Abstract
Background
Our recent study identified that human chemokine-like factor (CKLF)-like MARVEL transmembrane domain-containing family member 2 (CMTM2) was deregulated in hepatocellular carcinoma (HCC) tissues and posed as a potential tumor suppressor. However, the mechanism of CMTM2 in HCC occurrence and development has not been well elaborated.
Materials and Methods
The expression of CMTM2 was knocked-down by RNA interruption in Huh-7 and SMMC7721 cells. Cell proliferation ability was detected by CCK8 test and colony formation assay. The cell invasion and migration were measured by wound healing and Transwell assay.
Results
We found that the cell proliferation was significantly increased by interruption of CMTM2 expression, both in Huh-7 and SMMC7721 cells. Moreover, down-regulated CMTM2 could promote the invasion and migration ability of HCC cells through inducing the epithelial-mesenchymal transition (EMT) process. We further discovered that both the expression of CMTM2 and the EMT-associated marker E-cadherin were decreased in the same thirty cases of HCC tissues compared with the corresponding adjacent non-tumor tissues. Pearson correlation test showed that there was a significantly positive correlation between CMTM2 and E-cadherin in HCC tissues (P<0.05).
Conclusion
Based on the results of cell model and HCC tissues, our study suggests that down-regulated CMTM2 promotes HCC metastasis through inducing the EMT process.
Keywords: CMTM2, CMTM family members, HCC, metastasis, EMT
Introduction
Human chemokine-like factor (CKLF)-like MARVEL transmembrane domain-containing family (CMTM) members are novel proteins characterized by the MARVEL domain.1,2 There are nine members in the CMTM family, CMTM1, CMTM2, CMTM3, CMTM4, CMTM5, CMTM6, CMTM7, CMTM8, and CKLF. As a kind of cellular transmembrane protein and secretory protein, the CMTM family members implicate in many important physiological processes, such as male reproduction and body immunity. CMTM2 is highly expressed in male testes detected by immunohistochemistry (IHC).3 Knock-out of CMTM2 or CMTM4 in mice could lead to a significant decline in sperm production, decreased epididymal sperm motility, and increased abnormal sperms, showing an essential role of CMTM2 or CMTM4 in spermiogenesis and male fertility.4,5 Moreover, CMTM4 and CMTM6 were identified as critical protein regulators of PD-L1 by protecting it from ubiquitination. CMTM6 interference could cause deregulated PD-L1 expression in many cancer cells and primary human dendritic cells.6 In recent studies, some CMTM family members were found deregulated in tumors and acted as potential tumor suppressors. CMTM3 was found hyper-methylated in colorectal cancer, male laryngeal squamous cell carcinoma, and non-small cell lung cancer (NSCLC).7–9 CMTM3 could inhibit epidermal growth factor (EGF)-mediated migration and STAT3 signaling, suppress the expression of EGFR through promoting the degradation of EGFR in gastric cancer cells.10 In our previous studies, we have shown that CMTM4, CMTM6, and CMTM7 were down-regulated in hepatocellular carcinoma (HCC) tissues through IHC detection, and had a relationship with the poor prognosis of HCC patients after survival analysis, suggesting a tumor suppressor role of CMTM members in HCC carcinogenesis and progression.11–13
However, the role and mechanism of CMTM2 (alias name CKLFSF2) in tumors have not been well elucidated. Because of the high expression in human testes and spermatozoa, CMTM2 has been thought to be a pivotal protein for male reproduction.14 Most CMTM2 studies focused on explaining the relationship between CMTM2 expression and spermatogenesis through animal models. In these studies, deregulated CMTM2 expression was found to impair spermatogenesis, and spermatogenesis disorders could conversely lead to reduced CMTM2 expression. Patients with sertoli cell only syndrome (SCOS) had no expression of CMTM2 in the testicular tissues,15 and varicocele in rats caused a detrimental effect on CMTM2 levels and decreased spermatogonia cell number, seminiferous tubules diameter, and sperm indices.16 Apart from the wide expression in male reproductive systems, CMTM2 locates on chromosome 16q22.1 that harbors numerous tumor suppressor genes,3 implying a potential tumor-suppressive activity of CMTM2. Our recent study explored that CMTM2 was down-regulated in HCC tissues and had a correlation with the tumor grade of HCC patients.17 Furthermore, CMTM2 was found more highly expressed in metastatic salivary adenoid cystic carcinoma (SACC) tissues of patients without tumor recurrence or perineural invasion compared to those with tumor recurrence.18 These studies suggest that an important role of CMTM2 may play in the development of tumor metastasis.
To elaborate the role and mechanism of CMTM2 in HCC metastasis and progression, we knocked-down the expression of CMTM2 in HCC cell lines to measure the changes in cellular functions and epithelial-mesenchymal transition (EMT). We found that down-regulated CMTM2 promoted HCC metastasis through inducing the EMT process.
Materials and Methods
Tissue Samples
Thirty pairs of HCC and corresponding adjacent non-tumor tissues were collected at the Affiliated Hospital of Guilin Medical University between 2015 and 2016. All tissue samples were obtained from HCC patients underwent surgery without any radiotherapy or chemotherapy. The tissues were preserved in liquid nitrogen immediately after surgery. The tissue samples and clinicopathological information were collected under written informed consent from all patients. This study was conducted in accordance with the Declaration of Helsinki and with the approval from the Ethics Committee of Guilin Medical University.
Cell Lines and Plasmids
Human hepatocytes (L02) were purchased from the Cell Bank (Chinese Academy of Sciences, Shanghai, China), and hepatic tumor cell lines (Hep3B, Huh-7, SMMC7721) were purchased from ATCC (Manassas, VA, USA). The cell lines were all grown in a cell incubator at 37°C with a 5% CO2/95% air atmosphere.
Three shRNAs targeting CMTM2 were constructed from a GV102 vector by Genechem (Shanghai, China). The target sequence for KD-1# was 5ʹ-GGGCACGCTGAGATCAAGATT-3ʹ, the target sequence for KD-2# was 5ʹ-GGCCAGCAAAGGGAAAGAAAT-3ʹ, and the target sequence for KD-3# was 5ʹ-GACCTCTTCAACGACCTGATT-3ʹ.
Quantitative Reverse-Transcription Polymerase Chain Reaction (qRT-PCR)
Powdered tissues or cells were exposed to Trizol for total RNA extracting. After cDNA was produced from total RNA by reverse transcription reagents, the PCR reaction was started using SYBR Green Mixture (Tiangen, Beijing, China) according to the manufacture’s procedure. The primers for detecting are showed in Supplementary Table 1. The PCR reactions were: denaturation at 95°C for 10 min, 40 cycles of 95°C for 15 s, 65°C for 60 s. GAPDH was used as an internal control. The relative RNA expression was calculated using the 2−ΔΔCt method.
Western Blot
After extracted from cells using RIPA buffer and quantified, the soluble protein was resolved by SDS-PAGE and transferred to PVDF membranes for Western blot using ECL detection reagents. CMTM2 antibody (catalog No. NBP1-59439) was obtained from Novus (Littleton, CO, USA), EMT antibody sampler kit (catalog No. #9782) from Cell Signaling Technology (Danvers, MA, USA) and anti-β-actin antibody (catalog No. 21800) from Signalway Antibody LLC (College Park, Maryland, USA).
Cell Proliferation Test
The cell proliferation was detected by CCK8 assay using a kit from Beyotime (Shanghai, China). 5×103 cells/well were seeded in triplicate into 96-well plates. After 12 h, 24 h, 48 h, 72 h, and 96 h of culture, cells were treated with CCK8 and detected at OD values of 450 nm.
Colony Formation
Eight hundred cells/well were grown in a 6-well plate in triplicate and cultured for 2 weeks. After stained with crystal violet, the cell colony was photographed and counted under a microscope.
Cell Scratch Detection
Cells were grown in triplicate into 6-well plates. After culturing for 12 h, cells were scratched by 10 μL micropipette tips and added with fresh serum-free medium. After 0 h, 24 h, and 72 h from scratch, cells were photographed and measured for the migration length using Image J software. The migration length was determined as the wound width at 0 h minus the wound width at 24 h/72 h.
Cell Invasion and Migration Ability Test
Cell invasion and migration ability were detected by BD Transwell assay according to the previous published method.19
Statistical Analysis
All the data were shown as mean ± standard deviation and analyzed by SPSS software. Chi-square and t-tests were applied for the comparison of qualitative data and quantitative data between the two groups, respectively. P<0.05 was defined as statistically significant for all tests.
Results
Knock-Down of CMTM2 Promotes HCC Cell Proliferation
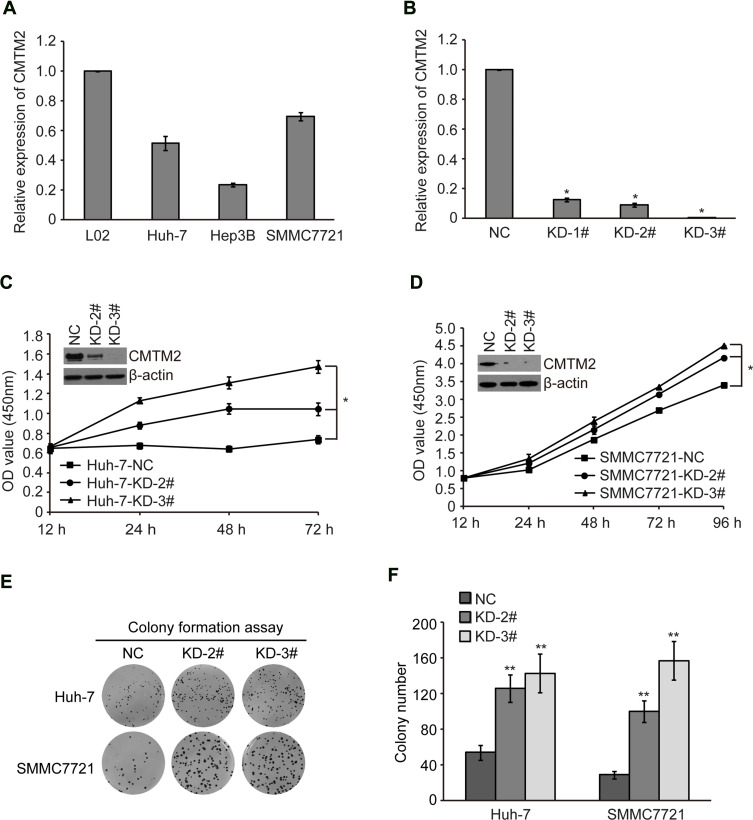
In order to explore the biological functions of CMTM2 in HCC cells, we first measured the mRNA expression of CMTM2 in several cell lines, SMMC7721, Huh-7, and Hep3B by qRT-PCR. As shown in Figure 1A, the mRNA expression of CMTM2 was lower in SMMC7721, Huh-7, and Hep3B cells than that in normal human liver L02 cells. Compared with L02 cells, there were about 69.68±2.64% of mRNA expression of CMTM2 in SMMC7721 cells, 51.66±4.62% in Huh-7 cells, and 23.49±1.32% in Hep3B cells. We then selected SMMC7721 and Huh-7 cell lines for subsequent CMTM2 knock-down assays. We purchased three shRNAs targeting CMTM2 from Genechem, named as KD-1#, KD-2#, and KD-3#. To find out the effect of these three shRNAs, we introduced the shRNAs into SMMC7721 cells. After 48 h of shRNAs transfection, we detected a significantly reduced mRNA expression of CMTM2 in SMMC7721 cells (P<0.05, Figure 1B). Because KD-2# and KD-3# had the more knock-down effect, they were chosen for the next cell function assays.
Figure 1.
Knock-down of CMTM2 promotes HCC cell proliferation.
Notes: (A) The mRNA expression of CMTM2 in L02 cell line and HCC cell lines as indicated was detected by qRT-PCR. (B) The mRNA expression of CMTM2 in SMMC7721 cells after introduction of CMTM2 shRNAs. NC means negative control shRNA and KD means shRNA targeting CMTM2. (C and D) Cell proliferation was detected by CCK-8 in Huh-7 and SMMC7721 cells after knocking-down of CMTM2. *P<0.05 compared to the control cells. (E and F) Cell proliferation was detected by colony formation assay in Huh-7 and SMMC7721 cells after knocking-down of CMTM2. **P<0.01 compared to the control cells.
Abbreviations: NC, negative control; KD, knock-down.
CMTM2 expression was then knocked-down by exogenous introduction of KD-2# and KD-3# in SMMC7721 and Huh-7 cell lines (named as SMMC7721-KD-2#, SMMC7721-KD-3#, Huh-7-KD-2#, and Huh-7-KD-3# cells). The control cells were introduced with a negative control plasmid (named as SMMC7721-NC and Huh-7-NC cells). After CMTM2 expression was knocked-down, both SMMC7721 and Huh-7 cells showed an increased cell proliferation, compared with the negative control cells (P<0.05, Figure 1C and D). Colony formation assay also showed that the cell proliferation ability was increased after knocking-down the expression of CMTM2 (P<0.01, Figure 1E and F). These results suggest that CMTM2 interference can promote the proliferation of HCC cells.
Knock-Down of CMTM2 Increases the Metastatic Ability of HCC Cells
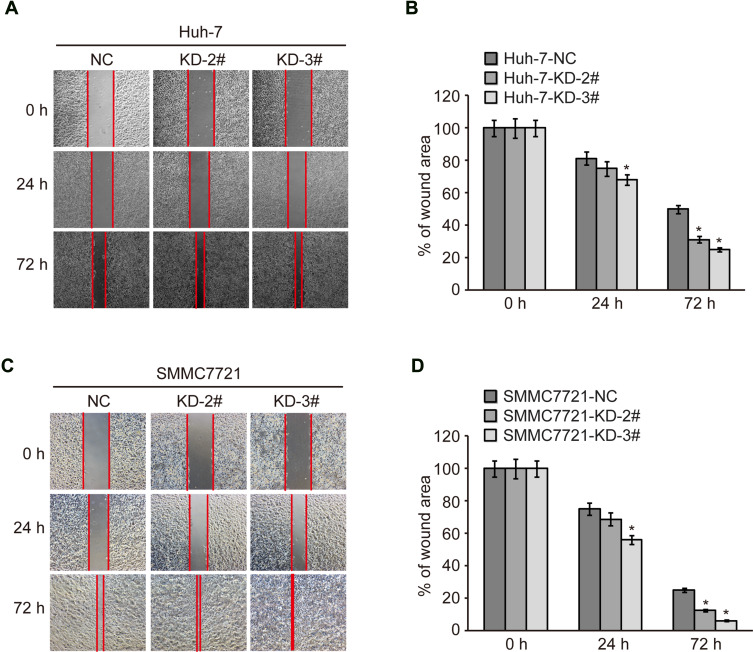
To find out whether CMTM2 plays a role in HCC cell metastasis, we performed a wound-healing assay to measure the metastatic ability of SMMC7721 and Huh-7 cells after knocking-down of CMTM2 expression (Figure 2A and C). As compared with Huh-7-NC cells, the metastatic ability of Huh-7-KD-2# and Huh-7-KD-3# cells was increased after 72 h of wound, especially Huh-7-KD-3# cells (P<0.05, Figure 2B). SMMC7721-KD-2# and SMMC7721-KD-3# cells also showed a wider migration distance than SMMC7721-NC cells after 72 h of wound (P<0.05, Figure 2D). The wound-healing assay indicates that CMTM2 interference can promote the metastasis of HCC cells.
Figure 2.
Knock-down of CMTM2 increases metastatic ability of HCC cells.
Notes: (A) Metastatic ability of Huh-7 cells was analyzed by wound healing after knocking-down of CMTM2. (B) Relative migration distance calculated after knocking-down of CMTM2 in Huh-7 cells. (C) Metastatic ability of SMMC7721 cells was analyzed by wound healing after knocking-down of CMTM2. (D) Relative migration distance calculated after knocking-down of CMTM2 in SMMC7721 cells. *P<0.05 compared to the control cells.
Abbreviations: NC, negative control; KD, knock-down.
Knock-Down of CMTM2 Promotes HCC Cell Invasion and Migration
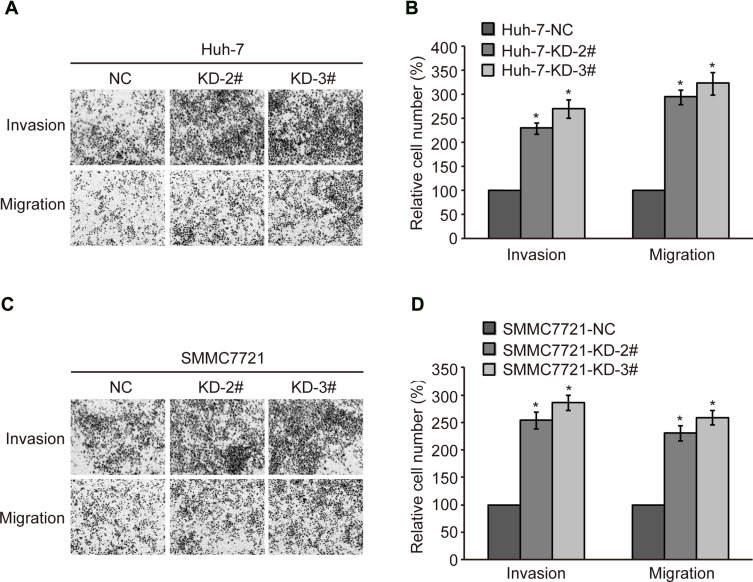
To further confirm the relationship between CMTM2 expression and HCC cell metastasis, a cell invasion and migration assay was conducted in SMMC7721 and Huh-7 cell lines. After CMTM2 expression was interrupted in SMMC7721 and Huh-7 cells, transwell assay showed an increased invasion and migration ability of CMTM2 knocked-down cells compared with SMMC7721-NC and Huh-7-NC cells (Figure 3A and C). As shown in Figure 3B and D, both KD-2# and KD-3# shRNAs could lead to an obvious up-regulation of invasion and migration in SMMC7721 and Huh-7 cells (P<0.05). These results show that decreased CMTM2 expression can promote the metastasis of HCC cells.
Figure 3.
Knock-down of CMTM2 promotes HCC cell invasion and migration.
Notes: (A) Invasion and migration of Huh-7 cells were analyzed by transwell assay after knocking-down of CMTM2. (B) Relative invasive and migrated cells calculated after knocking-down of CMTM2 in Huh-7 cells. (C) Invasion and migration of SMMC7721 cells were analyzed by wound healing after knocking-down of CMTM2. (D) Relative invasive and migrated cells calculated after knocking-down of CMTM2 in SMMC7721 cells. *P<0.05 compared to the control cells.
Abbreviations: NC, negative control; KD, knock-down.
Down-Regulated CMTM2 Promotes HCC Metastasis Through Inducing the EMT Process
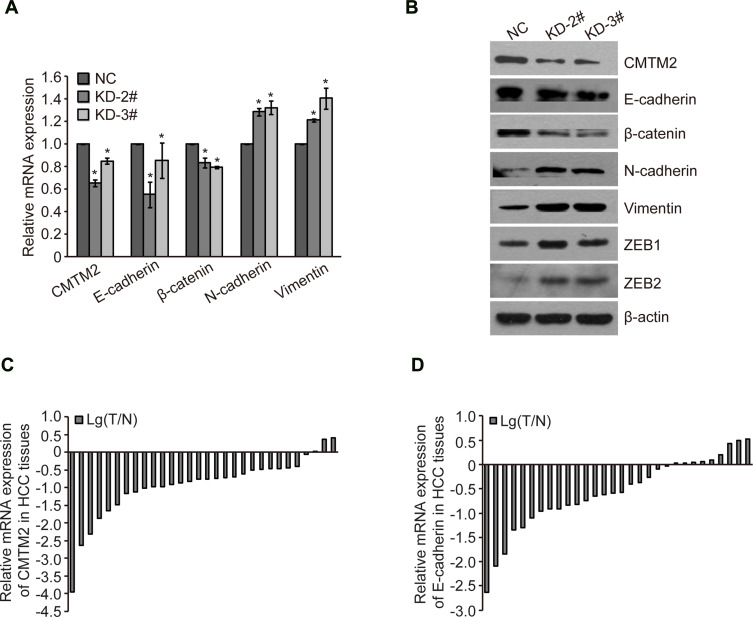
EMT is involved in the metastatic progression of various malignancies.20–22 We measured the expression of EMT-associated factors to determine the EMT process after knocking-down the expression of CMTM2. As compared to Huh-7-NC cells, the epithelial markers E-cadherin and β-catenin in Huh-7-KD-2# and Huh-7-KD-3# cells were decreased by 44.89±11.14%, 14.75±15.68%, 16.47±4.40%, and 20.72±1.03%, respectively. While the mesenchymal markers N-cadherin and Vimentin in Huh-7-KD-2# and Huh-7-KD-3# cells were increased by 28.64±3.24%, 32.49±5.82%, 21.38±1.30%, and 40.73±9.46%, respectively (Figure 4A). Western blot results also showed that epithelial markers E-cadherin and β-catenin were decreased, while mesenchymal markers N-cadherin, Vimentin, zinc-finger E-box binding protein 1 (ZEB1) and ZEB2 were increased after CMTM2 was knocked-down (Figure 4B). These results show down-regulated CMTM2 can induce the EMT process in HCC cells.
Figure 4.
Down-regulated CMTM2 promotes HCC metastasis through inducing EMT process.
Notes: (A) Relative gene expression after knocking-down of CMTM2 in Huh-7 cells. *P<0.05 compared to the control cells. (B) EMT protein expression after knocking-down of CMTM2 in Huh-7 cells. (C) Relative mRNA expression of CMTM2 in 30 HCC tissues. Lg(T/N) is logarithmic transformation of the CMTM2 expression in paired-HCC (T) and adjacent non-tumor tissues (N) by qRT-PCR. (D) Relative mRNA expression of E-cadherin in 30 HCC tissues. Lg(T/N) is logarithmic transformation of the E-cadherin expression in paired-HCC (T) and adjacent non-tumor tissues (N) by qRT-PCR.
Abbreviations: NC, negative control; KD, knock-down.
To further explore the relationship between CMTM2 and EMT process, we detected the expression of CMTM2 and E-cadherin in the same 30 paired HCC and adjacent non-tumor tissues. In consistent with our previous IHC results,17 the mRNA expression of CMTM2 was down-regulated in 90.0% (27/30) of HCC tissues by qRT-PCR (Figure 4C). In addition, CMTM2 had a relationship with metastasis in HCC patients after Fisher’s exact test (P<0.05, Table 1), which further supported the results in HCC cells. Furthermore, the mRNA expression of E-cadherin was also lower in 70.0% (21/30) of HCC tissues than that in the corresponding adjacent non-tumor tissues (Figure 4D). Although we did not find a significant correlation between the expression of E-cadherin and the clinicopathological features of HCC patients (P>0.05, Table 2), there was a significantly positive correlation between the expression of CMTM2 and E-cadherin in HCC tissues (r=0.437, P<0.05, Table 3). These results show that down-regulated expression of CMTM2 in HCC tissues can promote HCC metastasis through inducing the EMT process.
Table 1.
Correlation Between CMTM2 Expression and Clinic-Pathological Characteristics of HCC Patients
| Variables | Total | CMTM2 Expression | P value | |
|---|---|---|---|---|
| Low | High | |||
| Gender | ||||
| Male | 28 | 25 | 3 | 1.000 |
| Female | 2 | 2 | 0 | |
| Age-yr | ||||
| ≥50 | 18 | 18 | 0 | 0.054 |
| <50 | 12 | 9 | 3 | |
| Alcohol intake | ||||
| No | 20 | 19 | 1 | 0.251 |
| Yes | 10 | 8 | 2 | |
| HCC family history | ||||
| No | 27 | 24 | 3 | 1.000 |
| Yes | 3 | 3 | 0 | |
| HBV infection | ||||
| No | 5 | 5 | 0 | 1.000 |
| Yes | 25 | 22 | 3 | |
| Liver cirrhosis | ||||
| No | 12 | 10 | 2 | 0.548 |
| Yes | 18 | 17 | 1 | |
| AFP (ng/mL) | ||||
| ≥20 | 20 | 17 | 3 | 0.532 |
| <20 | 10 | 10 | 0 | |
| Tumor diameter (cm) | ||||
| ≥5 | 17 | 14 | 3 | 0.238 |
| <5 | 13 | 13 | 0 | |
| Tumor number | ||||
| 1 | 24 | 21 | 3 | 1.000 |
| >1 | 6 | 6 | 0 | |
| Tumor grade | ||||
| Ⅰ+Ⅱ | 14 | 13 | 1 | 1.000 |
| III | 16 | 14 | 2 | |
| Tumor stage | ||||
| Ⅰ+Ⅱ | 22 | 21 | 1 | 0.166 |
| III+Ⅳ | 8 | 6 | 2 | |
| Vascular invasion | ||||
| No | 21 | 20 | 1 | 0.234 |
| Yes | 9 | 7 | 2 | |
| Metastasis | ||||
| No | 26 | 25 | 1 | 0.039 |
| Yes | 4 | 2 | 2 | |
Notes: Bold value indicates significance; P value is based on Fisher’s exact test.
Abbreviation: AFP, α-fetoprotein.
Table 2.
Correlation Between E-Cadherin Expression and Clinic-Pathological Characteristics of HCC Patients
| Variables | Total | E-Cadherin Expression | P value | |
|---|---|---|---|---|
| High | Low | |||
| Gender | ||||
| Male | 28 | 8 | 20 | 0.517 |
| Female | 2 | 1 | 1 | |
| Age-yr | ||||
| ≥50 | 18 | 6 | 12 | 0.704 |
| <50 | 12 | 3 | 9 | |
| Alcohol intake | ||||
| Yes | 10 | 3 | 7 | 1.000 |
| No | 20 | 6 | 14 | |
| HCC family history | ||||
| Yes | 3 | 1 | 2 | 1.000 |
| No | 27 | 8 | 19 | |
| HBV infection | ||||
| No | 5 | 1 | 4 | 1.000 |
| Yes | 25 | 8 | 17 | |
| Liver cirrhosis | ||||
| No | 12 | 9 | 3 | 0.704 |
| Yes | 18 | 13 | 5 | |
| AFP (ng/mL) | ||||
| ≥20 | 20 | 8 | 12 | 0.204 |
| <20 | 10 | 1 | 9 | |
| Tumor diameter (cm) | ||||
| ≥5 | 17 | 6 | 11 | 0.691 |
| <5 | 13 | 3 | 10 | |
| Tumor number | ||||
| 1 | 24 | 8 | 16 | 0.637 |
| >1 | 6 | 1 | 5 | |
| Tumor grade | ||||
| Ⅰ+Ⅱ | 14 | 3 | 11 | 0.440 |
| III | 16 | 6 | 10 | |
| Tumor stage | ||||
| Ⅰ+Ⅱ | 22 | 6 | 16 | 0.666 |
| III+Ⅳ | 8 | 3 | 5 | |
| Vascular invasion | ||||
| Yes | 9 | 4 | 5 | 0.389 |
| No | 21 | 5 | 16 | |
| Metastasis | ||||
| Yes | 4 | 3 | 1 | 0.069 |
| No | 26 | 6 | 20 | |
Note: P value is based on Fisher’s exact test.
Abbreviation: AFP, α-fetoprotein.
Table 3.
The Correlation Between CMTM2 and E-Cadherin in HCC Tissues
| Correlation | CMTM2 | r value | P value | ||
|---|---|---|---|---|---|
| Low | High | ||||
| E-cadherin | Low | 20 | 1 | 0.437 | 0.016 |
| High | 7 | 2 | |||
Notes: Bold value indicates significance; P value is based on Pearson correlation test.
Discussion
Through knocking-down the expression of CMTM2 in HCC cells, we found that down-regulation of CMTM2 could promote cell proliferation and metastasis, further supporting the suppressive role of CMTM2 in HCC pathogenesis and progression. Moreover, we discovered a significantly positive correlation between the expression of CMTM2 and E-cadherin. both in HCC cells and tumor tissues, suggesting a novel mechanism of CMTM2 in HCC metastasis.
Cancer is one of the major diseases that seriously endanger human health in the world. Now the incidence of cancer is getting higher and younger. Many studies have found that metastasis is the main cause of high cancer mortality, and about 90% of cancer-related deaths are due to metastasis.23,24 Metastasis is also an important risk factor for cancer recurrence after clinical surgery, chemotherapy, or radiotherapy.25,26 Because of high recurrence and high metastasis, HCC is one of the malignant tumors with the highest mortality rate. The identification of mechanism in HCC metastasis is in a need for HCC treatment and prognosis.
Accumulated evidences indicate that EMT, an early sign of tumor metastasis, plays a vital role in HCC progression.27,28 EMT refers to the conversion of epithelial cells to mesenchymal cells under normal physiological or specific pathological conditions. EMT occurs in the initial stage of tumor metastasis, endows tumor cells with an invasion and migration ability to break through the basement membrane and enter the circulatory system, and then forms a life-threatening tumor distal metastasis.29 Molecular biological characteristics of EMT include the down-regulation of epithelial markers (such as E-cadherin and β-catenin) and up-regulation of mesenchymal markers (such as N-cadherin and Vimentin).30 The decreased expression of β-catenin might result from its nuclear accumulation or activated form.31,32 While E-cadherin could be silenced epigenetically by ZEB1 to recruit multiple chromatin enzymes of E-cadherin at its promoter.33 In HCC cells, we found that the epithelial markers E-cadherin and β-catenin were decreased, while the mesenchymal markers N-cadherin, Vimentin, ZEB1 and ZEB2 were increased after knocking-down the expression of CMTM2. Moreover, both the expression of E-cadherin and CMTM2 were down-regulated in HCC tissues. Our results showed that down-regulated CMTM2 could induce the EMT process to promote HCC metastasis.
CMTM family members are recently identified as tumor suppressors in many kinds of cancers. They are deregulated by genetic or epigenetic modifications in various cancers. Recurrent mutations in CMTM2 were found in diffuse-type gastric cancer (DGC) and pivotal in the progression of DGC.34 CMTM5 single-nucleotide polymorphism (SNP) rs3811178 and CMTM6 SNP rs164207 were thought to contribute to HCC susceptibility in the southern Chinese population.35 CMTM3 protein was absent in gastric cancer and colorectal cancer, which was well correlated with its methylation status.36 The promoter of CMTM5 can also be methylated to exhibit its tumor suppressor activity in oral squamous cell carcinoma.37 In addition, miR-135b-5p was reported to promote gastric cancer progression by targeting CMTM3.38 Our results showed that CMTM2 had a down-regulated expression in HCC cells and tissues. Whether there is copy number variation, mutation, promoter methylation, or microRNAs regulation of CMTM2 expression in HCC needs to be elaborated in future work.
Apart from CMTM2, some other CMTM family members have also been investigated to have a relationship with the EMT process. Yuan W et al showed that CMTM3 suppressed metastasis and EMT of gastric cancer via the STAT3/Twist1/EMT signaling pathway.39 Zhang W et al reported that knock-down of CMTM8 expression could acquire EMT features in HepG2 cells and increase invasive and migratory ability. They also found that the epithelial marker E-cadherin was reduced after CMTM8 knocking-down.40 When liver cancer SK-Hep-1 cells were over-expressed with CMTM7, the expression of E-cadherin protein was increased and N-cadherin protein was decreased, indicating CMTM7 had a migration-suppressive ability.41 The positive correlation between the expression of CMTM2 and E-cadherin in our study further confirms the role of CMTM members in cancer metastasis.
Conclusion
In conclusion, we report for the first time that down-regulated CMTM2 promotes the metastasis of HCC via inducing the EMT process. The mechanism of CMTM2 in HCC metastasis explored in this study will provide more potential targets for HCC therapy.
Acknowledgments
This work was supported by the National Natural Science Foundation of China (81860586, 81860602), Natural Science Foundation of Guangxi Province (2018GXNSFAA281054, 2018GXNSFBA281216) and Key Science and Technology Research and Development Program Project of Guangxi (AB17292074).
Disclosure
The authors report no conflicts of interest in this work.
References
- 1.Han W, Lou Y, Tang J, et al. Molecular cloning and characterization of chemokine-like factor 1 (CKLF1), a novel human cytokine with unique structure and potential chemotactic activity. Biochem J. 2001;357(1):127–135. doi: 10.1042/bj3570127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Han W, Ding P, Xu M, et al. Identification of eight genes encoding chemokine-like factor superfamily members 1-8 (CKLFSF1-8) by in silico cloning and experimental validation. Genomics. 2003;81(6):609–617. doi: 10.1016/S0888-7543(03)00095-8 [DOI] [PubMed] [Google Scholar]
- 3.Shi S, Rui M, Han W, et al. CKLFSF2 is highly expressed in testis and can be secreted into the seminiferous tubules. Int J Biochem Cell Biol. 2005;37(8):1633–1640. doi: 10.1016/j.biocel.2004.04.028 [DOI] [PubMed] [Google Scholar]
- 4.Zhang XW, Yin HQ, Li Q, et al. CMTM2 is involved in spermiogenesis in mice. Beijing Da Xue Xue Bao. 2019;51(2):228–293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Liu F, Liu X, Liu X, et al. Integrated analyses of phenotype and quantitative proteome of CMTM4 deficient mice reveal its association with male fertility. Mol Cell Proteomics. 2019;18(6):1070–1084. doi: 10.1074/mcp.RA119.001416 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mezzadra R, Sun C, Jae LT, et al. Identification of CMTM6 and CMTM4 as PD-L1 protein regulators. Nature. 2017;549(7670):106–110. doi: 10.1038/nature23669 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Li J, Chen C, Bi X, et al. DNA methylation of CMTM3, SSTR2, and MDFI genes in colorectal cancer. Gene. 2017;630:1–7. doi: 10.1016/j.gene.2017.07.082 [DOI] [PubMed] [Google Scholar]
- 8.Shen Z, Chen X, Li Q, et al. Elevated methylation of CMTM3 promoter in the male laryngeal squamous cell carcinoma patients. Clin Biochem. 2016;49(16–17):1278–1282. doi: 10.1016/j.clinbiochem.2016.08.002 [DOI] [PubMed] [Google Scholar]
- 9.Zhang Y, Wang R, Song H, et al. Methylation of multiple genes as a candidate biomarker in non-small cell lung cancer. Cancer Lett. 2011;303(1):21–28. doi: 10.1016/j.canlet.2010.12.011 [DOI] [PubMed] [Google Scholar]
- 10.Yuan W, Liu B, Wang X, et al. CMTM3 decreases EGFR expression and EGF-mediated tumorigenicity by promoting Rab5 activity in gastric cancer. Cancer Lett. 2017;386:77–86. doi: 10.1016/j.canlet.2016.11.015 [DOI] [PubMed] [Google Scholar]
- 11.Bei C, Zhang Y, Wei R, et al. Clinical significance of CMTM4 expression in hepatocellular carcinoma. Onco Targets Ther. 2017;10:5439–5443. doi: 10.2147/OTT.S149786 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zeng W, Zhu X, Luo W, Chen Q, Zhang Y, Tan S. Clinical significance of expression of CMTM7 in hepatocellular carcinoma. Shijie Huaren Xiaohua Zazhi. 2016;34:4568–4575. [Google Scholar]
- 13.Zhu X, Qi G, Li C, et al. Expression and clinical significance of CMTM6 in hepatocellular carcinoma. DNA Cell Biol. 2019;38(2):193–197. doi: 10.1089/dna.2018.4513 [DOI] [PubMed] [Google Scholar]
- 14.Liu G, Xin ZC, Chen L, et al. Expression and localization of CKLFSF2 in human spermatogenesis. Asian J Androl. 2007;9(2):189–198. doi: 10.1111/j.1745-7262.2007.00249.x [DOI] [PubMed] [Google Scholar]
- 15.Zhang XD, Yin LL, Zheng Y, Lu L, Zhou ZM, Sha JH. Expression of a novel beta adaptin subunit mRNA splice variant in human testes. Asian J Androl. 2005;7(2):179–188. doi: 10.1111/j.1745-7262.2005.00025.x [DOI] [PubMed] [Google Scholar]
- 16.Zhang XW, Dun YJ, Tang X, et al. Expression of chemokine like factor-like myelin and lymphocyte and related proteins for vesicle trafficking and membrane link transmembrane domain-containing protein 2 in rats with varicocele. Beijing Da Xue Xue Bao. 2016;48(4):579–583. [PubMed] [Google Scholar]
- 17.Guo X, Zhang S, Tan S, et al. Downregulated CMTM2 poses potential clinical significance in hepatocellular carcinoma. DNA Cell Biol. 2020;39(4):683–689. doi: 10.1089/dna.2019.5237 [DOI] [PubMed] [Google Scholar]
- 18.Mays AC, Feng X, Browne JD, Sullivan CA. Chemokine and chemokine receptor profiles in metastatic salivary adenoid cystic carcinoma. Anticancer Res. 2016;36(8):4013–4018. [PubMed] [Google Scholar]
- 19.Luo W, Zhu X, Liu W, et al. MYC associated zinc finger protein promotes the invasion and metastasis of hepatocellular carcinoma by inducing epithelial mesenchymal transition. Oncotarget. 2016;7(52):86420–86432. doi: 10.18632/oncotarget.13416 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hodorogea A, Calinescu A, Antohe M, et al. Epithelial-mesenchymal transition in skin cancers: a review. Anal Cell Pathol. 2019;2019:3851576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gurzu S, Kobori L, Fodor D, Jung I. Epithelial mesenchymal and endothelial mesenchymal transitions in hepatocellular carcinoma: a review. Biomed Res Int. 2019;2019:2962580. doi: 10.1155/2019/2962580 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Shakib H, Rajabi S, Dehghan MH, Mashayekhi FJ, Safari-Alighiarloo N, Hedayati M. Epithelial-to-mesenchymal transition in thyroid cancer: a comprehensive review. Endocrine. 2019;66(3):435–455. doi: 10.1007/s12020-019-02030-8 [DOI] [PubMed] [Google Scholar]
- 23.Rossetto A, De Re V, Steffan A, et al. Carcinogenesis and metastasis in liver: cell physiological basis. Cancers. 2019;11(11):1731. doi: 10.3390/cancers11111731 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yilmaz M, Christofori G, Lehembre F. Distinct mechanisms of tumor invasion and metastasis. Trends Mol Med. 2007;13(12):535–541. doi: 10.1016/j.molmed.2007.10.004 [DOI] [PubMed] [Google Scholar]
- 25.Mills MN, Figura NB, Arrington JA, et al. Management of brain metastases in breast cancer: a review of current practices and emerging treatments. Breast Cancer Res Treat. 2020;180(2):279–300. doi: 10.1007/s10549-020-05552-2 [DOI] [PubMed] [Google Scholar]
- 26.Zeeshan R, Mutahir Z. Cancer metastasis − tricks of the trade. Bosn J Basic Med Sci. 2017;17(3):172–182. doi: 10.17305/bjbms.2017.1908 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Scheau C, Badarau IA, Costache R, et al. The role of matrix metalloproteinases in the epithelial-mesenchymal transition of hepatocellular carcinoma. Anal Cell Pathol. 2019;2019:9423907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Giannelli G, Koudelkova P, Dituri F, Mikulits W. Role of epithelial to mesenchymal transition in hepatocellular carcinoma. J Hepatol. 2016;65(4):798–808. doi: 10.1016/j.jhep.2016.05.007 [DOI] [PubMed] [Google Scholar]
- 29.Blackwell RH, Foreman KE, Gupta GN. The role of cancer-derived exosomes in tumorigenicity & epithelial-to-mesenchymal transition. Cancers. 2017;9(8):105. doi: 10.3390/cancers9080105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lamouille S, Xu J, Derynck R. Molecular mechanisms of epithelial-mesenchymal transition. Nat Rev Mol Cell Biol. 2014;15(3):178–196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yang Y, Zhang N, Zhu J, et al. Downregulated connexin32 promotes EMT through the Wnt/β-catenin pathway by targeting snail expression in hepatocellular carcinoma. Int J Oncol. 2017;50(6):1977–1988. doi: 10.3892/ijo.2017.3985 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ye Y, Long X, Zhang L, et al. NTS/NTR1 co-expression enhances epithelial-to-mesenchymal transition and promotes tumor metastasis by activating the Wnt/β-catenin signaling pathway in hepatocellular carcinoma. Oncotarget. 2016;7(43):70303–70322. doi: 10.18632/oncotarget.11854 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zhang Y, Xu L, Li A, Han X. The roles of ZEB1 in tumorigenic progression and epigenetic modifications. Biomed Pharmacother. 2019;110:400–408. doi: 10.1016/j.biopha.2018.11.112 [DOI] [PubMed] [Google Scholar]
- 34.Choi JH, Kim YB, Ahn JM, et al. Identification of genomic aberrations associated with lymph node metastasis in diffuse-type gastric cancer. Exp Mol Med. 2018;50(4):6. doi: 10.1038/s12276-017-0009-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bei C, Tan C, Zhu X, Wang Z, Tan S. Association between polymorphisms in CMTM family genes and hepatocellular carcinoma in Guangxi of China. DNA Cell Biol. 2018;37(8):691–696. doi: 10.1089/dna.2018.4274 [DOI] [PubMed] [Google Scholar]
- 36.Wang Y, Li J, Cui Y, et al. CMTM3, located at the critical tumor suppressor locus 16q22.1, is silenced by CpG methylation in carcinomas and inhibits tumor cell growth through inducing apoptosis. Cancer Res. 2009;69(12):5194–5201. doi: 10.1158/0008-5472.CAN-08-3694 [DOI] [PubMed] [Google Scholar]
- 37.Zhang H, Nan X, Li X, et al. CMTM5 exhibits tumor suppressor activity through promoter methylation in oral squamous cell carcinoma. Biochem Biophys Res Commun. 2014;447(2):304–310. doi: 10.1016/j.bbrc.2014.03.158 [DOI] [PubMed] [Google Scholar]
- 38.Lu M, Huang Y, Sun W, Li P, Li L, Li L. miR-135b-5p promotes gastric cancer progression by targeting CMTM3. Int J Oncol. 2018;52(2):589–598. doi: 10.3892/ijo.2017.4222 [DOI] [PubMed] [Google Scholar]
- 39.Yuan W, Li T, Mo X, et al. Knockdown of CMTM3 promotes metastasis of gastric cancer via the STAT3/Twist1/EMT signaling pathway. Oncotarget. 2016;7(20):29507–29519. doi: 10.18632/oncotarget.8789 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zhang W, Mendoza MC, Pei X, et al. Down-regulation of CMTM8 induces epithelial-to-mesenchymal transition-like changes via c-MET/extracellular signal-regulated kinase (ERK) signaling. J Biol Chem. 2012;287(15):11850–11858. doi: 10.1074/jbc.M111.258236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Huang ZM, Li PL, Yang P, et al. Overexpression of CMTM7 inhibits cell growth and migration in liver cancer. Kaohsiung J Med Sci. 2019;35(6):332–340. doi: 10.1002/kjm2.12058 [DOI] [PMC free article] [PubMed] [Google Scholar]