Abstract
Fusion technology is widely used in protein-drug development to increase activity, stability, and bioavailability of protein therapeutics. Fusion proteins, like any other type of biopharmaceuticals, need to remain stable during production and storage. Due to the high complexity and additional intramolecular interactions, it is not possible to predict the behavior of fusion proteins based on the behavior the individual proteins. Therefore, understanding the stability of fusion proteins on the molecular level is crucial for the development of biopharmaceuticals. The current study on the albumin-neprilysin (HSA-NEP) fusion protein uses a combination of thermal and chemical unfolding with small angle X-ray scattering and molecular dynamics simulations to show a correlation between decreasing stability and increasing repulsive interactions, which is unusual for most biopharmaceuticals. It is also seen that HSA-NEP is not fully flexible: it is present in both compact and extended conformations. Additionally, the volume fraction of each conformation changes with pH. Finally, the presence of NaCl and arginine increases stability at pH 6.5, but decreases stability at pH 5.0.
Subject terms: Molecular modelling, SAXS, High-throughput screening
Introduction
Stability and efficacy of protein-drugs are essential to achieve desirable biopharmaceutical applications. One of the strategies in optimization of protein-therapeutics is fusion technology, which consists of linking a target protein to a more stable protein. This approach has shown to improve catalytic efficiency, activity, stability, and solubility of protein-drugs1. Moreover, the fusion-protein approach is used to prevent fast renal clearance by connecting a target protein to a protein with a longer half-life. Human serum albumin (HSA), which is used in this study, and immunoglobulin Gs are proteins widely used in fusion technology due to their long half-life.
HSA is a highly abundant and well-studied serum protein with a half-life around 19–22 days2. Currently, multiple albumin fusion proteins are under clinical trials, and two are already accepted by the Food and Drug Administration (FDA)3. One of them is albiglutide: an albumin fusion protein connected to a glucagon like peptide-1 receptor agonist4. This therapeutic is administered for treatment of type 2 diabetes. The second album fusion drug is albutrepenonacog alfa, linked to the recombinant coagulation factor IX, which is used for treatment of hemophilia B5. In both therapeutics, the presence of HSA contributes to a significant increase in half-life: from 1.5–5 min6 up to 3.6–8 days for albiglutide, and from 17–34 h up to 92 h for albutrepenonacog alfa5,7.
As mentioned above, fusion technology can be used to address a variety of problems in protein-drug development. However, as any other type of biopharmaceutical, fusion proteins require special conditions (formulation) that will preserve their stability during production and storage. As it is not yet possible to predict the behavior of different proteins under different conditions, formulation remains a long and expensive process in protein-drug production. Formulation is particularly challenging for fusion proteins7, as additional intramolecular interactions lead to a change in stability, and therefore it is hard to predict the behavior of fusion proteins based on stability of individual proteins.
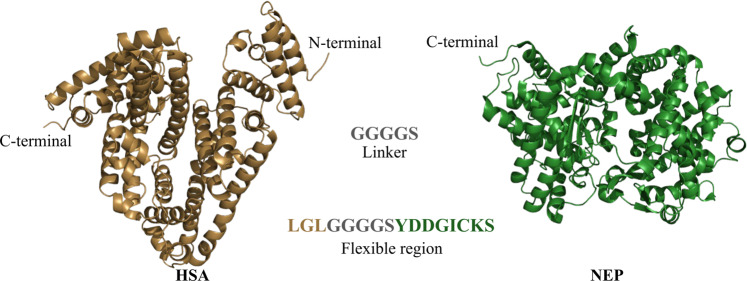
The aim of this study is to provide a better understanding on fusion protein stability and relate it to conformational changes. Specifically, we are investigating the stability of albumin fused to neprilisyn (HSA-NEP) (see Fig. 1). NEP is widely distributed in mammalian tissues and is involved in the inactivation of a variety of signaling peptides8,9. Additionally, it is involved in the degradation of amyloid β peptides, which makes it an attractive candidate as a protein-drug for treatment of Alzheimer’s disease10.
Figure 1.
HSA N-terminus fused with the NEP C-terminus via GGGGS linker (figure created using PyMOL25).
HSA-NEP was studied together with other proteins in PIPPI library11, and it was clear that the behavior of this protein was different from other flexible proteins, such as monoclonal antibodies.
In this study, the overall stability of HSA-NEP was investigated by thermal and chemical denaturation by varying pH and buffer composition, complemented by small angle X-ray scattering (SAXS) and molecular dynamics (MD) simulations. In combination, the stability studies and SAXS show a correlation between increasing protein repulsion and decreasing conformational stability. This contradicts conclusions from a previous study on a similar fusion protein: albumin fused with human growth hormone (HSA-hGH)12, where repulsion is concluded to have a stabilizing effect at certain conditions. Additionally, combining SAXS results with MD simulation results provided a molecular understanding of the determinants that cause the HSA-NEP conformational changes.
Results
The overall stability of HSA-NEP was analyzed by thermal and chemical denaturation using nano differential scanning fluorimetry (nanoDSF) and isothermal chemical denaturation (ICD). Initially, ICD was performed using two different denaturants: urea and guanidine hydrochloride (GuHCl). GuHCl is a strong denaturant that starts to unfold HSA-NEP at low concentrations. Therefore, urea was used for subsequent studies, in order to obtain well-defined denaturation curves. The initial analysis was performed as a function of pH (5–9) and NaCl concentration (0, 70, and 140 mM), reaching ionic strength of 150 mM. In this way, pH and ionic strength range covered most of the formulation conditions of biopharmaceuticals.
pH dependence
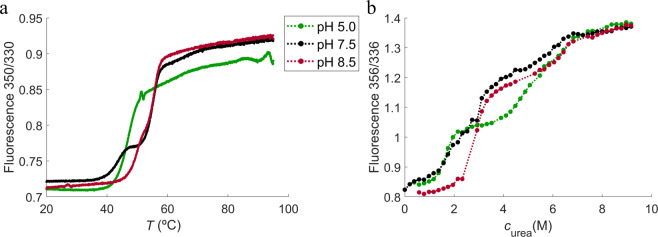
The denaturation curves from thermal unfolding using nanoDSF are shown in Fig. 2a. HSA-NEP has a single two-state unfolding (from folded to unfolded state) at pH 5.0, which is shifted towards a three-state unfolding with increasing pH (with the presence of the intermediate state). However, from pH 7.5 to pH 8.5 the thermal unfolding shifts back to a two-state (see Fig. 2a). Chemical denaturation results in a multi-state unfolding (with two intermediate states), which is shifted towards a three-state unfolding with increasing pH (see Fig. 2b). This means that the first intermediate state has a clear plateau at pH 5.0, less clear at pH 7.5, and not apparent at pH 8.5. The second intermediate state is not well-defined at pH 5.0, but more pronounced at pH 7.5 and 8.5. Hence, thermal and chemical denaturation of HSA-NEP show different unfolding mechanisms. It is difficult to speculate why, but one of the possible explanations is that the presence of urea in chemical denaturation inhibits aggregation, while thermal denaturation does not.
Figure 2.
NanoDSF (a) and ICD (b) curves at 10 mM histidine pH 5.0 (green), 10 mM histidine pH 7.5 (black), and 10 mM tris pH 8.5 (red).
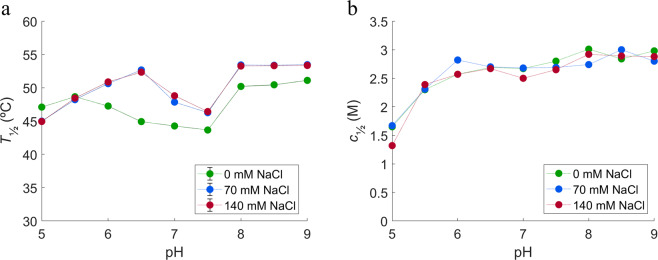
The temperature of unfolding (T½) and the denaturant needed to unfold 50% of the protein (c½) are shown in the Fig. 3. At 0 mM NaCl, T½ decreases from pH 5.5 to 7.5, with the higher values at lower (pH 5.0 and 5.5) and higher pH (from pH 8.0 to pH 9.0) (see Fig. 3a). The c½ increases from pH 5.0 to pH 6.0, where it reaches a plateau. It increases again around pH 8.0 (see Fig. 3b).
Figure 3.
Initial stability studies performed using (a) NanoDSF and (b) ICD at different pH and ionic strengths. In green: 0 mM NaCl, in blue: 70 mM NaCl, and in red: 140 mM NaCl.
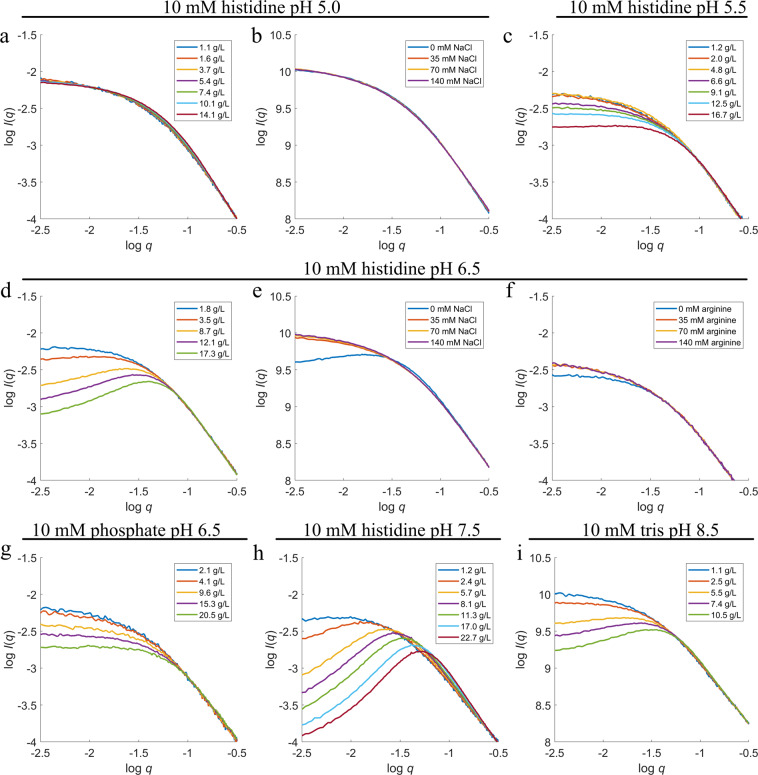
In order to study the associated conformational changes, SAXS concentration series data were collected at 10 mM histidine at pH 5.0, 5.5, 6.5, and 7.5, and 10 mM tris at pH 8.5 with 0 mM NaCl. All scattering curves and SAXS data analysis can be seen in supplementary information (SI) (see Table S.2 and Figure S.1). The intensity at low q-values decreases with increasing HSA-NEP concentration at all pH values (except pH 5.0) indicating a repulsive system (see Fig. 4). Moreover, repulsive interactions increase from pH 5.5 to 7.5 and decrease from pH 7.5 to 8.5, which correlates with the observed changes in T½ (see Fig. 3a).
Figure 4.
SAXS scattering curves for concentration series at (a) 10 mM histidine pH 5.0; (c) 10 mM histidine pH 5.5; (d) 10 mM histidine pH 6.5; (g) 10 mM phosphate pH 6.5; (h) 10 mM histidine pH 7.5; (i) 10 mM tris pH 8.5. SAXS scattering curves varying cNaCl at (b) 10 mM histidine pH 5.0 and (e) 10 mM histidine pH 6.5, with cHSA-NEP around 5.5–6 g/L; and varying carginine at (f) 10 mM histidine pH 6.5 with cHSA-NEP around 2 g/L.
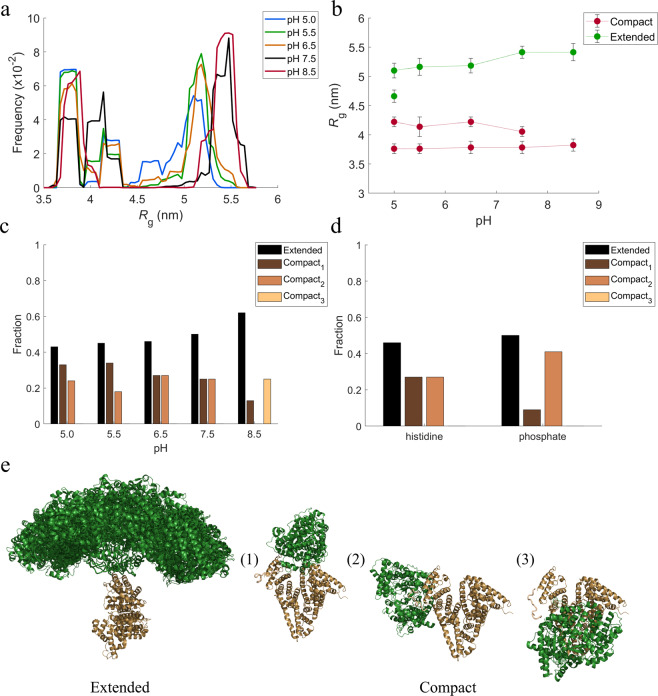
The Kratky plots of HSA-NEP show increase of the scattering at higher angles, which is characteristic for flexible systems13 (see Fig. S.2 in SI). Therefore, conformational changes of HSA-NEP were studied using Ensemble Optimization Method (EOM)14,15, which accounts for flexibility. This means that the relative position of HSA and NEP was allowed to vary under the constrains of the linker. HSA (PDB ID: 6EZQ16) and NEP (PDB ID: 6GID17) crystal structures were used as template structures for HSA and NEP. The EOM results were analyzed by looking at the resulting high-resolution structures, their probability (volume fractions) and radius of gyration (Rg) distribution. At all the investigated pH values, the Rg distributions shown in Fig. 5a,b, indicate the presence of two overall populations: a more compact conformation with Rg around 4 nm and a more extended conformation with Rg around 5 nm.
Figure 5.
Analysis from EOM. (a,b) Rg distributions, (c) volume fractions of different conformations in 10 mM histidine at pH 5.0, 5.5, 6.5, and 7.5 and 10 mM tris pH 8.5, (d) volume fractions of different conformations at pH 6.5 in histidine and phosphate buffers. (e) high resolution models for extended and compact conformations (figures created using PyMOL25).
The Rg distribution around 4 nm shows multiple peaks, indicating the presence of multiple compact conformations. The detailed SAXS analysis of the high-resolution models shows the presence of three different compact conformations (see Fig. 5e), whose distribution is pH dependent (see Fig. 5c).
The volume fraction of the extended conformation, as well as Rg, increase with increasing pH. The analysis of the high-resolution models shows many different possible conformations, which is an indication of high flexibility of the extended conformation. This is illustrated in Fig. 5e, where HSA is kept fixed, and the position of NEP is seen to vary amongst the suggested structures.
NaCl dependence
With the addition of NaCl, the thermal denaturation studies show an increase in T½ above pH 5.5 which points to an increase in thermal stability (see Fig. 3a), while ICD studies do not show a clear trend for NaCl effect. The SAXS experiments were performed in the presence of 0, 35, 70, and 140 mM NaCl at 10 mM histidine pH 5.0 and 6.5 (see Fig. 4b,e). At pH 5.0, the scattering curves do not change with the presence of NaCl. However, at pH 6.5 the intensity at low q-values increases with addition of NaCl and calculated molecular weight (MW) shifts from 136 to 150 kDa (see Table S.3 in SI), which is closer to the real MW of HSA-NEP. These results show that addition of NaCl screens the repulsive interactions present at pH 6.5, 0 mM NaCl.
Buffer and excipients dependence
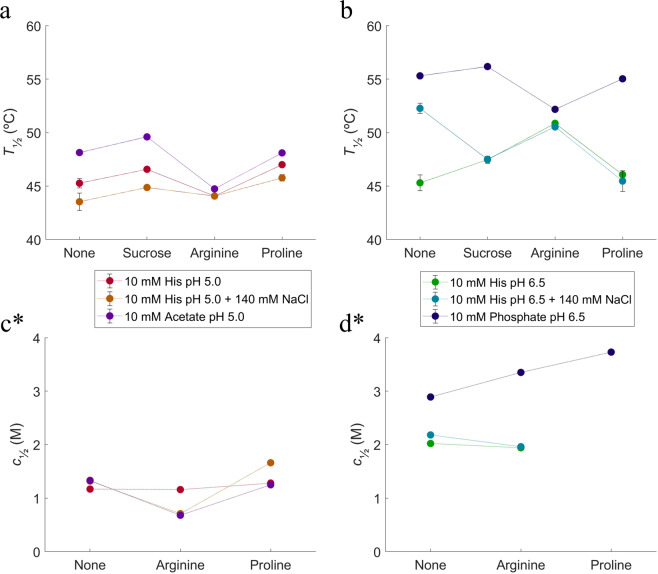
Relative to the histidine buffer, both the acetate and phosphate buffers give rise to higher T½ and c½ (see Fig. 6), which points to a higher conformational stability of the protein. Sucrose, arginine, and proline were selected as excipients and tested in the different buffers.
Figure 6.
NanoDSF and ICD stability studies using different buffers and excipients. (a) changes in T½ at histidine (0 and 140 mM NaCl) and acetate pH 5.0; (b) changes in T½ at histidine (0 and 140 mM NaCl) and phosphate pH 6.5; (c) changes in c½ at histidine (0 and 140 mM NaCl) and acetate pH 5.0; (d) changes in c½ at histidine (0 and 140 mM NaCl) and phosphate pH 6.5. Purple: 10 mM acetate pH 5.0, red: 10 mM histidine pH 5.0, orange: 10 mM histidine pH 5.0 with 140 mM NaCl, blue: 10 mM phosphate pH 6.5, green: 10 mM histidine pH 6.5, cyan: 10 mM histidine pH 6.5 with 140 mM NaCl. *It was not possible to perform ICD studies at pH 5.0 with sucrose and pH 6.5 with sucrose and proline, due to the crystallization of solution in high concentrations of urea.
At pH 5.0, both sucrose and proline have a weak positive effect on the HSA-NEP thermal stability, while the effect of arginine is negative and more pronounced. In acetate buffer, the presence of arginine is destabilizing, as it causes a decrease in T½ and c½ (see Fig. 6a,c, and Table 1). In histidine pH 5.0, arginine does not have a significant effect on the thermal stability, but the ICD studies show a decrease in c½ by around 1 M in the presence of NaCl.
Table 1.
Overview of the effect of excipients deduced from nanoDSF (T½) and ICD (c½) data analyses. Suc: sucrose, Arg: arginine-HCl, Pro: proline.
| 10 mM acetate pH 5.0 | 10 mM histidine pH 5.0 | ||||||||
|---|---|---|---|---|---|---|---|---|---|
| +140 mM NaCl | |||||||||
| Suc | Arg | Pro | Suc | Arg | Pro | Suc | Arg | Pro | |
| T½ | + | −− | 0 | + | − | + | + | + | ++ |
| c½ | + | −− | 0 | 0 | 0 | 0 | x | −− | + |
| 10 mM phosphate pH 6.5 | 10 mM histidine pH 6.5 | ||||||||
| +140 mM NaCl | |||||||||
| Suc | Arg | Pro | Suc | Arg | Pro | Suc | Arg | Pro | |
| T½ | + | −− | − | ++ | +++ | + | −− | − | −−− |
| c½ | x | + | ++ | x | 0 | x | x | − | X |
++++/−−−− ΔT½ > 10 °C Δc½ > 1.5 M.
+++/−−−5 °C < ΔT½ < 10 °C 1 M < Δc½ < 1.5 M.
++/−−2 °C < ΔT½ < 5 °C 0.5 M < Δc½ < 1.0 M.
+/− 0.5 °C < ΔT½ < 2 °C 0.2 M < Δc½ < 0.5 M.
0 ΔT½ < 0.5 °C Δc½ < 0.2 M.
Reference point for excipients: respective buffer without excipient.
+ stabilizes.
− destabilizes.
x – data not acquired.
The phosphate buffer at pH 6.5 was selected for SAXS measurements, as it has a clear positive effect in both ICD and nanoDSF experiments (see Fig. 6b,d and Table 1). Like in the histidine buffer, the intensity curves are decreasing at low q-values, with increasing cHSA-NEP (see Fig. 4d,g), which points to the presence of repulsive interactions. However, this decrease is less pronounced in phosphate buffer, which means that the system is less repulsive. At ~3 g/L, the calculated MWHSA-NEP is lower in phosphate buffer (166 kDa) than in histidine buffer (180 kDa) (see Table S.3 in SI), which points to a lower amount of larger species/aggregates in phosphate buffer. Moreover, the volume fractions of the compact conformations differs significantly: in histidine buffer both conformations, compact1 and compact2, are present in equal amounts (0.27), while in phosphate buffer, the amount of compact1 has decreased to 0.13 and the amount of compact2 has increased to 0.41 (see Fig. 5d).
At pH 6.5, the three excipients affect the system differently. Sucrose is slightly stabilizing in phosphate and histidine buffers, but in combination with NaCl it has a destabilizing effect, decreasing T½ by 4.8 °C (see Fig. 6b and Table 1). Proline has some stabilizing effect in histidine buffer, but in combination with NaCl it decreases T½ by 6.8 °C, which points to a decrease in stability. In phosphate buffer, both proline and arginine have a different effect in thermal and chemical denaturation studies: they have negative effect on thermal stability, but seem to protect HSA-NEP from chemical denaturation (see Fig. 6b,d, and Table 1). In histidine buffer at pH 6.5 arginine increases T½ by 5.4 °C, which means an increase in HSA-NEP stability. Additionally, SAXS data show a decrease in the intermolecular repulsion (see Fig. 4f). Addition of arginine in combination with NaCl has no effect.
Molecular dynamics simulations
In order to understand conformational changes of HSA-NEP under different physicochemical conditions, the interface between HSA and NEP in extended and compact conformations were studied using MD simulations, using high-resolution models based on SAXS data generated by EOM. Subsequently, changes in the electrostatic surface potentials at the protein-protein interface were investigated to understand conformational preference with pH.
Extended
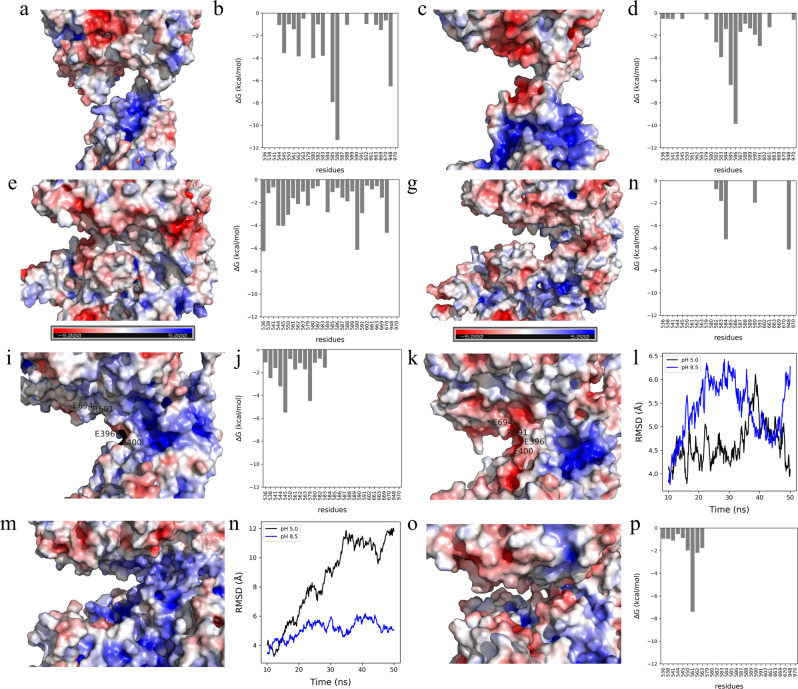
The electrostatic surface and interaction energy for inter-domain interface of the extended conformation are shown in Fig. 7a–d. The electrostatic surface at pH 5.0 shows less prominent positive and negative patches when compared to pH 8.5 (see Fig. 7a,c). Furthermore, the free energy of interaction at the interface region is similar as they do not share a large interface in extended conformation. Due to the neutral patches around the interface, HSA and NEP tend to be closer in space at pH 5.0 (Rg of 45.97 ± 0.22 Å). Contrary to this, both proteins remain far from each other at pH 8.5 (Rg of 50.20 ± 0.73 Å), due to repulsive negative charge-charge interactions.
Figure 7.
Surface coloring of the representative structure from MD trajectory clustering based on the electrostatic potentials at pH 5.0 for (a) extended conformation, (e) compact1, (i) compact2, and (m) compact3; at pH 8.5 for (c) extended conformation, (g) compact1, (k) compact2, and (o) compact3 (color-scale: red, white, and blue indicates negative, close to neutral, and positive potentials. Figures were created using PyMOL25). HSA is located in the bottom and NEP in the top. Interaction energy at pH 5.0 for (b) extended conformation, (f) compact1, and (j) compact2; at pH 8.5 for (d) extended conformation, (h) compact1 and (p) compact3. RMSD plot for (l) compact2, and (n) compact3 at pH 5.0 (in black) and 8.5 (in blue).
Compact1
The electrostatic surface and interaction energy for compact1 inter-domain interface are shown in Fig. 7e–h. With increasing pH, residues at the interface have a lower contribution to the free energy of interaction (see Fig. 7f,h). This is in agreement with the analysis of intermolecular interactions, which points to the presence of three salt bridges (E505-R665, D563-K665, and K564-D659) and two hydrogen bonds (R114-R633 and N503-Q610) at pH 5.0, and only one salt bridge (E565-K665) at pH 8.5. This change in the interactions from pH 5.0 to 8.5 leads to increase in Rg from 37.58 (±0.23 Å) to 39.67 (±0.48 Å) resulting in less compact conformation.
Compact2
The electrostatic surface and interaction energy for compact2 inter-domain interface are shown in Fig. 7i–k. In the compact2 conformation, the electrostatics change with pH around the interface region, which include residues E694, R691, D1044, E396, and E400 (see Fig. 7i,k). With increasing pH, the interface region becomes more hydrophilic, resulting in negative charge-charge repulsion. Moreover, solvent accessible surface area (SASA) plots as a function of simulation time shows that compact2 has increasing interface SASA with increasing pH (data not shown). Additionally, the root mean square deviation (RMSD) shows that compact2 is unstable at pH 8.5 (see Fig. 7l).
Compact3
The electrostatic surface and interaction energy for compact3 inter-domain interface are shown in Fig. 7m–p. At pH 5.0, compact3 conformation has a strong positive patch in the interface region, which makes HSA and NEP prone to repulsive behavior (see Fig. 7m). With increasing pH, the interface becomes neutral (see Fig. 7o), which makes compact3 conformation more favorable at pH 8.5. This is in agreement with analysis of intermolecular interactions: at pH 5.0 there are no interactions predicted, while at pH 8.5 they are three hydrogen bonds (K475-N634, D494-Q610, and E495-Q610). Additionally, SASA of the interface increases (100 Å) with decreasing pH and the RMSD shows that compact3 is unstable at pH 5.0 (see Fig. 7n). The overall interface energy at pH 8.5 favors compact3 in tris buffer, decreasing from −17.8 to −24.0 kCal/mol.
Denaturation process
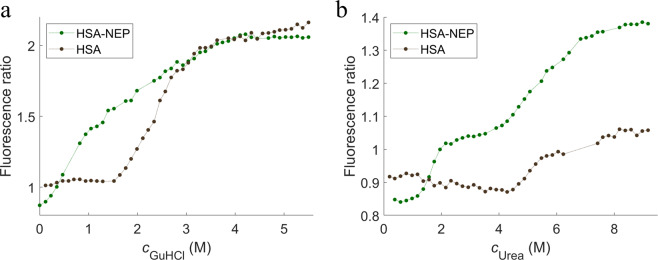
Both urea and GuHCl were used as denaturants in the chemical denaturation studies, and both show a multi-state unfolding (see Fig. 8a,b). However, they point to different unfolding mechanisms: in the presence of urea, the first two transitions are well separated with well-defined intermediate states, which is not the case with GuHCl. Unlike HSA-NEP, HSA alone has a simple two-state unfolding mechanism and requires higher concentrations of urea (~5 M) and GuHCl (~2.3 M) to unfold. It is seen in Fig. 8b that HSA alone starts to unfold with addition of around 4 M urea, which corresponds to the beginning of the second transition of HSA-NEP. This suggests that in the presence of urea, the NEP domain unfolds first.
Figure 8.
Chemical unfolding curves for HSA-NEP (in green) and HSA (in black) in histidine at pH 5.0 with (a) GuHCl and (b) urea.
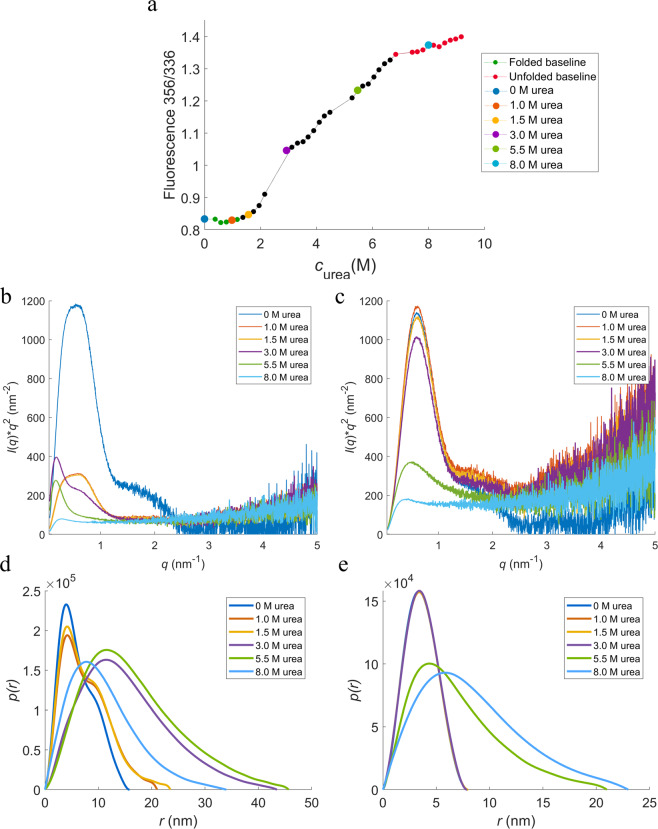
In order to confirm the previous statement and follow conformational changes during unfolding, SAXS experiments were performed at different concentrations of urea in 10 mM histidine at pH 5.5, where the transitions are more well-defined (see Fig. 9a,b). Without denaturant, HSA-NEP has a Kratky plot with a well-defined maximum and pair distribution (p(r)) function with a double peak, which is characteristic for multidomain proteins. In the presence of 1 and 1.5 M of urea, the shape of the peak changes, pointing to small conformational changes. By increasing the concentration up to 3 M, the shape of the peak shifts to a single peak, which indicates significant conformational changes. The maximum intensity in the Kratky plot decreases with increasing concentration of urea, due to decreasing contrast. Finally, in the presence of 8 M urea, the Kratky plot has a plateau instead of a peak, meaning that HSA-NEP is fully unfolded.
Figure 9.
HSA-NEP chemical denaturation study. (a) HSA-NEP (1 g/L) chemical unfolding curve. Kratky plots for (b) HSA-NEP (5 g/L) at different concentrations of urea, and for (c) HSA (5 g/L) at different concentrations of urea. p(r) functions for (d) HSA-NEP (5 g/L) at different concentrations of urea, and for (e) HSA (5 g/L) at different concentrations of urea. Urea concentrations are given in the insets.
The same experiment was performed with HSA alone (see Fig. 9c). With addition of 1, 1.5, and 3 M urea, the shape of the peak in p(r) function remains the same. By adding 5.5 M urea the peak becomes broader and the intensity increases at high q-values, which is characteristic for partially unfolded proteins. At the maximum concentration of urea, the Kratky plot of HSA (as well as HSA-NEP) has a plateau instead of a peak, which is characteristic of a fully unfolded protein.
Discussion
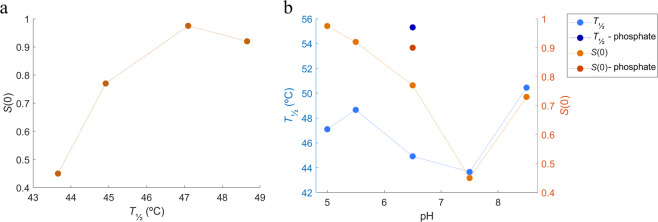
From the nanoDSF and SAXS data analyses it is clear that there seem to be a correlation between T½ and repulsive interactions in the system (see Fig. 10a,b). An increase in repulsion, seen as a decrease in the structure factor, S(0), in the SAXS data, is followed by a decrease in T½, which could be indicative of a destabilizing effect of repulsive interactions. The correlation seems to be dependent also on the choice of buffer, as seen in Fig. 10b. This finding contradicts the findings of Cordes et al.12, where repulsive interactions in 10 mM acetate at pH 5.0 show a positive effect on the HSA-hGH fusion protein stability12. Due to the similar pKa values for hGH and NEP (5.12 for hGH18 and 5.47 for NEP), similar non-specific protein-protein interactions would be expected, however the specific interactions must be responsible for these differences.
Figure 10.
Correlation between thermal stability and repulsive interactions with pH. (a) correlation between S0 and T½, (b) correlation between structure factor and T½ in histidine buffer (pH 5.0–7.5), phosphate buffer (pH 6.5), and tris buffer (pH 8.5). Thermal stability represented as T½ (in blue); and repulsive interactions represented as structure factors at cHSA-NEP around 10 g/L (in orange).
The increase in repulsion seen in histidine buffer from pH 5.5 to 7.5 can be explained by the pI values of the individual proteins. HSA has pI of 5.67 and NEP has pI of 5.47, which means that both proteins are negatively charged at pH> 6. Therefore, increasing pH leads to an increase in repulsive interactions, which is also in agreement with MD simulations (see Fig. 7). The increase in repulsion between the artificially connected HSA and NEP proteins could induce internal stress, which, according to the SAXS data, is followed by an increased volume fraction of the extended conformation. One possible scenario is that in order to minimize the internal repulsive interactions in the protein, both domains keep as far away from each other as possible, which leads to an increase in Rg (see Fig. 5a,b). This results in increased flexibility and that HSA-NEP is more easily unfolded. Additionally, from two possible conformations, HSA-NEP will prefer the extended conformation, where HSA and NEP are more separated in space (see Fig. 5c).
Furthermore, it is seen that at pH 6.5, where the protein clearly shows repulsive interactions and low thermal stability, the addition of NaCl to HSA-NEP increases T½ (see Fig. 3a). At the same time, SAXS data indicates that NaCl screens the repulsive interactions (see Fig. 4e), leading to increase in stability.
NanoDSF and ICD studies show that not only pH but also the buffer type influences HSA-NEP stability. Changing histidine buffer for phosphate buffer at pH 6.5 increases both T½ and c½ (see Fig. 6b,d), decreases repulsion (see Fig. 4e,g), and changes the volume fractions of compact1 and compact2 (see Fig. 5d). One of the reasons might be the significantly higher ionic strength in 10 mM phosphate buffer at pH 6.5 (0.013 M vs 0.005 M for 10 mM histidine buffer), which contributes to the screening of repulsive interactions. The changes in volume factions for the compact conformations might happen due to specific interactions between the phosphate buffer and HSA-NEP. At pH 5.0, HSA and NEP do not have significant intramolecular interactions in histidine buffer (see Fig. 4a). However, replacing it by acetate buffer leads to an increase in T½ and c½, which also might happen due to specific interaction between acetate and HSA-NEP.
Among all tested additives, arginine has a pronounced effect on the HSA-NEP stability. At pH 5.0, it generally destabilizes, for both chemical and thermal denaturation, with an exception in the thermal denaturation in histidine with 140 mM NaCl. In general, the effect of addition of arginine seems to be comparable to the addition of NaCl for thermal denaturation. However, the chemical denaturation curves indicate that arginine may have another role in this system: helping unfolding at pH 5.0 and hampering unfolding at pH 6.5 in phosphate. Proline seems to have a clear stabilizing effect at pH 6.5 in phosphate buffer (chemical denaturation), and a clear destabilizing effect in histidine buffer +140 mM NaCl (thermal denaturation).
As already mentioned, HSA-NEP is present in different compact conformations: compact1, compact2, and compact3 (see Fig. 5e). For better understanding of conformational changes, the interface between HSA and NEP was studied by MD simulations. Molecular understanding about the preference of the different conformation in varying pH was explored combining the electrostatics surface study and free energy of interaction at the HSA-NEP interface.
The SAXS data shows that up to pH 7.5 only the compact1 and compact2 conformations are present. According to the analysis of the MD simulations, compact3 is not present below pH 7.5 due to highly unfavorable repulsive interactions in the interface region (see Fig. 7m). Moreover, EOM analysis shows that up to pH 7.5, the volume fraction of compact1 decreases, while the volume fraction for extended conformations increases (see Fig. 5c). Analysis of the MD simulations shows that compact1 becomes less favorable due to increasing positive charge-charge repulsion and decreasing interaction energy between HSA and NEP.
Finally, SAXS data shows that at pH 8.5 HSA-NEP is present in compact1, compact3, and extended conformations. According to MD simulation results, tris as a buffering system enhances the stability of compact3, binding in the interface making it more a favorable conformation along with the extended conformations (see Fig. 5c). From MD simulations at pH 8.5, compact1 has less pronounced charge-charge repulsion when compared to compact2, which means that compact1 is more likely to be present at pH 8.5.
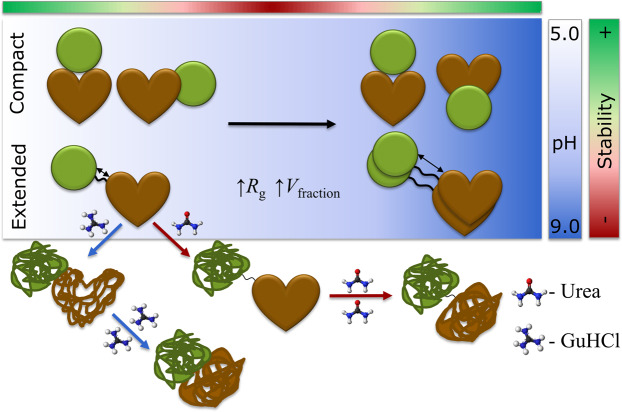
In combination, ICD and SAXS provide better understanding of the HSA-NEP unfolding process. HSA-NEP unfolding starts with NEP unfolding, which is followed by unfolding of HSA. Additionally, the GuHCl and urea denaturation curves point to different unfolding mechanisms (see Fig. 8). In the presence of urea, the first two transitions are well separated by well-defined intermediate state, which suggests presence of a stable intermediate, where HSA is folded and NEP is unfolded. In the presence of GuHCl, HSA-NEP also has multi-state unfolding, but the transitions between different states are not well-defined. This suggests that with GuHCl, unfolding of NEP might affect the integrity of HSA, which leads to the absence of a well-defined intermediate state (see Fig. 11).
Figure 11.
Schematic overview of HSA-NEP stability.
Moreover, multiple transitions in ICD curves (see Fig. 9) can also be explained by the presence of different HSA-NEP conformations. Compact conformations have multiple interactions at the interface, which might stabilize NEP. However, in the extended conformation, the NEP domain is more exposed, resulting in a faster unfolding.
Conclusion
From this study, we show how to explain behavior and stability of fusion proteins in general based on biophysical and molecular characterization. Our study shows that we cannot rely only on the knowledge we already have on the individual proteins. While the HSA-NEP protein shows increasing stability with decreasing repulsion, HSA alone shows the opposite19, as do most stable proteins. Furthermore, it is also not even possible to generalize the stability of albumin fusion proteins, as the MD simulation results clearly show the importance of specific interactions with the tris molecule indicating that for each system consisting of both protein and buffer a separate analysis is needed.
Unlike most proteins, HSA-NEP is less stable with increasing repulsion, due to intradomain repulsion. Moreover, interactions between HSA and NEP do not allow for full flexibility: HSA-NEP is present in an extended and multiple compact conformations. The compact conformations are mainly stabilized by salt bridges in the interface between HSA and NEP and are therefore more rigid, while the extended conformation is more flexible. Changes in pH induce changes at the interface, which shifts the equilibrium between different compact and extended conformations.
Methods
Dialysis and formulation
Dialysis and formulation procedure was performed according to the protocol described in Kulakova et al.20 HSA-NEP was provided by AstraZeneca in 49 g/L solution and was dialyzed for pH/NaCl and buffer/excipients screening. After dialysis, HSA-NEP was diluted to 20 g/L with dialysis buffer, followed by 1:20 dilution with final formulation buffer within pH of ±0.520. The concentration was determined with a NanodropTM 1000 (Thermo Fisher Scientific, Waltham, USA) (see Table S.1 in supplementary information (SI)).
Isothermal chemical denaturation
Chemical denaturation studies was on Unchained Labs HUNK system - AVIA ICD 2304 (Unchained Labs, Pleasanton, USA) according to the protocol described in Kulakova et al.20 using 1134 min of additional incubation time. Both urea and guanidine hydrochloride (GuHCl) were used as denaturants for pH/NaCl screening, while urea was selected for the buffer/excipients. Data analysis was performed using Formulator software v3.02 (Unchained Labs, Pleasanton, USA https://www.unchainedlabs.com/hunky/). For the native protein, the emission wavelength was selected from the data of the first point of the gradient, which corresponded to 336 nm. For the unfolded state, data of the last point of the gradient was considered and the maximum wavelength was 356 nm. The ratio 356/336 was plotted against denaturant concentration to monitor the unfolding process. The denaturation curve pointed to the presence of a multi-state unfolding process, therefore, both 3- and 4-state models were used for the data fitting. Additionally, secondary fits were performed for each NaCl concentration combining different pH. Free energy of unfolding (ΔGunfold), c½, and m-values were calculated for both transitions. HSA-NEP stability was analyzed by monitoring the change in c½ from the first transition, where the protein starts to unfold.
Thermal denaturation
Thermal stability studies were performed in triplicate with the Prometheus NT.48 (NanoTemper Technologies, Munich, Germany) according to the protocol described in Kulakova et al.20. HSA-NEP stability was analyzed by monitoring the change in T½ from the first transition. All measurements were done in triplicates and data analysis was performed using PR.Control v1.12.2 software (NanoTemper Technologies, Munich, Germany).
Small angle X-ray scattering
Data collection was performed at the P12 beamline at the Petra III storage ring (DESY, Hamburg DE)21 (see Table S.1 in SI for experimental details). Radius of gyration (Rg) and maximum dimension (Dmax) were derived from the experimental data with the graphical data analysis program PRIMUSQT22.
Ensemble Optimization Method (EOM) was used to analyze conformational polydispersity of HSA-NEP. EOM consists of two programs RANCH and GAJOE that were used separately. RANCH was used to generate a large pool of 10000 random conformations (genes) by using HSA (pdbid: 6EZQ16) and NEP (pdbid: 6GID17) from homology modelling. Flexible regions in C-terminal of HSA (LGLG) and N-terminal of NEP (YDDGICKS) were removed from the structures, increasing the length of the linker. GAJOE was used to select ensembles of conformations, such that the average structure fits to the experimental data. Experimental curves acquired at different HSA-NEP concentrations were merged in order to remove the noise. Additionally, first 80 points were removed due to the presence of repulsive interactions. The output files contain fit to experimental data, distribution of volume fractions, and information about Rg and Dmax distribution.
Molecular dynamics simulations
Initially, HSA (PDB ID: 6EZQ18) and NEP (PDB ID: 6GID19) crystal structures were aligned to the most representative structures from SAXS modelling (that are shown in Fig. 5e) to get the two crystal structures in the right orientation. This structure was taken further for homology modelling to model the linker region (GGGGS) using MODELLER9.2023 program.
Minimization was performed on the generated models to account for structure optimization. Using PDB2PQR24 plugin in PyMOL25, structures were prepared at different physicochemical conditions (pH 5, 6.5, and 8.5). Subsequently, these structures were taken for all-atom classical constant pH MD simulation of 50 ns in explicit solvent utilizing ff99SB26 force-field for proteins. In total, the solvated system contained approximately 60000 water molecules. Each system was neutralized with either sodium or chloride depending on the overall charge of the protein. The complete protocol used to setup MD simulations is described in our previous work27. HSA-NEP interaction interface is defined as follows: consider HSA, residues belonging to NEP within 5 Å of HSA’s protein surface, and vice versa is defined as interface residues for HSA-NEP interaction. The interface for each of the conformations is different, hence the interface was found separately for each conformation (see Table S.6 in SI). The titratable residues such as Asp, Glu, His, Lys, in the interface were subjected to titration during constant pH MD simulations to account for change in protonation state upon protein structure dynamics. Using the MM-GBSA28,29 free energy method, the free energy of interaction at the interface was calculated. The most representative structure found throughout the simulation using hierarchical clustering approach30,31 was taken for MM-GBSA calculations. Analyses were performed with CPPTRAJ32 in Amber 16, and VMD 1.9.333. The electrostatic surface potential was calculated for the most representative structure from the hierarchical clustering. Interface SASA was calculated by subtracting the SASA for the interface residues in HSA-NEP from the individual protein domains: HSA and NEP, respectively. All simulations were performed once for 50 ns each.
Additionally, the compact3 conformation was simulated independently in 10 mM and 150 mM tris to understand the effect of tris on the conformational stability. In total, 11 and 157 tris molecules corresponding to 10 mM and 150 mM tris were added to the solvated system containing approximately 60000 water molecules. The simulations were performed in duplicates for 50 ns starting froma random seed number to estimate the statistical uncertainty of the results. Tris was obtained from Zinc Database34. These molecules were prepared at pH 8.5 using the Ligprep tool in Schrödinger release 2016–3 (Schrödinger, LLC, New York, NY, USA)35. Parameter file for the tris molecule was prepared using the antechamber36 module in Amber 16 at pH 8.5 and applying the AM1-BCC37 charge method. Furthermore, an interaction score per residue (P(Iscore))27 was calculated to estimate the binding capacity of tris to the protein surface. The amount of tris accumulating at the HSA-NEP interaction surface was calculated by summing the number of contacts formed between the compact3 interface residues and tris molecules, further normalized by the number of tris molecules in the simulated system, which is defined as average normalized contact score.
The intermolecular interactions in the interface between HSA and NEP were analyzed for each compact conformation using NCONT from the CCP4 software38.
Supplementary information
Acknowledgements
This work was supported by European Union’s Horizon 2020 research and innovation program (grant agreement nr 675074 PIPPI). We thank the Danish Agency for Science, Technology, and Innovation for funding the instrument center DanScatt. The synchrotron SAXS data was collected at beamline P12 operated by EMBL Hamburg at the PETRA III storage ring (DESY, Hamburg, Germany). We would like to thank Stefano Da Vela for the assistance in using the beamline. AstraZeneca is acknowledged for kindly providing us with HSA-NEP fusion protein and Albumedix Ltd. for kindly providing us with Recombumin® native sequence HSA.
Author contributions
A.K. collected SAXS, nanoDSF, and ICD data and performed formal analysis on these. She wrote original draft and edited the manuscript. S.I. performed molecular dynamic simulations and wrote that part of the manuscript. P.S. contributed with data interpretation for small angle X-ray scattering. S.M. collected ICD data for HSA. W.S. supervised stability studies. G.P. supervised MD simulation studies. P.H. contributed with funding acquisition, conceptualization, and supervision. Commented and edited the manuscript.
Data availability
ICD, nanoDSF and SAXS data is deposited in PIPPI Data Base https://pippi-data.kemi.dtu.dk/ EOM models obtained from SAXS data analysis can be found in SASBDB database.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
is available for this paper at 10.1038/s41598-020-67002-9.
References
- 1.Yang H, Liu L, Xu F. The promises and challenges of fusion constructs in protein biochemistry and enzymology. Appl. Microbiol. Biotechnol. 2016;100:8273–8281. doi: 10.1007/s00253-016-7795-y. [DOI] [PubMed] [Google Scholar]
- 2.Peters, T. The Albumin Molecule in All About Albumin 9–75 (Elsevier, 1995).
- 3.Weimer, T., Metzner, H. J. & Schulte, S. Recombinant Albumin Fusion Proteins. in Fusion Protein Technologies for Biopharmaceuticals (ed. Schmidt, S.R.) 163–178 (John Wiley & Sons, Inc., 2013)
- 4.Trujillo JM, Nuffer W. Albiglutide. Ann. Pharmacother. 2014;48:1494–1501. doi: 10.1177/1060028014545807. [DOI] [PubMed] [Google Scholar]
- 5.Lyseng-Williamson KA. Coagulation Factor IX (Recombinant), Albumin Fusion Protein (Albutrepenonacog Alfa; Idelvion®): A Review of Its Use in Haemophilia B. Drugs. 2017;77:97–106. doi: 10.1007/s40265-016-0679-8. [DOI] [PubMed] [Google Scholar]
- 6.Hui H, Farilla L, Merkel P, Perfetti R. The short half-life of glucagon-like peptide-1 in plasma does not reflect its long-lasting beneficial effects. Eur. J. Endocrinol. 2002;146:863–869. doi: 10.1530/eje.0.1460863. [DOI] [PubMed] [Google Scholar]
- 7.Schmidt, S. R. Fusion Proteins: Applications and Challenges in Fusion Protein Technologies for Biopharmaceuticals (ed. Schmidt, S.R.) 1-24 (John Wiley & Sons, Inc., 2013).
- 8.Roques BP, Noble F, Dauge V, Fournié-Zaluski MC, Beaumont A. Neutral Endopeptidase 24.11: Structure, Inhibition, and Experimental and Clinical Pharmacology. Pharmacol. Rev. 1993;45:87–146. [PubMed] [Google Scholar]
- 9.Erdös EG, Skidgel RA. Neutral endopeptidase 24.11 (enkephalinase) and related regulators of peptide hormones. FASEB J. 1989;3:145–151. doi: 10.1096/fasebj.3.2.2521610. [DOI] [PubMed] [Google Scholar]
- 10.Howell S, Nalbantoglu J, Crine P. Neutral endopeptidase can hydrolyze β-amyloid(1–40) but shows no effect on β-amyloid precursor protein metabolism. Peptides. 1995;16:647–652. doi: 10.1016/0196-9781(95)00021-B. [DOI] [PubMed] [Google Scholar]
- 11.Gentiluomo L, et al. Advancing Therapeutic Protein Discovery and Development through Comprehensive Computational and Biophysical Characterization. Mol. Pharm. 2020;17:426–440. doi: 10.1021/acs.molpharmaceut.9b00852. [DOI] [PubMed] [Google Scholar]
- 12.Cordes AA, Platt CW, Carpenter JF, Randolph TW. Selective Domain Stabilization as a Strategy to Reduce Fusion Protein Aggregation. J. Pharm. Sci. 2012;101:1400–1409. doi: 10.1002/jps.23049. [DOI] [PubMed] [Google Scholar]
- 13.Svergun, D. I., Koch, M. H. J., Timmins, P. A. & May, R. P. Monodisperse systems in Small Angle X-Ray and Neutron Scattering from Solutions of Biological Macromolecules. 93–151 (Oxford University Press, 2013).
- 14.Tria G, Mertens HDT, Kachala M, Svergun DI. Advanced ensemble modelling of flexible macromolecules using X-ray solution scattering. IUCrJ. 2015;2:207–217. doi: 10.1107/S205225251500202X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bernadó P, Mylonas E, Petoukhov MV, Blackledge M, Svergun DI. Structural Characterization of Flexible Proteins Using Small-Angle X-ray Scattering. J. Am. Chem. Soc. 2007;129:5656–5664. doi: 10.1021/ja069124n. [DOI] [PubMed] [Google Scholar]
- 16.Wenskowsky L, et al. Identification and Characterization of a Single High-Affinity Fatty Acid Binding Site in Human Serum Albumin. Angew. Chemie Int. Ed. 2018;57:1044–1048. doi: 10.1002/anie.201710437. [DOI] [PubMed] [Google Scholar]
- 17.Moss S, Subramanian V, Acharya KR. High resolution crystal structure of substrate-free human neprilysin. J. Struct. Biol. 2018;204:19–25. doi: 10.1016/j.jsb.2018.06.004. [DOI] [PubMed] [Google Scholar]
- 18.Ettori C, et al. Purification of recombinant human growth hormone by isoelectric focusing in a multicompartment electrolyzer with immobiline membranes. J. Biotechnol. 1992;25:307–318. doi: 10.1016/0168-1656(92)90163-4. [DOI] [PubMed] [Google Scholar]
- 19.Sønderby P, et al. Self-Interaction of Human Serum Albumin: A Formulation Perspective. ACS Omega. 2018;3:16105–16117. doi: 10.1021/acsomega.8b02245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kulakova A, et al. Small angle X-ray scattering and molecular dynamic simulations provide molecular insight for stability of recombinant human transferrin. J. Struct. Biol. X. 2020;4:100017. doi: 10.1016/j.yjsbx.2019.100017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Blanchet CE, et al. Versatile sample environments and automation for biological solution X-ray scattering experiments at the P12 beamline (PETRA III, DESY) J. Appl. Crystallogr. 2015;48:431–443. doi: 10.1107/S160057671500254X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Petoukhov MV, et al. New developments in the ATSAS program package for small-angle scattering data analysis. J. Appl. Crystallogr. 2012;45:342–350. doi: 10.1107/S0021889812007662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Šali A, Blundell TL. Comparative Protein Modelling by Satisfaction of Spatial Restraints. J. Mol. Biol. 1993;234:779–815. doi: 10.1006/jmbi.1993.1626. [DOI] [PubMed] [Google Scholar]
- 24.Dolinsky TJ, Nielsen JE, McCammon JA, Baker NA. PDB2PQR: an automated pipeline for the setup of Poisson-Boltzmann electrostatics calculations. Nucleic Acids Res. 2004;32:W665–W667. doi: 10.1093/nar/gkh381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.DeLano, W. L. The PyMOL Molecular Graphics System (2002) on the World Wide Web (http://www.pymol.org).
- 26.Lindorff-Larsen K, et al. Improved side-chain torsion potentials for the Amber ff99SB protein force field. Proteins Struct. Funct. Bioinforma. 2010;78:1950–1958. doi: 10.1002/prot.22711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Indrakumar S, et al. Conformational Stability Study of a Therapeutic Peptide Plectasin Using Molecular Dynamics Simulations in Combination with NMR. J. Phys. Chem. B. 2019;123:4867–4877. doi: 10.1021/acs.jpcb.9b02370. [DOI] [PubMed] [Google Scholar]
- 28.Srinivasan J, Cheatham TE, Cieplak P, Kollman PA, Case DA. Continuum solvent studies of the stability of DNA, RNA, and phosphoramidate-DNA helices. J. Am. Chem. Soc. 1998;120:9401–9409. doi: 10.1021/ja981844+. [DOI] [Google Scholar]
- 29.Gohlke H, Case DA. Converging free energy estimates: MM-PB(GB)SA studies on the protein-protein complex Ras-Raf. J. Comput. Chem. 2004;25:238–250. doi: 10.1002/jcc.10379. [DOI] [PubMed] [Google Scholar]
- 30.Tan, P.-N., Steinbach, M. & Kumar, V. Chapter 8: Cluster Analysis: Basic Concepts and Algorithms. in Introduction to Data Mining 487-568 (Pearson Education, Inc. 2006).
- 31.Wolf A, Kirschner KN. Principal component and clustering analysis on molecular dynamics data of the ribosomal L11·23S subdomain. J. Mol. Model. 2013;19:539–549. doi: 10.1007/s00894-012-1563-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Roe DR, Cheatham TE. PTRAJ and CPPTRAJ: Software for Processing and Analysis of Molecular Dynamics Trajectory Data. J. Chem. Theory Comput. 2013;9:3084–3095. doi: 10.1021/ct400341p. [DOI] [PubMed] [Google Scholar]
- 33.Humphrey W, Dalke A, Schulten K. VMD: Visual molecular dynamics. J. Mol. Graph. 1996;14:33–38. doi: 10.1016/0263-7855(96)00018-5. [DOI] [PubMed] [Google Scholar]
- 34.Irwin JJ, Sterling T, Mysinger MM, Bolstad ES, Coleman RG. ZINC: A Free Tool to Discover Chemistry for Biology. J. Chem. Inf. Model. 2012;52:1757–1768. doi: 10.1021/ci3001277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Madhavi Sastry G, Adzhigirey M, Day T, Annabhimoju R, Sherman W. Protein and ligand preparation: parameters, protocols, and influence on virtual screening enrichments. J. Comput. Aided. Mol. Des. 2013;27:221–234. doi: 10.1007/s10822-013-9644-8. [DOI] [PubMed] [Google Scholar]
- 36.Wang J, Wolf RM, Caldwell JW, Kollman PA, Case DA. Development and testing of a general amber force field. J. Comput. Chem. 2004;25:1157–1174. doi: 10.1002/jcc.20035. [DOI] [PubMed] [Google Scholar]
- 37.Jakalian A, Jack DB, Bayly CI. Fast, efficient generation of high-quality atomic charges. AM1-BCC model: II. Parameterization and validation. J. Comput. Chem. 2002;23:1623–1641. doi: 10.1002/jcc.10128. [DOI] [PubMed] [Google Scholar]
- 38.Winn MD, et al. Overview of the CCP4 suite and current developments. Acta Crystallogr. D Biol. Crystallogr. 2011;67:235–242. doi: 10.1107/S0907444910045749. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
ICD, nanoDSF and SAXS data is deposited in PIPPI Data Base https://pippi-data.kemi.dtu.dk/ EOM models obtained from SAXS data analysis can be found in SASBDB database.