Abstract
Lack of insight into the identity of the cells that initiate metastasis hampers the development of antimetastatic therapies. Only a tiny fraction of tumor cells termed metastasis-initiating cells (MICs) are able to successfully seed metastases, causing recurrence and therapeutic resistance. Using metastasis models, we describe a subpopulation of MIC derivates from lung metastases that do not have proliferation advantages, express high levels of the PDGF receptors and EMT/stemness-related genes, and are unique in their ability to initiate metastasis. PDGF factors specifically boost the metastatic potential of MIC populations in a PDGFR-dependent manner. However, PDGFR inhibition preferentially suppresses lung metastases, but does not reduce the primary tumor burden. Thus, we found that PDGFR inhibition blocks AKT activation, whereas SGK1, which shares high-similarity kinase domain and overlap substrates with AKT overexpression remains active in MICs. SGK1 and PDGF signaling act in concert to promote metastatic formation, and SGK1 inhibition confers vulnerability to PDGFR inhibitors, also eliciting a powerful antitumor effect. In vivo, SGK1 inhibitors sensitize xenograft tumors to PDGFR-targeted therapies by reducing primary tumor growth and lung metastasis. Consequently, dual inhibition of PDGFR and SGK1 exhibited strong antitumor activities in established breast cancer cell lines in vitro and in vivo. Therefore, this approach not only provides insight into MIC transformation but also aids the design of improved therapeutic strategies for advanced breast cancer.
Subject terms: Cancer stem cells, Metastasis
Introduction
Metastases are the cause of ~90% of all cancer-related deaths [1]. It is estimated that <0.02% of disseminated tumor cells succeed in forming secondary tumors [2, 3], and these specialized cancer cells are termed metastasis-initiating cells (MICs) [4]. Similar to tumor-initiating cells (TICs), MICs can utilize normal stem cell pathways to increase their stemness and plasticity, but harbor additional abilities that allow them to survive and function as TICs in an unfamiliar microenvironment that is distinctively altered from the primary tumor microenvironment. Once MICs expand their daughter cells at secondary locations and form overt metastatic lesions, many of the currently available therapies often fail.
The origin of MICs remains elusive. Recently, whole-exome sequencing analyses of multiple metastasis models showed that the competency of metastasis is derived from the selection of pre-existing mutations [5–7], supporting the possibility of the presence of MICs at the primary tumor site. A recent study by using highly sensitive fluorescence-actived cell sorting and single-cell sequencing analysis concluded that primary tumor contains a rare population of stem-like cells, and that the percentage of these cells correlates with metastatic potential [8]. Pascual et al. also revealed that in oral squamous cell carcinoma cell lines, fatty acid receptor CD36 effectively marked a subpopulation of tumor cells with high metastasis-initiating potential in CD44+ TICs [9]. Using human mammary carcinoma-derived tumorspheres as a functional readout for an aggressive breast cancer phenotype, we performed an in vivo screen for MIC-enriched cell populations. This strategy yielded cell populations that possess phenotypic and molecular features of MICs. With this model, we provide evidence that platelet-derived growth factor (PDGF) signaling is a key regulator of MICs. By driving the function of MICs, PDGF activates downstream signaling pathways that ultimately promote tumorigenesis, stemness, invasion, and metastasis. Blocking the PDGF pathway preferentially decreased lung metastases, but unfortunately did not reduce primary tumor burden.
A number of mechanisms have been proposed to mediate resistance to targeted therapies, including activation of a bypass signaling pathway. Hyperactivation of the downstream phosphoinositide 3-kinase (PI3K)–AKT pathway is the best characterized of these bypass pathways [10, 11]. In this study, we sought to develop a clinically relevant model of breast cancer MICs that could be used to uncover the driving factors and mechanisms of metastasis formation. We also aimed to validate findings from this model in cell lines and xenografts and to translate our results into a therapeutic strategy that could be rapidly evaluated in clinical trials.
Materials and methods
Cell culture
Fresh surgical breast cancer tissues were minced and digested immediately with collagenase (300 units/ml, Sigma-Aldrich) and hyaluronidase (100 units/ml, Sigma-Aldrich) on an orbital shaker at 37 °C. After 4 h, the mixture was filtered through a 100- μm filter (BD Biosciences) and then resuspended in Dulbecco’s modified Eagle’s medium (DMEM)/F12 medium supplemented with penicillin and streptomycin before centrifugation at 400 rcf. The pellet was resuspended in DMEM/F12 basic medium and centrifuged again at 400 rcf. The resulting cells were propagated in vitro in DMEM/F12 medium (Gibco) supplemented with B27 (1:50), basic fibroblast growth factor (20 ng/ml, R&D), epidermal growth factor (20 ng/ml, R&D), R-Spondin 1 (250 ng/ml Peprotech), Noggin (100 ng/ml, Peprotech), and Y-27632 (5 μM, Abmole) at 37 °C with 5% CO2 unless otherwise specified. Their human strain of origin was confirmed by short tandem repeat analysis (Bioassay Methods Group, NIST). Other breast cancer cell lines used in this study were obtained from the ATCC and authenticated by short tandem repeat testing. In some experiments, cells were stably transfected with a luciferase-expressing lentiviral vector. All cell lines were routinely tested for mycoplasma contamination.
Animal studies
All animal experiments were approved by the Animal Ethics Committee of Sun Yat-sen University and conducted in the Animal Centre of Sun Yat-sen University. Female NOD or NOD-SCID mice (5–7 weeks old) were obtained from Beijing Vital River Laboratory Animal Technology Company Limited, and were acclimated for 1 week prior to experimentation.
For in vivo selection, a limited number (1 × 104) of parental cells were suspended in PBS with 50% Matrigel (356231, BD Biosciences) and injected orthotopically into the mammary fat pads of NOD-SCID mice. The resulting tumors were excised, minced, and digested immediately with collagenase (300 units/ml, Sigma-Aldrich) and hyaluronidase (100 units/ml, Sigma-Aldrich) at 37 °C. After 120 min, the mixture was filtered through a 40 -µm strainer (BD Biosciences) and then resuspended in DMEM/F12 medium supplemented with penicillin and streptomycin. The resulting cells were propagated in vitro to yield TE1 derivatives, which were then subjected to another round of in vivo selection (TE2 derivatives). A third round of in vivo selection yielded TE3 derivatives, which were injected into the mammary fat pads of NOD-SCID mice to generate lung metastatic nodules. These nodules were dissociated into single-cell suspensions that were propagated in vitro to yield LM derivatives. LM1 and LM2 are two single-cell-derived lung metastatic clones from the LM populations.
For orthotopic tumor initiation assays, cells were suspended in PBS with 50% Matrigel and injected (102, 103, or 104 cells per injection) into the mammary fat pads of NOD-SCID mice. Mice were killed at the observation time point, and necropsy was performed. For quantification, n is the number of independent mice.
For in vivo bioluminescence analysis, 1 × 105 luciferase-labeled LM cells were implanted into the mammary fat pads of NOD mice, which were monitored for lung metastasis by bioluminescence imaging. Mice were anesthetized with isoflurane, intraperitoneally administered 100 mg/kg D-luciferin (Promega), and imaged with an IVIS imaging system (Caliper Life Sciences) to record the bioluminescence signal, which was quantified with Living Image software (PerkinElmer).
For in vivo drug treatment experiments, tumor-bearing mice were randomly assigned into the respective treatment groups once the tumor was palpable (~100 mm3).
Paclitaxel, CP-673451 and either GSK-650394 or PEG-400 (as vehicle) were stocked in DMSO and freshly prepared in PBS. Mice were treated via intraperitoneal injection of GSK-650394 (150 μg/day, subcutaneous administration), CP-673451 (50 mg/kg/day, intraperitoneal administration), or both for 5 days a week. Paclitaxel was intraperitoneally administered at a dose of 10 mg/kg every 4 days. Necropsies were performed either after 4 weeks of treatment and 8 weeks of observation, or when the mean tumor volumes were greater than 2500 mm3. At the experimental endpoint, primary tumors were harvested, and macroscopic metastatic lesions were scored.
Tumorsphere assays
Tumorsphere assays were performed as previously described [12]. Briefly, single-cell suspensions containing 1000 cells were plated on ultralow attachment 24-well plates (Corning Costar) in the stem cell culture medium. The media were replaced every 48 h with fresh media. Cells were cultured for 4–7 days, and tumorspheres with a diameter of >50 mm were counted. For serial passages, tumorspheres were collected and dissociated with trypsin for 2 min at 37 °C to obtain single-cell suspensions.
3D Matrigel on-top culture
The Matrigel on-top culture was performed as described elsewhere [13]. Briefly, cells were resuspended as a single-cell suspension (104 cells/ml) in culture medium (DMEM/F12 medium supplemented with 5% FBS) containing 2% Matrigel, plated on 96-well culture plates pre-coated with 15 μl/well of 100% Matrigel (BD Biosciences), and cultured at 37 °C for 6–14 days.
RNA-seq
RNA samples were extracted from parental and LM cells, as well as their derived primary tumors and lung metastatic nodules with TRIzol (ThermoFisher), and submitted to Guangzhou SAGENE Biotechnology Company Limited for RNA-seq analysis. A detailed depiction of the GSEA methodology is provided at http://www.broadinstitute.org/gsea/doc/GSEAUserGuideFrame.html.
Statistical analysis
Statistical analyses were performed with GraphPad Prism 7 software. Comparisons between two groups were performed with an unpaired two-sided t test. All P-values <0.05 were considered significant. Statistical significance is indicated as follows: **P < 0.01, and *P < 0.05; ‘n.s.’ indicates not significant.
Results
Establishment of a breast cancer model of MICs
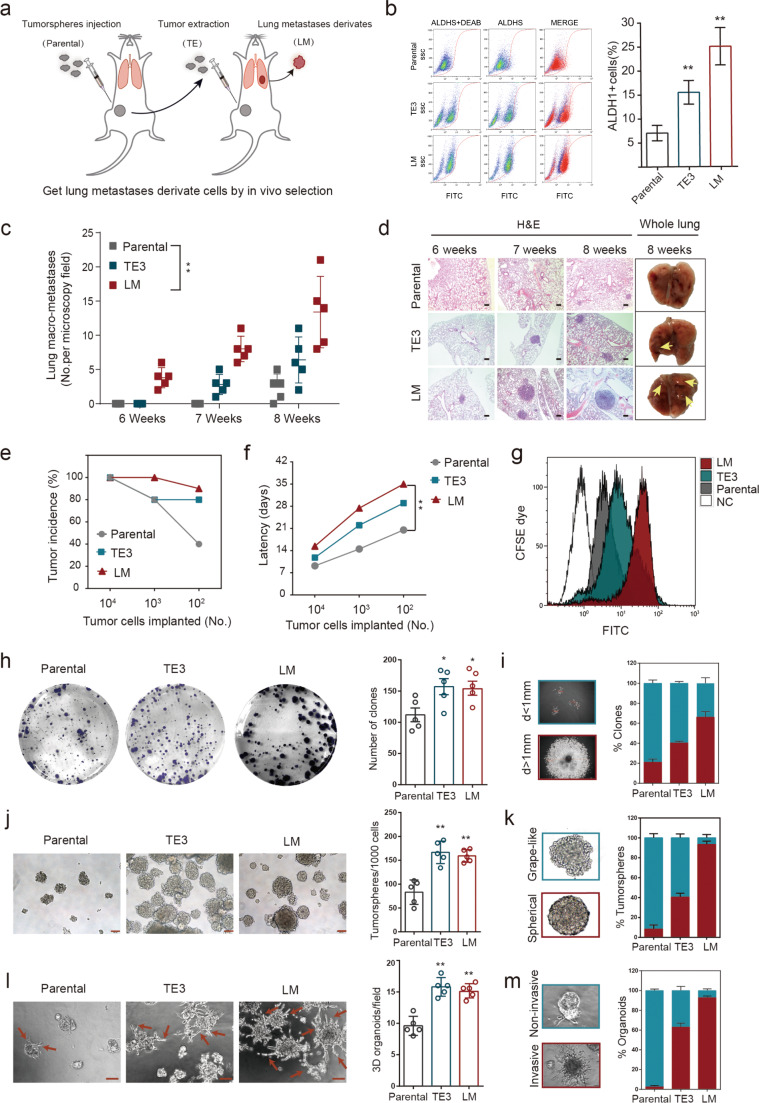
To investigate the key molecular determinants governing MICs, we employed an in vivo selection strategy in mice to select subpopulations of patient-derived cells with enhanced metastasis-initiating capacity (Fig. 1a). We first isolated and propagated tumorspheres in vitro from fresh breast cancer clinical samples that were diagnosed as triple-negative breast cancer by biopsy. Tumorspheres that can expand for more than five passages in vitro were adopted for further examination in vivo (Supplementary Fig. 1a). While most of the tumorspheres either failed to form primary tumors by low-dose (103–104) injection or were unable to colonize locoregional lymph nodes or disseminate to the lung, only clone 409 (Supplementary Fig. 1b) was able to disseminate to the lung and was termed the parental cell line. After three rounds of in vivo selection, a tumor-extracted (TE) tumorsphere derivative, TE3, demonstrated enhanced lung metastasis (Supplementary Fig. 1c). Finally, we obtained a polyclonal metastatic mammary carcinoma cell line, LM, derived from a spontaneous lung metastasis in a TE3 tumor burden mouse (Fig. 1a). LM derivatives present enrichment of ALDH1 marker profiles relative to those of their parental populations (Fig. 1b). When injected intravenously, LM cells gave rise to a greater number of macroscopic colonies in the lungs relative to the parental and TE3 cell populations (Fig. 1c, d). Therefore, LM cells were more effective at re-initiating tumors in a metastatic organ relative to their parental populations.
Fig. 1. Establishment of an MIC model.
a Schematic of the in vivo selection strategy used to establish the MIC model. Breast cancer cells cultured with sphere culture medium were injected at low cell numbers into the mammary fat pads of NOD-SCID mice, and the resulting tumors were dissociated into single-cell suspensions, cultured in vitro, and re-injected over multiple rounds. TE: tumor extraction; LM: lung metastases. b Left: representative ALDEFLUOR analyses of parental, TE3, and LM cells. DEAB, a specific inhibitor of ALDH1, was used as a control. Right: quantification of the ALDEFLUOR assays (n = 3). Data are presented as the mean ± SD. c Parental, TE3, or LM cells were injected intravenously (i.v., tail vein) (5 × 105 cells per mouse), and macro-metastases (macro-mets) >100 µm were quantified from microscopy tissue sections from four lung lobes per animal to calculate the metastasis count at the indicated time point. Data are presented as the mean ± SD. d H&E images of lungs from mice shown in (c). Scale bars, 200 µm. e, f Cells were admixed with Matrigel and inoculated into the mammary fat pad of NOD-SCID mice at the indicated numbers (1 × 104 cells, n = 5 per group; 1 × 103 cells, n = 10 per group; 1 × 102 cells, n = 10 per group). Incidence of tumor formation (e) and tumor latency at the indicated cell number (f). g Flow cytometry analysis of CFSE dye retention by parental, TE3, and LM cells after 3 days. NC: negative control. h, i Colony-formation assays of parental, TE3, and LM cells. h Cells were cultured for 14 days followed by staining (left) and quantification (right, n = 5). Representative photographs are shown. i Representative images (left) and quantification (right) of small clones (d < 1 mm) (teal) and large and compacted clones (d > 1 mm) (red). Data are presented as the mean ± SEM. j, k Sphere formation assays of parental, TE3, and LM cells. j Representative bright-field images (left) and quantification (right, n = 5) of tumorsphere formation after 6 days of culture. Scale bars, 100 µm. k Representative images (left) and quantification (right) of grape-like (teal), and spherical (red) tumorspheres. Data are presented as the mean ± SEM. l, m 3D Matrigel on-top assays of parental, TE3, and LM cells. l Representative phase-contrast images (left) and quantification (right, n = 5) of 3D organoid formation after 10 days of culture. Arrows indicate filopodium-like protrusions. Scale bars, 100 µm. m Representative images (left) and quantification (right) of invasive organoid (red) and mass organoid (teal). Data are presented as the mean ± SEM.
MICs possess pre-metastatic capacity independent of proliferation ability
LM cells showed an increased ability to seed tumors in vivo, but were slow to do so in the early stages (Fig. 1e, f). This effect was evident when limited numbers (1 × 104, 1 × 103, and 1 × 102) of tumor cells were injected (Fig. 1e, f; Supplementary Fig. 2a). Upon administering CFSE dye and monitoring the cells’ bioluminescence, we found a higher percentage of label-retaining cells (LRCs) in the LM populations after 3 days of culture (Fig. 1g) and after a prolonged period (14 days) of in vitro culture (Supplementary Fig. 2b).
Subsequent in vitro experiments revealed that although LM cells generated more clones in standard adherent cell culture conditions, more tumorspheres in suspension cultivation and more organoids in 3D-Matrigel culture than did the parental cells, LM populations, and TE3 populations did not exhibit significant differences (Fig. 1h, j, l). Notably, the LM populations showed distinct differences in cell morphology. Single clones of LM cells were larger and rounder than those of parental and TE3 cells (Fig. 1h, i). The LM-derived tumorspheres were compact and spherical, whereas parental-derived tumorspheres were irregular grape-like clusters (Fig. 1j, k). In 3D on-top assays, LM populations displayed abundant filopodium-like protrusions (Fig. 1l, m). Taken together, these data indicate that LM cells possess key features of MICs, including the capacity to retain metastasis-initiating and tumorigenic potential and the maintenance of a stem-like phenotype.
LM populations express high levels of PDGFRα/β
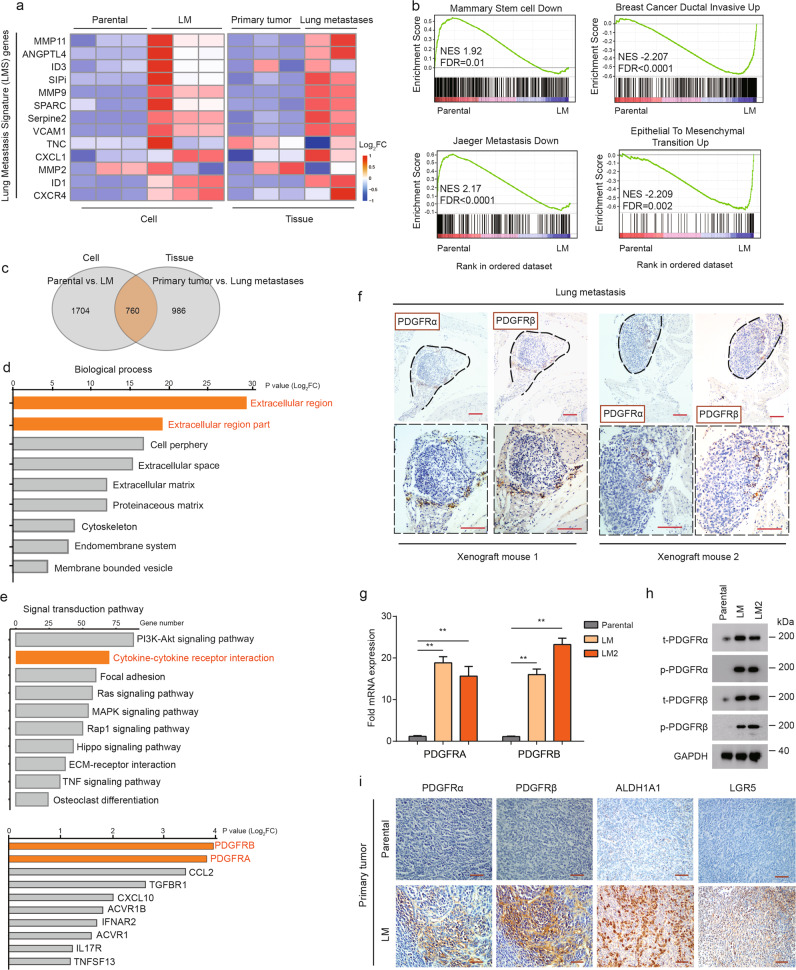
Having established a breast cancer model of MICs, we next examined whether LM populations have molecular signatures similar to those of MICs. With tumorspheres and tumor tissues, RNA-seq analysis of LM populations and their parental populations was performed. LM populations showed higher expression of genes of the lung metastasis signature (LMS) than did the parental populations, both in vitro and in vivo (Fig. 2a). The KEGG enrichment map revealed a significant upregulation of the MIC molecular signature in LM cells, including stemness, metastasis, breast cancer ductal invasiveness, and epithelial–mesenchymal transition (EMT) (Fig. 2b).
Fig. 2. PDGFRα and PDGFRβ are overexpressed in LM populations.
a Heatmap representation of the expression of lung metastasis signature (LMS) genes in indicated cell populations and tumor tissues. Data are presented as rows normalized. b KEGG enrichment maps of upregulated MIC-associated gene sets of LM cells versus paternal cells. NES normalized enrichment score, FDR false discovery rate. c Venn diagram showing the overlap of the differentially expressed genes of the parental and LM populations between in vitro and in vivo analyses. Signatures are generated by including genes with fold changes higher or lower than +1 and −1, respectively. d Top represents biological process categories upregulated in LM cells and tissues. e Overrepresented signal transduction pathways and top overrepresented genes in the cytokine–cytokine receptor interaction in LM populations. f Representative IHC images of PDGFRα and PDGFRβ staining in serially sectioned mouse lung metastases formed from LM cells. Images below (black boxes) represent 2× enlargement of the image above. Scale bars, 100 µm. g PDGFRA/B mRNA levels in parental, LM, and LM2 cells. The levels were normalized to those of GAPDH. Data are presented as the mean ± SD. h Western blot analysis of PDGFRα and PDGFRβ expression in parental, LM, and LM2 cells. i Representative IHC images of PDGFRα, PDGFRβ, ALDH1A1, and LGR5 staining in serially sectioned mouse primary tumor formed from LM cells. Scale bars, 100 µm.
Further analysis from gene expression profiles showed that there were 760 overlapping differentially expressed genes between the LM and parental populations both in vitro and in vivo (Fig. 2c). Gene ontology analysis revealed that the overlapping genes were strongly enriched in the processes of “extracellular region” and “extracellular region part” (Fig. 2d). On the other hand, signal transduction pathway analyses revealed that these overlapping genes were strongly enriched in metastasis-related pathways (Fig. 2e). Among these pathways, the top-ranked cytokine–cytokine receptor pathway in the process of “extracellular region” was chosen as the focus of subsequent analyses. PDGFRB and PDGFRA, which were the top two overrepresented genes in the cytokine–cytokine receptor pathway, were selected for further study (Fig. 2e). We validated the differential expression of several of these genes by quantitative PCR with reverse transcription (RT–qPCR) in LM and its two single-cell-derived lung metastatic clones (LM1 and LM2, Supplementary Fig. 3a, b). This differential expression was confirmed in LM2 tumor line (Fig. 2f). For further confirmation, QPCR and western blot analyses were performed, and the results showed that PDGFRα and PDGFRβ were highly expressed in LM1 and LM2 cells (Fig. 2g, h). In vivo, the LM-derived tumors continued to express PDGFRα/β and high levels of the cancer stem cell markers ALDH1 and LGR5, but the parental-derived tumors did not (Fig. 2i). Accordingly, lung metastatic lesions from patients exhibited higher PDGFRα/β than their primary tumors (Supplementary Fig. 4a, b). Interestingly, PDGFRα and PDGFRβ were also co-expressed in lung macro-metastasis nodules (Fig. 2i). Collectively, these results suggest that LM populations possess molecular signatures of MICs and that PDGFRα/β are overexpressed in MICs.
PDGFR inhibition preferentially suppresses lung metastases
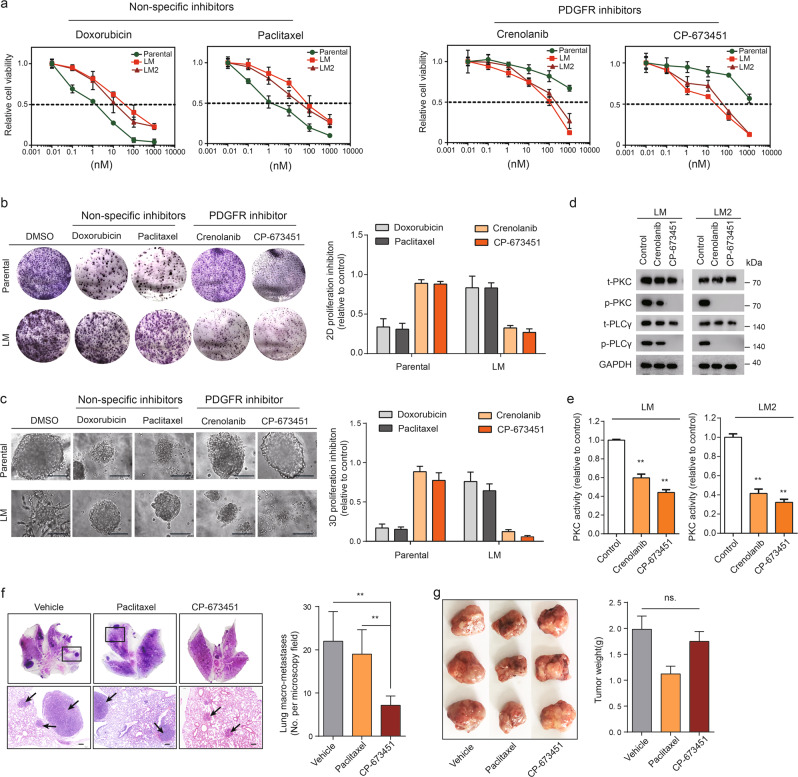
To investigate whether PDGFR inhibition preferentially affects MIC-enriched LM1 and LM2 cells, we utilized two antagonists, crenolanib and CP-673451, that are specific to PDGFRα/β, and found that LM1 and LM2 cells were more sensitive to PDGFR inhibition relative to parental cells. In contrast, LM1 and LM2 cells were more resistant to the nonspecific inhibitors paclitaxel and doxorubicin than were the parental cells (Fig. 3a). While chemotherapy with doxorubicin or paclitaxel increased the inhibition of clonogenic and 3D clump formation in parental cells, crenolanib or CP-673451 increased the inhibition of clonogenic and organoid formation in LM cells (Fig. 3b, c). Exposure of LM1 and LM2 cells to either crenolanib or CP-673451 led to a significant reduction in the levels of p-PKC, which is a known downstream effector of PDGFR, and p-PLCγ [14], which is known to transduce signals from PDGFR to PKC (Fig. 3d). Likewise, pharmacologic inhibition of PDGFR in LM1 and LM2 cells reduced PKC activity (Fig. 3e). Together, these data suggest the preferential potency of PDGFR inhibitors to MICs in vitro. In vivo, paclitaxel resulted in modest suppression of lung macro-metastases in LM tumor-bearing mice, whereas treatment with CP-673451 led to increased attenuation of lung macro-metastases compared with that observed with paclitaxel (Fig. 3f). However, treatment with CP-673451 was unable to inhibit primary tumor growth (Fig. 3g). Together, these data suggest that PDGFR inhibitors preferentially suppress lung metastases of MICs, but have little impact on primary tumors.
Fig. 3. Preferential anti-metastasis efficiency of PDGFR inhibition.
a Cell viability of parental, LM1, and LM2 cells upon treatment with increasing concentrations of doxorubicin, paclitaxel, crenolanib, or CP-673451. b Colony-formation assays of parental or LM cells treated with DMSO, doxorubicin (50 nM), paclitaxel (10 nM), crenolanib (1 μM), or CP-673451 (1 μM). Cells were cultured for 8 days followed by staining (left) and quantification (right). Scale bars, 100 µm. c 3D-Matrigel on-top assays of parental or LM cells treated with DMSO, doxorubicin (50 nM), paclitaxel (10 nM), crenolanib (1 μM), or CP-673451 (1 μM). Representative images (left) and quantification (right) of 3D-organoid formation after 10 days of culture. Scale bars, 100 µm. d LM1 and LM2 cells were treated with DMSO, crenolanib (1 µM), or CP-673451 (1 μM) for 48 h, and changes in the total PKC, p-PKC, total PLCγ, and p-PLCγ protein levels were analyzed by western blot. GAPDH served as a loading control. e Levels of total PKC kinase activity in LM1 and LM2 cells treated with crenolanib (1 μM) or CP-673451 (1 μM) relative to DMSO-treated cells are shown. f Representative H&E staining (left) and quantification (right) of lung macro-metastases of LM tumor-bearing mice treated with vehicle, paclitaxel (10 mg/kg), or CP-673451 (50 mg/kg). Scale bars, 200 µm. g Photographs of harvested tumors (left) and quantification (right) of primary tumor weight of LM tumor-bearing mice treated with vehicle, paclitaxel (10 mg/kg), or CP-673451 (50 mg/kg).
PDGF ligands drive the development of metastasis of MICs
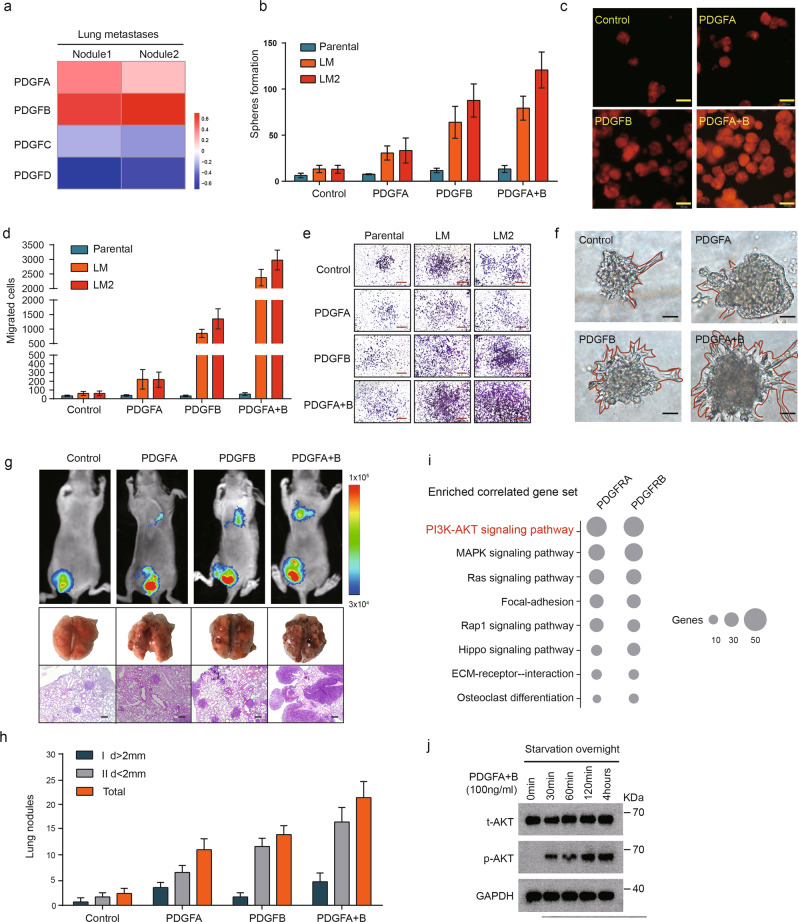
We next asked whether the activation of PDGF signaling can promote the metastasis of parental and LM cells. RNA-seq analysis showed that lung metastasis nodules expressed high levels of PDGFA and PDGFB (Fig. 4a). We speculated that the overexpression of two PDGF ligands in lung metastases is necessary for PDGFR activation and MIC colonization. When either PDGFA or PDGFB protein was added, the numbers of spheres formed from the LM1 and LM2 populations were significantly increased, whereas the number of spheres formed from the parental cell line remained the same (Fig. 4b, c). LM1 and LM2 cells generated higher numbers of migratory cells in response to PDGFA or PDGFB than to DMSO (Fig. 4d, e). Notably, LM cells treated with PDGFB formed disorganized structures that invaded the extracellular matrix in 3D culture (Fig. 4f). Remarkably, these effects were further strengthened by the co-treatment with PDGFA. Next, we tested the effects of PDGF on metastasis-initiating function by admixing luciferase-labeled LM cells with Matrigel containing either PDGF factor or PBS control (Fig. 4g). The LM tumors had developed significantly fewer lung macro-metastases than did the LM + PDGF factor tumors. Importantly, the addition of both PDGFA and PDGFB resulted in a further increase in the number of lung macro-metastases (Fig. 4g, h). This phenotype did not seem to stem from faster proliferation, since primary tumors derived from cells cultured in growth factor-containing medium did not grow faster than LM cells cultured in PBS-containing medium in vivo, as evident from the similar tumor size (Supplementary Fig. 5a) and weight (Supplementary Fig. 5b). These findings suggested that PDGF factors specifically promote breast cancer metastasis formation by enhancing MIC ability.
Fig. 4. The development of MIC metastasis is sufficiently driven by PDGFα/β.
a Heatmap representation of the expression of PDGFA, PDGFB, PDGFC, and PDGFD in tissues from two lung metastatic nodules. Data are presented as rows normalized. b The number of tumorspheres formed by parental, LM1, or LM2 cells treated with DMSO, PDGFA (100 ng/ml), PDGFB (100 ng/ml), or both (100 ng/ml each, PDGFA:PDGFB = 1:1) for 7 days. Data are presented as the mean ± SD. c Representative images of tumorsphere formation of LM cells treated with DMSO, PDGFA (100 ng/ml), PDGFB (100 ng/ml), or both (100 ng/ml each, PDGFA:PDGFB = 1:1). Scale bars, 50 µm. d The number of migratory cells of parental, LM1, and LM2 cells treated with DMSO, PDGFA (100 ng/ml), PDGFB (100 ng/ml), or both (100 ng/ml each, PDGFA:PDGFB = 1:1) based on the migration assay. Data are presented as the mean ± SD. e Representative images of the migration assays of (d). Scale bars, 100 µm. f Bright-field images of LM cells treated with PDGFA (100 ng/ml), PDGFB (100 ng/ml), or both (100 ng/ml each, PDGFA:PDGFB = 1:1). Scale bars, 100 µm. g (Top) Bioluminescence imaging of LM tumor-bearing mice treated with vehicle, PDGFA (100 ng/ml), PDGFB (100 ng/ml), or both (100 ng/ml each, PDGFA:PDGFB = 1:1). (Bottom) Photographs of excised lungs and representative H&E staining of the lung sections are shown. Scale bars, 200 µmk. h Quantification of the metastatic lung nodules formed with the indicated treatment. i Pathways that correlated with the expression of PDGFRA/PDGFRB in LM cells. j Western blot analyses of total AKT, p-AKT in LM cells treated with PDGFA and PDGFB (100 ng/ml each, PDGFA:PDGFB = 1:1) at different time points. GAPDH served as a loading control.
To understand how PDGF signaling confers an MIC signature, we used GSEA and found that the expression of PDGFRA or PDGFRB was significantly correlated with AKT gene signatures (Fig. 4i). The addition of PDGFA resulted in an increase in p-PDGFRα, while the addition of PDGFB resulted in an increase in both p-PDGFRα and p-PDGFRβ, and the presence of both factors increased the levels of phosphorylated AKT (Fig. 4j). These data highlight specific signaling pathways downstream of PDGF activation in MICs.
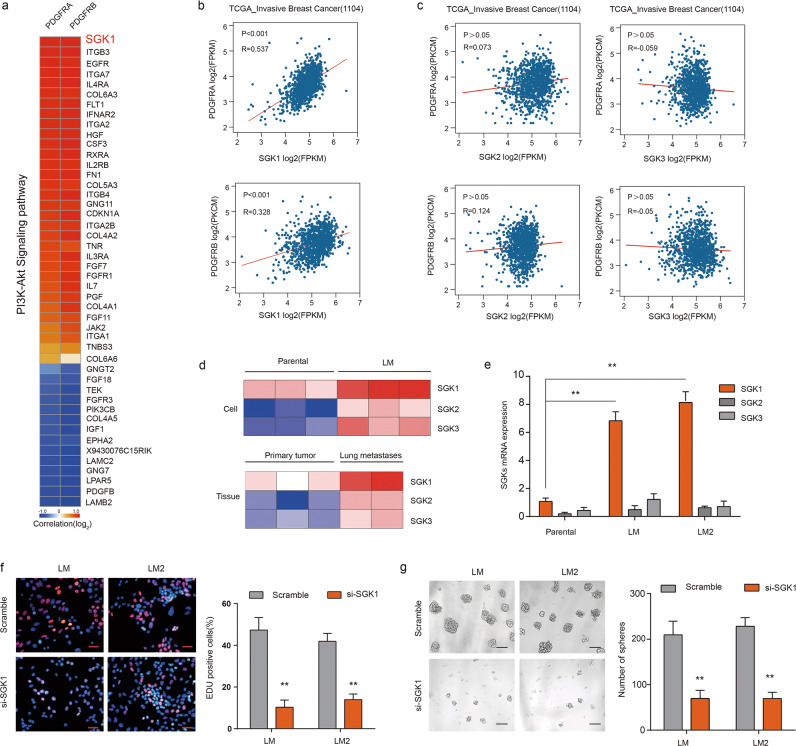
Relevance of SGK1 and PDGFRA/PDGFRB expression to the growth of MICs
We next asked whether the genes in the PI3K–AKT pathway positively correlated with the expression of PDGFRA/PDGFRB. Using RNA-seq data from LM cells, we found that the top gene positively co-expressed with PDGFRA and PDGFRB was serum and glucocorticoid-induced kinase 1 (SGK1) (Fig. 5a). SGK1 is a kinase that belongs to the SGK family (including SGK1–3), has a high similarity with the kinase domain of AKT, and functions as a mediator of PI3K signaling [15, 16]. We sought to determine whether SGK1 serves as a functional kinase in MICs. Analyses of TCGA-invasive breast carcinoma cohorts [17] also yielded a significant association between the expression of PDGFRA/B and SGK1 (Fig. 5b), but not its family members SGK2 and SGK3 (Fig. 5c). In the Oncomine database, we found that SGK1 expression was higher in invasive cancer than in normal tissue or ductal breast cancer (Supplementary Fig. 6). Consistent with these biostatistics, we also observed increased SGK1 expression in the LM population compared with the parental population by RNA-seq analysis, both in cell cultures and tumor tissues (Fig. 5d). We also confirmed with RT-PCR that SGK1 showed significantly enhanced expression in LM1 and LM2 cells compared with the parental cells (Fig. 5e). These results prompted us to investigate whether SGK1 functionally regulates MIC function and metastasis. Knockdown of SGK1 via small interfering RNAs (siRNAs) led to a significant decrease in the number of EdU-positive cells and spheres derived from LM1 or LM2 cells (Fig. 5f, g). These findings, together with the established role of SGK1 in cell migration and invasion [18, 19], suggest that SGK1 plays an important role in MICs to form metastases.
Fig. 5. SGK1 and PDGFRA/PDGFRB are co-expressed in MICs.
a Heatmap of genes of PI3K–AKT pathway that correlated with the expression of PDGFRA and PDGFRB based on RNA-seq data of LM cells. b The gene expression levels of SGK1 and PDGFRA/B were measured in the TCGA_invasive breast cancer cohort (1104) data sets. Correlation values and P-values were determined as indicated. c The gene expression levels of SGK2/3 and PDGFRA/B were measured in the TCGA_invasive breast cancer cohort (1104) data sets. Gene expression is reported as log2 values. d Heatmap representation of the expression of SGK1, SGK2, and SGK3 in the indicated cell populations and tumor tissues. Data are presented as rows normalized. e RT-PCR analyses of SGK1, SGK2, and SGK3 expression in parental, LM1, and LM2 cell populations. The levels were normalized to those of GAPDH. Data are presented as the mean ± SD. f Representative images (left) and quantification (right) of EdU-positive LM1 and LM2 cells transfected with the indicated siRNAs in the EdU assay. Scale bars, 20 µm. g Representative images (left) and quantification (right) of spheres formed from LM1 and LM2 cells transfected with the indicated siRNAs. Scale bars, 50 µm.
SGK1 inhibition sensitizes MICs to PDGFR inhibition
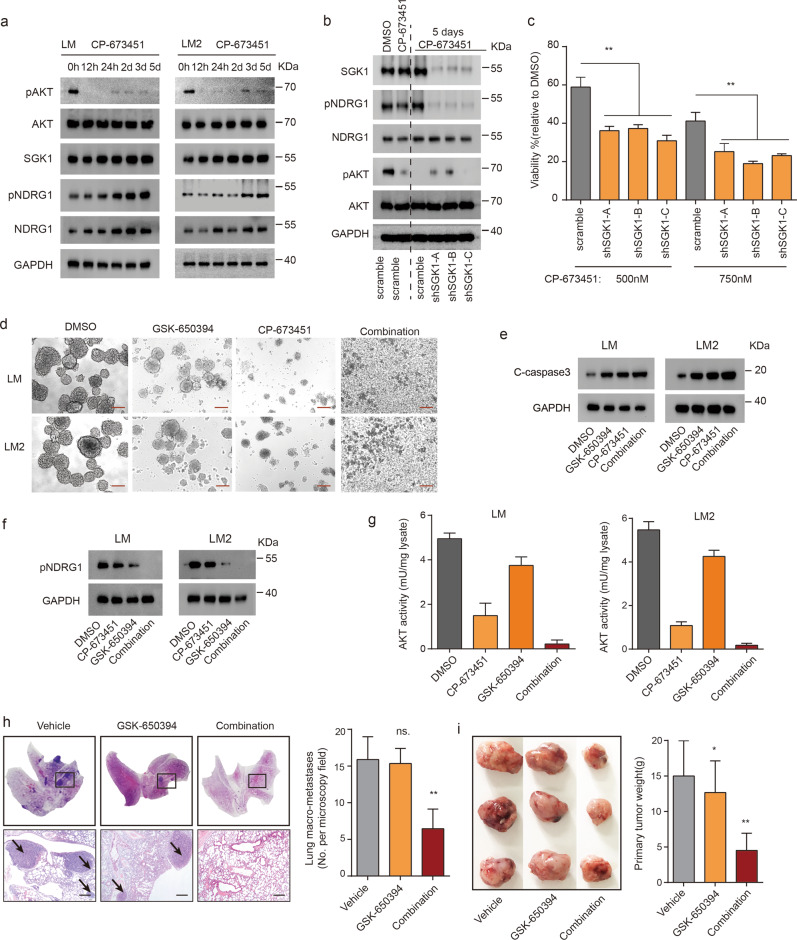
To determine whether additional kinases are involved in the acquisition of drug resistance, we treated LM1 and LM2 cells with the PDGFR inhibitor CP-673451 for a prolonged period, and immunoblot analysis confirmed a reduction in the levels of phosphorylated AKT (Fig. 6a). Given that SGK1 shares high similarity with AKT at the catalytic domain and some of its downstream targets, we reasoned that SGK1 is involved in restraining the response to PDGF signaling inhibition in lieu of AKT. Consistent with our speculation, NDRG1, a major substrate of SGK1, was phosphorylated in LM1 and LM2 cells whenever AKT kinase activity was blocked after CP-673451 treatment. Although SGK1 expression correlates with p-NDRG1 levels [20], AKT can also phosphorylate NDRG1 in the absence of SGK1 [21]. To underscore the role of SGK1 in this setting, we used three shRNAs to knock down SGK1 protein and found that this knockdown prevented NDRG1 phosphorylation in LM cells subjected to prolonged PDGFR inhibitor treatment (Fig. 6b). Accordingly, we observed a decrease in cell viability in the SGK1-knockdown cells that was enhanced in the presence of CP-673451 (Fig. 6c). This is indicative of compensatory signaling between AKT and SGK1, and that only the simultaneous combination of PDGFR and SGK1 inhibitors can block the activity of the endogenous enzymes in MICs.
Fig. 6. SGK1 inhibition sensitizes MICs to PDGF inhibition.
a LM1 and LM2 cells were treated with CP-673451 (1 μM) at the indicated times, and changes in p-AKT, total AKT, SGK1, p-NDRG1, and total NDRG1 protein levels were analyzed by western blot. GAPDH served as a loading control. b LM cells were transfected with scramble siRNA or three SGK1-targeted siRNAs and treated with DMSO or CP-673451 for 1 h or 5 days at the indicated times, and changes in SGK1, p-NDRG1, NDRG1, p-AKT, and AKT protein levels were analyzed by western blot. GAPDH served as a loading control. c Cell viability of LM cells with stable SGK1-knockdown cells (shSGK1-A/B/C) treated with CP-673451 (0.5 or 0.75 μM). d Representative bright-field images of tumorsphere formation of LM1 and LM2 cells treated with DMSO, GSK-650394 (2 μM), CP-673451 (1 μM), or a combination of both agents. Scale bars, 50 µm. e Western blot analyses of cleaved caspase-3 expression in LM1 or LM2 cells treated with DMSO, GSK-650394 (2 μM), CP-673451 (1 μM), or a combination of both agents. GAPDH served as a loading control. f Western blot analysis of p-NDRG1 expression in LM1 and LM2 cells treated with DMSO, GSK-650394 (2 μM), CP-673451 (1 μM), or a combination of both agents for 4 h. g Endogenous kinase assay for AKT activity in LM1 and LM2 cells treated with DMSO, GSK-650394 (2 μM), CP-673451 (1 μM), or a combination of both agents for 4 h. h LM tumor-bearing mice were treated with GSK-650394 (150 µg/day, 5 days/week, subcutaneous administration), CP-673451 (50 mg/kg/day, 5 days/week, intraperitoneal administration), or a combination of both agents. Representative H&E staining (right) and quantification (left) of lung macro-metastases. Data are presented as the mean ± SD. Scale bars, 200 µm. i Photographs of harvested primary tumors (right) and quantification (left). Data are presented as the mean ± SD.
Treatment with either SGK1 inhibitor GSK-650394 or PDGFR inhibitor CP-673451 alone inhibited tumorsphere formation in the LM1 and LM2 populations (Fig. 6d). Notably, the combination of GSK-650394 and CP-673451 led to superior inhibition of tumorsphere formation (Fig. 6d). Apoptosis detection by western blot showed that the level of cleaved caspase-3 was increased upon GSK-650394 and CP-673451 treatment (Fig. 6e). We also found that only the combination of GSK-650394 and CP-673451 was able to decrease the levels of phosphorylated NDRG1 in LM1 and LM2 cells (Fig. 6f). The combination of both drugs is required to effectively inhibit the activity of AKT (Fig. 6g).
To test whether SGK1 or/and PDGFR antagonism would inhibit pulmonary metastasis and primary tumors in vivo, we evaluated the effects of GSK-650394 alone and in combination with CP-673451 in mice engrafted with LM tumors. In LM tumor-bearing mice, GSK-650394 alone resulted in modest suppression of lung macro-metastases; notably, treatment with GSK-650394 and CP-673451 led to further attenuation of lung macro-metastases compared with that observed with GSK-650394 alone (Fig. 6h). Compared with no treatment, GSK-650394 alone also resulted in a modest reduction in primary tumor weight in LM tumor-bearing mice (Fig. 6i). Remarkably, mice treated with the combination of GSK-650394 and CP-673451 experienced further inhibition of primary tumor growth (Fig. 6i). The above data suggest the potent efficacy of the combination of a PDGFR inhibitor and an SGK1 inhibitor in the treatment of MIC populations.
The combination of CP-673451 and GSK-650394 is a potential treatment for breast cancer
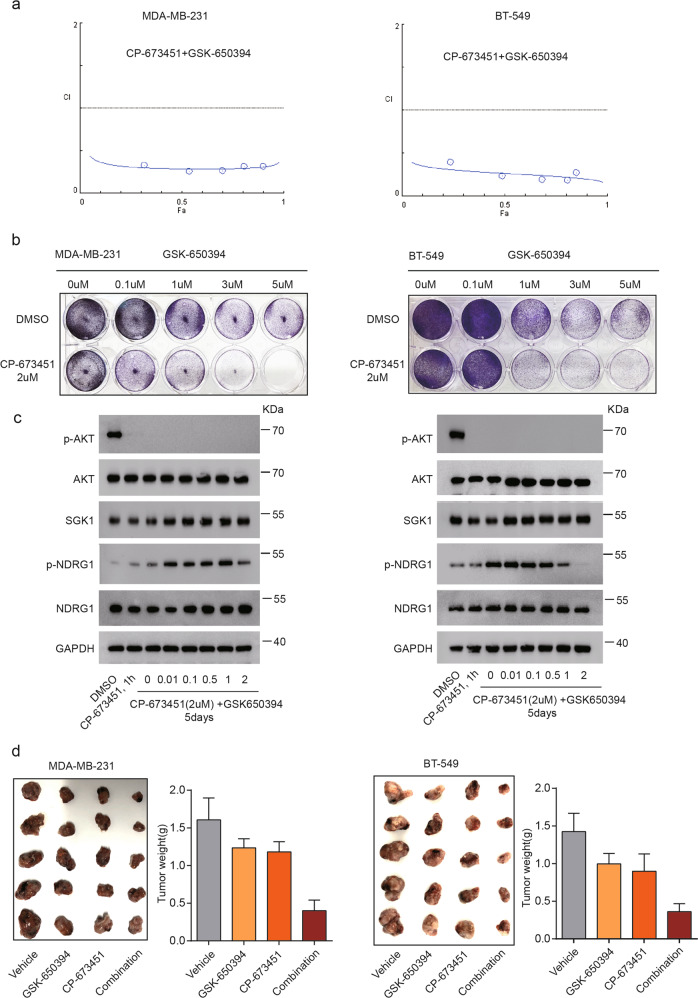
We further tested whether the combined PDGFR and SGK1 inhibition could extend to well-established breast cancer cell lines. Using the breast cancer cell lines MDA-MB-231 and BT-549, which express elevated levels of PDGFR and SGK1, we found that inhibition of SGK1 with GSK-650394 had a synergistic effect with PDGFR inhibition with CP-673451 in both cell lines (Fig. 7a). Co-treatment with CP-673451 and GSK-650394 led to a strong reduction in colony formation in MDA-MB-231 and BT-549 cells in a concentration-dependent manner (Fig. 7b). Central to our work, the combination of CP-673451 and GSK-650394 was able to decrease the levels of phosphorylated NDRG1 and AKT in both cell lines (Fig. 7c). Finally, we tested the effect of combined CP-673451 and GSK-650394 administration in xenotransplantation breast cancer models. In both MDA-MB-231 and BT-549 tumor-bearing mice, compared with the vehicle group, all treatment groups displayed a slightly decreased tumor formation (Fig. 7d). Compared with the single-agent treatment group, the combination treatment group (CP-673451 and GSK-650394) showed significantly decreased tumor formation (Fig. 7d). Together, these data suggest the therapeutic potential of CP-673451 and GSK-650394 in breast cancer.
Fig. 7. Combined PDGF and SGK1 inhibition is a potential treatment for breast cancer.
a Combination index (CI) calculation for CP-673451 and GSK-650394 treatment in MDA-MB-231 (left) and BT-549 cells (right). b Colony-formation assays of MDA-MB-231 (left) and BT-549 (right) cells treated with DMSO or CP-673451 (2 μM) with increasing concentrations of GSK-650394. c MDA-MB-231 (left) and BT-549 (right) cells were treated with DMSO or CP-673451 for 1 h or a combination of CP-673451 (2 μM) and increasing doses of GSK-650394 for 5 days. Changes in p-AKT, total AKT, SGK1, p-NDRG1, and NDRG1 protein levels were analyzed by western blot. GAPDH served as a loading control. d Photographs of harvested tumors and quantification of MDA-MB-231 (left) and BT-549 (right) tumor-bearing mice treated with vehicle, GSK-650394 (150 µg/day, 5 days/week, subcutaneous administration), CP-673451 (50 mg/kg/day, 5 days/week, intraperitoneal administration), or a combination of both agents. Data are presented as the mean ± SD.
Discussion
Lack of insight into the mechanisms behind metastasis initiation and progression poses challenges to the development of antitumor therapeutics. Here, we describe a scenario in which the activation of PDGF signaling leads to metastatic initiation and progression. Notably, SGK1 is co-expressed with PDGFR in MICs and contributes to the maintenance of residual AKT downstream activity and consequent cell survival. Suppression of both PDGFR and SGK1 prevents AKT and NDRG activation and results in strong antitumor activities. This work provides a rationale for the use of a combination treatment for metastatic breast cancer.
The presumed features of MICs include cellular plasticity, resistance to anoikis and apoptosis, evasion of the immune system response, metabolic reprogramming, and the ability to enter and exit dormancy [4]. In our study, LM cells are highly predisposed to metastasize to the lungs and are enriched for genes of the LMS [22, 23]. A defining trait of LM cells is their stem-like phenotype. LM cells formed larger clones than parental cells, but this phenotype did not seem to stem from more rapid cycling, suggesting that MICs harbor a greater percentage of slow-cycling long-term cells that can grow into metastatic lesions in a distant organ. This observation is in line with previous reports that subpopulations of cancer stem cells (CSCs) undergo EMT, which is associated not only with a migratory phenotype but also with a slow proliferative state that confers resistance to antiproliferative drugs [24, 25]. Our results suggest that PDGF signaling activation can drive metastatic success by promoting several key traits of MICs. Our present findings demonstrate that PDGF signaling is a central regulatory node in the MIC population.
Indeed, many efforts have been made to exploit strategies to inhibit PDGF signaling in cancer. However, currently approved cancer therapies for the inhibition of kinase activity of PDGFRs are based on multi-targeted small molecules [26], making it difficult to separately assess the function of these signaling pathways. Our results showed that compared with chemotherapy, treatment with inhibitors specific to PDGFRα/β led to significantly increased growth inhibition of MICs. The metastatic heterogeneity substantially contributes to the current inability to successfully eradicate or prevent metastatic diseases. Considering the heterogeneity of cancer cells within tumors, it is therefore reasonable to propose a combination rationale for patients with breast cancer. In fact, our preliminary results indicate that the combination of a PDGFR inhibitor and an SGK1 inhibitor led to a further inhibition of pulmonary metastases and primary breast tumors. As most agents used in adjuvant therapies of breast cancer are designed to target proliferative cancer cells rather than cells with a metastasis-initiating phenotype [27], our results yield clues regarding how combination therapy can treat and prevent metastases arising from breast cancer.
Importantly, an emerging characteristic of SGK1 is that it may not only represent a therapeutic target in itself but also could be exploited to improve the efficacy of existing therapies. For example, SGK1 promotes resistance of breast cancer cells to AKT- and PI3Kα-directed agents [21, 28]. Growing evidence has indicated that SGK1 is a stress-induced survival factor and that its expression is promptly induced under pathophysiological conditions such as exposure to growth factors (such as EGF and IGF), cytokines, and various cellular stresses such as oxidative stress and irradiation [29]. However, much remains unclear about SGK1 function. We showed that in the presence of a PDGFR inhibitor, SGK1, but not AKT remains active; conversely, upon GSK-650394 treatment, SGK1, but not AKT, is fully inhibited. The single activity of either of these kinases seems to be sufficient to propagate downstream pro-survival signaling. This is also confirmed by the fact that the combination of both agents elicited a powerful antitumor effect.
In conclusion, our findings show that SGK1 mediates the reduction of the pharmacodynamic effect of PDGFR inhibitors while playing a key role in PDGF signaling in MICs. SGK1 inhibition can sensitize MICs to PDGFR inhibition, and combined PDGFR and SGK1 inhibition exhibited strong antitumor activities in established breast cancer cell lines. Further studies are necessary to evaluate the therapeutic efficacy of a PDGF inhibitor and an SGK1 inhibitor in a clinical setting. This study highlights the importance of understanding the underlying mechanisms of kinase regulation in uncovering critical nodes for pharmacological intervention and improving the therapeutic options for metastatic cancers.
Supplementary information
Acknowledgements
This study was supported by the National Natural Science Foundation of China (Nos. 81672598, 81872152, 81772961, and 81702620).
Compliance with ethical standards
Conflict of interest
The authors declare that they have no conflict of interest.
Footnotes
Edited by H.-U. Simon
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
These authors contributed equally: Lu Yang, Ning Li, Zhicheng Xue
Contributor Information
Hailin Tang, Email: tanghl@sysucc.org.cn.
Xiaoming Xie, Email: xiexm@sysucc.org.cn.
Supplementary information
The online version of this article (10.1038/s41418-019-0485-4) contains supplementary material, which is available to authorized users.
References
- 1.Spano D, Heck C, De Antonellis P, Christofori G, Zollo M. Molecular networks that regulate cancer metastasis. Semin Cancer Biol. 2012;22:234–49. doi: 10.1016/j.semcancer.2012.03.006. [DOI] [PubMed] [Google Scholar]
- 2.Husemann Y, Geigl JB, Schubert F, Musiani P, Meyer M, Burghart E, et al. Systemic spread is an early step in breast cancer. Cancer Cell. 2008;13:58–68. doi: 10.1016/j.ccr.2007.12.003. [DOI] [PubMed] [Google Scholar]
- 3.Cameron MD, Schmidt EE, Kerkvliet N, Nadkarni KV, Morris VL, Groom AC, et al. Temporal progression of metastasis in lung: cell survival, dormancy, and location dependence of metastatic inefficiency. Cancer Res. 2000;60:2541–6. [PubMed] [Google Scholar]
- 4.Celia-Terrassa T, Kang Y. Distinctive properties of metastasis-initiating cells. Genes Dev. 2016;30:892–908. doi: 10.1101/gad.277681.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Yachida S, Jones S, Bozic I, Antal T, Leary R, Fu B, et al. Distant metastasis occurs late during the genetic evolution of pancreatic cancer. Nature. 2010;467:1114–7. doi: 10.1038/nature09515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Jacob LS, Vanharanta S, Obenauf AC, Pirun M, Viale A, Socci ND, et al. Metastatic competence can emerge with selection of preexisting oncogenic alleles without a need of new mutations. Cancer Res. 2015;75:3713–9. doi: 10.1158/0008-5472.CAN-15-0562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Campbell PJ, Yachida S, Mudie LJ, Stephens PJ, Pleasance ED, Stebbings LA, et al. The patterns and dynamics of genomic instability in metastatic pancreatic cancer. Nature. 2010;467:1109–13. doi: 10.1038/nature09460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lawson DA, Bhakta NR, Kessenbrock K, Prummel KD, Yu Y, Takai K, et al. Single-cell analysis reveals a stem-cell program in human metastatic breast cancer cells. Nature. 2015;526:131–5. doi: 10.1038/nature15260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Pascual G, Avgustinova A, Mejetta S, Martin M, Castellanos A, Attolini CS, et al. Targeting metastasis-initiating cells through the fatty acid receptor CD36. Nature. 2017;541:41–45. doi: 10.1038/nature20791. [DOI] [PubMed] [Google Scholar]
- 10.Straussman R, Morikawa T, Shee K, Barzily-Rokni M, Qian ZR, Du J, et al. Tumour micro-environment elicits innate resistance to RAF inhibitors through HGF secretion. Nature. 2012;487:500–4. doi: 10.1038/nature11183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Goel S, Wang Q, Watt AC, Tolaney SM, Dillon DA, Li W, et al. Overcoming therapeutic resistance in HER2-positive breast cancers with CDK4/6 inhibitors. Cancer Cell. 2016;29:255–69. doi: 10.1016/j.ccell.2016.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yang L, Tang H, Kong Y, Xie X, Chen J, Song C, et al. LGR5 promotes breast cancer progression and maintains stem-like cells through activation of Wnt/beta-catenin signaling. Stem Cells. 2015;33:2913–24. doi: 10.1002/stem.2083. [DOI] [PubMed] [Google Scholar]
- 13.Shibue T, Brooks MW, Weinberg RA. An integrin-linked machinery of cytoskeletal regulation that enables experimental tumor initiation and metastatic colonization. Cancer Cell. 2013;24:481–98. doi: 10.1016/j.ccr.2013.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fais P, Leopizzi M, Di Maio V, Longo L, Della Rocca C, Tagliaro F, et al. Phosphoinositide-specific phospholipase C in normal human liver and in alcohol abuse. J Cell Biochem. 2019;120:7907–17. [DOI] [PubMed]
- 15.Pelzl L, Tolios A, Schmidt EM, Alesutan I, Walker B, Munzer P, et al. Translational regulation of the serum- and glucocorticoid-inducible kinase-1 (SGK1) in platelets. Biochem Biophy Res Commun. 2012;425:1–5. doi: 10.1016/j.bbrc.2012.07.026. [DOI] [PubMed] [Google Scholar]
- 16.Orlacchio A, Ranieri M, Brave M, Arciuch VA, Forde T, De Martino D, et al. SGK1 is a critical component of an AKT-independent pathway essential for PI3K-mediated tumor development and maintenance. Cancer Res. 2017;77:6914–26. doi: 10.1158/0008-5472.CAN-17-2105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Li JH, Liu S, Zhou H, Qu LH, Yang JH. starBase v2.0: decoding miRNA-ceRNA, miRNA-ncRNA and protein-RNA interaction networks from large-scale CLIP-Seq data. Nucleic Acids Res. 2014;42(Database issue):D92–97. doi: 10.1093/nar/gkt1248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Greenawalt EJ, Edmonds MD, Jain N, Adams CM, Mitra R, Eischen CM. Targeting of SGK1 by miR-576-3p Inhibits Lung Adenocarcinoma Migration and Invasion. Mol Cancer Res. 2019;17:289–98. doi: 10.1158/1541-7786.MCR-18-0364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Liu W, Wang X, Wang Y, Dai Y, Xie Y, Ping Y, et al. SGK1 inhibition-induced autophagy impairs prostate cancer metastasis by reversing EMT. J Exp Clin Cancer Res. 2018;37:73. doi: 10.1186/s13046-018-0743-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Murray JT, Campbell DG, Morrice N, Auld GC, Shpiro N, Marquez R, et al. Exploitation of KESTREL to identify NDRG family members as physiological substrates for SGK1 and GSK3. Biochemical J. 2004;384(Pt 3):477–88. doi: 10.1042/BJ20041057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sommer EM, Dry H, Cross D, Guichard S, Davies BR, Alessi DR. Elevated SGK1 predicts resistance of breast cancer cells to Akt inhibitors. Biochemical J. 2013;452:499–508. doi: 10.1042/BJ20130342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gupta GP, Perk J, Acharyya S, de Candia P, Mittal V, Todorova-Manova K, et al. ID genes mediate tumor reinitiation during breast cancer lung metastasis. Proc Natl Acad Sci USA. 2007;104:19506–11. doi: 10.1073/pnas.0709185104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Minn AJ, Gupta GP, Siegel PM, Bos PD, Shu W, Giri DD, et al. Genes that mediate breast cancer metastasis to lung. Nature. 2005;436:518–24. doi: 10.1038/nature03799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Oshimori N, Oristian D, Fuchs E. TGF-beta promotes heterogeneity and drug resistance in squamous cell carcinoma. Cell. 2015;160:963–76. doi: 10.1016/j.cell.2015.01.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Feng XH. The changing faces of cancer cells. Nat Rev Mol cell Biol. 2010;11:466. doi: 10.1038/nrm2923. [DOI] [PubMed] [Google Scholar]
- 26.Papadopoulos N, Lennartsson J. The PDGF/PDGFR pathway as a drug target. Mol Asp Med. 2018;62:75–88. doi: 10.1016/j.mam.2017.11.007. [DOI] [PubMed] [Google Scholar]
- 27.Hensel JA, Flaig TW, Theodorescu D. Clinical opportunities and challenges in targeting tumour dormancy. Nat Rev Clin Oncol. 2013;10:41–51. doi: 10.1038/nrclinonc.2012.207. [DOI] [PubMed] [Google Scholar]
- 28.Castel P, Ellis H, Bago R, Toska E, Razavi P, Carmona FJ, et al. PDK1-SGK1 signaling sustains AKT-independent mTORC1 activation and confers resistance to PI3Kalpha inhibition. Cancer Cell. 2016;30:229–42. doi: 10.1016/j.ccell.2016.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lang F, Gawaz M, Borst O. The serum- & glucocorticoid-inducible kinase in the regulation of platelet function. Acta Physiol. 2015;213:181–90. doi: 10.1111/apha.12334. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.