Abstract
The molecular and genetic basis of tumor recurrence is complex and poorly understood. RIPK3 is a key effector in programmed necrotic cell death and, therefore, its expression is frequently suppressed in primary tumors. In a transcriptome profiling between primary and recurrent breast tumor cells from a murine model of breast cancer recurrence, we found that RIPK3, while absent in primary tumor cells, is dramatically reexpressed in recurrent breast tumor cells by an epigenetic mechanism. Unexpectedly, we found that RIPK3 knockdown in recurrent tumor cells reduced clonogenic growth, causing cytokinesis failure, p53 stabilization, and repressed the activities of YAP/TAZ. These data uncover a surprising role of the pro-necroptotic RIPK3 kinase in enabling productive cell cycle during tumor recurrence. Remarkably, high RIPK3 expression also rendered recurrent tumor cells exquisitely dependent on extracellular cystine and undergo necroptosis upon cystine deprivation. The induction of RIPK3 in recurrent tumors unravels an unexpected mechanism that paradoxically confers on tumors both growth advantage and necrotic vulnerability, providing potential strategies to eradicate recurrent tumors.
Subject terms: Cancer metabolism, Cancer models, Oncogenes, Tumour biomarkers
Introduction
While enormous progress has been made for the diagnosis and treatment of primary breast tumors, the emergence of recurrent tumors still poses significant challenges. Recurrent breast tumors are generally incurable and unresponsive to the treatments [1]. However, the mechanisms leading to tumor recurrence remain largely unknown. To study the mechanism, genetically engineered mouse (GEM) models of recurrent breast cancers have been established using the doxycycline-inducible expression of oncogenes in the mammary gland [2–6]. In the GEM models of recurrent breast tumors, mammary adenocarcinomas can be induced by doxycycline and regressed after doxycycline withdrawal. Importantly, recurrent tumors will eventually emerge in most mice after the withdrawal of the oncogene [2–6] which bear significant similarities to human recurrent breast cancer in several ways: (1) tumor recurrence occurs over a timeframe, similar to the timing in human breast cancer; (2) the formation of recurrent tumors is independent from the initial tumor-driving oncogenes; (3) similar molecular pathways and clinical courses between mouse and human recurrent tumors [2–6]. Unfortunately, most recurrent-enriched pathways are not readily amenable to therapeutic intervention, necessitating novel therapeutic approaches to target recurrent tumor cells.
One relative unexplored aspect of recurrent tumor is the metabolic reprogramming and nutrient addictions [7, 8]. Previously, we demonstrated that renal cell carcinomas and triple-negative breast cancer cells are highly susceptible to cystine deprivation or inhibitors of cystine transporter (xCT) [9, 10]. In addition, we have identified ATM [11], MESH1 [12], KEAP1 glycosylation [13], and TAZ [14–16] as novel determinants of cystine addiction of cancer cells. Cystine is the limiting precursor of glutathione (GSH), a crucial antioxidant to decrease reactive oxygen species (ROS) [17]. Therefore, cystine deprivation results in GSH depletion, unopposed ROS surge, and triggering cell death [18]. Cystine deprivation activates the receptor-interacting serine/threonine kinase 1 (RIPK1), which promotes RIPK3 activation and phosphorylation/polymerization of mixed lineage kinase domain like pseudokinase (MLKL), resulting in membrane rupture and necroptosis [19, 20]. Accordingly, RIPK3 expression is often silenced in primary tumors due to the promoter methylation as a part of death-evading strategy [21, 22].
Here we report that the RIPK3 is highly expressed in the recurrent breast tumors and required for productive proliferation. However, this exaggerated reexpression of RIPK3 also renders the recurrent tumor cells vulnerable to the cell death triggered by cystine deprivation. Thus, RIPK3-depenent proliferation of recurrent tumor cells creates the collateral vulnerability to cystine deprivation that can serve as a novel therapeutic strategy for recurrent tumors.
Results
Exaggerated expression of RIPK3 in the recurrent tumor cells
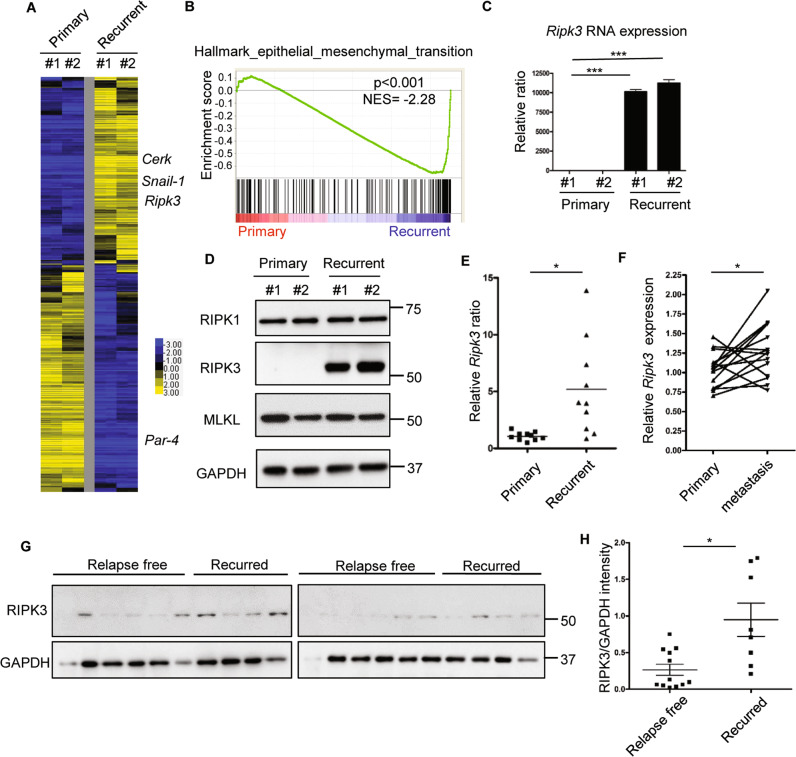
To investigate the phenotypic differences between primary and recurrent tumors, tumor cells were isolated and expanded from HER2 driven murine MTB/TAN model before the oncogenic withdrawal (primary tumors) and after the recurrence (recurrent tumors) [4]. To identify differentially expressed genes, we performed microarrays to compare the transcriptome differences between primary and recurrent tumor cells (GEO: GSE116513) [23]. This comparison validates the reported upregulation of Ceramide Kinase (Cerk) [24] and downregulation of prostate apoptosis response-4 [25] in the recurrent tumor cells (Fig. 1a). Also, recurrent tumor cells expressed a higher level of epithelial–mesenchymal transition (EMT)-driving snail family transcriptional repressor 1 [5] (Fig. 1a), consistent with EMT by gene set enrichment analysis (GSEA) (Fig. 1b). Therefore, these data confirm distinct gene expression patterns reported previously.
Fig. 1. Transcriptome profiling of primary and recurrent tumor cells revealed RIPK3 upregulation in recurrent cells.
a Heatmap of the transcriptional difference between two primary and two recurrent cell lines. Color scale indicates log2-fold-change. b GSEA analysis showed the enrichment of EMT gene set in the recurrent tumor cells. c Ripk3 was highly expressed in recurrent tumor cells by RT-PCR. n = 3 independent repeats. d Western blot showed a robust RIPK3 protein expression only in recurrent tumor cell lines. e Comparison of Ripk3 RNA expression between ten primary and ten recurrent mouse tumors showed an overall increase in recurrent tumors. f Comparison of RIPK3 expression between primary breast cancer and matching lymph node metastasis in human dataset (GSE61723). g Western blot showed that RIPK3 protein expression is higher in human primary breast tumors with a distant recurrence when compared with primary tumors with long-term (>10 years) disease-free survival. h Quantification of relative RIPK3 protein in g by normalizing to the intensity of loading control (GAPDH) using Image J software. Bars show standard error of the mean. *p < 0.05, ***p < 0.001, two-tailed Student’s t test.
When we examined the expression of genes involved in the necroptosis [19], we noted a robust overexpression of Ripk3 in the recurrent tumors cells (Fig. 1a). RT-PCR validated the dramatically increased Ripk3 mRNA expression in the recurrent tumor cells (Fig. 1c). Western blots revealed that RIPK3 protein, while absent in the primary tumor cells, was abundantly expressed in the recurrent tumor cells (Fig. 1d). In comparison, RIPK1 and MLKL proteins, the canonical regulator and target of RIPK3, respectively, were found to be expressed at similar levels in the primary and recurrent tumor cells (Fig. 1d). Similar Ripk3 mRNA overexpression is noted in a panel of mouse recurrent breast tumors, when compared with primary breast tumors (Fig. 1e). The absence of RIPK3 protein expression in primary tumor cells was previously noted and assumed to be an evolutionary strategy of tumors to escapes necroptosis as part of the cancer hallmarks [21, 26]. Therefore, the absence of RIPK3 in primary tumor cells is consistent with these reports. However, the reexpression of RIPK3 in the recurrent tumor cells was unexpected.
This observation from the murine recurrent breast tumor model is supported by two human dataset of gene expression comparison between primary breast cancer and matching lymph node metastasis [27, 28]. First, RIPK3 mRNA was significantly increased by 2.08-fold in metastatic tumors when compared with primary human tumors (Supplementary Table 1) [27]. Another human dataset (GSE61723) [28] that compared 16 pairs of primary breast cancer and matching lymph node metastasis, also showed an increase in RIPK3 mRNA expression in 11 out of 16 pairs with an overall significant upregulation (Fig. 1f). Collectively, these data indicate the upregulation of RIPK3 expression occurs in recurrent tumors in both a mouse model and two human studies.
To further validate the association between RIPK3 protein and metastasis, we compared a series of human primary breast tumors for which periodic follow-up was available through the Duke Tumor Registry and Duke electronic records. By comparing with either long-term (>10 years) disease free or a clear distant recurrence (Fig. 1g), we found that primary breast tumors with a distant metastasis showed higher RIPK3 protein expression (Fig. 1h). These data suggest that higher RIPK3 expression may render primary tumors more resistant to the therapy and recurred.
Epigenetic regulation of Ripk3 in the primary versus recurrent tumor cells
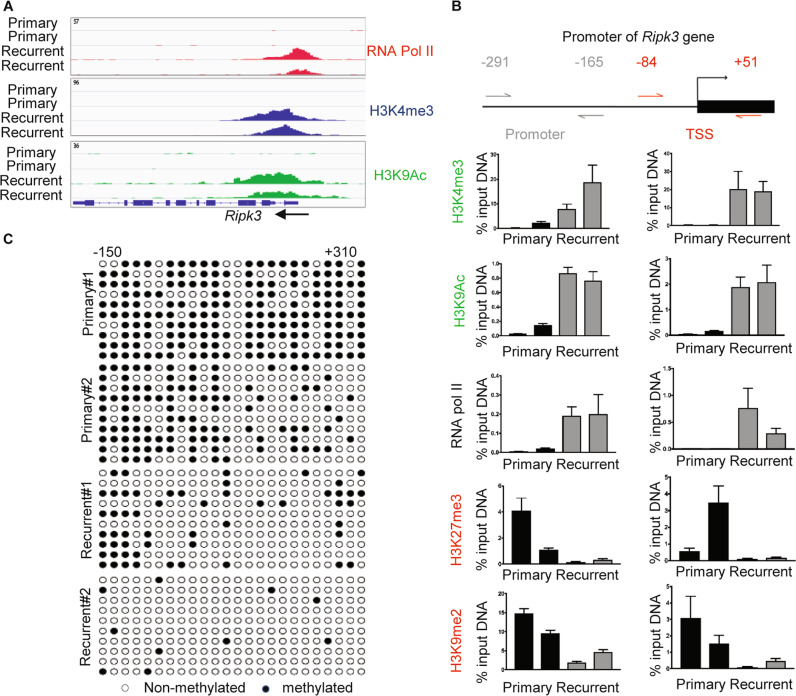
To understand the basis of the Ripk3 mRNA upregulation in the recurrent tumor cells, we investigated the epigenetic landscape of the regulatory regions of Ripk3 locus in the primary and recurrent tumor cells by ChIP-sequencing. Consistent with the recurrent-specific expression of Ripk3, RNA polymerase II dramatically occupied the regulatory regions of Ripk3 gene in the recurrent tumor cells, but not in the primary tumor cells (Fig. 2a). Next, we compared the ChIP-Seq data of activating epigenetic histone markers, H3K9Ac and H3K4me3 and found them highly enriched in the regulatory regions of Ripk3 gene adjacent to the RNA polymerase II binding site in the recurrent tumor cells, but not in the primary tumor cells (Fig. 2a).
Fig. 2. Epigenetic landscape of the regulatory regions of Ripk3 in the primary versus recurrent tumor cells.
a ChIP-Seq data showed the occupancy of RNA Pol II, H3K4me3, and H3K9ac in regulatory regions of Ripk3 of recurrent tumor cells. b ChIP-qPCR analysis of H3K4me3, H3K9ac, RNA pol II, H3K27me3, and H3K9me2 enrichment at two indicated regions in the promoter of the Ripk3 genes in two primary and two recurrent tumor cell lines. Data are presented as the percentage of input DNA. c The cytosine methylation of CpG dinucleotides (circles) within the Ripk3 promoter and gene body (−150 to +310) for two primary and two recurrent tumor cell lines. Bisulfite-treated DNA was transformed into bacteria and ten replicate colonies were sequenced (rows). Open circles denote unmethylated CpG dinucleotides, while closed circles denoted methylated CpG dinucleotides.
Next, we designed two sets of primers that cover the promoters (−291 to −165), transcriptional start site (TSS, −84 to +51) (Fig. 2b) to measure the epigenetic changes of Ripk3 locus by ChIP-PCR. We found that the promoter and TSS of Ripk3 gene are marked by the activation markers (H3K4me3 and H3K9Ac) and RNA polymerase II occupancy only in the recurrent tumor cells (Fig. 2b). Reciprocally, we found that these Ripk3 regulatory regions in the primary tumor cells are marked by the silencing markers (H3K27me3 and K3K9me2) (Fig. 2b).
We further performed bisulfite sequencing to measure the degree of DNA methylation of the CpG island in the Ripk3 regulatory regions (−150 to +310) (Fig. 2c). We found that most of the cytosines in the Ripk3 CpG island are methylated in the primary tumor cells, but unmethylated in the recurrent tumor cells (Fig. 2c). Together, these data indicate that epigenetic changes in the histone modification and DNA methylations are likely responsible for the silencing of Ripk3 in the primary tumor cells and robust expression in the recurrent tumor cells.
Ripk3 knockdown triggers mitotic defects and p53 activation
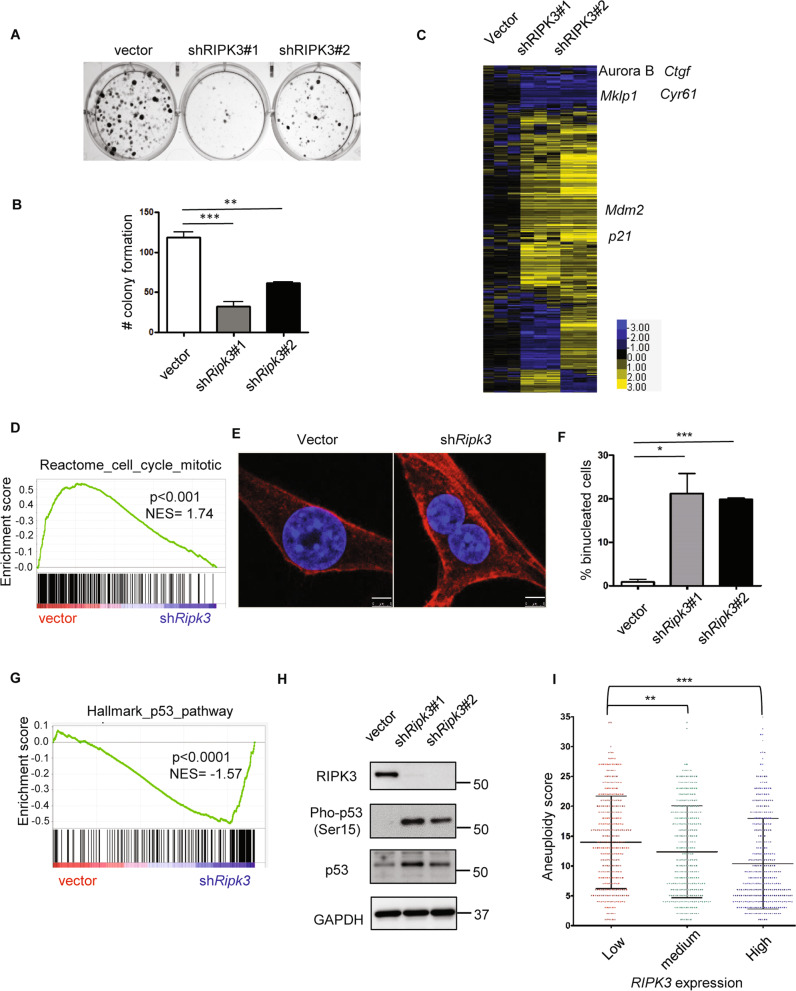
Given the unexpected robust expression of RIPK3 in the recurrent tumors, we investigated its functional role. First, we performed the clonogenic assay to determine whether Ripk3 silencing affects their capacity to proliferate and form colonies (Fig. 3a, b, Supplementary Fig. 1A, B). We found that Ripk3 knockdown by two independent shRNAs significantly reduced colony formation in the recurrent tumor cells (Fig. 3a, b), but not in the primary tumor cells (Supplementary Fig. 1A, B), indicating the crucial role of RIPK3 for the proliferation and survival of recurrent tumor cells.
Fig. 3. Ripk3 knockdown triggers p53 signaling and mitotic defects.
a Ripk3 silencing decreased colony formation. The clonogenic assay was performed by plating 500 recurrent tumor cells to six-well plates. After 10 days of incubation, cells were fixed with paraformaldehyde (4%), and stained with crystal violet. b Quantification of number of colony formation. c Heatmap of the transcriptional response to Ripk3 silencing in recurrent cells with several affected genes indicated. d GSEA analysis showed depletion of reactome cell cycle mitosis upon Ripk3 silencing. e Ripk3 silencing dramatically increased binucleated cells. Recurrent cells were stained with DAPI (nucleus) and Alexa Flour 594 Phalloidin (F-actin). Scale bar, 5 μm. f Quantification of binucleated cells under Ripk3 silencing. g GSEA analysis showed that Ripk3 silencing enriched p53 signaling pathway. h Ripk3 silencing led to the accumulation of p53 and increased Serine 15 phosphorylation. i Lower RIPK3 expression in human breast cancers is associated with increased amount of aneuploidy. *p < 0.05; **p < 0.01; ***p < 0.001, two-tailed Student’s t test. b n = 4 and f n = 3 independent repeats. Bars show standard error of the mean. i n = 1024.
MLKL is the downstream effector of RIPK3 during necroptosis [20]. We found that Mlkl silencing in recurrent tumor cells recapitulated the effect of Ripk3 knockdown and suppressed colony formation (Supplementary Fig. 1C, D). Consistently, an MLKL inhibitor, necrosulfonamide (NSA) [20, 29], also reduced colony formation of recurrent tumor cells (Supplementary Fig. 1E, F). These data show that the canonical necroptosis-driving RIPK3-MLKL signaling axis is required for cell proliferation and clonogenic growth in recurrent breast tumor cells.
To understand the mechanisms by which Ripk3 knockdown reduced clonogenic capacity of recurrent tumor cells, we investigated the transcriptional responses to Ripk3 knockdown by RNA-seq (GEO: GSE124634) (Fig. 3c). We found downregulation of several mitotic regulators in Ripk3 knockdown cells, including Aurora B and Mklp1 (Fig. 3c), as well as the depletion of the reactome to mitosis gene set by GSEA (Fig. 3d). Ripk3 knockdown also dramatically increased the number of binucleated cells by ~20-folds (Fig. 3e, f). These data suggest that Ripk3 affects the proper execution of mitosis in recurrent tumor cells. Binucleated/multinucleated cells generally result from cytokinesis failure [30]. Previous study showed that cytokinesis failure can lead to the activation of tumor suppressor p53 [31]. Consistently, GSEA revealed the enrichment of genes in p53 signaling pathway (Fig. 3g) and confirmed the upregulation of p53 target genes, Mdm2 and p21, in Ripk3 knockdown cells (Fig. 3c). Indeed, p53 protein is phosphorylated at Ser15 upon Ripk3 silencing (Fig. 3h), expected to weaken the interaction between p53 and its negative regulator Mdm2 [32] and stabilize p53 accumulation (Fig. 3h). Collectively, these data suggest that robust Ripk3 expression in recurrent tumor cells determines proper mitotic progression and cell proliferation.
Given that Ripk3 knockdown increased binucleated cells, which cause genomic instability [33], we speculated that Ripk3 knockdown may lead to aneuploidy. A recent report has made the scores of aneuploidy available in a pan-cancer TCGA dataset [34]. Therefore, we correlated the level of RIPK3 expression with its aneuploidy score in breast cancer patients (Fig. 3i). Our results indicate that low levels of RIPK3 mRNA expression are significantly associated with higher aneuploidy in breast cancers (Fig. 3i). These data in human breast tumors further support that RIPK3 is critical in preventing chromosome instability and aneuploidy in recurrent tumor cells.
Ripk3 knockdown represses YAP/TAZ
Cytokinesis failure can activate Hippo tumor suppressor pathway [31] and inactivate the two Hippo pathway effectors, YAP (Yes-associated protein 1) and TAZ (transcriptional coactivator with PDZ-binding motif). These proteins are coactivators of TEAD family transcription factors mediating the expression levels of proliferative and oncogenesis genes [35]. When Hippo is on, YAP/TAZ is inactivated by phosphorylation and exclusion from the nucleus. When Hippo is off, YAP/TAZ is localized in the nucleus and able to interact with TEAD and leads to downstream gene expression [35]. We found that Ripk3 silencing in recurrent tumor cells led to a depletion of YAP/TAZ signature by GSEA (Fig. 4a). RT-PCR confirmed that two canonical YAP/TAZ target genes: Ctgf and Cyr61, were dramatically repressed upon Ripk3 knockdown (Figs. 3c and 4b). In addition, we examined how Ripk3 silencing affects the subcellular localization of YAP and TAZ by nuclear/cytosol fractionation (Fig. 4c). While Ripk3 silencing slightly reduced nuclear YAP, it significantly depleted nuclear TAZ (to ~18%) with a corresponding increase in the cytosolic TAZ (Fig. 4c). Confocal microscopy further confirmed the reduced nuclear YAP/TAZ upon Ripk3 silencing (Fig. 4d). Thus, we speculated that the depletion of YAP/TAZ in the nucleus under Ripk3 silencing may contribute to low efficiency of colony formation and cell proliferation. To test this hypothesis, we expressed constitutively active mutants of YAP/TAZ, YAP S127A, and TAZ S89A [36, 37] in Ripk3 knockdown cells (Fig. 4e, f). We observed a complete rescue by TAZ S89A expression under Ripk3 knockdown whereas YAP S127A partially rescued colony formation (Fig. 4e, f). These data suggest that reduced nuclear YAP/TAZ levels and activities, especially TAZ, contribute significantly to reduced clonogenic formation induced by Ripk3 silencing in recurrent tumor cells.
Fig. 4. Ripk3 knockdown abolishes YAP/TAZ-dependent cell growth.
a GSEA analysis showed the depletion of YAP/TAZ transcriptional target gene set upon Ripk3 silencing in recurrent cells. b RT-PCR validated the downregulation of Ctgf and Cyr61, two canonical YAP/TAZ target genes upon Ripk3 knockdown. c Nuclear/cytosol fractionation showed the depletion of TAZ upon RIPK3 knockdown. α-tubulin: cytosolic marker; Lamin A/C: nuclear marker. Relative YAP/TAZ ratio was determined by normalizing YAP/TAZ intensity to Lamin A/C using Image J. d Confocal microscopy confirmed the depletion of YAP/TAZ upon RIPK3 knockdown. Scale bar, 10 μm. e Overexpression of constitutively active YAP S127A and TAZ S89A rescued the low colony formation upon Ripk3 knockdown as quantified in f. *p < 0.05; **p < 0.01; ***p < 0.001, two-tailed Student’s t test. n = 3 independent repeats. Bars show standard error of the mean.
Recurrent breast tumor cells are uniquely addicted to exogenous cystine
Cystine is imported into mammalian cells in exchange of the export of glutamate via the xCT transporter [38], which can be blocked by xCT inhibitor, erastin [39–41]. We have found that the cystine deprivation can trigger extensive cell death in renal cell carcinomas [9] and triple-negative breast cancer [10]. Given the robust RIPK3 expression in recurrent tumor cells, we investigated whether recurrent tumor cells are vulnerable to cystine deprivation.
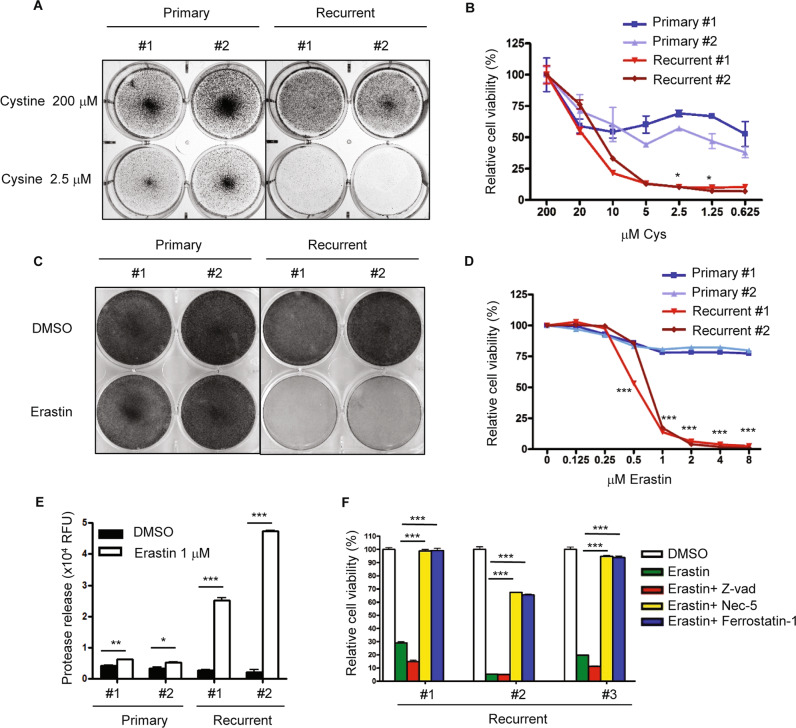
We subjected two primary and two recurrent cell lines to normal or cystine-deprived media and determined the cell viability. We found that cystine deprivation eliminated most of recurrent tumor cells, but only had modest effects on primary tumor cells (Fig. 5a). Under varying degrees of cystine deprivation, the recurrent cells were largely eliminated under 5 µM of cystine (Fig. 5b) as the primary tumor cells still maintained ~50% viability at 0.625 µM of cystine (Fig. 5b). Collectively, recurrent tumor cells are more sensitive to cystine deprivation.
Fig. 5. Recurrent tumor cells are more sensitive to cystine deprivation and erastin-induced death.
a Recurrent tumor cells, when compared with primary tumor cells, were more sensitive to cystine deprivation. Two primary and two recurrent cell lines were incubated in full media (200 µM) or cystine-deprived media (2.5 µM) for 16 h. The cells were then fixed with paraformaldehyde (4%) for crystal violet staining. b Recurrent tumor cells, when compared with primary tumor cells, were more sensitive to cell death under cystine deprivation. Primary and recurrent cells were incubated with decreasing level of cystine for 16 h. The viability was then measured by ATP level using the Celltiter-Glo assay. c Recurrent tumor cells are more sensitive to erastin treatment than primary tumor cells. Two primary and two recurrent cell lines were incubated in erastin (1 µM) or DMSO for 18 h. The cells were then fixed for crystal violet staining. d Primary and recurrent tumor cells were treated with increasing indicated doses of erastin for 18 h and the viability was measured by the Celltiter-Glo assay. e Erastin induced more cell rupture and protease release in recurrent cells. Primary and recurrent cells were treated with 1 µM of erastin for 16 h. The media was then harvested for protease measurement. f Erastin-induced cell death was rescued by Nec-5 and Ferrostatin-1. Erastin (2 µM) were treated at the same time with DMSO, Z-vad (20 µM), Nec-5 (5 µM), and Ferrostatin-1 (1 µM) in recurrent cell lines for 18 h. The cell viability was then determined by the Celltiter-Glo assay. b, d p < 0.0001, Two-way ANOVA, *p < 0.05, ***p < 0.001. Bonferroni post hoc tests. e, f *p < 0.05; **p < 0.01; ***p < 0.001, two-tailed Student’s t test. n = 3 independent repeats. Bars show standard error of the mean.
Next, we compared their sensitivity to erastin and found recurrent tumor cells were much more sensitive by crystal violet staining (Fig. 5c) and the CellTiter-Glo assay (Fig. 5d). While recurrent tumor cells were largely eliminated between 0.5 and 1 µM erastin, the primary cells survived more than 8 µM of erastin with ~75% viability (Fig. 5d). Such recurrent-specific erastin sensitivities are further confirmed by the higher levels of protease release upon cell membrane breakage (Fig. 5e).
Thus, we further analyzed the protein expression of RIPK3 and MLKL in primary and recurrent tumor cell lines after erastin treatment. We found that RIPK3 protein is only expressed in recurrent tumor cells and modestly elevated by erastin treatment (Supplementary Fig. 2A). An upregulated base level of phosphorylated MLKL was noted in recurrent tumor cells (Supplementary Fig. 2A). Moreover, Ripk3 silencing abolished MLKL phosphorylation (Supplementary Fig. 2B). Therefore, the elevated RIPK3 proteins and constitutive MLKL phosphorylation may prime the recurrent tumor cells to cell death triggered by erastin or cystine deprivation.
Erastin is considered to trigger to cell death by ferroptosis, a programmed cell death distinct from apoptosis and necroptosis [40]. However, at low dose of erastin, we have previously found that necroptosis pathway and RIPK3 is also required for programmed cell death [9]. Given the elevated RIPK3 in recurrent tumor cells, we further determined the cell death mechanisms. In addition to low dose of erastin, we used different inhibitors to define the cell death mechanisms caused by erastin. We found that the apoptosis inhibitor Z-Vad did not rescue the erastin-induced death. In contrast, both ferroptosis inhibitor (ferrostatin-1) [42] and necroptosis inhibitor (necrostatin-5) [43] robustly rescued the cell death, suggesting the potential role of RIPK3 in the cell death triggered by erastin (Fig. 5f). While the requirement for RIPK3 may not be generally applicable to all erastin-induced cell death, RIPK3 may be particularly critical in the recurrent tumor cells with high level of RIPK3 expression and constitutive MLKL phosphorylation.
Ripk3 expression contribute to cystine addiction in both primary and recurrent tumor cells
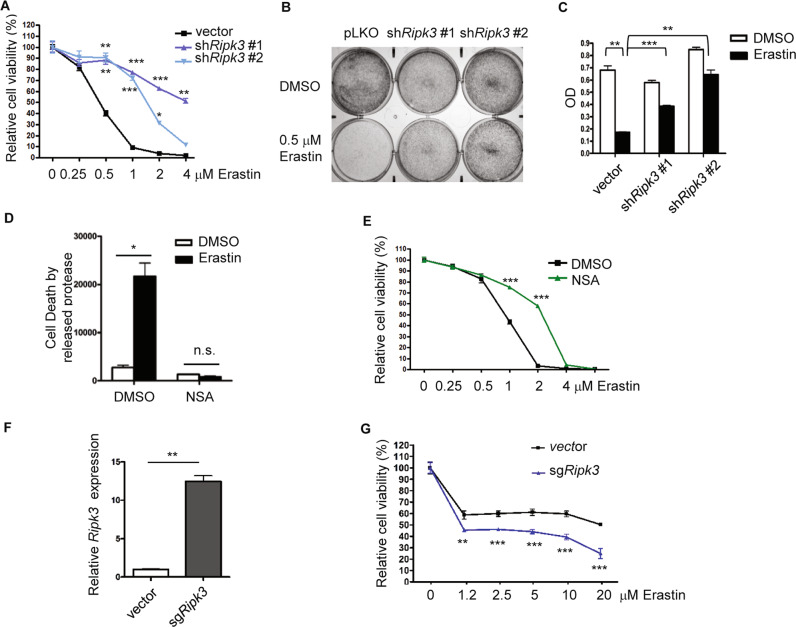
To examine whether high Ripk3 expression in recurrent tumor cells contributes to its vulnerability to cystine deprivation, we knocked down Ripk3 by two independent shRNAs and found a significant reduction of erastin-induced cell death with ~70–80% of viability (Fig. 6a), while erastin (1 µM) eliminated the control cells to less than 10% cell viability (Fig. 6a). Similar results were also obtained by crystal violet staining and follow-up quantification (Fig. 6b, c). Therefore, the robust Ripk3 expression of recurrent tumor cells contributes to the cystine addiction phenotypes.
Fig. 6. Ripk3 expression modulates cystine addiction in tumor cells.
a Ripk3 knockdown mitigated the erastin-induced cell death in recurrent tumor cells. Recurrent tumor cells transduced with control or two Ripk3 shRNAs were treated with increasing dose of erastin for 16 h. Cell viability was then measured by the Celltiter-Glo assay. b, c Recurrent cells transduced with control or two Ripk3 shRNAs were treated with 0.5 µM of erastin for 16 h before assessing their viability by crystal violet staining (b), as quantified in c. d, e MLKL phosphorylation by RIPK3 contributed to the erastin-induced cell death. Inhibiting MLKL by compound inhibitor (NSA, 5 µM) protected recurrent tumor cells from cell death under erastin treatment (0.5 µM) when measured by protease release (d) or the Celltiter-Glo assay (e). f, g CRISPR-a upregulated Ripk3 RNA expression in primary tumor cells and sensitized primary tumor cells to indicated doses of erastin. Primary tumor cells transduced with control or Ripk3 sgRNA with synergistic activation mediators were validated by RT-PCR (f) and examined its sensitivity to increasing dose of erastin by the Celltiter-Glo assay after 48 h of treatment (g). a, e, g p < 0.0001, Two-way ANOVA, *p < 0.05, **p < 0.01, ***p < 0.001, Bonferroni post hoc tests. c, d, f *p < 0.05; **p < 0.01; ***p < 0.001, two-tailed Student’s t test. n = 3 independent repeats. Bars show standard error of the mean.
Since the MLKL phosphorylation by RIPK3 is required for MLKL oligomerization and activation of necroptosis, we further inhibited MLKL oligomerization by NSA [20, 29] in recurrent tumor cells. We found NSA rescued the erastin-induced cell death using the protease release assay (Fig. 6d) and the CellTiter-Glo assay (Fig. 6e). Therefore, the exaggerated RIPK3 expression and MLKL phosphorylation in recurrent tumor cells both enable recurrent tumor cells to proliferate and pose the cells to necroptosis-inducing signaling triggered by cystine deprivation or erastin.
Next, we tested whether HDAC inhibitors and/or hypomethylating agents reactivate the epigenetic silencing of Ripk3 expression in primary tumor cells. We found that treatment of pan-HDAC inhibitor (SAHA), EZH2 inhibitor (EPZ6438), DNMT (5-Aza-DC) alone, or in combination treatments could only slightly increase the Ripk3 expression (Supplementary Fig. 3A). Even extended combination treatment of pan-HDAC and DNMT inhibitor failed to induce Ripk3 upregulation (Supplementary Fig. 3B). Thus, we employed dCas9 for CRISPR activation [44] to bring transcription activator VP64 to the promoter region of Ripk3. We found CRISPR activation led to ~12-fold of increase in Ripk3 expression in primary tumor cells (Fig. 6f). Importantly, this Ripk3 upregulation by CRISPR activation also rendered primary tumor cells sensitive to erastin (Fig. 6g). Taken together, these data suggest the manipulation of Ripk3 expression level in both primary and recurrent tumor cells can modulate its sensitivity to erastin-induced cell death.
Discussion
Global changes in epigenetic landscapes are a hallmark of cancer. While RIPK3 expression is found in most of the normal tissue, the promoter region of RIPK3 usually becomes highly methylated in cancer cells, which leads to almost absence of RIPK3 expression in several cancer cell types [21, 22, 26, 45, 46]. Besides necroptosis, RIPK3 has been reported in regulating cytokine production, inflammation, and antitumor immunity [47–49]. Therefore, RIPK3 is suggested as a tumor suppressor gene and absence in RIPK3 expression can be considered as adaptation process for tumor cells to evade death from various necroptosis-triggering signals and chemotherapy [21, 50]. In this study, primary tumor cell indeed showed low Ripk3 expression (Fig. 1c–f). However, after withdrawing of oncogene, the recurrent tumor cells showed exaggerating amount of RIPK3 that triggers constitutive MLKL phosphorylation (Fig. 1c–f and Supplementary Fig. 2A). Given the potential risk of Ripk3 upregulation in triggering necroptosis, we speculated an alternative function of Ripk3 to provide recurrent tumor cells’ fitness advantages after oncogene withdrawal. Previous study showed increased RIPK3 promote vascular permeability and extravasation in lung, which enables metastatic tumor formation [50]. Knockdown of RIPK3 in MDA-MB-231 breast cancer cells also contribute to arrest in in vivo tumor growth [51]. Similarly, the RIPK1/RIPK3 are highly expressed in pancreatic cancers and the in vivo deletion of these necroptosis proteins delayed oncogenic progression [46]. In this study, we found that both Ripk3 and Mlkl silencing rendered recurrent tumor cells inefficient in colony formation (Fig. 3a, b and Supplementary Fig. 1C, D), which support the role of RIPK3-MLKL axis in supporting cell growth during tumor recurrence. We also found Ripk3 silencing increased cytokinesis failure, p53 stabilization, and inactivation of YAP/TAZ signaling pathway [31], which in term leading to DNA damage and mitotic defects. Interestingly, RIPK1, which activates RIPK3 during activated necroptosis, is also reported to maintain chromosome stability by interacting with caspase-8 [52]. In our study, we have found similar critical role of the RIPK3 in the maintaining the integrity and proper progression of mitosis. More interestingly, RIPK3 together with RIPK1, caspase-8, and PLK is also found in the mitosis-associated ripoptosome [52]. While RIPK3 was not shown to play a critical role in the previous study, the exaggerated expression of RIPK3 in the recurrent tumor cells may be required to ensure the proper mitosis and cytokinesis under the altered properties of recurrent tumor cells.
Our studies also have significant therapeutic implications. Erastin can induce a nonapoptotic form of cell death named ferroptosis [40]. Although ferroptosis is considered to be regulated by GSH peroxidase 4 [53], our previous study has shown that low dose of erastin can induce necroptosis [9], which is supported by another study [54]. Consistent with our data, in the recurrent tumor cells with exaggerated RIPK3 expression, cell death induced by erastin can be mitigated by knockdown of Ripk3 (Fig. 6). Furthermore, both Nec-5 and ferrostatin-1 rescued erastin-induced cell death (Fig. 5f). These data indicate that high RIPK3 expression may contribute to its sensitivities to cell death induced by cystine deprivation. While tumor recurrence is usually considered incurable, this finding suggests that the collateral vulnerability to cystine deprivation or xCT inhibitors may hold therapeutic potential. In vivo cystine removal using recombinant cyst(e)inase [55] and inhibitors of cystine importer xCT [56] are being developed for clinical translation. Our data suggest that the recurrent tumors expressing high RIPK3 may be uniquely sensitive to these therapeutic approaches.
Methods
Cell culture
Primary and recurrent MTB/TAN tumor cells described previously [5] were cultured in Dulbecco’s modified Eagle’s medium (DMEM; GIBCO-11995) supplemented with 10% fetal bovine serum and 1 × antibiotics (penicillin, 10,000 UI/ml and streptomycin, 10,000 UI/ml) in a humidified incubator at 37 °C and 5% CO2. For primary cells, 10 ng/ml EGF, 5 µg/ml insulin, 1 µg/ml hydrocortisone, 5 µg/ml prolactin, 1 µM progesterone, and 2 µg/ml doxycycline were added to the media to maintain HER2/neu expression. For recurrent cells, 10 ng/ml EGF and 5 µg/ml insulin were added to the media. These cell lines have been tested to be mycoplasma free using the LookOut mycoplasma PCR detection kit (Sigma, MP0035).
Constructs and lentivirus infections
ShRNA targeting mouse Ripk3 RNA were purchase from Sigma (TRCN0000022536 and TRCN0000424625). ShRNAs targeting mouse Mlkl were purchase from Sigma (TRCN0000022599 and TRCN000022602). Lentivirus expressing RIPK3 shRNA was generated by transfecting HEK-293T cells in six-well plate with a 1:0.1:1 ratio of pMDG2:pVSVG:pLKO.1 with TransIT-LT1 transfection reagent (Mirus). After filtering through 0.45 µm of cellulose acetate membrane (VWR, 28145–481), lentivirus (250 µl) were added to a 60 mm dish of recurrent cells with polybrene (8 µg/ml) and selected with puromycin (5 µg/ml). For CRISPR activation, lenti dCAS-VP64_Blast, lentiMPH v2, and lenti sgRNA (MS2) puro backbone were gifts from Feng Zhang (Addgene plasmids #61425, #89308, and #73795). sgRNA targeting mouse Ripk3 (guide sequence: CTAAGGGGTCCAAGAGAGAC) was cloned into lenti sgRNA(MS2)_puro backbone by Golden Gate reaction.
Cell viability and cytotoxicity
Cell viability assay was performed by using the CellTiter-Glo luminescent cell viability assay (Promega) following manufacturer’s protocol. Cytotoxicity was determined by membrane rupture and protease release using the CellTox Green cytotoxicity assay (Promega) following manufacturer’s protocol.
Cell/tumor lysate and western blots
Cells were washed with ice cold PBS, harvested, and then resuspended in NP-40 buffer with protease and phosphatase inhibitors and lysed by incubating in at 4 °C with constant vortex for 30 min, then spun down at 13000 rpm for 10 min at 4 °C. Supernatant was transferred to another tube, and protein concentration was measured by the BCA protein assay kit (#23225, ThermoFisher). Frozen human breast cancer specimens were collected, stored, and identity retained under a Duke approved IRB protocol (eIRB Pro00012025). Tissue specimens were from primary breast cancers resected at Duke University Medical Center. We compared a series of primary tumors for which periodic follow-up was available through the Duke Tumor Registry and Duke electronic health records. We selected cases that had either experienced a clear distant recurrence or were long-term (>10 years) disease free. Tumor contents of the thus selected specimen was evaluated by an H&E stained section to have >50% tumor nuclei contents before protein extraction. Protein was extracted by placing frozen tissue in lysing matrix A tubes (cat. # 6910–100 from MP Biomedical formerly Biogene) with lysis buffer containing protease and phosphatase inhibitors. Tissue was homogenized in a mini-bead beater BIO 101 in the coldroom for 30 s at setting 6, spun at 14 K RPM for 2 min. The supernatant was passed through a 20-g needle, spun at 14 K RPM for 20 min at 4 °C. The supernatant was removed, and protein concentration determined by the Bradford method. Protein was stored at −80 °C until use.
Western blotting was performed as previously described [57]. Nuclear and cytoplasmic extraction for YAP/TAZ was performed by following manufacturer’s protocol (#78835, ThermoFisher). Quantification of YAP/TAZ was performed by Image J software and normalized to Lamin A/C protein level. Around 20 µg of protein was loaded on 8% SDS-PAGE gels, transferred to PDVF membrane, blocked with 5% nonfat milk in 1× TBST, incubated with primary antibodies overnight at 4 °C. Primary antibodies: RIPK1 (1:1000, 610458, BD biosciences); RIPK3 (1:1000, sc-374639, Santa Cruz); GAPDH (1:2000, sc-25778, Santa Cruz); Phospho-MLKL (Ser345) (1:1000, #62233, Cell signaling); MLKL (1:1000, #28640, Cell signaling); Pho-p53-S15 (1:1000, #92845, Cell signaling); Lamin A/C (1:1000, #4777T, Cell signaling); TAZ (1:1000, 560235, BD biosciences); YAP (1:1000, sc376830, Santa Cruz).
Quantitative real-time PCR
RNA from the samples was extracted by the RNeasy Mini Kit (Qiagen) following the manufacturer’s protocol. RNA was reverse transcribed to cDNA by random hexamers and SuperScript II (Invitrogen). Quantitative real-time PCR was performed following the manufacturer’s protocol by using Power SYBR Green PCR Mix (Applied Biosystems) and StepOnePlus Real-time PCR system (Applied Biosystems). Samples were biologically triplicated for mean ± SEM. Data were representative of three independent repeats. Mouse beta-actin (reference gene) primers: sense, 5′-GGC TGT ATT CCC CTC CAT CG-3′, antisense, 5′-CCA GTT GGT AAC AAT GCC ATG T-3′; Mouse RIPK3 primers: sense, 5′-TCT GTC AAG TTA TGG CCT ACT GG-3′, antisense, 5′-GGA ACA CGA CTC CGA ACC C-3′. Mouse CTGF primers: sense, 5′- GCC TAC CGA CTG GAA GAC AC-3′, antisense, 5′-GGA TGC ACT TTT TGC CCT TCT TA-3′. Mouse CYR61 primers: sense, 5′-CTG CGC TAA ACA ACT CAA CGA-3′, antisense, 5′-GCA GAT CCC TTT CAG AGC GG-3′.
ChIP-Seq and ChIP-PCR
ChIP-Seq and ChIP-PCR were performed as described previously [23]. Tumor cells were crosslinked in 1% formaldehyde (Sigma) for 10 min, prior to quenching with 250 mM glycine. DNA was sonicated to an average shear length of ~250–450 bp length. Lysates were precleared with protein A/G beads and immunoprecipitated with 5 μg of H3K9ac, H3K4me3, H3K27me3, H3K9me2, and RNApol2 antibodies purchased from Abcam. DNA was sequentially washed with wash buffers. DNA was eluted from washed beads and reverse crosslinked with concentrated NaCl overnight. After reverse cross-linking, proteins were digested with Proteinase K and chelated with EDTA. DNA was purified using PCR purification columns (Qiagen) according to manufacturer instructions. All qPCR reactions were carried out with SYBR green (Bio-Rad) Primers for promoter region of RIPK3: sense, 5′-CTT GGA CCC CTT AGC TCC AC-3′, antisense, 5′-GTA CCT GGC CCA AGA CAA CC-3′. Primers for TSS region of RIPK3: sense, 5′-CCC GGA CTT TGA ATG AGC GA-3′, antisense, 5′-CTC GGG TGG AAG CAG TTT CA-3′. Ct values were normalized to input DNA.
Immunoprecipitated DNA was sequenced on Illumina HiSeq 4000 sequencer with 50 bp single reads at an approximate depth of 55 million reads per sample. Sequencing reads underwent strict quality control processing with the TrimGalore package and were mapped to the mm10 genome using Bowtie aligner. Alignment files were converted to bigwig files by binning reads into 100-bp segments. H3K4me3, H3K9ac, and RNA pol II tracks were visualized for the Ripk3 promoter by IGV desktop viewer (Broad Institute).
Bisulfite sequencing
Bisulfite sequencing was performed using the EpiTect Bisulfite Kit (Qiagen) according to manufacturer instructions. The Ripk3 promoter region was PCR amplified using primers designed following previous study [58]. Sense, 5′-AGA GAA TTC GGA TCC TGG AGT TAA GGG GTT TAA GAG AGA T-3′, antisense, 5′-CTT CCA TGG CTC GAG CTT TAT CCC CTA CCT CAA AAA AAA C-3′. Amplified DNA was gel purified and transformed into competent bacteria. Ten independent bacterial colonies were sequenced for Ripk3, and DNA sequences were aligned with DNASTAR MegAlign software.
Immunofluorescence microscopy
Recurrent tumor cells were washed once with PBS and fixed in 4% paraformaldehyde for 15 min, followed by permeabilization and blocking with 0.2% Triton X-100 and 2% BSA for 15 min. Primary antibodies were incubated with the cells for 1 h. Immunofluorescence microscopy were performed using EVOS FL cell imaging system (ThermoFisher) or confocal microscope (880, Zeiss). Antibody: Alexa Fluor 594 Phalloidin (1:100, A12381, ThermoFisher); TAZ (1:100, 560235, BD biosciences); pho-histone H2AX-S139 (1:100, GTX628996, GeneTex).
RNA-seq and GESA
TrimGalore toolkit is used to process RNA-seq data. It employs Cutadapt to trim low-quality bases and Illumina sequencing adapters from the 3′ end of the reads. Reads (>20 nt) after trimming were kept for further analysis. By using the STAR RNA-seq alignment tool, reads were mapped to the GRCm38v73 version of the mouse genome and transcriptome. If reads were mapped to a single-genomic location, it were kept for subsequent analysis. Quantification of read counts of genes were performed using HTSeq. Only genes that had more than ten reads in any given library were further analyzed. DESeq2 Bioconductor package with the R statistical programming environment were applied for differential analysis to compare recurrent tumor cells with control or shRIPK3 silencing. The false discovery rate was calculated to control for multiple hypothesis testing. GSEA was performed to identify gene ontology terms and pathways associated with altered gene expression for the comparisons between control and recurrent cells with shRIPK3 silencing.
Statistical analysis
All measurements in biochemical assays were conducted in either triplicates or quadruplets to obtain mean and standard deviations. Randomization were conducted by assigned random order for processing. Data represent the mean ± the standard error of the mean. p values were determined by two ANOVA test with Bonferroni post hoc tests or a two-tailed Student’s t test in Graphpad. Error bars represent SEM, and significance between samples is denoted as *p < 0.05; **p < 0.01; and ***p < 0.001.
Supplementary information
Acknowledgements
We are grateful for technical support from the members of the Chi lab; Dr David Corcoran for technical assistance with RNA-seq. We are also grateful for the help of Drs Kelly Kiejda, Ricardo Vilain, and Rodney Scott for samples and data access. We acknowledge the financial support in part by DOD grants (W81XWH-17–1–0143, W81XWH-15–1–0486, and W81XWH-19–1–0842), NIH grants (GM124062, 1R01NS111588–01A1), the Duke Bridge Fund and Duke Cancer Institute (DCI) pilot fund.
Data availability
RNA-seq for recurrent cells with shRIPK3 silencing has been deposited in the NCBI Genome Expression Omnibus (GEO, GSE124634). All data and reagents supporting the findings of this study are available from the authors upon reasonable request.
Compliance with ethical standards
Conflict of interest
The authors declare that they have no conflict of interest.
Footnotes
Edited by J. Silke
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
These authors contributed equally: James V. Alvarez, Jen-Tsan Chi
Supplementary information
The online version of this article (10.1038/s41418-020-0499-y) contains supplementary material, which is available to authorized users.
References
- 1.Kimbung S, Loman N, Hedenfalk I. Clinical and molecular complexity of breast cancer metastases. Semin Cancer Biol. 2015;35:85–95. doi: 10.1016/j.semcancer.2015.08.009. [DOI] [PubMed] [Google Scholar]
- 2.D’Cruz CM, Gunther EJ, Boxer RB, Hartman JL, Sintasath L, Moody SE, et al. c-MYC induces mammary tumorigenesis by means of a preferred pathway involving spontaneous Kras2 mutations. Nat Med. 2001;7:235–9. doi: 10.1038/84691. [DOI] [PubMed] [Google Scholar]
- 3.Gunther EJ, Moody SE, Belka GK, Hahn KT, Innocent N, Dugan KD, et al. Impact of p53 loss on reversal and recurrence of conditional Wnt-induced tumorigenesis. Genes Dev. 2003;17:488–501. doi: 10.1101/gad.1051603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Moody SE, Sarkisian CJ, Hahn KT, Gunther EJ, Pickup S, Dugan KD, et al. Conditional activation of Neu in the mammary epithelium of transgenic mice results in reversible pulmonary metastasis. Cancer Cell. 2002;2:451–61. doi: 10.1016/s1535-6108(02)00212-x. [DOI] [PubMed] [Google Scholar]
- 5.Moody SE, Perez D, Pan TC, Sarkisian CJ, Portocarrero CP, Sterner CJ, et al. The transcriptional repressor Snail promotes mammary tumor recurrence. Cancer Cell. 2005;8:197–209. doi: 10.1016/j.ccr.2005.07.009. [DOI] [PubMed] [Google Scholar]
- 6.Boxer RB, Jang JW, Sintasath L, Chodosh LA. Lack of sustained regression of c-MYC-induced mammary adenocarcinomas following brief or prolonged MYC inactivation. Cancer Cell. 2004;6:577–86. doi: 10.1016/j.ccr.2004.10.013. [DOI] [PubMed] [Google Scholar]
- 7.Keenan MM, Chi JT. Alternative fuels for cancer cells. Cancer J. 2015;21:49–55. doi: 10.1097/PPO.0000000000000104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tang X, Keenan MM, Wu J, Lin CA, Dubois L, Thompson JW, et al. Comprehensive profiling of amino acid response uncovers unique methionine-deprived response dependent on intact creatine biosynthesis. PLoS Genet. 2015;11:e1005158. doi: 10.1371/journal.pgen.1005158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tang X, Wu J, Ding CK, Lu M, Keenan MM, Lin CC, et al. Cystine deprivation triggers programmed necrosis in VHL-deficient renal cell carcinomas. Cancer Res. 2016;76:1892–903. doi: 10.1158/0008-5472.CAN-15-2328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tang X, Ding CK, Wu J, Sjol J, Wardell S, Spasojevic I, et al. Cystine addiction of triple-negative breast cancer associated with EMT augmented death signaling. Oncogene. 2017;36:4379. doi: 10.1038/onc.2017.192. [DOI] [PubMed] [Google Scholar]
- 11.Chen PH, Wu J, Ding CC, Lin CC, Pan S, Bossa N, et al. Kinome screen of ferroptosis reveals a novel role of ATM in regulating iron metabolism. Cell Death Differ. 2019. 10.1038/s41418-019-0393-7. [DOI] [PMC free article] [PubMed]
- 12.Ding C-KC, Rose J, Wu J, Sun T, Chen K-Y, Chen P-H, et al. Mammalian stringent-like response mediated by the cytosolic NADPH phosphatase MESH1. 2018. https://www.biorxiv.org/content/10.1101/325266v1.full.
- 13.Chen PH, Smith TJ, Wu J, Siesser PF, Bisnett BJ, Khan F, et al. Glycosylation of KEAP1 links nutrient sensing to redox stress signaling. EMBO J. 2017;36:2233–50. doi: 10.15252/embj.201696113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yang WH, Ding CKC, Sun T, Hsu DS, Chi JT. The hippo pathway effector TAZ regulates ferroptosis in renal cell carcinoma. Cell Rep. 2019;28:2501–8. e2504. doi: 10.1016/j.celrep.2019.07.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yang W-H, Chi J-T. Hippo pathway effectors YAP/TAZ as novel determinants of ferroptosis. Mol Cell Oncol. 2020;7:1. 10.1080/23723556.2019.1699375. [DOI] [PMC free article] [PubMed]
- 16.Yang WH, Huang Z, Wu J, Ding C-KC, Murphy SK, Chi J-T. A TAZ-ANGPTL4-NOX2 axis regulates ferroptotic cell death and chemoresistance in epithelial ovarian cancer. Mol Cancer Res. 2019;2019:molcanres.0691. doi: 10.1158/1541-7786.MCR-19-0691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lu SC. Regulation of glutathione synthesis. Mol Asp Med. 2009;30:42–59. doi: 10.1016/j.mam.2008.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Garcia-Ruiz C, Fernandez-Checa JC. Redox regulation of hepatocyte apoptosis. J Gastroenterol Hepatol. 2007;22:S38–42. doi: 10.1111/j.1440-1746.2006.04644.x. [DOI] [PubMed] [Google Scholar]
- 19.Newton K. RIPK1 and RIPK3: critical regulators of inflammation and cell death. Trends Cell Biol. 2015;25:347–53. doi: 10.1016/j.tcb.2015.01.001. [DOI] [PubMed] [Google Scholar]
- 20.Wang H, Sun L, Su L, Rizo J, Liu L, Wang LF, et al. Mixed lineage kinase domain-like protein MLKL causes necrotic membrane disruption upon phosphorylation by RIP3. Mol Cell. 2014;54:133–46. doi: 10.1016/j.molcel.2014.03.003. [DOI] [PubMed] [Google Scholar]
- 21.Koo GB, Morgan MJ, Lee DG, Kim WJ, Yoon JH, Koo JS, et al. Methylation-dependent loss of RIP3 expression in cancer represses programmed necrosis in response to chemotherapeutics. Cell Res. 2015;25:707–25. doi: 10.1038/cr.2015.56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Geserick P, Wang J, Schilling R, Horn S, Harris PA, Bertin J, et al. Absence of RIPK3 predicts necroptosis resistance in malignant melanoma. Cell Death Dis. 2015;6:e1884. doi: 10.1038/cddis.2015.240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mabe NW, Fox DB, Lupo R, Decker AE, Phelps SN, Thompson JW, et al. Epigenetic silencing of tumor suppressor Par-4 promotes chemoresistance in recurrent breast cancer. J Clin Investig. 2018;128:4413–28. doi: 10.1172/JCI99481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Payne AW, Pant DK, Pan TC, Chodosh LA. Ceramide kinase promotes tumor cell survival and mammary tumor recurrence. Cancer Res. 2014;74:6352–63. doi: 10.1158/0008-5472.CAN-14-1292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Alvarez JV, Pan TC, Ruth J, Feng Y, Zhou A, Pant D, et al. Par-4 downregulation promotes breast cancer recurrence by preventing multinucleation following targeted therapy. Cancer Cell. 2013;24:30–44. doi: 10.1016/j.ccr.2013.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Karami-Tehrani F, Malek AR, Shahsavari Z, Atri M. Evaluation of RIP1K and RIP3K expressions in the malignant and benign breast tumors. Tumour Biol. 2016;37:8849–56. doi: 10.1007/s13277-015-4762-7. [DOI] [PubMed] [Google Scholar]
- 27.Vecchi M, Confalonieri S, Nuciforo P, Vigano MA, Capra M, Bianchi M, et al. Breast cancer metastases are molecularly distinct from their primary tumors. Oncogene. 2008;27:2148–58. doi: 10.1038/sj.onc.1210858. [DOI] [PubMed] [Google Scholar]
- 28.Mathe A, Wong-Brown M, Morten B, Forbes JF, Braye SG, Avery-Kiejda KA, et al. Novel genes associated with lymph node metastasis in triple negative breast cancer. Sci Rep. 2015;5:15832. doi: 10.1038/srep15832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sun L, Wang H, Wang Z, He S, Chen S, Liao D, et al. Mixed lineage kinase domain-like protein mediates necrosis signaling downstream of RIP3 kinase. Cell. 2012;148:213–27. doi: 10.1016/j.cell.2011.11.031. [DOI] [PubMed] [Google Scholar]
- 30.Normand G, King RW. Understanding cytokinesis failure. Adv Exp Med Biol. 2010;676:27–55. doi: 10.1007/978-1-4419-6199-0_3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ganem NJ, Cornils H, Chiu SY, O’Rourke KP, Arnaud J, Yimlamai D, et al. Cytokinesis failure triggers hippo tumor suppressor pathway activation. Cell. 2014;158:833–48. doi: 10.1016/j.cell.2014.06.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Shieh SY, Ikeda M, Taya Y, Prives C. DNA damage-induced phosphorylation of p53 alleviates inhibition by MDM2. Cell. 1997;91:325–34. doi: 10.1016/s0092-8674(00)80416-x. [DOI] [PubMed] [Google Scholar]
- 33.Giam M, Rancati G. Aneuploidy and chromosomal instability in cancer: a jackpot to chaos. Cell Div. 2015;10:3. doi: 10.1186/s13008-015-0009-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Taylor AM, Shih J, Ha G, Gao GF, Zhang X, Berger AC, et al. Genomic and functional approaches to understanding cancer aneuploidy. Cancer Cell. 2018;33:676–89 e673. doi: 10.1016/j.ccell.2018.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zanconato F, Cordenonsi M, Piccolo S. YAP/TAZ at the roots of cancer. Cancer Cell. 2016;29:783–803. doi: 10.1016/j.ccell.2016.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zhao B, Wei X, Li W, Udan RS, Yang Q, Kim J, et al. Inactivation of YAP oncoprotein by the Hippo pathway is involved in cell contact inhibition and tissue growth control. Genes Dev. 2007;21:2747–61. doi: 10.1101/gad.1602907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yang Z, Nakagawa K, Sarkar A, Maruyama J, Iwasa H, Bao Y, et al. Screening with a novel cell-based assay for TAZ activators identifies a compound that enhances myogenesis in C2C12 cells and facilitates muscle repair in a muscle injury model. Mol Cell Biol. 2014;34:1607–21. doi: 10.1128/MCB.01346-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lewerenz J, Klein M, Methner A. Cooperative action of glutamate transporters and cystine/glutamate antiporter system Xc- protects from oxidative glutamate toxicity. J Neurochem. 2006;98:916–25. doi: 10.1111/j.1471-4159.2006.03921.x. [DOI] [PubMed] [Google Scholar]
- 39.Dolma S, Lessnick SL, Hahn WC, Stockwell BR. Identification of genotype-selective antitumor agents using synthetic lethal chemical screening in engineered human tumor cells. Cancer Cell. 2003;3:285–96. doi: 10.1016/s1535-6108(03)00050-3. [DOI] [PubMed] [Google Scholar]
- 40.Dixon SJ, Lemberg KM, Lamprecht MR, Skouta R, Zaitsev EM, Gleason CE, et al. Ferroptosis: an iron-dependent form of nonapoptotic cell death. Cell. 2012;149:1060–72. doi: 10.1016/j.cell.2012.03.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Gout PW, Buckley AR, Simms CR, Bruchovsky N. Sulfasalazine, a potent suppressor of lymphoma growth by inhibition of the x(c)- cystine transporter: a new action for an old drug. Leukemia. 2001;15:1633–40. doi: 10.1038/sj.leu.2402238. [DOI] [PubMed] [Google Scholar]
- 42.Skouta R, Dixon SJ, Wang J, Dunn DE, Orman M, Shimada K, et al. Ferrostatins inhibit oxidative lipid damage and cell death in diverse disease models. J Am Chem Soc. 2014;136:4551–6. doi: 10.1021/ja411006a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wang K, Li J, Degterev A, Hsu E, Yuan J, Yuan C. Structure-activity relationship analysis of a novel necroptosis inhibitor, Necrostatin-5. Bioorg Med Chem Lett. 2007;17:1455–65. doi: 10.1016/j.bmcl.2006.11.056. [DOI] [PubMed] [Google Scholar]
- 44.Konermann S, Brigham MD, Trevino AE, Joung J, Abudayyeh OO, Barcena C, et al. Genome-scale transcriptional activation by an engineered CRISPR-Cas9 complex. Nature. 2015;517:583–8. doi: 10.1038/nature14136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hockendorf U, Yabal M, Herold T, Munkhbaatar E, Rott S, Jilg S, et al. RIPK3 restricts myeloid leukemogenesis by promoting cell death and differentiation of leukemia initiating cells. Cancer Cell. 2016;30:75–91. doi: 10.1016/j.ccell.2016.06.002. [DOI] [PubMed] [Google Scholar]
- 46.Seifert L, Werba G, Tiwari S, Giao Ly NN, Alothman S, Alqunaibit D, et al. The necrosome promotes pancreatic oncogenesis via CXCL1 and Mincle-induced immune suppression. Nature. 2016;532:245–9. doi: 10.1038/nature17403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Najjar M, Saleh D, Zelic M, Nogusa S, Shah S, Tai A, et al. RIPK1 and RIPK3 kinases promote cell-death-independent inflammation by toll-like receptor 4. Immunity. 2016;45:46–59. doi: 10.1016/j.immuni.2016.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Yatim N, Jusforgues-Saklani H, Orozco S, Schulz O, Barreira da Silva R, Reis e Sousa C, et al. RIPK1 and NF-kappaB signaling in dying cells determines cross-priming of CD8(+) T cells. Science. 2015;350:328–34. doi: 10.1126/science.aad0395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Moriwaki K, Balaji S, McQuade T, Malhotra N, Kang J, Chan FK. The necroptosis adaptor RIPK3 promotes injury-induced cytokine expression and tissue repair. Immunity. 2014;41:567–78. doi: 10.1016/j.immuni.2014.09.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Hanggi K, Vasilikos L, Valls AF, Yerbes R, Knop J, Spilgies LM, et al. RIPK1/RIPK3 promotes vascular permeability to allow tumor cell extravasation independent of its necroptotic function. Cell Death Dis. 2017;8:e2588. doi: 10.1038/cddis.2017.20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Liu X, Zhou M, Mei L, Ruan J, Hu Q, Peng J, et al. Key roles of necroptotic factors in promoting tumor growth. Oncotarget. 2016;7:22219–33. doi: 10.18632/oncotarget.7924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Liccardi G, Ramos Garcia L, Tenev T, Annibaldi A, Legrand AJ, Robertson D, et al. RIPK1 and caspase-8 ensure chromosome stability independently of their role in cell death and inflammation. Mol Cell. 2019;73:413–28. e417. [DOI] [PMC free article] [PubMed]
- 53.Yang WS, SriRamaratnam R, Welsch ME, Shimada K, Skouta R, Viswanathan VS, et al. Regulation of ferroptotic cancer cell death by GPX4. Cell. 2014;156:317–31. doi: 10.1016/j.cell.2013.12.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Chen MS, Wang SF, Hsu CY, Yin PH, Yeh TS, Lee HC, et al. CHAC1 degradation of glutathione enhances cystine-starvation-induced necroptosis and ferroptosis in human triple negative breast cancer cells via the GCN2-eIF2alpha-ATF4 pathway. Oncotarget. 2017;8:114588–602. doi: 10.18632/oncotarget.23055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Cramer SL, Saha A, Liu J, Tadi S, Tiziani S, Yan W, et al. Systemic depletion of L-cyst(e)ine with cyst(e)inase increases reactive oxygen species and suppresses tumor growth. Nat Med. 2017;23:120–7. doi: 10.1038/nm.4232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Yoshikawa M, Tsuchihashi K, Ishimoto T, Yae T, Motohara T, Sugihara E, et al. xCT inhibition depletes CD44v-expressing tumor cells that are resistant to EGFR-targeted therapy in head and neck squamous cell carcinoma. Cancer Res. 2013;73:1855–66. doi: 10.1158/0008-5472.CAN-12-3609-T. [DOI] [PubMed] [Google Scholar]
- 57.Lin CC, Kitagawa M, Tang X, Hou MH, Wu J, Qu DC, et al. CoA synthase regulates mitotic fidelity via CBP-mediated acetylation. Nat Commun. 2018;9:1039. doi: 10.1038/s41467-018-03422-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Yang Z, Jiang B, Wang Y, Ni H, Zhang J, Xia J, et al. 2-HG inhibits necroptosis by stimulating DNMT1-dependent hypermethylation of the RIP3 promoter. Cell Rep. 2017;19:1846–57. doi: 10.1016/j.celrep.2017.05.012. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
RNA-seq for recurrent cells with shRIPK3 silencing has been deposited in the NCBI Genome Expression Omnibus (GEO, GSE124634). All data and reagents supporting the findings of this study are available from the authors upon reasonable request.