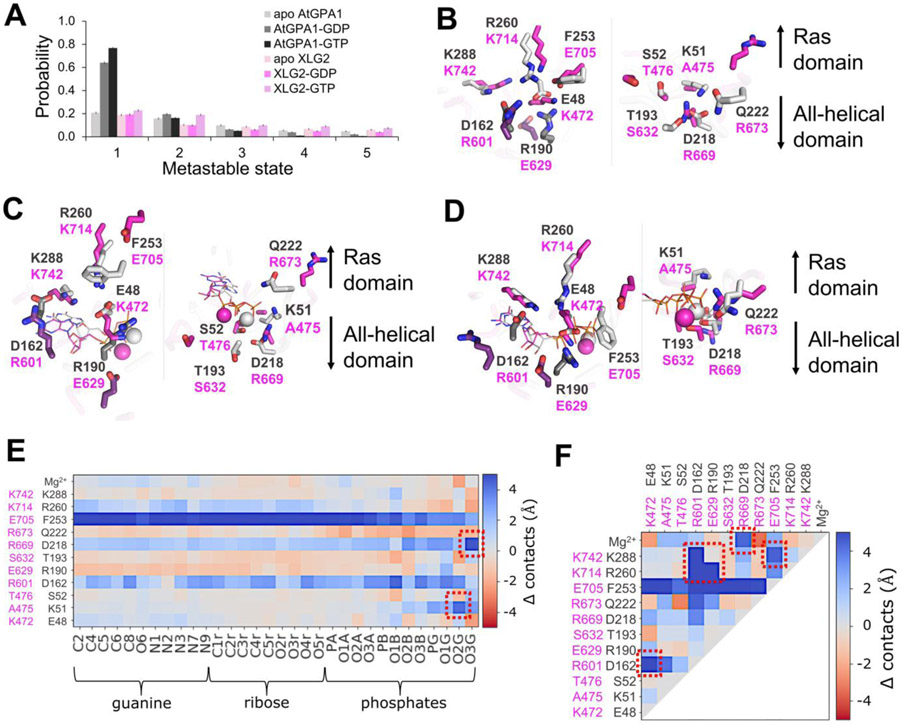
In preparation for MD simulations, the structure of XLG2 obtained by homology modeling, was subjected to molecular mechanics minimization following several protocols as described in the Methods section in order to avoid unnatural clashes between atoms resulting from homology modeling. To understand the overall dynamics of the proteins, we analyzed RMS fluctuations per residue and calculated RMSD using all C-alpha atoms of the proteins (Fig. S4, S5), which showed that the general fold of AtGPA1 was more stable and the amino-acid residues displayed lower mobility compared to XLG2. We then sought to understand the dynamics of the nucleotide binding site and explored the key differences in the interactions formed within the binding site. First, we visualized the binding sites of the two proteins to explore the main differences in terms of the amino-acid residue composition (Fig. 5A). The following differences in the similarly-positioned, binding-site residues were determined between AtGPA1 and XLG2: E48 to K472, D162 to R601, R190 to E629, F253 to E705, R260 to K714, K288 to K742 in guanine and ribose binding sites, and K51 to A475, S52 to T476, T193 to S632, D218 to R669, Q222 to R673 in Mg2+ and phosphates binding sites (Fig. 1, 5).
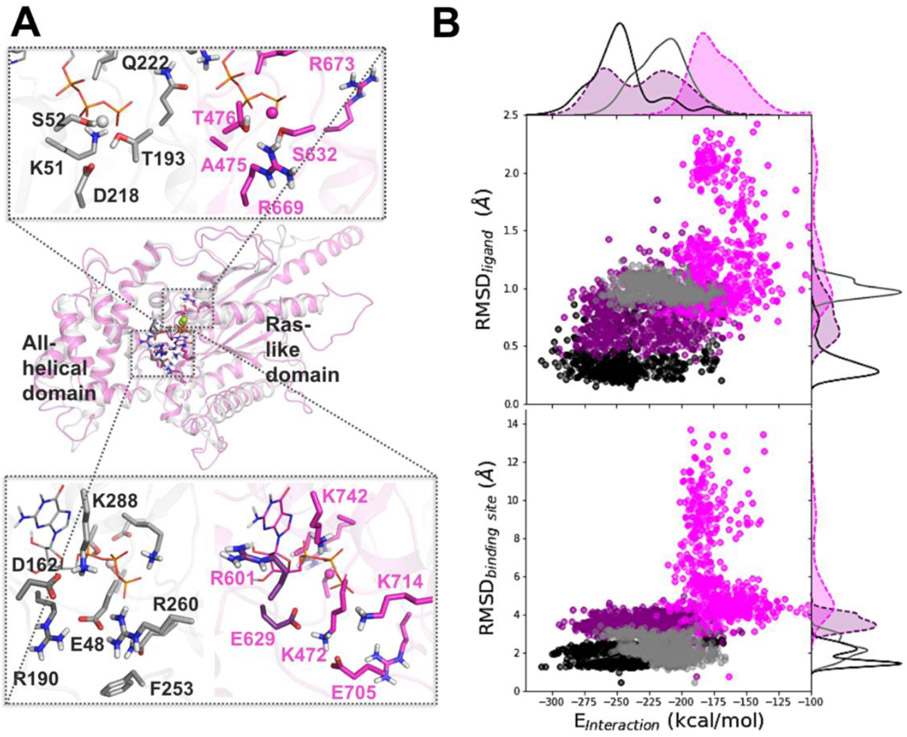
Figure 5. Difference in dynamics between the nucleotide-bound AtGPA1 and XLG2: Insights from MD simulations.
Homology modeling and MD simulations reveal the main differences in amino-acid residue composition and the nucleotide binding site dynamics of AtGPA1 (grey) and XLG2 (magenta) (A) Aligned minimized AtGPA1 crystal structure (PDB ID 2xtz) and the homology modeled XLG2. The zoomed in plots separately display phosphate and Mg2+ binding site (top) and guanine and ribose binding site (bottom) on the example of GTP-bound complexes, highlighting the most prominent differences in the amino-acid residues. (B) The relationship between the nucleotide-protein interaction energies (designated as Einteraction on the scatter plots, and calculated as the sum of the Coulomb and LJ terms) and mobility of the nucleotide (RMSDligand) and binding site (RMSDbinding site) display substantial separation among AtGPA1-GTP (black points and solid line), XLG2-GTP (purple points and dashed line), AtGPA1-GDP (grey points and solid line), and XLG2-GDP (pink points and dashed line) complexes (Fig. S6-8).
To understand the differences in the binding site dynamics, we first calculated RMSD of the heavy atoms of residues located in the binding sites (Fig. 5B and Fig. S6), which we defined as the protein residues within 4 Å from GTP (see Methods). We observed that AtGPA1 and XLG2 nucleotide binding sites differed in conformational dynamics and had distinctly different configurations as elaborated in the following paragraph. Next, we aimed to understand the impact of the difference in dynamics of the binding site residues on the nucleotide mobility and nucleotide binding preferences (Fig. 5B & Figs. S7, S8). Through exploring the relationship between the nucleotide-protein interaction energy (calculated as the sum of intermolecular Coulomb and LJ terms of the molecular mechanics energy of the nucleotide-protein complexes) and mobilities of the binding site and ligand, we observed substantial differences across the four complexes formed when AtGPA1 and XLG2 bound to both GDP and GTP. The molecular systems occupied distinct regions on each of these two landscapes. Importantly the ranking order of the means of two parameters, (i) the nucleotide mobility in the pocket as characterized by the RMSD of the nucleotide (from smallest to largest), and subsequently (ii) the nucleotide-protein interaction energies characterized as the sum of all LJ and Coulomb terms of the nucleotide-protein interactions (from more negative to less negative), agreed with the ranking order in terms of our experimental binding affinities (Kd +/− StDev) as follows: 1st) GPA1-GTP (0.021 +/− 0.018 μM), 2nd) XLG2-GTP (2.4 +/− 0.5 μM), 3rd) GPA1-GDP (28 +/−12 μM), 4th) XLG2-GDP (177 +/−33 μM) (Table 1).
This result added confidence to our structural and simulations-derived interpretations of the molecular complex formations. We would like to emphasize, however, that such calculations of the intermolecular interaction energies are merely estimates of the relative strengths of ligand-protein interactions in the bound state, which by no means is equivalent to the assessment of the change in Gibbs free energy of binding (Fujitani et al., 2005; Ganotra & Wade, 2018; Henriksen, Fenley, & Gilson, 2015; Mobley & Gilson, 2017).
Cluster analysis of the generated MD trajectories (Abramyan, Snyder, Thyparambil, Stuart, & Latour, 2016; Daura et al., 1999; Shao, Tanner, Thompson, & Cheatham, 2007 ) (see Methods for details) revealed metastable states with distinct configurations of the binding sites linked to the experimental nucleotide binding preferences (Fig. 6). For the most populated metastable state of each molecular complex, we explored the specific intra-protein chemical interactions which directly impact the dynamics of the active sites, and their impact on the nucleotide-binding interactions. The results show that nucleotide-bound AtGPA1 achieves stable dominant (i.e., frequently visited) conformations, whereas XLG2 complexes tend to transition between conformationally-diverse states with lower probabilities. Such low frequency populations of the top clusters are associated with a more dynamic binding pocket in XLG2. Interestingly, both apo-proteins assume multiple states with low probabilities across the top five clusters (Fig. S6A, S9, Supplemental Movies 1-6).
Figure 6. Apo and nucleotide-bound proteins obtain distinct configurations defining the nucleotide binding preferences of AtGPA1 and XLG2.
(A) The top five most populated metastable states of the nucleotide binding site obtained in cluster analysis of MD trajectories indicate that nucleotide-bound AtGPA1 obtains a stable frequently visited conformational state, whereas XLG2 complexes tend to transition between conformationally diverse states with lower probabilities. Interestingly, both apo proteins obtain multiple states with equivalently low probabilities (Fig. S6,S9). Aligned centroids of the largest metastable states are presented in panels B-D. (B) The most populous apo states show stable E48-R190-R260 and D162-K288 salt bridge networks in the guanine binding site (left image) of AtGPA1 (grey), and a more destabilized salt bridge network between similarly positioned residues in XLG2 (magenta) primarily contributed by R601-K742 electrostatic repulsion. K51-D218 salt bridge in the phosphate and Mg2+ binding sites (right image) enables a more structures AtGPA1, while the neutral A475 and a repulsion between R669 and R673 cause a more disintegrated XLG2. (C) GDP- and (D) GTP-bound complexes retain the strong salt bridge network in AtGPA1 and less stable electrostatic interactions in XLG2. K51 reorients and forms an additional bond with phosphates in AtGPA1, which is prevented by the equivalently positioned neutral A475 in XLG2. K472 breaks its bonds with E629 and re-arranges to interact with closer located phosphates. The absence of γ-phosphate in GDP makes the nucleotide more mobile, losing the frequency of its contacts. R673 in GTP-bound XLG2, however, forms a relatively stable bond with the γ-phosphate seemingly increasing the nucleotide binding affinity. The residues shown in darker shades in panels B-D (D162 and R190 of GPA1; R601 and E629 of XLG2) make the key intra-protein interactions defining the binding site shape. The differences in the frequency of the aforementioned interactions, on the example of GTP-bound complexes, are clearly seen through heatmaps of Δ contacts (minimum distances) (E) between the non-hydrogen atoms of the nucleotide and binding site residues, and (F) within the binding site residues. 100 states of the most populated clusters were used to generate the heatmaps. The red squares highlight the most prominent changes in the interactions stipulating the importance of the residues to the stability of the active sites and to the interactions with the nucleotide.
The most populous states of the apo-protein show a stable salt bridge network in AtGPA1, but no network and fewer coherent salt bridges are found in XLG2-1. In the guanine binding site of AtGPA1, salt bridges formed between E48 of the P-loop, R190 of Switch I, and R260 of Switch III, as well as between D162 and K288, while a more destabilized salt bridge network appeared between similarly positioned residues in XLG2-1, namely: E629 of Switch I, K472 of P-loop, K714 of Switch III and E705 (Fig. 6B, S9-12). In XLG2-1, a positive charge at K714 (position equivalent to R260 in AtGPA1) is not capable of forming a salt bridge with K472 (position equivalent to E48 in AtGPA1). In the phosphate and Mg2+ binding sites, the K51-D218 salt bridge enables a more structured apo-AtGPA1, while for XLG2, the neutral sidechain of A475 (position equivalent to K51 in AtGPA1) together with a repulsion between R669 and R673 precludes a stabilizing salt bridge. In general, AtGPA1 salt bridges formed by the two loop residues D162 and R190 (and equivalently placed E629 in XLG2) drawing the two domains closer together to subsequently increase the number of interactions between the all-helical domain and the nucleotide.
The nucleotide-bound complexes retain the aforementioned strong salt bridge network in AtGPA1 and weak electrostatic interactions in XLG2 (Fig. 6C,D, Fig. S9-12). In the bound state, the K51 sidechain of AtGPA1 was re-arranged to form an additional bond with phosphates, which is an additional salt bridge lacking by the neutral A475 in XLG2-1. In the GTP-bound XLG2-1 model, E705 lost its K472 interaction to γ-phosphate which also attracted R673 for further stabilization. Moreover, E48 in AtGPA1 avoided the negatively-charged phosphates which further promoted electrostatic interactions with both R190 and R260 to stabilize this cavity. In the GDP-occupied state, due to the lack of the γ-phosphate, GDP is more mobile and loses a number of its contacts with the active site residues in both Gα proteins, but more so in XLG2-1 due its structurally-unstable binding site.
The heatmaps of Δ contacts from experiments determining the difference in the minimum distances between the two Gα proteins and (i) the atoms of the nucleotide and binding site residues, and (ii) intra-protein interactions within the binding site residues, clearly show the contrast between the interaction frequencies within these two proteins. As highlighted in Fig 6E and F, the most prominent changes in the interactions important to the stability of the active sites and to the interactions with the nucleotide, in addition to the previously indicated interactions, reside with F253 in AtGPA1 which was overall closer to both the nucleotide and the binding site residues compared with similarly positioned E705 in XLG2-1. This aromatic residue makes frequent pi-cation interactions with R190 in both apo- and ligand-bound protein and occasional pi-cation interactions with the Mg2+ in the ligand-bound state.
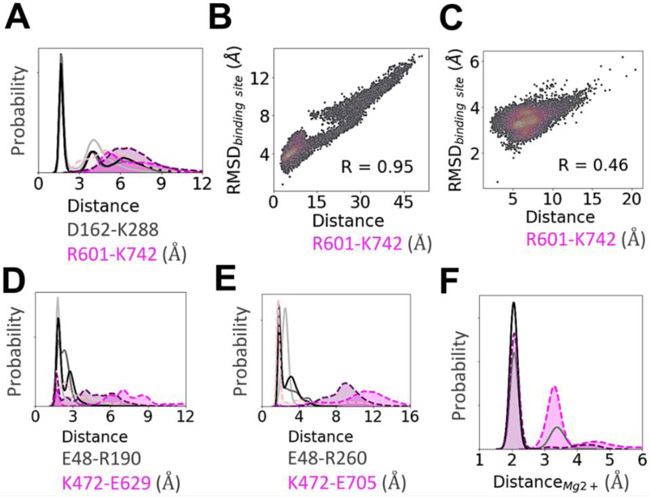
Our analyses shows that the D162-K288 salt bridge (Fig. 7A) is one of the key interactions maintaining the shape of the apo-AtGPA1 nucleotide-binding site. In contrast in XLG2-1, two positively charged residues, R601 and K742, situated in positions equivalent to D162 and K288 of AtGPA1 caused electrostatic repulsion, pushing away the all-helical domain of the protein from the Ras-like domain, resulting in an increased mobility and a less structured nucleotide binding site in XLG2-1. The R601-K742 distance in XLG2-1 was highly correlated with the fluctuations of the binding site in XLG2-GDP and to a lesser extent in XLG2-GTP (Fig. 7B and C). The reason for the former is the lack of an extra anchor in terms of γ-phosphate in GDP to enable the electrostatic repulsion between R601 and K742 to be the main contributor to the instability of XLG2-GDP binding pocket. This agrees with the experimentally observed poor binding affinity of GDP to XLG2. Although this correlation still exists in XLG2-GTP, it is less pronounced due to the presence of the extra phosphate in GTP. In XLG2, K714 makes ionic bonds with phosphates, however, this does not seem to be sufficient to retain the binding site integrity distorted by the aforementioned repulsion.
Figure 7. Key intra-protein distances responsible for the experimental nucleotide binding affinities as determined from MD simulations.
Distribution of the minimum distances between the residues emphasized in Fig. 6 and text. In general, a stronger salt bridge network in AtGPA1 maintains the shape of its nucleotide-bound binding site (see text for more details). (A) Distance between D162 and K288 and the equivalently placed R601 and K742 in XLG2. (B) The repulsion between R601 and K742 is highly correlated (R=0.95) with the binding site RMSD in XLG2-GDP and (C) to a lesser extent in XLG2-GTP (R=0.46). The 2D correlation plots were constructed using kernel-density estimation with Gaussian kernels. (D) Distance between E48 and R190 in AtGPA1 and similarly positioned K472 and E629. (E) Distance between E48 and R260 in AtGPA1 and K472 and E705 (aligned with F253 of AtGPA1) in XLG2. The distributions show that both of the salt bridges (panels E and D) are dominant in the apo proteins, and are less persistent in nucleotide-bound XLG2. (F) The distributions of the minimum distance between Mg2+ and γ-phosphate binding site residues (D218, S52, T193, and D218 in GPA1; T476, S632, R669 in XLG2) and Mg2+ counterion. For clarity in panels A, D, E, and F the values of probabilities on the y-axis are hidden. Apo AtGPA1 is plotted with solid light grey lines, AtGPA1-GDP—solid grey, AtGPA1-GTP—solid black, apo XLG2—dotted pink, XLG2-GDP—dotted magenta, XLG2-GTP—dotted purple; the probability densities for XLG2 are shaded for contrast.
Another distinction between the two proteins is in the Mg2+ binding site (Fig. 7F & Fig. S13) discussed above. In AtGPA1, D218 formed an H-bond with S52 (which interacts with Mg2+) and a Coulomb interaction with Mg2+, whereas the ‘bulkier’ and positively charged sidechain of R669 in XLG2 (position equivalent to D218 in AtGPA1) did not form stable interactions with either T476 or S632 (which interact with Mg2+) and caused an electrostatic repulsion with Mg2+. The distribution of the minimum distance between the Mg2+ binding site residues (S52, T193, D218, and Q222 in AtGPA1; T476, S632, R669, and R673 in XLG2) and the Mg2+ counterion clearly explain this effect.
To interpret the observed equivalent AtRGS1 binding capability of the two Gα proteins (Fig. 3, 4, Table 1), we estimated the structural stability of the specific regions that are involved in AtRGS1 binding (Fig. S14). We showed that the three equivalently placed AtRGS1 binding site residues of apo-AtGPA1 and apo-XLG2-1 similarly maintained their structural integrity over the course of our simulations. Such conformationally-preserved regions in the apo-proteins position them to bind AtRGS1 when it is tethered close to either protein.
Conclusion
XLG2 binds GTP in vitro poorly such that at the estimated concentration of GTP in plant cells, XLG2 is not expected to be nucleotide bound. However, XLG2 binds regulatory partners, AtRGS1 and Gβγ. Therefore, XLG2 negatively regulates signaling by two ways. One is sequestering the Gβγ dimer directly and the other is by promoting AtGPA1 interacting with Gβγ indirectly through freeing AtGPA1 from the AtRGS1::AtGPA1 complex. While this concept shares similarities for control of G signaling by dominant negative mutations of canonical G protein in animals (Brandy Barren & Nikolai O. Artemyev, 2007), it is unique in that the negative control is provided in trans by a genetically-encoded, atypical G protein.
Taken together, our modeling data provide credible interpretations for the experimentally observed strengths of guanine nucleotide binding to AtGPA1 and XLG2. Several key intra-protein and nucleotide-protein interactions in AtGPA1 were shown to be attributed to the higher structural stability of the binding site of the protein and to more persistent contacts of the protein with the nucleotide and magnesium. We show mechanistically that among the chief intra-protein interactions preserving the stability of the binding site in both apo- and nucleotide-bound-states of AtGPA1 include the following ionic bonds: D162-K288, R190-E48-R260, and K51-D218. Because XLG2 is important for disease resistance and development (X. Liang et al., 2016; X. Liang, et al., 2018; Y. Liang, Gao, & Jones, 2017), engineering these equivalent residues may lead to improvements in crop performance.
MATERIALS AND METHODS
Protein expression and purification
AtGPA1 and XLG2 proteins were expressed and purified as described previously (Li et al., 2018; Y. Liang, et al., 2017). For protein expression, proteins were transformed into ArcticExpress RP cells (Agilent Technologies). In large scale culture, 0.5 L LB medium in 2.5 L flasks were incubated at 37 °C, 225 rpm. At OD600 = 0.6 to 0.8, protein expression was induced using 0.5 mM IPTG at 12 °C for 16 hours. All purifications were performed at 4 °C.
Purification of AtGPA1 and XLG2
For quantification of interaction between AtGPA1 and AtRGS1 and AGB1/AGG1, Twinstep-GPA1 was used. Twinstrep-AtGPA1-pDEST17 (His tag removed) was transformed into ArcticExpress RP cells (Agilent Technologies). Protein expression was induced by 0.5 mM IPTG at OD600 0.75 and cultured at 12 °C for 16 hours. Cell pellets were resuspended in extraction buffer (25 mM Tris-HCl pH 8.0, 150 mM NaCl, 2 mM MgCl2, 20 μM GDP, 5 mM 2-mercaptoethanol, 1 mM PMSF, 0.25 mg/mL Lysozyme, 0.1% Thesit (Sigma, 88315), 1 X protease inhibitor cocktail, 10% glycerol) and mixed for 30 min at 4 °C. The suspension was sonicated (Sonic Dismembrator, Model 550, Fisher Scientific, power level 5, 0.50/0.50 off for 1 min, 2 cycles) to disrupt the cells. The lysate was centrifuged at 30,000 X g for 40 mins, and then the soluble fraction was collected and incubated with strep-tactin sepharose (50% suspension, cat no. 2-1201-010, IBA) for 30min at 4 °C. Then resin was washed with washing buffer (= extraction buffer above except with neither lysozyme nor thesit) and eluted with elution buffer (Washing buffer supplemented with 2.5 mM desthibiotin (Sigma-Aldrich)). The eluted proteins were run on size exclusion column (Superdex 200 10/300 GL, GE Healthcare) with running buffer (20 mM Tris-HCl, pH 7.5, 50 mM NaCl, 10 mM MgCl2, 50 μM GDP, 1 mM DTT, and 10% Glycerol). Aliquoted protein samples were snap frozen by liquid nitrogen and store at −80 °C.
For quantification of interaction between AtGPA1 and guanine nucleotides, polyHis-tagged-AtGPA1 was used. For polyHis-tagged AtGPA1 production, the cells were resuspended in N1 buffer (25 mM Tris-HCl pH 7.6, 20 μM GDP, 100 mM NaCl, 5% glycerol, 10 mM imidazole, 10 mM MgCl2, 12.5 mM 2- mercaptoethanol, 1 mM PMSF, 10 mM leupeptin, 0.25 mg/mL Lysozyme, 0.1% Thesit (Sigma, 88315)) and mixed for 30 min. After sonication (Sonic Dismembrator, Model 550, Fisher Scientific, power level 5, 0.50/0.50 off for 1 min, 2 cycles), the concentration of NaCl was raised to 300 mM and the lysate was mixed for another 30 min. The soluble fraction was separated by centrifugation at 30,000 × g for 45 min, then the supernatant was incubated with TALON Metal Affinity Resin (50 μL 50% slurry per 1 g cell pellet) for 1.5 h. The resin was washed with washing buffer (25 mM Tris-HCl pH 7.6, 300 mM NaCl, 5% glycerol, 10 mM imidazole, 10 mM MgCl2, 10 mM leupeptin) and eluted with elution buffer (50 mM Tris-HCl pH 7.6, 300 mM NaCl, 5% glycerol, 300 mM imidazole, 10 mM MgCl2). The eluate was dialyzed against dialysis buffer (20 mM Tris-HCl pH 7.6, 50 mM NaCl, 1 mM MgCl2, 1 mM DTT) overnight.
The purification procedure for XLG2 C terminal Gα domain (residues from 435 to 861) was similar to His-tagged AtGPA1 purification except the buffer was pH 7.5 and the polyHis tag was cleaved by TEV protease to generate untagged XLG2.
His-and GST-tagged RGS+Ct (AtRGS1, residues from 284 to 459) were cloned into pDEST17 or pDEST15 destination vector as previously described (Li et al., 2016; Urano et al., 2012). The procedure for expression and purification was similar to His-tagged AtGPA1 purification except in the absence of GDP. The His-GST tag was cleaved by TEV protease. This recombinant TEV enzyme had a 100 percent cutting efficiency.
Gβγ Protein Purification
AGB1 and AGG1 were purified as described previously (Jones, Temple, Jones, &. Dohlman, 2011). For expression of Gβγ in Sf9 cells with the baculovirus expression system, N-terminal 6XHis tagged AGB1 and 6XHis tagged AGG1 were subcloned into the pFastBacDual vector (Invitrogen) at the BamHI/PstI and XhoI/KpnI sites, respectively. AGB1/AGG1 recombinant proteins were expressed by P2 baculoviral stock (titer ~ 2X 108 pfu/ml) infection of sf9 cells at multiplicity of infection (MOI) of 1 for 48 hours. Purification steps were performed at 4°C. Pellets were resuspended in extraction buffer (25 mM Tris, pH 8, 200 mM NaCl, 20 mM imidazole, 1 mM 2-mercaptoethanol, 1 mM PMSF, 0.25 mg/mL Lysozyme, 0.1% thesit (Sigma, 88315), 1X protease inhibitor cocktail). The cells were disrupted by sonication (Sonic Dismembrator, Model 550, Fisher Scientific, power level 5, 0.50/0.50 off for 1 min, 2 cycles). Sonicated lysate was centrifugated at 50,000 X g for 30 mins to obtain the soluble fraction, and then the soluble fraction was incubated with Ni-NTA Agarose (Qiagen, Mat. No, 1018244) for 1 hour. The Ni-NTA Agarose was washed with washing buffer (same as extraction buffer with no lysozyme and thesit) and eluted with elution buffer (25 mM Tris, pH 8, 200 mM NaCl, 250 mM imidazole, 1 mM 2-mercaptoethanol, 1 mM PMSF, 1Xprotease inhibitor cocktail, 20% glycerol). The eluate was run on a size exclusion column (Superdex 200 10/300 GL, GE Healthcare) with running buffer (20 mM Tris-HCl, pH 8, 50 mM NaCl, 1 mM DTT, and 10% Glycerol). Aliquoted protein samples were snap frozen in liquid nitrogen and store at −80 °C. Purified proteins were examined by SDS-PAGE gel with both Coomassie Blue staining and anti-His tag western blot. Aliquoted protein samples were snap frozen by liquid nitrogen and store at −80 °C.
Quantitative binding analyses by microscale thermophoresis (MST)
Measurements for equilibrium binding were performed using 50 nM fluorescently labelled protein using Monolith NTTM Protein Labeling Kit RED-NHS (Nanotemper Technologies). The dye/protein ratio used was 10:1. Experiments were conducted at 25°C and in ‘MST Buffer’ [PBS pH 7.4, 0.05% Tween-20]. Protein sample was centrifuged for 30 min (21,000 × g, 4°C) before experiment. For thermophoresis measurements, ligand and labeled protein sample was mixed 1:1 with each of the ligand dilution series. After 10-min incubation at room temperature, each dilution was filled into Monolith NTTM MST Premium-coated capillaries (Nanotemper Technologies). A capillary scan was performed with 40% LED power. Binding curves were fitted to two sets of replicates. For binding experiments, Kd values were calculated via the MO. Affinity Analysis V2.3 software. This software performs a quality control based on the levels of starting fluorescence, and the distribution of the signal within the capillaries, an indicator of non-specific binding. A signal to noise ration above 5 provides high confidence of the data.
XLG2 model building
To compare the protein structures between XLGs and canonical Gα, we created high-quality models of the Gα homology domains of XLG1, XLG2 and XLG3 using MODELLER and the aligned sequences shown in Fig. 1. The human RGS4 and Giα1 transition state (Ligand: AlF4 and GDP) complex (PDB [1AGR]) was used as template to generate the models of XLG2. Five models (XLG2-1 to XLG2-5) were created using the Automodel script based on the template of human Giα1 (PDB [1AGR]). For evaluation and selection of the best model, we calculated the objective function (molpdf), Discrete Optimized Protein Energy (DOPE) score and GA341 assessment score between the model and the template (Figure S2). The first model XLG2-1 was selected given the relatively low value of the molpdf and overall DOPE assessment scores. In addition, DOPE scores were calculated per-residue and the template and the models were compared using Gnuplot (Figure S2).
Molecular dynamics simulations
The crystal structure of Arabidopsis AtGPA1 (PDB ID 2XTZ) (J. C. Jones, J. W. Duffy, et al., 2011) and the homology model structure of Arabidopsis XLG2 were used as the starting structures in the molecular dynamics (MD) simulations. Upon the alignment of the two proteins, the nucleotide from the 2XTZ structure of AtGPA1 protein was copied into the XLG2 protein. Histidine residues were protonated using the Interactive Optimizer in the H-bond assignment section of the Protein Preparation Wizard module available through Maestro (Maestro Schrödinger software (release 2018-4, Schrödinger, LLC: New York, NY)). The protein structures were then energy-minimized using OPLS3 force field (FF) and the Optimal scheme available in the Macromodel Minimization module of Maestro, which used PRCG method with 3-point line searcher since the number of unfixed atoms was more than 1,000. The molecular systems were then prepared to perform MD simulations in Gromacs 2018.2 simulation package using CHARMM36 protein forcefiled (Berendsen, Vanderspoel, & Vandrunen, 1995). End caps were added to both termini of each protein. The protein complex was minimized in vacuum using the steepest decent algorithm for 5,000 steps or until the maximum force of 1,000 kJ*mol-1*nm-1 was reached. The molecular systems were then solvated in TIP3P water (Jorgensen, 1982), counterions were added for system neutrality, and NaCl was added by replacing water molecules in order to mimic 0.15 M physiological conditions. The total system sizes were ~80,000 atoms for GPA1 systems and ~130,000 atoms for XLG2 systems. Solvent energy-minimization was then performed, followed by a two-step equilibration, during which all heavy atoms of the system, excluding those of water and counterions, were restrained: 0.1 ns in NVT ensemble using the modified Berendsen thermostat (Berendsen, Postma, Vangunsteren, Dinola, & Haak, 1984) set at constant 310 K, and 1 ns in NPT ensemble at constant 1 atm and 310 K using the Parinello-Rahman pressure coupling (Parrinello & Rahman, 1981). All simulations were conducted using the Leapfrog integrator in periodic boundary conditions. The 6-12 Lennard-Jones potential was used to describe the vdW interactions, and the nonbonded cutoff distance was set at 0.1 nm. The particle mesh Ewald algorithm (Darden, York, & Pedersen, 1993) controlled the long-range electrostatic interactions. Bonds involving hydrogen atoms were constrained using the linear constraint solver algorithm (LINCS) (Hess, Bekker, Berendsen, & Fraaije, 1997). The production simulations were conducted in NPT ensemble with all atoms free to move. The volta GPU nodes on UNC Longleaf supercomputer cluster were used, and each simulation was performed on a combination of 1 GPU and 4 associated CPUs. Each of the six molecular systems were subjected to three independent 1,000 ns long MD runs totaling 18 μs cumulative simulation time. Gromacs’s trajectory analysis tools, MDTraj (McGibbon et al., 2015) along with in house bash and python scripts were used for data analysis, and matplotlib and seaborn were employed for plotting. Molecular visualization and generation of graphics were performed in MacPyMOL v1.8.6.2 (The PyMOL Molecular Graphics System, Version 1.8.6.2, Schrödinger, LLC). Cluster analysis was performed with gmx cluster available in Gromacs (Daura, et al., 1999) using the RMSD cutoff of 2.0 Å. The following binding site residues were used to calculate RMSD of the binding site and for clustering: GPA1: 47-53, 162-163, 187-193, 218-222, 253, 260, 287-288, 290-291, 354-356; XLG2: 471-477, 601-602, 606, 626-632, 669-673, 705, 714, 741-742, 744-745, 817-819. Additionally, for clustering of the nucleotide-bound complexes the ligand and magnesium were also included. For Fig. 6A the binomial standard deviations σ of the state probabilities were calculated through
where P is the probability of the cluster and N is the total number of observations.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by NIGMS (R01GM065989) and NSF (MCB-0718202) awarded to Alan. M. Jones. The deep computational analyses were supported by the resources of the UNC Longleaf cluster (https://its.unc.edu/research-computing/longleaf-cluster/)
REFERENCES
- Abramyan TM, Snyder JA, Thyparambil AA, Stuart SJ, & Latour RA (2016). Cluster analysis of molecular simulation trajectories for systems where both conformation and orientation of the sampled states are important. Journal of Computational Chemistry, 37(21), 1973–1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashihara H, & Nygaard P (1989). Purine nucleotide and RNA synthesis in suspension cultured cells of carrot. Physiologia Plantarum, 75, 31–36. [Google Scholar]
- Barren B, & Artemyev NO (2007). Mechanisms of dominant negative G-protein alpha subunits. J Neurosci Res, 85(16), 3505–3514. [DOI] [PubMed] [Google Scholar]
- Berendsen HJC, Postma JPM, Vangunsteren WF, Dinola A, & Haak JR (1984). Molecular-Dynamics with Coupling to an External Bath. Journal of Chemical Physics, 81(8), 3684–3690. [Google Scholar]
- Berendsen HJC, Vanderspoel D, & Vandrunen R (1995). Gromacs - a Message-Passing Parallel Molecular-Dynamics Implementation. Computer Physics Communications, 91(1-3), 43–56. [Google Scholar]
- Berman DM, Wilkie TM, & Gilman AG (1996). GAIP and RGS4 are GTPase-activating proteins for the Gi subfamily of G protein alpha subunits. Cell, 86(3), 445–452. [DOI] [PubMed] [Google Scholar]
- Bosch DE, Willard FS, Ramanujam R, Kimple AJ, Willard MD, Naqvi NI, et al. (2012). A P-loop mutation in Galpha subunits prevents transition to the active state: implications for G-protein signaling in fungal pathogenesis. PLoS Pathog, 8(2), e1002553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chakravorty D, Gookin TE, Milner MJ, Yu Y, & Assmann SM (2015). Extra-Large G proteins expand the repertoire of subunits in Arabidopsis heterotrimeric G protein signaling. Plant Physiology, 169(1), 512–529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coleman DE, Berghuis AM, Lee E, Linder ME, Gilman AG, & Sprang SR (1994). Structures of active conformations of Giα1 and the mechanism of GTP hydrolysis. Science, 265, 1405–1412. [DOI] [PubMed] [Google Scholar]
- Darden T, York D, & Pedersen L (1993). Particle Mesh Ewald - an N.Log(N) Method for Ewald Sums in Large Systems. Journal of Chemical Physics, 98(12), 10089–10092. [Google Scholar]
- Daura X, Gademann K, Jaun B, Seebach D, van Gunsteren WF, & Mark AE (1999). Peptide folding: When simulation meets experiment. Angewandte Chemie International Edition, 38(1-2), 236–240. [Google Scholar]
- Fujitani H, Tanida Y, Ito M, Jayachandran G, Snow CD, Shirts MR, et al. (2005). Direct calculation of the binding free energies of FKBP ligands. J Chem Phys, 123(8), 084108. [DOI] [PubMed] [Google Scholar]
- Ganotra GK, & Wade RC (2018). Prediction of Drug-Target Binding Kinetics by Comparative Binding Energy Analysis. ACS Med Chem Lett, 9(11), 1134–1139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilman AG (1987). G proteins: transducers of receptor-generated signals. Annu Rev Biochem, 56, 615–649. [DOI] [PubMed] [Google Scholar]
- Henriksen NM, Fenley AT, & Gilson MK (2015). Computational Calorimetry: High-Precision Calculation of Host-Guest Binding Thermodynamics. J Chem Theory Comput, 11(9), 4377–4394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heo JB, Sung S, & Assmann SM (2012). Ca2+-dependent GTPase, Extra-large G Protein 2 (XLG2), promotes activation of DNA-binding protein related to Vernalization 1 (RTV1), leading to activation of floral integrator genes and early flowering in Arabidopsis. Journal of Biological Chemistry, 287(11), 8242–8253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hess B, Bekker H, Berendsen HJC, & Fraaije JGEM (1997). LINCS: A linear constraint solver for molecular simulations. Journal of Computational Chemistry, 18(12), 1463–1472. [Google Scholar]
- Iiri T, Herzmark P, Nakamoto JM, van Dop C, & Bourne HR (1994). Rapid GDP release from Gs alpha in patients with gain and loss of endocrine function. Nature, 371(6493), 164–168. [DOI] [PubMed] [Google Scholar]
- Johnston CA, Taylor JP, Gao Y, Kimple AJ, Grigston JC, Chen J-G, et al. (2007). GTPase acceleration as the rate-limiting step in Arabidopsis G protein-coupled sugar signaling. Proceedings of the National Academy of Sciences, 104(44), 17317–17322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnston CA, Willard MD, Kimple AJ, Siderovski DP, & Willard FS (2008). A sweet cycle for Arabidopsis G-proteins: Recent discoveries and controversies in plant G-protein signal transduction. Plant Signal Behav, 3(12), 1067–1076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones JC, Duffy JW, Machius M, Temple BR, Dohlman HG, & Jones AM (2011). The crystal structure of a self-activating G protein alpha subunit reveals its distinct mechanism of signal initiation. Sci Signal, 4(159), ra8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones JC, Jones AM, Temple BR, & Dohlman HG (2012). Differences in intradomain and interdomain motion confer distinct activation properties to structurally similar Galpha proteins. Proc Natl Acad Sci U S A, 109(19), 7275–7279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones JC, Temple BRS, Jones AM, & Dohlman HG (2011). Functional reconstitution of an atypical G protein heterotrimer and regulator of G protein signaling protein (RGS1) from Arabidopsis thaliana. Journal of Biological Chemistry, 286(15), 13143–13150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jorgensen WL (1982). Quantum and Statistical Mechanical Studies of Liquids .24. Revised Tips for Simulations of Liquid Water and Aqueous-Solutions. Journal of Chemical Physics, 77(8), 4156–4163. [Google Scholar]
- Kleuss C, Raw AS, Lee E, Sprang SR, & Gilman AG (1994). Mechanism of GTP hydrolysis by G-protein alpha subunits. Proc Natl Acad Sci U S A, 91(21), 9828–9831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lambert NA (2008). Dissociation of heterotrimeric g proteins in cells. Sci Signal, 1(25), re5. [DOI] [PubMed] [Google Scholar]
- Lee E, Taussig R, & Gilman AG (1992). The G226A mutant of Gs alpha highlights the requirement for dissociation of G protein subunits. J Biol Chem, 267(2), 1212–1218. [PubMed] [Google Scholar]
- Li B, Makino S.-i., Beebe ET, Urano D, Aceti DJ, Misenheimer TM, et al. (2016). Cell-free translation and purification of Arabidopsis thaliana regulator of G signaling 1 protein. Protein Expression and Purification, 126, 33–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li B, Tunc-Ozdemir M, Urano D, Jia H, Werth EG, Mowrey DD, et al. (2018). Tyrosine phosphorylation switching of a G protein substrate. J. Biol Chem, 293(13), 4752–4766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang X, Ding P, Lian K, Wang J, Ma M, Li L, et al. (2016). Arabidopsis heterotrimeric G proteins regulate immunity by directly coupling to the FLS2 receptor. [JOUR]. eLife, 5, e13568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang X, Ma M, Zhou Z, Wang J, Yang X, Rao S, et al. (2018). Ligand-triggered de-repression of Arabidopsis heterotrimeric G proteins coupled to immune receptor kinases. Cell Research, 28(5), 529–543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang Y, Gao Y, & Jones AM (2017). Extra large G-Protein interactome reveals multiple stress response function and partner-dependent XLG subcellular localization. [Original Research]. Frontiers in Plant Science, 8(1015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malinski JA, Zera EM, Angleson JK, & Wensel TG (1996). High affinity interactions of GTPγS with the heterotrimeric G Protein, transducin: Evidence at high and low protein concentrations J Biol Chem, 271(22), 12919–12924. [DOI] [PubMed] [Google Scholar]
- Maruta N, Trusov Y, Brenya E, Parekh U, & Botella JR (2015). Membrane-localized extra-large G proteins and Gβγ of the heterotrimeric G proteins form functional complexes engaged in plant immunity in Arabidopsis. Plant Physiology, 167(3), 1004–1016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGibbon Robert T., Beauchamp Kyle A., Harrigan Matthew P., Klein C, Swails Jason M., Hernández Carlos X., et al. (2015). MDTraj: A modern open library for the analysis of molecular dynamics trajectories. Biophysical Journal, 109(8), 1528–1532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer R, & Wagner KG (1985). Analysis of the nucleotide pool during growth of suspension cultured cells of Nicotiana tabacum by high performance liquid chromatography. Physiologia Plantarum, 65(4), 439–445. [Google Scholar]
- Mobley DL, & Gilson MK (2017). Predicting binding free energies: Frontiers and Benchmarks. Annu Rev Biophys, 46, 531–558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noel JP, Hamm HE, & Sigler PB (1993). The 2.2 A crystal structure of transducin-alpha complexed with GTP gamma S. Nature, 366(6456), 654–663. [DOI] [PubMed] [Google Scholar]
- Parrinello M, & Rahman A (1981). Polymorphic Transitions in Single-Crystals - a New Molecular-Dynamics Method. Journal of Applied Physics, 52(12), 7182–7190. [Google Scholar]
- Posner BA, Mixon MB, Wall MA, Sprang SR, & Gilman AG (1998). The A326S mutant of Gialpha1 as an approximation of the receptor-bound state. J Biol Chem, 273(34), 21752–21758. [DOI] [PubMed] [Google Scholar]
- Raw AS, Coleman DE, Gilman AG, & Sprang SR (1997). Structural and biochemical characterization of the GTPgammaS-, GDP.Pi-, and GDP-bound forms of a GTPase-deficient Gly42 --> Val mutant of Gialpha1. Biochemistry, 36(50), 15660–15669. [DOI] [PubMed] [Google Scholar]
- Saraste M, Sibbald PR, & Wittinghofer A (1990). The P-loop--a common motif in ATP- and GTP-binding proteins. Trends Biochem Sci, 15(11), 430–434. [DOI] [PubMed] [Google Scholar]
- Seeburg PH, Colby WW, Capon DJ, Goeddel DV, & Levinson AD (1984). Biological properties of human c-Ha-ras1 genes mutated at codon 12. Nature, 312(5989), 71–75. [DOI] [PubMed] [Google Scholar]
- Shao J, Tanner S, Thompson N, & Cheatham T (2007. ). Clustering molecular dynamics trajectories: 1. Characterizing the performance of different clustering algorithms. J Chem Theory Comput., 3(6), 2312–2334. [DOI] [PubMed] [Google Scholar]
- Sprang S (1997). G proteins, effectors and GAPs: structure and mechanism. Current Opinion in Structural Biology, 7(6), 849–856. [DOI] [PubMed] [Google Scholar]
- Sprang SR (1997). G protein mechanisms: insights from structural analysis. Annu Rev Biochem, 66, 639–678. [DOI] [PubMed] [Google Scholar]
- Sprang SR (2016). Invited review: Activation of G proteins by GTP and the mechanism of Galpha-catalyzed GTP hydrolysis. Biopolymers, 105(8), 449–462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Temple BR, Jones CD, & Jones AM (2010). Evolution of a signaling nexus constrained by protein interfaces and conformational States. PLoS Comput Biol, 6(10), e1000962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thung L, Trusov Y, Chakravorty D, & Botella J (2012). Gγ1 + Gγ2 + Gγ3 = Gβ: The search for heterotrimeric G-protein γ subunits in Arabidopsis is over. J. Plant Physiol, 169, 542. [DOI] [PubMed] [Google Scholar]
- Ullah H, Chen J-G, Temple B, Boyes D, & Alonso J (2003). The β subunit of the Arabidopsis G protein negatively regulates auxin-induced cell division and affects multiple developmental processes. Plant Cell, 15, 393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urano D, Chen J-G, Botella JR, & Jones AM (2013). Heterotrimeric G protein signalling in the plant kingdom. Open Biology, 3(3). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urano D, & Jones AM (2014). Heterotrimeric G protein–coupled signaling in plants. Annual Review of Plant Biology, 65(1), 365–384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urano D, Maruta N, Trusov Y, Stoian R, Liang Y, Jaiswal D, et al. (2016). Saltatory evolution of the heterotrimeric G protein signaling mechanisms in the plant kingdom. Science Signaling, 9(446), ra93. [DOI] [PubMed] [Google Scholar]
- Urano D, Miura K, Wu Q, Iwasaki Y, Jackson D, & Jones AM (2016). Plant morphology of heterotrimeric G protein mutants. Plant and Cell Physiology, 57(3), 437–445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urano D, Phan N, Jones JC, Yang J, Huang J, Grigston J, et al. (2012). Endocytosis of the seven-transmembrane RGS1 protein activates G-protein-coupled signalling in Arabidopsis. [10.1038/ncb2568]. Nat Cell Biol, 14(10), 1079–1088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin Y, Katahira R, & Ashihara H (2014). Metabolism of purine nucleosides and bases in suspension-cultured Arabidopsis thaliana cells. Eur. Chem. Bull, 3(9), 925–934. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.