Abstract
The CRISPR-Cas9 system from Streptococcus pyogenes has been exploited as a programmable RNA-guided DNA targeting and editing platform. This evolutionary tool enables diverse genetic manipulations with unprecedented precision and ease. Cas9 is an allosteric enzyme, which is allosterically regulated in conformational activation, target recognition, and DNA cleavage. Here, we outline the underlying allosteric control over the Cas9 complex assembly and targeting specificity. We further review the strategies for mitigating intrinsic Cas9 off-target effects through allosteric modulations and the advances in engineering controllable Cas9 systems that are responsive to external allosteric signals. Future development of highly specific, tunable CRISPR-Cas9 systems through allosteric modulations would greatly benefit applications that require both conditional control and high precision.
Keywords: CRISPR-Cas9, allosteric regulation
Graphical abstract
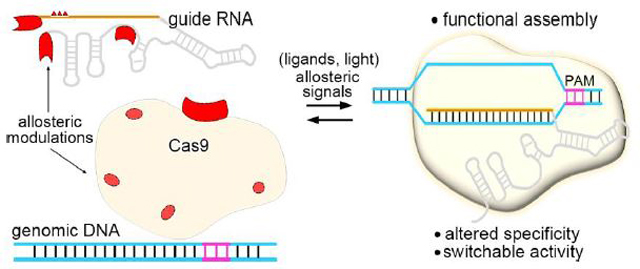
Introduction
The prokaryotic Clustered Regularly Interspaced Short Palindromic Repeats associated protein 9 (CRISPR-Cas9) system has been adapted as a powerful and versatile DNA targeting platform that enables control, analysis and engineering of genomes with an unprecedented ease [1,2].
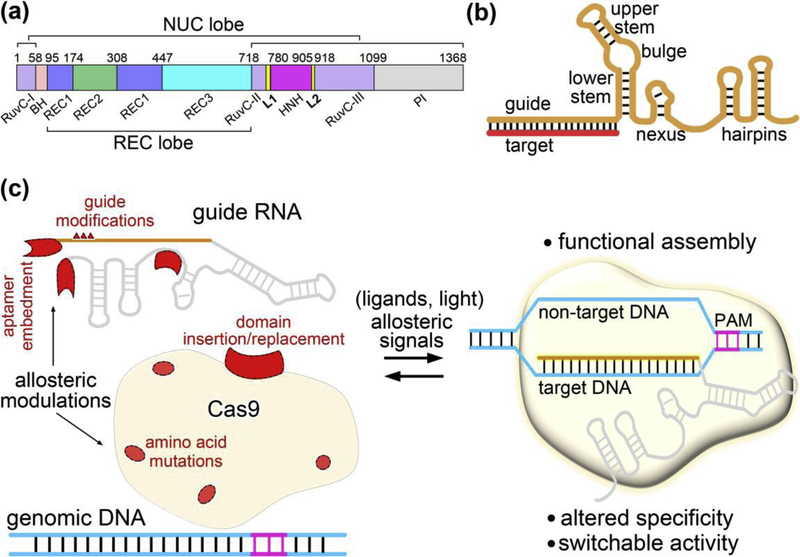
The most widely used Cas9 from Streptococcus pyogenes (SpCas9) is a dual RNA-guided, multidomain endonuclease (Figure 1a) [3,4]. Unless otherwise noted, we designate SpCas9 as Cas9 hereafter. Cas9 is complexed with a chimeric single guide RNA (sgRNA) (Figure 1b) to perform its double stranded DNA (dsDNA) targeting and cleavage function. By programming the 20-nucleotide guide sequence at the 5’ end of sgRNA, Cas9 can be targeted to any genomic site flanked by a short protospacer adjacent motif (PAM) [5–7]. The apo-Cas9 structure adopts a bi-lobed architecture and comprises an alpha-helical recognition (REC) lobe and a nuclease (NUC) lobe connected by an arginine-rich bridge helix (BH) (Figure 1a). The REC lobe is further divided into three domains (REC1, REC2 and REC3), and the NUC lobe consists of a PAM-interacting (PI) domain and two Mg2+-dependent nuclease domains (HNH and RuvC). HNH and RuvC function as scissors, cutting the target strand (that is complementary to the sgRNA 5’ end) and the nontarget strand of a dsDNA target, respectively.
Figure 1.
Allosteric regulation of CRISPR-Cas9. (a) Domain organization of Cas9. (b) Schematic representation of the Cas9 sgRNA base pairing with target DNA. The sgRNA functional modules are defined as in ref. [12]. (c) Diverse allosteric modulations leading to CRISPR-Cas9 with altered properties. Cas9 is subjected to allosteric regulation at different assembly stages following guide RNA loading and DNA binding, and its DNA targeting specificity and activity can be allosterically modulated through various RNA or protein engineering approaches. Allosteric modulators are shown in red.
Allostery is the structural and functional response at one site of a macromolecule to a perturbation at a distal site [8,9]. Cas9 is a typical allosteric enzyme, which undergoes a sequence of precise conformational rearrangements toward target recognition and cleavage [10,11]. This process involves multiple layers of exquisite allosteric regulation to ensure its functional activity and targeting precision [2,10,11].
Allostery also plays an essential role in the development of strategies to modulate Cas9 function. Beyond its utility of genome editing, the Cas9-based toolbox has been extended to transcriptional regulation, epigenetic modification, and base editing that rely on nuclease-dead Cas9 (dCas9) or Cas9 nickase [1,2]. All of these applications benefit from allosteric modulation approaches, which enhance the target specificity of Cas9 and control precision over Cas9-guide RNA function across dimensions of dose, time and space.
In this review, we first describe multilevel allosteric regulation underlying Cas9 effector complex assembly, followed by reviewing the recent advances in allosteric modulation of CRISPR-Cas9 targeting specificity and allosteric switch of CRISPR-Cas9 activity via protein and RNA engineering (Figure 1c).
Allosteric assembly of Cas9 complex
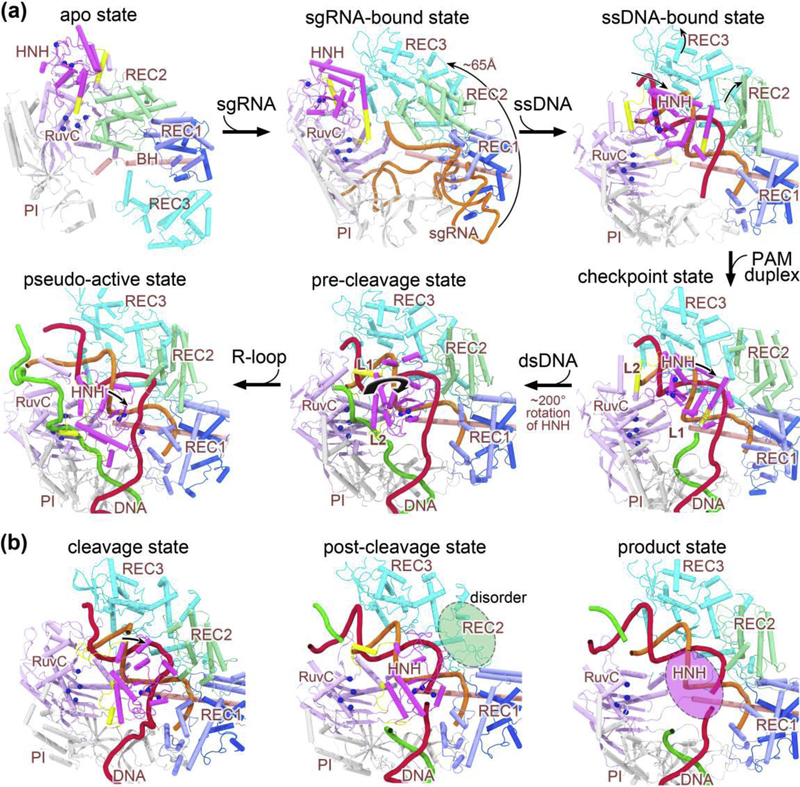
The structures of Cas9 in different bound forms have been successively reported since 2014. They represent the apo state [4], sgRNA-bound pre-targeting state [13], incomplete DNA-bound intermediate state [14,15], dsDNA-bound pre-cleavage state [16], R-loop-bound pseudo-active state [17], cleavage state [18•], post-cleavage state [19••], and product state [19••]. With these structures, we can now delineate a complete conformational transition pathway along which Cas9 achieves its nuclease activation (Figure 2). The conformational changes of Cas9 from the apo state to the pre-cleavage state have been well documented in previous reviews (Figure 2a) [10,11]. Most recently, two papers have reported the structures of Cas9 bound to DNA target in the cleavage, post-cleavage, and product states (Figure 2b) [18•,19••], thereby completing the last steps along the Cas9 reaction cycle. Importantly, both studies confirmed that the HNH domain uses the catalytic triad D839-H840-N863 for target DNA cleavage. The corresponding HNH conformation is hence referred to as the N863-IN conformation. This conformation is distinct from the most frequently observed N863-OUT conformation with N863 being oriented outside the catalytic center, which is incompatible with catalysis. The N863-OUT to N863-IN state transition was suggested to be collectively driven by the Mg2+ ions and target DNA [18•]. In the following part, we will focus on the role of allosteric effects on Cas9 conformational activation following target DNA binding.
Figure 2.
Stepwise conformational changes in Cas9 leading to cleavage activation. (a) The X-ray and cryo-EM structures of Cas9 reported from years 2014 to 2017 that highlight major conformational changes at each stage of assembly. Along the conformational activation pathway, these structures (shown clockwise) represent the apo state (PDB code: 4CMP), the sgRNA-bound pre-targeting state (PDB code: 4ZT0), the ssDNA-bound state (PDB code: 4OO8), the checkpoint state (PDB code: 4UN3), the pre-cleavage state (PDB code: 5F9R), and the pseudo-active state (PDB code: 5Y36), respectively. (b) The computational model and cryo-EM structures of Cas9 ternary complex presented in 2019 capturing the cleavage state (left), post-cleavage (middle; PDB code: 6O0Y), and product state (right; PDB code: 6O0X). The protein domains are color-coded as in Figure 1a, and the sgRNA, target DNA strand, non-target DNA strand are represented in orange, red, and green cartoon forms, respectively. Note that the split parts of REC1 are colored light and dark blue, respectively. For sgRNA, only the targeting region at its 5’ end is displayed for clarity (except 4ZT0). The alpha-carbon atoms of the active residues in the two nuclease domains (i.e. HNH and RuvC) are depicted as blue spheres. Major domain rearrangements of Cas9 are indicated with curved arrows. Note that the HNH and REC2 domains display alternate disorder transitions (highlighted with translucent ovals) in the post-cleavage and product complexes. Abbreviations: BH, bridge helix; PI, PAM-interacting domain; sgRNA, single guide RNA; ssDNA, single-stranded DNA; dsDNA, double-stranded DNA.
The conformational state of nucleic acid-bound Cas9 is primarily defined by that of the HNH domain [20–23]. Upon target DNA binding, the Cas9 HNH domain undergoes significant rigid body translation and rotation toward the cleavage state (Figure 2). A conformational checkpoint has been identified during this process, which determines whether Cas9 cuts its bound DNA target [21]. Single-molecule Förster resonance energy transfer (FRET) studies suggested that the HNH domain tends to be trapped in this checkpoint intermediate when the number of guide RNA-target DNA mismatches at the PAM-distal end exceeds a threshold [21]. After exploiting this unique allosteric control in Cas9, Chen et al. [24••] rationally designed a hyper-accurate Cas9 variant (HypaCas9) bearing amino acid mutations on the PAM-distal REC3 domain to elevate the energy barrier underlying the HNH domain reorientation. Noticeably, the Cas9 REC2 domain has been proposed to act as an allosteric transducer that mediates the signal propagation from the PAM-distal end to the HNH domain [24••,25]. Intriguingly, a recent FRET work by Sung el al. [26•] showed that the REC2 domain also allosterically destabilizes the displaced nontarget DNA strand in the Cas9/R-loop complex through the positive charges on its surface. This finding seems to be in contrast to the previous notion that the positively charged residues on Cas9 surface stabilize the unwound conformation of DNA [27], which warrants further investigation on the allosteric role of REC2 domain.
Furthermore, the activated HNH domain in turn exerts an allosteric control over the RuvC nuclease activity. It was discovered in structural studies that the two linker regions (named L1 and L2) connecting the HNH and RuvC nuclease domains undergo a remarkable folding-unfolding transition upon dsDNA binding, which is in concert with the large scale reorientation of the HNH domain [13–15,28]. These two linkers have thus been proposed to function as allosteric transducers to mediate concerted cleavage of both DNA strands [11,24], which was supported by biochemical assays [20]. Meanwhile, allosteric crosstalk between the REC lobe, HNH and RuvC domains to facilitate assembly of the Cas9 effector complex has also been well characterized through molecular dynamics (MD) simulations [25,29].
Magnesium ion (Mg2+), beyond its catalytic role, also plays an essential role in activating the Cas9 conformation via a cryptic allosteric mechanism. The FRET studies by Dagdas et al. [21] revealed that the Cas9 HNH domain is always trapped in the checkpoint intermediate and unable to undergo a reorientation docking onto the target DNA without Mg2+ ions. Upon addition of Mg2+ ion or other divalent ions that do not support cleavage, most of the Cas9 complexes populated the docking state [21]. However, it remains perplexing as to how the divalent cations facilitate such a striking structural rearrangement as well as the local conformational transition of N863-OUT to N863-IN [18•].
Allosteric modulation of Cas9 targeting specificity
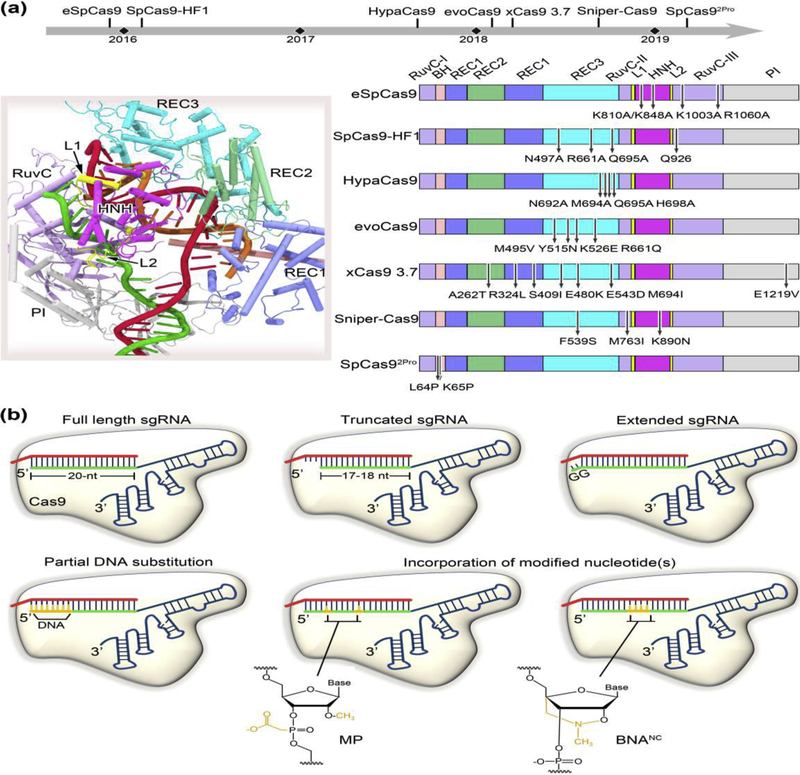
Off-target effects constitute a major bottleneck in the development of transformative therapeutic applications of CRISPR-Cas9 [30]. Extensive efforts to increase the CRISPR-Cas9 targeting specificity have resulted in the development of various high-fidelity Cas9 variants that contain amino-acid substitutions, viz., eSpCas9 [27], SpCas9-HF1 [31], HypaCas9 [24••], evoCas9 [32], Sniper-Cas9 [33], xCas9 3.7 [34], and SpCas92Pro [35•] (Figure 3a). Notably, eSpCas9, SpCas9-HF1, HypaCas9 and SpCas92Pro have been engineered through structure-guided rational approaches, while evoCas9, Sniper-Cas9 and xCas9 3.7 have been identified by directed evolution. All of these Cas9 variants involve amino-acid mutations to allosterically modulate Cas9 function for specificity improvement.
Figure 3.
Allosteric modulation of Cas9 targeting accuracy through protein or sgRNA engineering. (a) Specificity enhanced Cas9 mutants obtained through the strategies of structure-guided rational design or directed evolution. The upper panel shows the timeline of various high-fidelity Cas9 variants reported to date. The PDB structure 5F9R is used for illustration of individual Cas9 domains where the mutations are located (left panel). The mutation positions for each Cas9 variant are marked in the right panel. eSpCas9, SpCas9-HF1, HypaCas9 and SpCas92Pro have been designed by rational approaches, and evoCas9, xCas9 3.7, and Sniper-Cas9 screened through directed evolution. (b) Schematic diagram of sgRNA modifications conferring improved specificity. Most of these modifications occur at the 5’ end of the sgRNA, including truncation of two or three nucleotides, extension of two guanine nucleotides, and partial DNA replacement. The incorporation of chemically modified nucleotides, such as 2’-O-methyl-3’-phosphonoacetate (MP) and bridged nucleic acids (2’,4’-BNANC[N-Me]) at specific locations to the 5’ end or within the central region of the sgRNA guide has also been demonstrated to broadly reduce off-target cleavage.
The initial design rationale for eSpCas9 and SpCas9-HF1 is established on the “excess energy” hypothesis that the Cas9-guide RNA complex possesses more energy than is needed for its effective on-target recognition, thereby enabling cleavage at mismatched sites [27,31]. In other words, weakening the binding affinity between Cas9 and the target DNA or guide RNA could in principle improve the Cas9 specificity. However, subsequent studies discovered that the binding affinities of the two Cas9 variants for on-target and PAM-distal mismatched targets are similar to the wild type enzyme, suggesting a distinct mechanism existing for cleavage specificity improvement of these variants [24••]. A conformational proofreading mechanism was then proposed, which hypothesized that Cas9 targeting specificity could be governed through allosteric communication between the HNH domain and the PAM-distal interactions [24••]. In this mechanism, the PAM-distal region in Cas9 (such as REC3 and RuvC-III) acts as an allosteric effector that senses the RNA-DNA heteroduplex to allow for HNH domain transition to the active state. Mutations in this region could raise conformational threshold of HNH domain activation, thereby promoting off-target discrimination. This mechanism can well account for the enhanced specificity on eSpCas9, SpCas9-HF1, HypaCas9 and evoCas9, as these Cas9 mutants bear most or all of the mutations near the PAM-distal end (Figure 3a). However, the precise mode of communication from the PAM-distal region, e.g. REC3, to the HNH domain remain to be elucidated. REC2 appears to serve as a signal relay during this process, as this domain sterically permits or occludes HNH domain access to the target DNA strand scissile phosphate from the Cas9 ternary structures [13–15,18•,19••,28]. Additionally, it is worth noting that a recent cryo-EM study has uncovered a patch of positively charged residues in a RuvC loop that interact with the distal DNA duplex [19••]. This region could be an alternative target for future mutational test, as in developing eSpCas9 [27].
By contrast, xCas9 3.7, Sniper-Cas9 and SpCas92Pro modulate Cas9 DNA targeting specificity in a different fashion. Specifically, xCas9 3.7 contains seven amino acid substitutions (six in REC lobe plus one in PI domain) (Figure 3a) and achieves high editing efficiency, enhanced targeting specificity and broad PAM compatibility simultaneously [34]. A recent study by Guo et al. [36•] indicated that the substitution of E1219V determines relaxed PAM recognition, while only three of those mutations (i.e., R324L, S409I, and M694I) are important for specificity enhancement of xCas9 3.7. In xCas9 3.7, the central channel within the REC and NUC lobes opens widely at the PAM-distal end comparing to the wild type Cas9 [14], as characterized by ~10 Å displacement of the REC2 and REC3 domains away from the RNA-DNA heteroduplex. Further structural comparison suggests that the central channel expansion is allosterically induced by two remote mutations (i.e., R324L and M694I) on the REC1 domain, and might also be cooperatively facilitated by M694I-causing steric hindrance at the PAM-distal end. Nevertheless, this reduction in interactions between REC3 and the RNA-DNA heteroduplex at the PAM-distal end is in line with that observed for the above-mentioned Cas9 mutants, leading to a similar mechanism of conformational checkpoint [21,24••]. Interestingly, Sniper-Cas9 involves three amino acid substitutions (F539S, M763I and K890N), which are located on the REC3, RuvC and HNH domains, respectively (Figure 3a), while SpCas92Pro was derived by two proline substitutions (L64P/K65P) on the bridge helix of Cas9. Yet all of these residues are not in direct contacts with the bound nucleic acids. It remains elusive regarding how Sniper-Cas9 and SpCas92Pro allosterically modulate target DNA cleavage and mismatch tolerance. Future structural and computational studies are necessary to help to decipher the underlying mechanisms.
Cas9 specificity can also been improved through guide RNA modifications (Figure 3b). Interestingly, most of these modifications appear at or near the guide RNA 5’ end (i.e., the PAM-distal end), including truncation of two to three nucleotides [37], extension of two extra guanine nucleotides [38], and partial DNA replacement [39]. The other guide RNA variants have unnatural chemical modifications (i.e. 2’-O-methyl-3’-phosphonoacetate and bridged nucleic acids) incorporated at specific sites near to the 5’ end or within the central part of the guide RNA targeting region [40,41]. Obviously, these diverse modifications can interfere with the RNA-DNA heteroduplex formation at or near the PAM-distal end. As a result, Cas9 cleavage activity is more sensitive to mismatched DNA targets due to increased threshold crossing the conformational checkpoint, as detailed above.
Allosteric switch of Cas9 activity
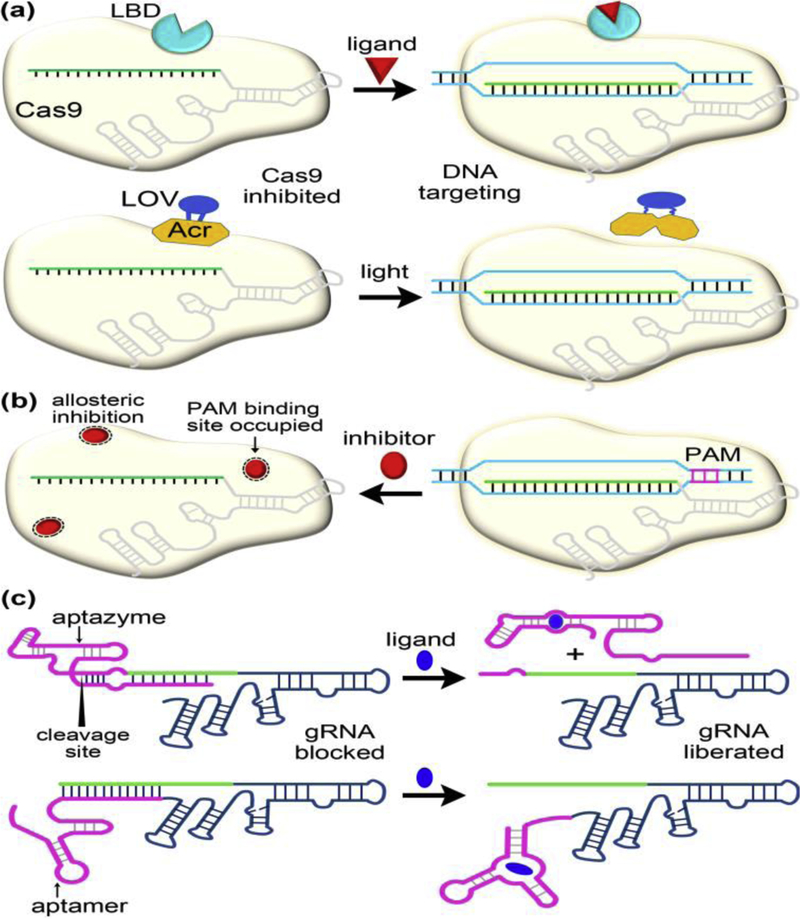
Recent years have seen broad interest in developing strategies for conditional control of CRIPSR-Cas9-based tools across multiple dimensions, including dose, time, space, and orthogonal modulation of different genes [42–45, 46••]. The conditional control is generally achieved at the post-translational level by modifying either the Cas9 protein itself or its programmable guide RNA molecules (Figure 4a,4c). The activities of these engineered Cas9-sgRNA variants are often subject to allosteric switch by external stimuli (such as small molecules and light).
Figure 4.
Selected case studies demonstrating allosteric control over Cas9-guide RNA function through protein or guide RNA engineering (a,c) or through direct modulation by small molecule inhibitors (b). These engineered systems enable conditional regulation of genome editing and transcription. (a) Insertion of a ligand binding domain from human estrogen receptor to Cas9 or insertion of a light-oxygen-voltage photosensor domain to AcrIIA4 (an anti-CRIPSR protein inhibiting Cas9) leads to switchable Cas9 systems by small molecule ligands (e.g. 4-hydroxytamoxifen) or light. (b) Cas9-DNA binding disrupted by small molecule inhibitors (e.g. the newly identified BRD0539). (c) Extending sgRNA to include small molecule-responsive aptazymes or aptamers imparts allosteric switch in Cas9 by regulating the accessibility of the sgRNA guide region to target DNA. The aptazymes or aptamers are modified to append an antisense sequence that base pairs with the sgRNA guide region in the absence of small molecules. Addition of a cognate ligand triggers ribozyme self-cleavage or riboswitch conformational change, promoting the release of the blocking sequence (i.e. antisense strand) from the guide RNA. Abbreviations: LBD, ligand binding domain; LOV, light-oxygen-voltage; Acr, anti-CRIPSR protein.
The approaches of domain insertion, fusion and replacement have been popularly employed to engineer small-molecule-controlled Cas9 systems. Using domain insertion profiling, Oakes et al. [47] identified a specific insertion site in the Cas9 REC2 domain and constructed an allosterically regulated Cas9 (arCas9) by embedding the ligand binding domain of human estrogen receptor-α (ER-LBD) (Figure 4a, upper panel). The function of this Cas9 construct is activated in the presence of 4-hydroxytamoxifen (4-HT). The underlying mechanism of activation is unclear. The ER-LBD incorporation likely causes steric block of the RNA-DNA binding channel within Cas9, and the 4-HT binding could induce REC2 and ER-LBD movements that open the channel. Another study [48] strategically developed a chemical inducible Cas9 variant (named ciCas9) as an intramolecular autoinhibitory switch by replacing the original REC2 domain with the BCL-xL protein and fusing an interacting BH3 peptide. The intramolecular interaction between BCL-xL and BH3 locks Cas9 in an inactive state. Disruption of the BCL-xL-BH3 interaction by addition of the small molecule A-385358 (A3) results in release of autoinhibition and allosteric activation of Cas9. ciCas9 serves as a rapidly inducible single-component system displaying temporal and dose-dependent control of nuclease activity with lower background and off-target activities. Notably, its on-target specificity can be further improved by introduction of the previously identified mutations for eSpCas9.
Another avenue for allosteric switch of Cas9 is leveraging phage-derived anti-CRISPR (Acr) proteins. Anti-CRISPR proteins are naturally occurring off-switches for CRISPR-Cas immunity [49,50]. However, Bubeck et al. [51•] inserted the LOV2 photosensor from Avena sativa to AcrIIA4, a potent SpCas9 inhibitor and generated an ArcIIA4 variant that enables optogenetic control of Cas9 activity (Figure 4a, lower panel). The N and C termini of LOV2 are in close proximity in the dark, thereby preserving the AcrIIA4 inhibitory conformation after LOV2 insertion. Light-induced unfolding of the LOV2 terminal helices, however, perturbs the AcrIIA4 structure, which results in allosteric unleashing of Cas9 ribonucleoprotein. Moreover, Bubeck et al. rationally designed several Acr-LOV smutants that exhibited enhanced Cas9 inhibition in the dark while retaining light activation. The Acr-LOV hybrid and its optimized mutants are also able to exert strong light regulation on xCas9.
A general issue with the protein engineering approaches is that all target genes are regulated in the same manner and the Cas9 activity does not depend on the cell state or exogenous ligand, limiting the therapeutic application of Cas9. As the function of Cas9 is directly governed by its sgRNA, a complementary strategy is to develop controllable sgRNAs that are responsive to different ligands. Such class of sgRNAs have the potential to facilitate differential regulation of multiple gene targets in multiplexing scenarios. Two groups [52,53] have first reported various ligand-responsive sgRNA variants by appending a modified aptamer and a modified aptazyme to the sgRNA at its 5’ and 3’ ends, respectively (Figure 4c). The sgRNA targeting region combines with a complementary, blocking sequence in the extended RNAs without ligands, which prevents Cas9 DNA targeting. Upon addition of cognate ligands, the aptamers undergo a conformational change, and the aptazymes undergo self-cleavage to promote the release of the blocking sequence that allosterically restores Cas9-guide RNA function. The aptazyme-integrated sgRNAs are irreversible for Cas9 regulation.
Most recently, Kundert et al. [54••] discovered an additional route for allosteric control of Cas9 activity using sgRNA. By structure-guided insertion of the aptamer into one of the sgRNA functional modules, i.e., the upper stem, nexus, and hairpin (Figure 1b), Kundert et al. obtained both ligand-activated and ligand-deactivated sgRNAs (termed ligRNA+ and ligRNA-, respectively). For the mechanism of ligRNA+, it has been suggested that ligand binding leads to strand displacements that stabilize the functional sgRNA conformation [54]. By contrast, it remains enigmatic regarding the mechanism underlying the ability of ligRNA- to deactivate Cas9 function in the presence of a ligand. A possible explanation is that ligand binding to the aptamer would disrupt the key interactions of R77 in Cas9 with U59 in ligRNA- as shown in the crystal structure of Cas9 ternary complex [15]. Nonetheless, the design of ligRNAs enables new applications that require differential and opposing temporal control of multiple genes.
In contrast to the variety of the protein and RNA engineering approaches for Cas9 regulation, a much-less explored strategy is to identify small molecule modulators that act directly on Cas9. In a latest work [46••], the researchers have successfully identified a potent inhibitor (BRD0539) blocking Cas9-DNA binding through a well-established high-throughput screening platform. In view of its structural and chemical properties, BRD0539 appears not to directly compete with the canonical NGG PAM for binding the PAM-interacting arginine residues. Rather, this inhibitor likely hinders the DNA-bound state formation via an unknown allosteric mechanism (Figure 4b), which warrants further structural studies.
Conclusions
The series of Cas9 structures along the reaction pathway have suggested the key roles that allostery plays in Cas9 complex assembly and Cas9 targeting specificity improvement. Other Cas9 “snapshots”, especially those between the checkpoint intermediate and the cleavage conformation, are likely to provide additional mechanistic insights into how the PAM-distal RNA-DNA base-pairing allosterically regulates the conformational dynamics of the Cas9 HNH domain [55,56]. The allosteric modulations have aided in engineering of more efficient Cas9 variants with reduced off-target effects. Allosteric switches are designed to improve the precise control of Cas9 function over multiple dimensions. We anticipate the further elucidated allosteric mechanism of Cas9 and the development of next-generation CRISPR-Cas9 tools through allosteric modulations in the near future.
Highlights.
Cas9 conformational activation are subjected to allosteric regulation.
Cas9 specificity can be allosterically enhanced by protein and RNA engineering.
Cas9 activity can be conditionally tuned through diverse allosteric switches.
Acknowledgements
This work was supported by Shanghai Municipal Education Commission under the program for Professor of Special Appointment at Shanghai Institutions of Higher Learning to Z. Z.; National Heart, Lung, and Blood Institute of the National Institutes of Health under award number R15HL147265 to J.L. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
Footnotes
Conflict of interest statement
The authors declare no conflict of interest.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References and recommended reading
Papers of particular interest, published within the period of review, have been highlighted as:
• of special interest
•• of outstanding interest
References
- 1.Knott GJ, Doudna JA: CRISPR-Cas guides the future of genetic engineering. Science 2018, 361:866–869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wilkinson RA, Martin C, Nemudryi AA, Wiedenheft B: CRISPR RNA-guided autonomous delivery of Cas9. Nat Struct Mol Biol 2019, 26:14–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Jinek M, Chylinski K, Fonfara I, Hauer M, Doudna JA, Charpentier E: A Programmable Dual-RNA-Guided DNA Endonuclease in Adaptive Bacterial Immunity. Science 2012, 337:816–821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Jinek M, Jiang F, Taylor DW, Sternberg SH, Kaya E, Ma E, Anders C, Hauer M, Zhou K, Lin S, et al. : Structures of Cas9 endonucleases reveal RNA-mediated conformational activation. Science 2014, 343:1247997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mali P, Yang LH, Esvelt KM, Aach J, Guell M, DiCarlo JE, Norville JE, Church GM: RNA-Guided Human Genome Engineering via Cas9. Science 2013, 339:823–826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cong L, Ran FA, Cox D, Lin SL, Barretto R, Habib N, Hsu PD, Wu XB, Jiang WY, Marraffini LA, et al. : Multiplex Genome Engineering Using CRISPR/Cas Systems. Science 2013, 339:819–823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Charpentier E, Doudna JA: Biotechnology: Rewriting a genome. Nature 2013, 495:50–51. [DOI] [PubMed] [Google Scholar]
- 8.Hayatshahi HS, Ahuactzin E, Tao P, Wang S, Liu J: Probing protein allostery as a residue-specific concept via residue response maps. J Chem Inf Model 2019, 10.1021/acs.jcim.9b00447. [DOI] [PubMed] [Google Scholar]
- 9.Greener JG, Sternberg MJ: Structure-based prediction of protein allostery. Curr Opin Struct Biol 2018, 50:1–8. [DOI] [PubMed] [Google Scholar]
- 10.Jiang F, Doudna JA: CRISPR-Cas9 Structures and Mechanisms. Annu Rev Biophys 2017, 46:505–529. [DOI] [PubMed] [Google Scholar]
- 11.Chen JS, Doudna JA: The chemistry of Cas9 and its CRISPR colleagues. Nat Rev Chem 2017, 1:0078. [Google Scholar]
- 12.Briner AE, Donohoue PD, Gomaa AA, Selle K, Slorach EM, Nye CH, Haurwitz RE, Beisel CL, May AP, Barrangou R: Guide RNA functional modules direct Cas9 activity and orthogonality. Mol Cell 2014, 56:333–339. [DOI] [PubMed] [Google Scholar]
- 13.Jiang F, Zhou K, Ma L, Gressel S, Doudna JA: A Cas9-guide RNA complex preorganized for target DNA recognition. Science 2015, 348:1477–1481. [DOI] [PubMed] [Google Scholar]
- 14.Anders C, Niewoehner O, Duerst A, Jinek M: Structural basis of PAM-dependent target DNA recognition by the Cas9 endonuclease. Nature 2014, 513:569–573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Nishimasu H, Ran FA, Hsu PD, Konermann S, Shehata SI, Dohmae N, Ishitani R, Zhang F, Nureki O: Crystal structure of Cas9 in complex with guide RNA and target DNA. Cell 2014, 156:935–949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Anders C, Bargsten K, Jinek M: Structural Plasticity of PAM Recognition by Engineered Variants of the RNA-Guided Endonuclease Cas9. Mol Cell 2016, 61:895902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Huai C, Li G, Yao RJ, Zhang Y, Cao M, Kong LL, Jia CQ, Yuan H, Chen HY, Lu DR, et al. : Structural insights into DNA cleavage activation of CRISPR-Cas9 system. Nature Communications 2017, 8:1375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zuo Z, Zolekar A, Babu K, Lin VJ, Hayatshahi HS, Rajan R, Wang YC, Liu J: Structural and functional insights into the bona fide catalytic state of Streptococcus pyogenes Cas9 HNH nuclease domain. Elife 2019, 8:e46500.[•] Using computational simulations and functional assays, this study reported two distinct conformations adopted by the Cas9 HNH domain. The HNH domain cleavage conformation was suggested to be collectively promoted by interactions with DNA substrate and Mg2+ ion.
- 19.Zhu X, Clarke R, Puppala AK, Chittori S, Merk A, Merrill BJ, Simonovic M, Subramaniam S: Cryo-EM structures reveal coordinated domain motions that govern DNA cleavage by Cas9. Nat Struct Mol Biol 2019, 26:679–685.[••] Three cryo-EM structures of Cas9-sgRNA-DNA complex along the last reaction steps were dertermined at a resolution of ~3.3 Å. The first structure captured the Cas9 in a state akin to the previously reported checkpoint conformation. The last two structures represent the postcatalytic and product sates, respectivley. The new interactions identified from these structures may aid in rational enginnering of new Cas9 variants with improved accuracy and efficiency through allosteric modulations.
- 20.Sternberg SH, LaFrance B, Kaplan M, Doudna JA: Conformational control of DNA target cleavage by CRISPR-Cas9. Nature 2015, 527:110–113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Dagdas YS, Chen JS, Sternberg SH, Doudna JA, Yildiz A: A Conformational Checkpoint Between DNA Binding And Cleavage By CRISPR-Cas9. Sci Adv 2017, 3:eaao0027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Osuka S, Isomura K, Kajimoto S, Komori T, Nishimasu H, Shima T, Nureki O, Uemura S: Real-time observation of flexible domain movements in CRISPR-Cas9. EMBO J 2018, 37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yang MY, Peng SJ, Sun RR, Lin JD, Wang N, Chen CL: The Conformational Dynamics of Cas9 Governing DNA Cleavage Are Revealed by Single-Molecule FRET. Cell Rep 2018, 22:372–382. [DOI] [PubMed] [Google Scholar]
- 24.Chen JS, Dagdas YS, Kleinstiver BP, Welch MM, Sousa AA, Harrington LB, Sternberg SH, Joung JK, Yildiz A, Doudna JA: Enhanced proofreading governs CRISPR-Cas9 targeting accuracy. Nature 2017, 550:407–410.[••] This work first discovered that the non-catalytic REC3 domain of Cas9 serves as a sensor of target complementarity to allosterically govern the conformational state of the HNH domain. Exploiting this allosteric regulation, Chen et al. rationally designed a new hyper-accurate Cas9 variant (HypaCas9) with alanine substitutions on the PAM-distal REC3 domain that raises the energy threshold for HNH domain reorientation.
- 25.Palermo G, Chen JS, Ricci CG, Rivalta I, Jinek M, Batista VS, Doudna JA, McCammon JA: Key role of the REC lobe during CRISPR–Cas9 activation by ‘sensing’, ‘regulating’, and ‘locking’ the catalytic HNH domain. Quart Rev Biophys 2018, 51:e9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sung K, Park J, Kim Y, Lee NK, Kim SK: Target Specificity of Cas9 Nuclease via DNA Rearrangement Regulated by the REC2 Domain. J Am Chem Soc 2018, 140:77787781.[•] Using single-molecule FRET approach, Sung et al. demonstrated that the positive residues on the REC2 domain of Cas9 have a role in allosterically destablizing the unwound conformation of the non-target DNA strand, thereby contributing to Cas9 target specificity. This finding offers a new route to minimizing Cas9 off-target effects.
- 27.Slaymaker IM, Gao L, Zetsche B, Scott DA, Yan WX, Zhang F: Rationally engineered Cas9 nucleases with improved specificity. Science 2016, 351:84–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jiang FG, Taylor DW, Chen JS, Kornfeld JE, Zhou KH, Thompson AJ, Nogales E, Doudna JA: Structures of a CRISPR-Cas9 R-loop complex primed for DNA cleavage. Science 2016, 351:867–871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Palermo G, Ricci CG, Fernando A, Basak R, Jinek M, Rivalta I, Batista VS, McCammon JA: Protospacer Adjacent Motif-Induced Allostery Activates CRISPR-Cas9. J Am Chem Soc 2017, 139:16028–16031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kim D, Luk K, Wolfe SA, Kim JS: Evaluating and Enhancing Target Specificity of Gene-Editing Nucleases and Deaminases. Annu Rev Biochem 2019, 88:191–220. [DOI] [PubMed] [Google Scholar]
- 31.Kleinstiver BP, Pattanayak V, Prew MS, Tsai SQ, Nguyen NT, Zheng Z, Joung JK: High-fidelity CRISPR-Cas9 nucleases with no detectable genome-wide off-target effects. Nature 2016, 529:490–495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Casini A, Olivieri M, Petris G, Montagna C, Reginato G, Maule G, Lorenzin F, Prandi D, Romanel A, Demichelis F, et al. : A highly specific SpCas9 variant is identified by in vivo screening in yeast. Nat Biotechnol 2018, 36:265–271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lee JK, Jeong E, Lee J, Jung M, Shin E, Kim YH, Lee K, Jung I, Kim D, Kim S, et al. : Directed evolution of CRISPR-Cas9 to increase its specificity. Nat Commun 2018, 9:3048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hu JH, Miller SM, Geurts MH, Tang W, Chen L, Sun N, Zeina CM, Gao X, Rees HA, Lin Z, et al. : Evolved Cas9 variants with broad PAM compatibility and high DNA specificity. Nature 2018, 556:57–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Babu K, Amrani N, Jiang W, Yogesha SD, Nguyen R, Qin PZ, Rajan R: Bridge Helix of Cas9 Modulates Target DNA Cleavage and Mismatch Tolerance. Biochemistry 2019, 58:1905–1917.[•] This work showed that two proline substitutions within the arginine-rich bridge helix of Cas9 impart a higher selectivity in DNA targeting, which is in contrast with previous practices targeting large Cas9 domains for specificity modulation. It was proposed that the substitutions may work by affecting the positioning of the endonuclease sites and the allosteric communication between these sites. Further studies are needed to decipher the underlying mechanism.
- 36.Guo M, Ren K, Zhu Y, Tang Z, Wang Y, Zhang B, Huang Z: Structural insights into a high fidelity variant of SpCas9. Cell Res 2019, 29:183–192.[•] A highly specific Cas9 varaint (xCas9 3.7) with broad PAM compatibility has been reported by Hu et al. in 2008 (see ref. 34). Guo et al. subsequently determined the crystal structures of xCas9 3.7 with two DNA targets bearing different, non-canonical PAMs. This study provides structural basis for enhanced specificity and relaxed PAM requirement of xCas9 3.7. Notably, the solved structures indicate that the amino acid mutations at the REC1 and REC2 interface of Cas9 could allosterically result in reduced interactions of the REC3 and REC2 domains with the RNA-DNA hybrid by broadening the central channel within Cas9 at the PAM-distal end. This observation is reminiscent of the mechanism of specificity improvement on SpCas9-HF1 and HypaCas9.
- 37.Fu Y, Sander JD, Reyon D, Cascio VM, Joung JK: Improving CRISPR-Cas nuclease specificity using truncated guide RNAs. Nat Biotechnol 2014, 32:279–284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kim D, Kim S, Kim S, Park J, Kim JS: Genome-wide target specificities of CRISPR-Cas9 nucleases revealed by multiplex Digenome-seq. Genome Res 2016, 26:406–415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Yin H, Song CQ, Suresh S, Kwan SY, Wu Q, Walsh S, Ding J, Bogorad RL, Zhu LJ, Wolfe SA, et al. : Partial DNA-guided Cas9 enables genome editing with reduced off-target activity. Nat Chem Biol 2018, 14:311–316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ryan DE, Taussig D, Steinfeld I, Phadnis SM, Lunstad BD, Singh M, Vuong X, Okochi KD, McCaffrey R, Olesiak M, et al. : Improving CRISPR-Cas specificity with chemical modifications in single-guide RNAs. Nucleic Acids Res 2018, 46:792–803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Cromwell CR, Sung K, Park J, Krysler AR, Jovel J, Kim SK, Hubbard BP: Incorporation of bridged nucleic acids into CRISPR RNAs improves Cas9 endonuclease specificity. Nat Commun 2018, 9:1448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Mills EM, Barlow VL, Luk LYP, Tsai YH: Applying switchable Cas9 variants to in vivo gene editing for therapeutic applications. Cell Biol Toxicol 2019, 10.1007/s10565-019-09488-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Gangopadhyay SA, Cox KJ, Manna D, Lim D, Maji B, Zhou Q, Choudhary A: Precision Control of CRISPR-Cas9 Using Small Molecules and Light. Biochemistry 2019, 58:234–244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Richter F, Fonfara I, Gelfert R, Nack J, Charpentier E, Moglich A: Switchable Cas9. Curr Opin Biotechnol 2017, 48:119–126. [DOI] [PubMed] [Google Scholar]
- 45.Nunez JK, Harrington LB, Doudna JA: Chemical and Biophysical Modulation of Cas9 for Tunable Genome Engineering. ACS Chem Biol 2016, 11:681–688. [DOI] [PubMed] [Google Scholar]
- 46.Maji B, Gangopadhyay SA, Lee M, Shi M, Wu P, Heler R, Mok B, Lim D, Siriwardena SU, Paul B, et al. : A High-Throughput Platform to Identify Small-Molecule Inhibitors of CRISPR-Cas9. Cell 2019, 177:1067–1079 e1019.[••] Maji et al. developed a suite of high-throughput assays that enables screening of small-molecule inhibitors of CRISPR-Cas9. Specifically, a potent Cas9 inhibitor (BRD0539) has been identified that is cell-permeable, non-toxic, and stable under physiological conditions. Presumably this compound disrupts the formation of Cas9 DNA-bound state by directly competing with the PAM for binding to the PAM-recognizing arginine residues or via an allosteric mode. This study represents the first development of synthetic small-molecule inhibitors of CRISPR-Cas9.
- 47.Oakes BL, Nadler DC, Flamholz A, Fellmann C, Staahl BT, Doudna JA, Savage DF: Profiling of engineering hotspots identifies an allosteric CRISPR-Cas9 switch. Nat Biotechnol 2016, 34:646–651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Rose JC, Stephany JJ, Valente WJ, Trevillian BM, Dang HV, Bielas JH, Maly DJ, Fowler DM: Rapidly inducible Cas9 and DSB-ddPCR to probe editing kinetics. Nat Methods 2017, 14:891–896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Pawluk A, Amrani N, Zhang Y, Garcia B, Hidalgo-Reyes Y, Lee J, Edraki A, Shah M, Sontheimer EJ, Maxwell KL, et al. : Naturally Occurring Off-Switches for CRISPR-Cas9. Cell 2016, 167:1829–1838 e1829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Hwang S, Maxwell KL: Meet the Anti-CRISPRs: Widespread Protein Inhibitors of CRISPR-Cas Systems. CRISPR J 2019, 2:23–30. [DOI] [PubMed] [Google Scholar]
- 51.Bubeck F, Hoffmann MD, Harteveld Z, Aschenbrenner S, Bietz A, Waldhauer MC, Borner K, Fakhiri J, Schmelas C, Dietz L, et al. : Engineered anti-CRISPR proteins for optogenetic control of CRISPR-Cas9. Nat Methods 2018, 15:924–927.[•] AcrIIA4 is an anti-CRISPR (Acr) protein that potently hampers SpCas9 DNA targeting function. Bubeck et al. strategically inserted a photosensor domain (LOV2) into to a solvent-exposed loop in AcrIIA4 and obtained several AcrIIA4 variants that enable optogenetic control of CRISPR-Cas9. In the absence of light, these AcrIIA4 variants retain most of the inhibitory activity for Cas9. Light-induced unfolding of LOV2 perburbs the fold of AcrIIA4 and inhibits its anti-CRISPR function, which allosterically liberates Cas9-guide RNA. This study contribute a novel avenue for controlling CRISPR-Cas9 without modifying the Cas9 protein itself or its programmable RNA.
- 52.Tang W, Hu JH, Liu DR: Aptazyme-embedded guide RNAs enable ligand-responsive genome editing and transcriptional activation. Nat Commun 2017, 8:15939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Liu Y, Zhan Y, Chen Z, He A, Li J, Wu H, Liu L, Zhuang C, Lin J, Guo X, et al. : Directing cellular information flow via CRISPR signal conductors. Nat Methods 2016, 13:938944. [DOI] [PubMed] [Google Scholar]
- 54.Kundert K, Lucas JE, Watters KE, Fellmann C, Ng AH, Heineike BM, Fitzsimmons CM, Oakes BL, Qu J, Prasad N, et al. : Controlling CRISPR-Cas9 with ligand-activated and ligand-deactivated sgRNAs. Nat Commun 2019, 10:2127.[••] This study reported the development of ligand-responsive sgRNA variants through insertion of RNA aptamers. In contrast to prior sgRNA-based designs, this approach can be used to allosterically activate and deactivate CRISPR-Cas9 function in response to a small molecule. Since the system acts directly on each target-specific sgRNA, it enables differential and opposing temporal control of multiple genes.
- 55.Zuo Z, Liu J: Structure and Dynamics of Cas9 HNH Domain Catalytic State. Sci Rep 2017, 7:17271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Taylor DW: The final cut: Cas9 editing. Nat Struct Mol Biol 2019, 26:669–670. [DOI] [PubMed] [Google Scholar]