Abstract
Orchid (Orchidaceae) is one of the largest families in angiosperms and presents exceptional diversity in lifestyle. Their unique reproductive characteristics of orchid are attracted by scientist for centuries. One of the synapomorphies of orchid plants is that their seeds do not contain endosperm. Lipids are used as major energy storage in orchid seeds. However, regulation and mobilization of lipid usage during early seedling (protocorm) stage of orchid is not understood. In this study, we compared transcriptomes from developing Phalaenopsis aphrodite protocorms grown on 1/2-strength MS medium with sucrose. The expression of P. aphrodite MALATE SYNTHASE (PaMLS), involved in the glyoxylate cycle, was significantly decreased from 4 days after incubation (DAI) to 7 DAI. On real-time RT-PCR, both P. aphrodite ISOCITRATE LYASE (PaICL) and PaMLS were down-regulated during protocorm development and suppressed by sucrose treatment. In addition, several genes encoding transcription factors regulating PaMLS expression were identified. A gene encoding homeobox transcription factor (named PaHB5) was involved in positive regulation of PaMLS. This study showed that sucrose regulates the glyoxylate cycle during orchid protocorm development in asymbiotic germination and provides new insights into the transcription factors involved in the regulation of malate synthase expression.
Subject terms: Plant embryogenesis, Plant molecular biology
Introduction
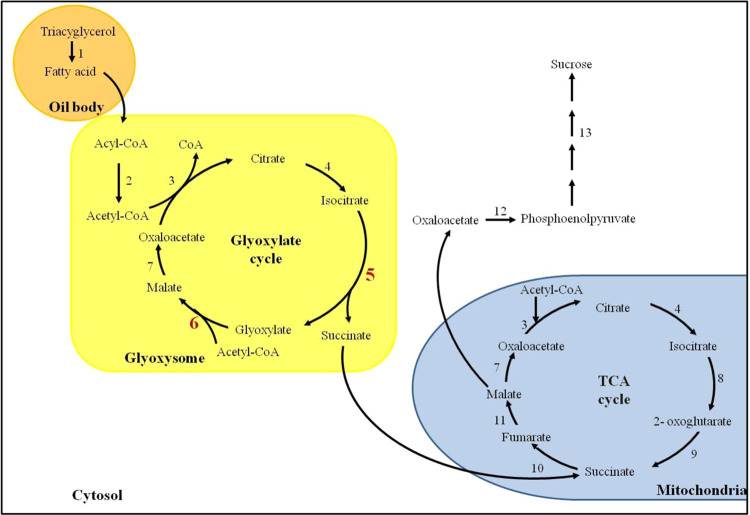
In many dicot seeds, such as in Arabidopsis thaliana, Glycine max, and Brassica napus, a large amount of lipids is stored in cotyledons, which are therefore called “oil seeds”. During oil seed germination, lipids cannot be directly used by seeds to provide energy. Therefore, the oil mobilization pathway, including β-oxidation, glyoxylate cycle, tricarboxylic acid (TCA) cycle and gluconeogenesis, is essential for converting storage lipids to soluble sugars that are used to fuel seedling growth (Fig. 1). In oil mobilization, the glyoxylate cycle plays a central role in use of seed storage oil. The cycle uptakes the two-carbon product acetyl-CoA and synthesizes the 4-carbon compound succinate, which subsequently forms a substrate for gluconegenesis to generate sucrose in the cytosol. These soluble sugars are then catabolized by glycolysis to support seedling growth1,2. The glyoxylate cycle involves two unique key enzymes: isocitrate lyase (ICL, EC 4.1.3.1), which catalyzes the lysis of isocitrate to glyoxylate and succinate, and malate synthase (MLS, EC 2.3.3.9), which synthesizes malate from glyoxylate and acetyl-CoA1,2. In Arabidopsis seedlings, the transcript level and enzyme activity of ICL and MLS are induced after imbibition and decrease after post-germinative growth. This expression pattern is strongly correlated with lipid breakdown1. Control of plant metabolism at the transcription level is most apparent when the intracellular nutritional status changes. In the regulation of the glyoxylate cycle, carbon catabolite repression has been well studied in cell culture, seedlings and mature plant tissue of cucumber3–5. The gene expression of ICL and MLS was induced in cucumber cell culture during sugar starvation1 and was down-regulated on treatment with different hexose sugars (sucrose, glucose, fructose and mannose). Deletion analysis of the promoter region of cucumber ICL and MLS revealed separate conserved sequence elements that are necessary for induction in response to a change in carbohydrate status3. Suppressed ICL enzyme activity was also found in the presence of glucose in Arabidopsis during post-germinative growth6.
Figure 1.
Oil mobilization pathway in oil seeds. 1, triacylglycerol lipase; 2, fatty acid β-oxidation; 3, citrate synthase; 4, aconitate hydratase; 5, isocitrate lyase; 6, malate synthase; 7, malate dehydrogenase; 8, isocitrate dehydrogenase; 9, 2-oxoglutarate dehydrogenase and succinyl-CoA synthetase; 10, succinate dehydrogenase; 11, fumarate hydratase; 12, phosphoenolpyruvate carboxykinase; 13, gluconeogenesis.
The seeds of orchid are often referred to as ‘dust seeds. The seed is very tiny and contains a globular-shape embryo without well development as in other flowering plants7,8. The seed development in orchids is unique as compared with most flowering plants. Once the ovules mature, a zygote and a polar chalazal complex could be formed after successful double fertilization7. However, the polar chalazal complex do not has ability to develop into an endosperm9. Thus, the mature orchid seeds do not contain endosperm. The lipid is the major energy storage found in orchid embryo.
Because of the minute size and limited stored nutrient reserves of orchid seeds, symbiosis with mycorrhizal fungus under natural conditions is essential for germination. Seed reserves are mobilized to provide nutrition for early seedling development before photosynthesis7,8. After the fungus is established in the orchid, growth generally occurs with carbon flux from fungi. The swollen embryos grow and form protocorms, a structure between the embryo and the seedling that lacks chlorophyll8. After germination, young seedlings have insufficient reserves to allow for ongoing growth without fungus-supplied carbon10. On realizing that the main function of the fungus is to provide a carbon source, sugar was added into culture medium to enable asymbiotic germination8. A wide range of sugars has been shown to support germination and growth of orchid seeds; they include mono-, di- and oligosaccharides (such as glucose, fructose, maltose and trehalose)11–13. This technique of asymbiotic orchid seed germination is useful for propagation for most orchids in the absence of mycorrhizal fungus7. However, how the carbon source affects the metabolic events of orchid seed storage oil during protocorm development remain the least studied and most poorly understood.
In this study, we compared the expression of both P. aphrodite ISOCITRATE LYASE (PaICL) and PaMLS genes regulated by sucrose at protocorm stage. We identified a positive transcription factor (TF) regulating PaMLS expression, thereby providing the basis for an expanded understanding of orchid seed storage-oil utilization.
Results
Distribution of storage products in P. aphrodite mature seeds
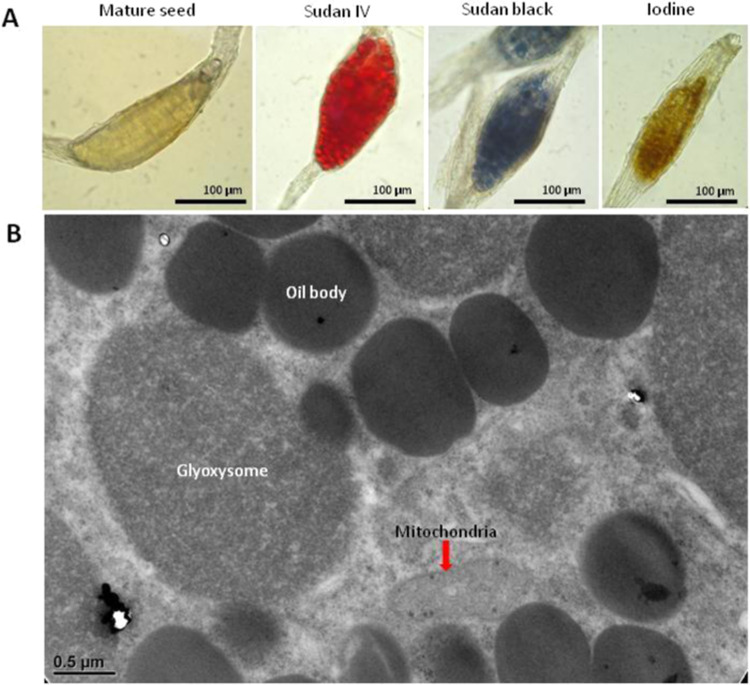
According to previous study, endosperm is absent in mature orchid seeds, but lipids are accumulated during the seed maturation stage. To verify the storage components in P. aphrodite mature seed, we used 0.3% Sudan IV solution and 0.3% Sudan black solution to stain for lipids (Fig. 2A). The entire proembryo of seeds was red with Sudan IV staining and dark-blue with Sudan black staining. Therefore, a large amount of lipids accumulated in the proembryo of mature P. aphrodite seed. However, the absence of starch was visualized as dark-brown staining with 5% iodine solution.
Figure 2.
Distribution of storage components in Phalaenopsis aphrodite mature seeds (A) P. aphrodite mature seeds and seeds stained with Sudan IV, Sudan black and iodine. Large amounts of lipids were visualized in red with Sudan IV and in dark-blue with Sudan black. Absence of starch deposition is seen as dark-brown on iodine staining. (B) Transmission electron micrograph of a section of a P. aphrodite protocorm at 0 DAI. DAI, days after incubation.
TEM of a section in a P. aphrodite protocorm at 0 DAI
In germinating seed, fatty acids are released from lipids stored in oil bodies, then imported into glyoxysomes to produce succinate via β-oxidation and the glyoxylate cycle. Succinate molecules are then transported to mitochondria to be converted to malate for gluconeogenesis. Therefore, the location of oil bodies, glyoxysomes and mitochondria are close in germinating seeds. To investigate the spatial distribution of the three organelles in germinating orchid seeds, P. aphrodite seeds were resuspended in 1/2 MS liquid medium to imbibe the seeds (0 DAI protocorms) and observed by TEM (Fig. 2B). Many oil bodies were found in protocorms at 0 DAI, shown in dark-grey, with glyoxysomes (light-grey organelles) located beside them. Mitochondria were in close proximity to glyoxysomes and oil bodies.
Effect of sucrose on protocorm development in P. aphrodite
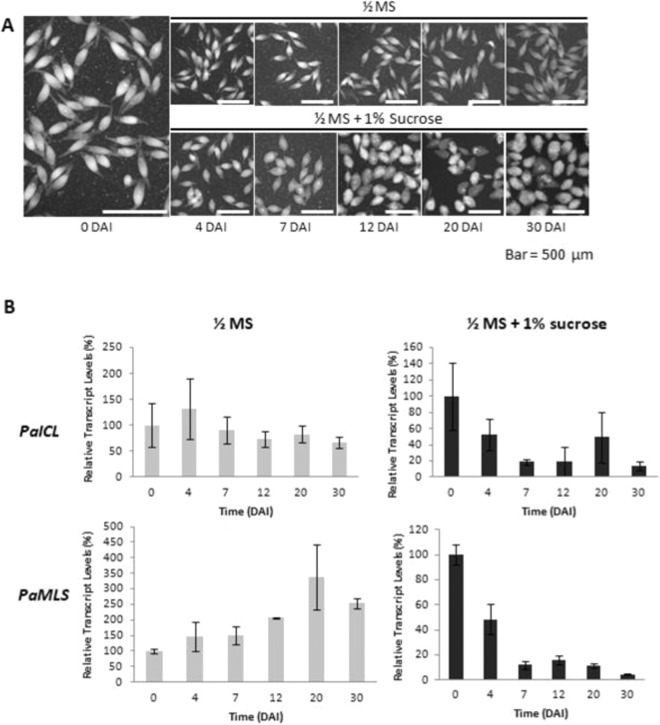
To examine the effect of sucrose on protocorm development, P. aphrodite mature seeds were grown in medium containing 1/2-strength MS salts with or without 1% (w/v) sucrose (Fig. 3A). In the presence of sucrose, spindle-like seeds swelled and greenish protocorms were observed at 7 DAI. Most protocorms were greenish and became round and a few protocorms were bleached from 12 to 20 DAI. At 30 DAI, many protocorms were bleached and some continue enlarged and turned to dark green. In contrast, seeds grown without sucrose did not swell and turn greenish until 30 DAI. These results suggest that sucrose is important to provide energy for supporting orchid protocorm development and/or germination.
Figure 3.
Phenotypes and expression profiles of PaICL and PaMLS at different developmental protocorm stages in P. Aphrodite (A) P. aphrodite seeds were resuspended in 1/2-strength MS (1/2 MS) liquid medium for 48 h, defined as 0-DAI protocorms, then incubated in 1/2 MS medium with or without 1% sucrose for 4 to 30 days. DAI, days after incubation. Bar = 500 μm. (B) Real-time quantitative RT-PCR analysis of PaICL and PaMLS expression normalized to Phalaenopsis Actin4 for each sample; experiments were performed in triplicate. Data are mean ± SEM.
Expression of genes involved in oil mobilization pathway
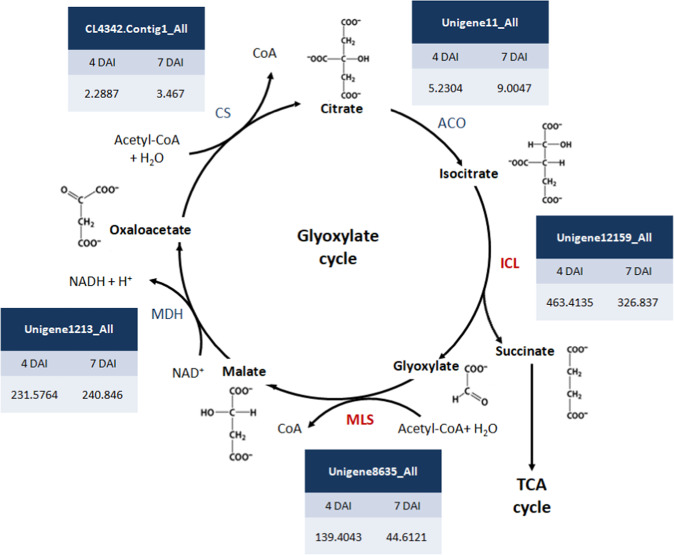
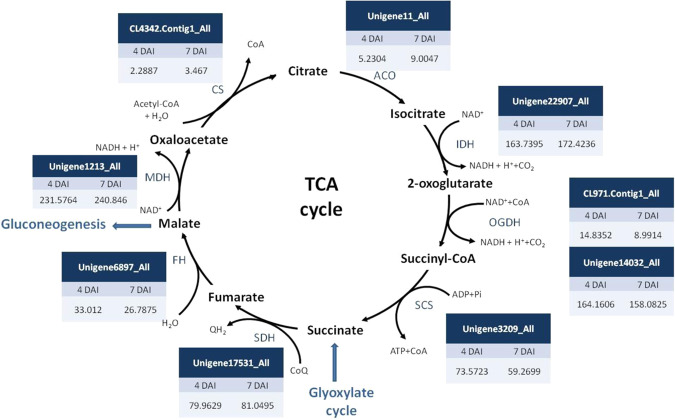
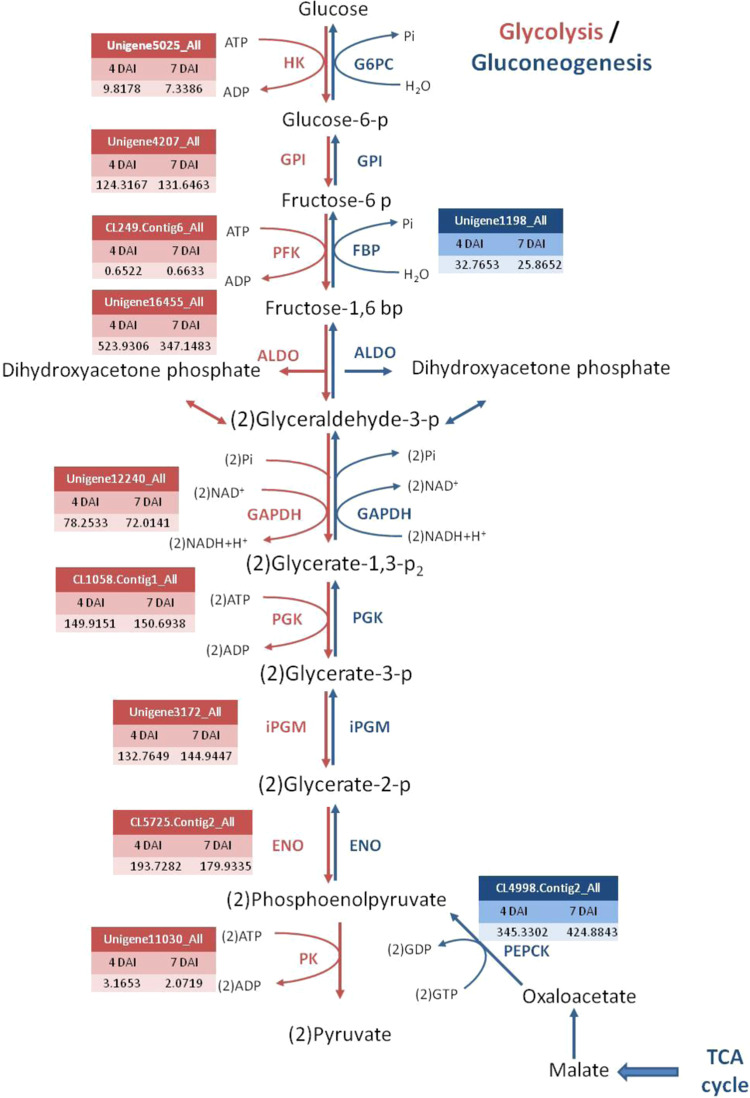
The expression of ICL and MLS involved in the glyoxylate cycle has been found correlated with lipid breakdown and regulated by the hexose sugars1. To further reveal whether sucrose affects the protocorm oil mobilization pathway, we compared transcriptomes from 4- and 7-DAI protocorms treated with sucrose. The expression of enzymes involved in the glyoxylate and TCA cycles and gluconeogenesis and glycolysis pathway were compared by differential expression analysis. Unigenes involved in each pathway were obtained by using genes sequences from the KEGG Pathway (http://www.genome.jp/kegg/pathway.html 14) to blast against protocorm transcriptomes. The expression levels were shown with FPKM in 4- and 7-DAI protocorm transcriptomes. In the glyoxylate cycle, MLS showed 3-fold down-regulation and ICL 1.4-fold down-regulation in 7-DAI protocorms. We found no significant changes in unigene hits for other enzymes (Fig. 4). However, sucrose did not significantly affect the expression of genes in the TCA cycle (Fig. 5) or gluconeogenesis and glycolysis pathway (Fig. 6): the expression of unigenes involved in these pathways did not significantly differ between 4- and 7-DAI protocorms. Interestingly, hexokinase, phosphofrucokinase, and pyruvate kinase genes involved in glycolysis showed low FPKM value at both 4 and 7 DAI.
Figure 4.
Expression of unigenes involved in glyoxylate cycle in 4- and 7-DAI protocorms The expression levels were derived from P. aphrodite protocorm transcriptomes. ACO: aconitate hydratase, ICL: isocitrate lyase, MLS: malate synthase, MDH: malate dehydrogenase, CS: citrate synthase.
Figure 5.
Expression of unigenes involved in tricarboxylic acid (TCA) cycle between 4- and 7-DAI protocorms. The expression levels were derived from P. aphrodite protocorm transcriptomes. CS: citrate synthase, ACO: aconitate hydratase, IDH: isocitrate dehydrogenase, OGDH: 2-oxoglutarate dehydrogenase, SCS: succinyl-CoA synthetase, SDH: succinate dehydrogenase, FH: fumarate hydratase, MDH: malate dehydrogenase.
Figure 6.
Expression of unigenes involved in glycolysis/gluconeogenesis between 4- and 7-DAI protocorms. The expression levels were derived from P. aphrodite protocorm transcriptomes. HK: hexokinase, G6PC: glucose-6-phosphatase, GPI: glucose-6-phosphate isomerase, PFK: 6-phosphofructokinase 1, FBPase: fructose-1,6-bisphosphatase I, ALDO: fructose-bisphosphate aldolase, GAPDH: glyceraldehydes 3-phosphate dehydrogenase, PGK: phosphoglycerate kinase, iPGM: 2,3-bisphosphoglycerate-independent phosphoglycerate mutase, ENO, enolase, PK: pyruvate kinase, PEPCK: phosphoenolpyruvate carboxykinase.
Identification and phylogenetic analysis of PaICL and PaMLS in P. aphrodite
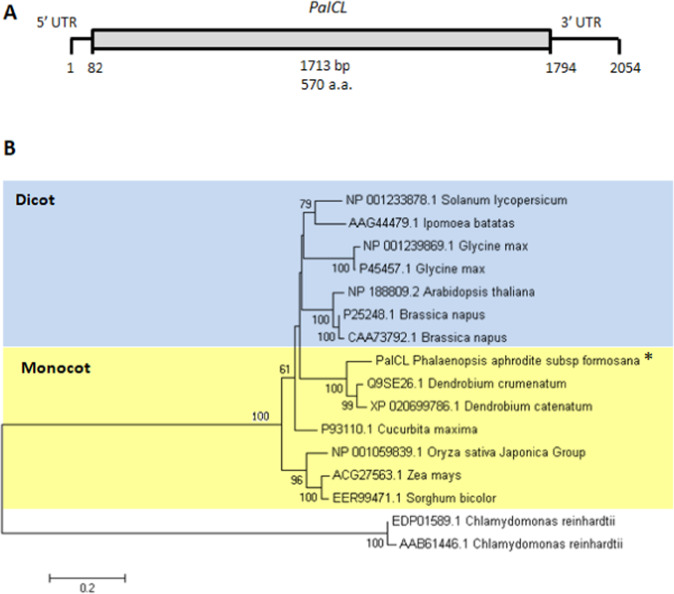
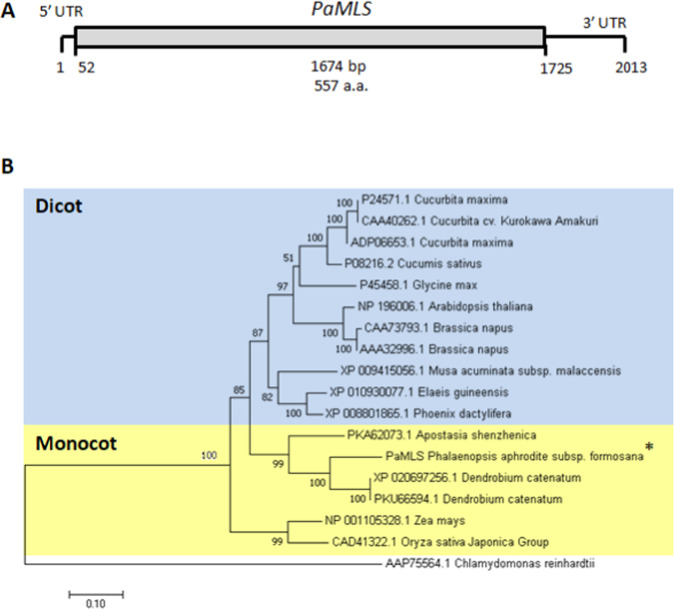
Full-length cDNA sequences of P. aphrodite ICL and MLS were obtained from protocorm cDNA at 0 DAI by 5′ RACE and were named PaICL and PaMLS, respectively. Full-length cDNA of PaICL was 2,054 bp with the coding region from 82 to 1,794 bp (Fig. 7A). Full-length cDNA of PaMLS was 2,013 bp with the coding region from 52 to 1,725 bp (Fig. 8A). Conceptual translation of the open reading frames encoded by genes yielded proteins 570 and 557 amino acids long for PaICL and PaMLS, respectively.
Figure 7.
Phylogenetic analysis of ICL. (A) Structure of PaICL full-length cDNA. (B) Phylogenetic tree created with MEGA 5.0 by the neighbor-joining method and the bootstrap test involved 1000 iterations.
Figure 8.
Phylogenetic analysis of MLS. (A) Structure of of PaMLS full-length cDNA. (B) Phylogenetic tree created with MEGA 5.0 by the neighbor-joining method and the bootstrap test involved 1000 iterations.
To determine the phylogenetic relationships of PaICL and PaMLS with other plant isocitrate-lyase and malate-synthase genes, we reconstructed the phylogenetic tree for these genes by using the coding regions of amino acid sequences retrieved from the National Center for Biotechnology Information (NCBI). Both PaICL (Fig. 7B) and PaMLS (Fig. 8B) were closely related to their respective orthologs in the primitive orchid Apostaisa15 and the other epiphytic orchid Dendrobium16. Both PaICL and PaMLS formed a clade with the homologs of monocots (Figs. 7B and 8B).
Temporal expression of PaICL and PaMLS during protocorm development
To verify the effects of sucrose on glyoxylate cycle during protocorm development in P. aphrodite, we used real-time quantitative PCR to confirm the temporal expression patterns of PaICL and PaMLS (Fig. 3B). Without sucrose in the medium, PaICL and PaMLS expressed in the entire protocorm developmental stages, from 0 to 30 DAI. In contrast, with sucrose, PaICL and PaMLS were substantially down-regulated during protocorm development. Thus, sucrose might inhibit lipid hydrolysis through the glyoxylate cycle during orchid protocorm development.
Comparison of sucrose-affected transcript profiles between protocorms at 4 and 7 DAI
To gain more insight into the genes related to the decreased expression of PaMLS in 7-DAI protocorms treated with sucrose, we used a 2-fold expression cutoff for selecting differentially expressed unigenes between protocorms at 4 and 7 DAI. In total, 3,830 and 7,648 genes showed differential expression in protocorms at 4 and 7 DAI, respectively.
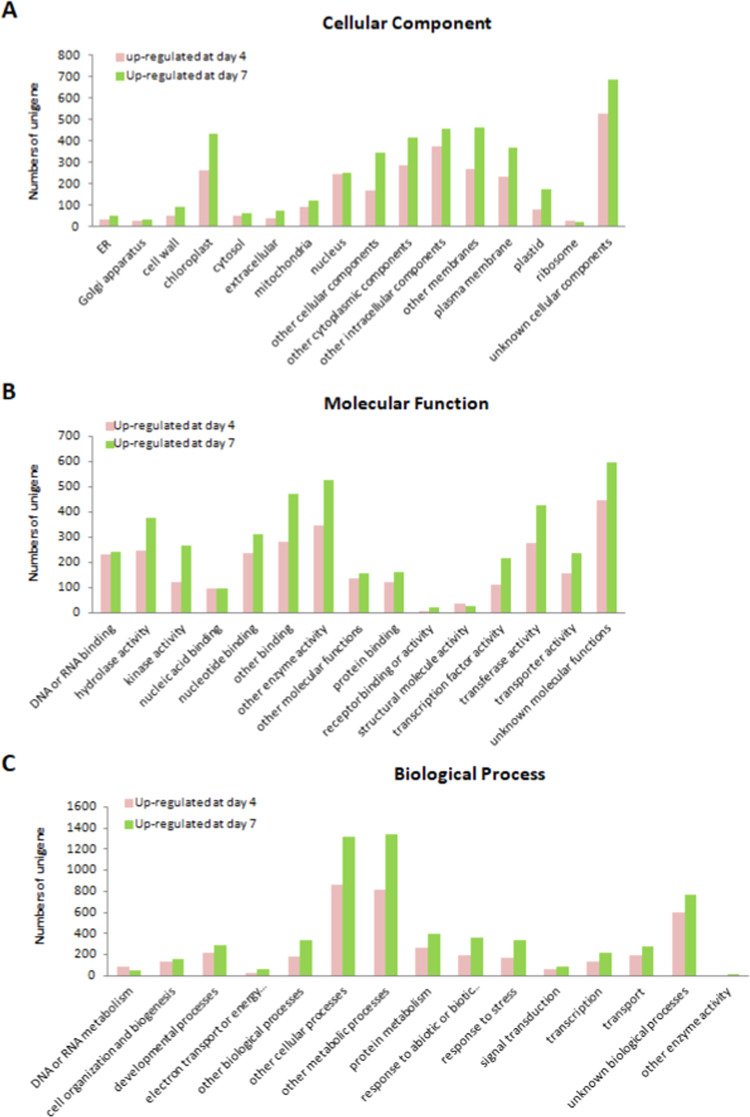
Gene Ontology (GO) classification system for plants developed at TAIR17 (https://www.arabidopsis.org/help/helppages/go_slim_help.jsp) was further adopted to characterize the possible function of developing stage-dominant unigenes (Fig. 9). For 4-DAI protocorm-dominant unigenes, 90% (3,451/3,830), 65% (2,510/3,830), 64% (2,467/3,830) were partitioned to biological processes, molecular functions and cellular components, respectively, and for 7-DAI protocorm-dominant unigenes, 72% (5,513/7,648), 49% (3,796/7,648), 49% (3,798/7,648) were assigned, respectively. The 7-DAI protocorm unigenes were highly represented in most GO categories. In biological processes, 7-DAI protocorm unigenes were involved in other cellular processes and other metabolic processes, but 4-DAI protocorm unigenes were involved in DNA or RNA metabolism. In molecular functions, 7 DAI protocorm unigenes were highly represented in hydrolase activity, kinase activity and TF activity, but 4-DAI protocorm unigenes were involved in nucleic acid binding and structural molecule activity. In cellular components, 7-DAI protocorm unigenes were overrepresented in chloroplasts, other cellular components and plastids as compared with 4-DAI protocorm unigenes. In contrast, 7-DAI protocorm unigenes were slightly underrepresented in ribosomes.
Figure 9.
Gene ontology (GO) assignments for 4- and 7-DAI dominant genes derived from P. aphrodite protocorm transcriptomes. Genes were filtered with absolute value of log2 ratio ≧1. The GO Slim Classification for Plants developed at TAIR was used to characterize the unigenes functionally. (A) Cellular component, (B) molecular function, (C) biological process.
Characterization of developing stage–dominant unigenes by KEGG pathway analysis
Stage–dominant unigenes were mapped to KEGG pathways (http://www.genome.jp/kegg/pathway.html) (Table 1). About 10.2% (392/3,830) of 4-DAI protocorm-dominant unigenes were mapped to 93 KEGG pathways, and 8.6% (656/7,648) of 7-DAI unigenes were mapped to 97 KEGG pathways. Among 392 hits of 4-DAI protocorm-dominant unigenes, 251 hits were assigned to metabolism, 104 to genetic information processing, 17 to organismal systems, 16 to cellular processes, and 4 to environmental information processing. 656 7-DAI protocorm-dominant unigenes were respectively classified into metabolism (521), genetic information processing (73), organismal systems (41), cellular processes (18), and environmental information processing (3). In metabolism, the number of unigenes associated with biosynthesis of other secondary metabolites, carbohydrate metabolism, and lipid metabolism was obviously increased in 7-DAI protocorms. In lipid metabolism, more genes were involved in α-linolenic acid metabolism in 7- than 4-DAI protocorms, which suggests that α-linolenic acids might be the main components in orchid seed-storage lipids. The significant increase in biosynthesis of secondary-metabolite genes indicated that protocorms may generate secondary metabolites to adapt to the environment.
Table 1.
Number of differentially expressed unigenes mapped in KEGG pathways.
| KEGG pathway | Sub-pathway of KEGG pathway | No. of hits for 4-DAI protocorm unigenes | No. of hits for 7-DAI protocorm unigenes |
|---|---|---|---|
| Metabolism | 251 | 521 | |
| Carbohydrate Metabolism | 50 | 105 | |
| Energy Metabolism | 7 | 50 | |
| Lipid Metabolism | 27 | 75 | |
| Nucleotide Metabolism | 30 | 14 | |
| Amino Acid Metabolism | 46 | 79 | |
| Metabolism of Other Amino Acids | 10 | 25 | |
| Glycan Biosynthesis and Metabolism | 7 | 18 | |
| Metabolism of Cofactors and Vitamins | 23 | 19 | |
| Metabolism of Terpenoids and Polyketides | 27 | 44 | |
| Biosynthesis of Other Secondary Metabolites | 24 | 92 | |
| Genetic Information Processing | 104 | 73 | |
| Transcription | 21 | 13 | |
| Translation | 17 | 13 | |
| Folding, Sorting and Degradation | 24 | 32 | |
| Replication and Repair | 42 | 15 | |
| Environmental Information Processing | 4 | 3 | |
| Signal Transduction | 4 | 3 | |
| Cellular Processes | 16 | 18 | |
| Transport and Catabolism | 16 | 18 | |
| Organismal Systems | 17 | 41 | |
| Environmental Adaptation | 17 | 41 | |
Transcription factors related to the regulation of PaMLS expression
To explore the TFs related to the regulation of PaMLS expression, unigenes with 10-fold differential expression between 4- and 7-DAI protocorms were screened and BLAST searched against rice TFs derived from plantTFDB (Supplementary Fig. S2). Furthermore, 2,000-bp upstream sequences of PaMLS translation start sits for the Phalaenopsis genome were retrieved18 and analyzed by using plantPAN to predict TF binding sites. In all, 14 unigenes with at least 10-fold differential expression and presenting a putative binding sequence at the PaMLS promoter were filtered. Eight genes corresponded to the 14 unigenes after alignment to the Phalaenopsis genome (Table 2). These genes were named PaHB5, PaANT, PaMADS2, PaMYB4, PaPIF3, PaRAV1-1, PaWRKY18 and PaWRKY71.
Table 2.
Identified putative transcription factors for PaMLS expression.
| Transcription factor | Unigene | Stage with higher expression | Fold change expression | Full length cDNA (bp) | Coding sequence (bp) | Protein length (a.a) |
|---|---|---|---|---|---|---|
| PaHB5 | CL2645.Contig2_All | 4 DAI | 10.5 | 816 | 60–686 | 208 |
| PaWRKY71 | CL696.Contig6_All | 7 DAI | 10.2 | 990 | 158–739 | 192 |
| PaPIF3 | Unigene23033_All | 7 DAI | 10.5 | 1161 | 446–751 | 100 |
| PaANT | Unigene4788_All | 7 DAI | 14.4 | 950 | 42–827 | 260 |
| PaMADS2 | Unigene23959_All | 7 DAI | 20.4 | 764 | 81–764 | 237 |
| PaMYB4 | Unigene23199_All | 7 DAI | 21.4 | 702 | 1–702 | 232 |
| PaWRKY18 | Unigene14862_All | 7 DAI | 49.2 | 661 | 26–538 | 171 |
| PaRAV1–1 | Unigene22378_All | 7 DAI | 42.8 | 579 | 1–579 | 191 |
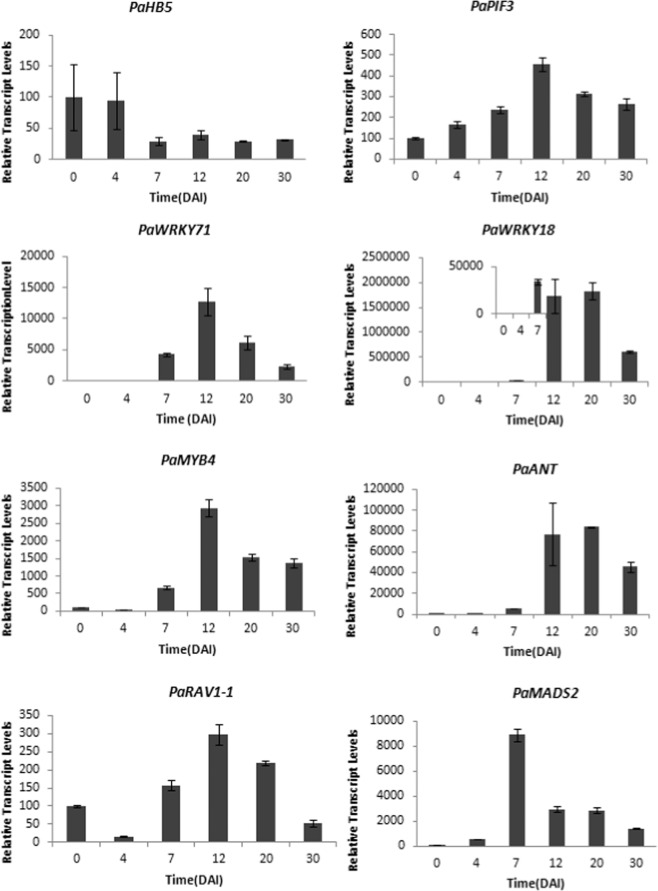
Temporal expression of 8 putative transcription factors during protocorm development
Real-time quantitative PCR were used to confirm the temporal expression of 8 putative TFs during developing protocorms treated with sucrose (Fig. 10). The predicted TF PaHB5 had higher expression at early protocorm developing stages (0 DAI and 4 DAI), and its expression was significantly decreased from 7 DAI. PaPIF3 transcripts continually increased and peaked at 12 DAI, then decreased to 30 DAI. PaWRKY18, PaWRKY71, PaMYB4, and PaANT showed a similar expression pattern but the expression was extremely low in 0- and 4-DAI protocorms. The expression patterns of PaRAV1-1 and PaPIF3 were similar, but PaRAV1-1 level was decreased in 4-DAI protocorms. PaMADS2 showed low expression in 0- and 4-DAI protocorms, and highly accumulated transcripts in 7-DAI protocorms, which was earlier than for other putative regulators.
Figure 10.
Real-time quantitative PCR analysis of putative transcription factor genes during P. aphrodite protocorm development. RNA was extracted from 0- to 30-DAI protocorms cultured with 1% sucrose. Quantification was normalized to Phalaenopsis Actin4 for each sample; experiments were performed in triplicate. Data are mean ± SEM.
Activation ability of PaHB5 on PaMLS promoter
To predict the putative PaHB5 binding sites, a 2,000-bp upstream sequence of PaMLS was analyzed by using the plantPAN database, with 22 putative binding sites obtained for PaHB5. Most binding sites were localized in the region of the 1,000-bp upstream regulatory sequence of PaMLS (19 binding sites) (Supplementary Table S2). Therefore, a 1-kb fragment of the upstream regulatory sequence of PaMLS was selected for dual luciferase assay (Fig. 11A). PaHB5 conferred 2.73-fold transactivation activity on the 1-kb promoter of PaMLS (Fig. 11B), which suggests that PaHB5 is a positive regulator of PaMLS.
Figure 11.
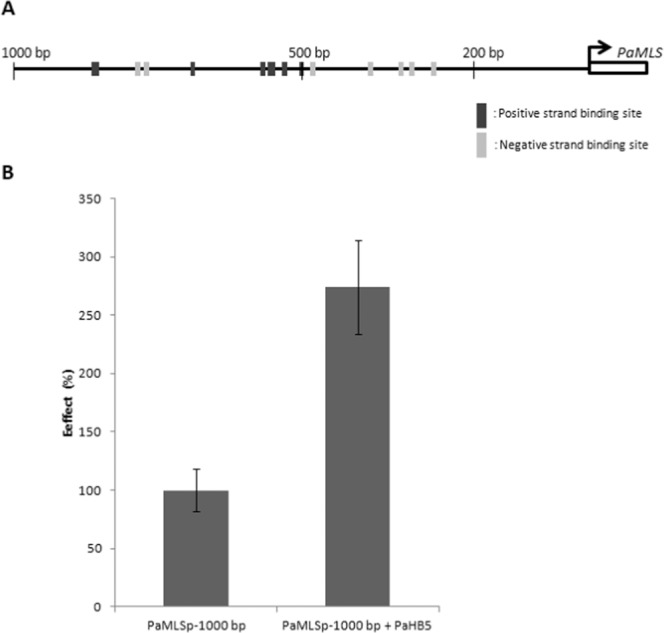
Activation ability of PaHB5 on the PaMLS promoter. (A) Diagram of 1-kb PaMLS promoter. Numbers represent the distance in nucleotides from the 5′ end of the promoter to the start codon of PaMLS. The dark-grey and light-grey blocks on the promoter represent the putative binding sites of ATHB5 predicted by promoter analysis with PlantPAN. (B) Effect of PaHB5 on PaMLS promoter shown by relative luciferase activity of different treatments.
Discussion
In the life cycle of angiospermss, seed germination and post-germinative growth are pivotal stages19. Previous studies have indicated that the glyoxylate cycle plays a crucial role in mobilization of storage oils during germination and post-germinative growth of oilseed20. In germinating oilseeds, glyoxylate cycle is located in the glyoxysome surrounded by a specialized single membrane1. In this paper, we found a large amount of lipids accumulated in the proembryo of mature P. aphrodite seeds while starch is absence (Fig. 2A). We also observed that glyoxysomes exist in the embryo cells of imbibed Phalaenopsis orchid seed. In addition, oil bodies and mitochondria could be found in close proximity to glyoxysomes (Fig. 2B). From the observation, it might imply that endogenous reserves of lipid could be used in the imbibed Phalaenopsis orchid seed. Interestingly, it has been reported that no glyoxysomes were found in Catteleya aurantiaca protocorms grown on medium with sucrose or without sucrose21. Another observation from some terrestrial orchids such as Disa, Disperis and Huttonaea indicated that glyoxysomes are absent in seeds incubated without exogenous sucrose, and glyoxysomes appear in the presence of sucrose22. The differences could be due to the situations that different species are studied. In fact, previous studies showed that all the glyoxylate cycle’s enzymes are located in the glyoxysome, with the exception of aconitase1,23. Detection of significant expression of PaICL and PaMLS supported that existence of glyoxysomes in Phalaenopsis imbibed seeds.
With sucrose in the medium, the expression of both PaICL and PaMLS was significantly down-regulated from 0 to 7 DAI during protocorm development (Fig. 3B). The expression of both PaICL and PaMLS may be sensitive markers of exogenous carbon source in orchid seed germination. It has been demonstrated that the fungal requirement of orchid seeds can be successfully bypassed in vitro with provision of sucrose in the culture medium24. These results suggested that sucrose is a key regulatory factor supplied by the symbiont for regulation of orchid seed lipid metabolism in natural environment. We deduced that at natural condition, if an orchid seed do not meet an appropriate symbiont, it do not geminate and slowly breaks down lipid for longer longevity in the wild. If an orchid seed meet an appropriate symbiont, it will use carbon source provided by the symbiont to geminate and suppress lipid usage. Thus, the minus-sucrose treatment in the medium could be considered similar to seeds which fail to seek out appropriate symbionts. Furthermore, plus-sucrose condition might be similar to the successful symbiotic seed germination in the natural environment.
In oilseeds or cucumber cell culture, the sugar-mediated repression of transcript level or enzyme activity is consistent for both ICL and MLS4,25. In Arabidopsis, lipid breakdown and hypocotyl elongation were strongly inhibited when Arabidopsis icl was grown in the dark, and the icl mutant is also deficient in conversion of 14C acetate into sugar. In mls mutants, inhibition of growth is less severe, and some carbon can be converted from acetate into sugar20. Therefore, the glyoxylate produced by ICL may be channeled into gluconeogenesis via the serine–glycine shuttle26, which was confirmed by the accumulation of serine and glycine residues in the mls mutant27. In culture of orchid asymbiotic seeds, sucrose provides the carbon source to protocorms and therefore might mediate the end-product inhibition of the glyoxylate cycle to block lipid breakdown. The repressed expression of PaICL and PaMLS by sucrose during protocorm development might serve to reserve the endogenous energy resource and give priority to utilize the exogenous available carbon source.
Eight putative TFs are predicted to bind on the PaMLS promoter, including one putative activator (PaHB5) and 7 putative repressors (PaWRKY71, PaPIF3, PaANT, PaMADS2, PaMYB4, PaWRKY18 and PaRAV1-1) (Table 2). The transcriptional activator PaHB5 showed positive transactivation activity on the PaMLS promoter, which suggests that PaHB5 has an important role in regulating the glyoxylate cycle for energy management during protocorm development. Moreover, several putative repressors responding to sucrose were identified to possibly regulate the expression of PaMLS. Thus, positive and negative regulation may coordinate the expression of PaMLS.
Previous studies showed one CArG box-like motif (CCA/T6GG) on the promoter of MLS in cucumber4. In addition, the expression of MLS has been detected in cucumber petal5. These results suggest that a floral homeotic MADS TF might have a regulatory role in the expression of MLS in cucumber. A number of MADS box proteins were identified as involved in proteome changes of Oncidium sphacelatum mycorrhizal protocorms at different developmental stages28. In this study, a MADS box TF, PaMADS2, a homolog of PeMADS2, which is involved in sepal and petal development in P. equestris29,30, was also predicted to suppress PaMLS expression (Table 2).
The MADS-box genes may directly control MLS expression through the sucrose signaling pathway during protocorm development. MYB and WRKY TFs have been reported to be involved in the sugar response. Two crucial TFs, MYBS1 and MYBGA, were found to integrate diverse nutrient-starvation and gibberellin signaling pathways during germination of cereal grains. Although MYBS1 synthesis is repressed by sugar but induced by sugar starvation in rice, MYB TFs were involved in the metabolic response31. Some MYB TFs were found to be glucose-inducible in Arabidopsis seedlings32. Several WRKY proteins responded to and were up-regulated by sugar starvation33,34. For example, the WRKY TF, SUSIBA2, is reported to bind to sugar-responsive elements of the isoamylase 1 promoter in crops35. Therefore, the putative MYB and WRKY TFs are potential candidates to regulate PaMLS transcription under metabolic changes during orchid protocorm development. Phytochrome-interacting factors, belonging to the Arabidopsis basic helix-loop-helix superfamily36, were reported to repress seed germination, promote seedling skotomorphogenesis and promote shade-avoidance by regulating the expression of thousands of genes. They are also required for sucrose-dependent growth promotion during post-germinative growth37. Thus, showing a negative correlation with the expression of PaMLS in response to sucrose treatment (Fig. 10), PaPIF3 may play a negative regulating role on the repression of PaMLS expression. In conclusion, this study provides the basis for understanding the regulation of the use of orchid seed-storage energy.
Conclusion
We observed that the glyoxysomes locate in close proximity to oil bodies and mitochondria in imbibed Phalaenopsis protocorms, suggesting storage oil could be catabolized to provide nutrition for the protocorm development and growth. The expression of the PaICL and PaMLS involved in glyoxylate cycle could be down-regulated by the exogenous sucrose. The transcriptional activator PaHB5 identified from transcriptome comparison presented positive transactivation activity on the PaMLS promoter, which indicates that PaHB5 play an important role in regulating the glyoxylate cycle for energy management during protocorm development. This study provides new insights into the regulation of glyoxylate cycle during early protocorm development of orchids.
Materials and Methods
Plant materials and growth conditions
Collection and growth of P. aphrodite seeds was as described by Balilashaki et al.38 with some modification. Mature seeds were collected from the capsules of P. aphrodite. Seeds were surface-sterilized with 1% (v/v) NaOCl solution for 15 min. After rinsing with sterilized water twice, seeds were re-suspended in half-strength Murashige and Skoog liquid medium to imbibe for 48 h, then applied to 0.85% (w/v) agar plates containing 1/2 MS salts with or without 1% (w/v) sucrose and grew in culture room at 23~25 °C. Protocorms were then collected at 0, 4, 7, 12, 20 and 30 days after incubation (DAI).
Histology
To reveal the content in the P. aphrodite mature seeds, seeds were stained with Sundan IV, Sudan black and iodine for 40 min at 50 °C, and then washed with 70% ethanol. The stained sections were observed by microscopy.
Transmission electron microscopy (TEM)
The P. aphrodite 0 DAI protocorms were prefixed in 4% (g/v) paraformaldehyde and 2.5% (v/v) glutaraldehyde in 67 mM phosphate buffer (NaH2PO4˙H2O and Na2HPO4) for 24 h. The samples were fixed in 1% (g/v) OsO4 in 0.067 M phosphate buffer, dehydrated through an acetone series (15–30–50–70–90–100%), embedded in Spurr’s resin and polymerized in a vacuum oven at 70 °C. Sections 100 nm thick were obtained and placed on grids, then counterstained with uranyl acetrate and lead nitrate, and observed by transmission electron microscopy (JEM-1400, JEOL).
RNA preparation
Total RNA was extracted as described29. Plant materials were immersed in liquid nitrogen and stored at −80 °C. Briefly, frozen tissue (0.5–1 g) was pulverized with liquid nitrogen by using a pestle and mortar and then homogenized in TRIZOL reagent. The dissolved RNA was extracted with chloroform. After centrifugation at 13000 rpm to remove insoluble material, total RNA was precipitated with 0.8 M sodium citrate and 1.2 M NaCl at −20 °C overnight, then precipitated again with 4 M LiCl, pelleted, and washed. The final RNA precipitate was dissolved in a suitable volume of sterilized DEPC-treated water. To remove the DNA contamination, total RNA was treated with RNase-free DNase I.
Expression analyses by RT-PCR and real-time quantitative RT-PCR
RNA was used as a template for cDNA synthesis with reverse transcriptase and the SuperScript II kit (Invitrogen). Transcripts of PaMLS, PaICL, and 8 candidate TF genes including PaANT, PaHB5, PaMADS2, PaMYB4, PaPIF3, PaRAV1-1, PaWRKY18 and PaWRKY71 were detected by RT-PCR and real-time quantitative PCR. The primer pairs are in Supplementary Table S1.
The methods of RT-PCR and real-time quantitative RT-PCR were as described39 with modification. The RT-PCR program was 94 °C for 5 min for denaturation, then 94 °C for 30 s, 72 °C for 30 s, and extension at 72 °C for 30 min. Annealing temperature and number of amplified cycles varied with different primer pairs (PaICL and PaMLS: 58 °C/25 cycles, candidate TF genes: 62 °C/33 cycles). The amplified products were analyzed on agarose gel and photographed. Only one amplified band with expected size was detected for each of PaICL and PaMLS (Supplementary Fig. S1).
For real-time quantitative RT-PCR analysis, the PCR program was incubation at 50 °C for 2 min, then 95 °C for 10 min, and thermal-cycling for 40 cycles (95 °C for 15 s and 60 °C for 1 min) by using the ABI 7500 Real-Time PCR instrument39. Triplicate experiments were performed for each sample. Sequencing Detection System v1.2.3 (Applied Biosystems) was adopted for data analysis.
5′ Rapid amplification of cDNA ends (5′ RACE) for PaICL and PaMLS
The 5′ RACE was performed as described29. Briefly, the full-length cDNAs were synthesized by extending the 5′ ends of cDNA by using the SMART RACE cDNA amplification kit (Clontech, Palo Alto, CA, USA). First-strand cDNAs were synthesized from 1 μg total RNA from P. aphrodite protocorms at 0 DAI following the manufacturer’s protocol. The cDNA containing the 5′ end for PaICL clones was obtained by PCR amplification with a 5′-specific universal primer (Clontech) and 3′ gene-specific primer sequences for PaICL, 5′-GCCCCGAGAATGAACTGGTGGTC-3′ and PaMLS, 5′-GTCCCTTCTTTTACCTCCCCAA-3′. The thermal cycling protocol was initial denaturation at 94 °C for 5 min, followed by 35 cycles at 94 °C for 30 s, 53 °C for 30 s and 72 °C for 30 s, and a final extension at 72 °C for 7 min. RACE-products were re-amplified with gene-specific nested primer sequences for PaICL, 5′- CCTCTTGCTGCTTGCGGTCGTGGTAG-3′ and PaMLS, 5′-GCTCCGTTCTCACTCTGCTGG-3′ and the nested universal primer provided in the RACE kit. The PCR protocol consisted of an initial denaturation at 94 °C for 5 min, followed by 35 cycles at 94 °C for 30 s, 53 °C for 30 s and 72 °C for 30 s and a final extension at 72 °C for 7 min. The PCR products were cloned into the pGEM-T Easy vector (Promega, Madison, WI, USA) and sequenced from both strands of six positive clones selected randomly.
Sequence alignments and phylogenetic analysis
The full-length proteins sequences of 34 genes retrieved from GenBank were aligned by using Clustal W with the default parameters. The neighbor-joining phylogenetic tree was conducted in MEGA540 with default settings. Bootstrap value was obtained by 1,000 replicate runs.
Comparison of transcriptomic profiling by RNA sequencing
RNA from P. aphrodite protocorms at 4 and 7 DAI grown in medium containing1/2 MS plus 1% (w/v) sucrose were collected as described above. Both 2 μg RNA samples were treated with Dnase I, and then sequenced by using Solexa/Illumina RNA-seq (Illumina Hiseq. 2000 platform, BGI Tech Solutions). Before assembly, high-quality reads were obtained by removing adaptor sequences, and low-quality reads were filtered by using TRIMMOMATIC41 from raw reads. The resulting high-quality reads were de novo assembled and annotated as described42. Transcript abundance was normalized by using the fragments per kilobase per million mapped reads (FPKM) method43.
Promoter analysis of PaMLS
A fragment of a 2000-bp upstream sequence of PaMLS was obtained from the corresponding genome sequence in the P. equestris genome18, and then analyzed by using PlantPAN44 (http://plantpan.mbc.nctu.edu.tw/) to predict putative TF binding sites on the PaMLS promoter.
Ten-fold up-regulated unigenes were screened by comparing gene expression of protocorm transcriptomes at 4 and 7 DAI. The selected unigenes were BLAST searched in the TF database of Oryza sativa subsp. japonica from PlantTFDB45 (http://planttfdb.cbi.pku.edu.cn/) to identify the putative TF genes. These putative unigenes were then BLAST searched in the database against putative TF genes binding on the PaMLS promoter.
Construction of transformed fusions
Genomic DNA was extracted from floral buds of P. aphrodite as described29. The 1-kb PaMLS promoter fragments were amplified from genomic DNA of floral buds of P. aphrodite by PCR-amplification with EX Taq DNA polymerase (Takara) by using forward primers (PaMLSp_1000_5′_BamH I) and a reverse primer (PaMLSp_3′_Nco I) (Supplementary Table S1). The PCR products were cloned into the pGM-T vector (GeneMark), and then digested with the restriction endonucleases BamH I and Nco I to obtain the promoter fragments. The promoter fragments were cloned into the pJD301 vector that contains a firefly luciferase gene.
The PaHB5 coding sequence was amplified by PCR from cDNA of protocorms of P. aphrodite at 4 DAI that were grown on medium plus sucrose by using primers for PaHB5_5′_Xba I and PaHB5_3′_BamH I (Supplementary Table S1). The PCR products were cloned into the pGM-T vector (GeneMark), then digested with restriction endonuclease BamH I and Xba I to obtain the PaHB5 fragment. The fragment was cloned into the multiple cloning sites downstream of the CaMV 35 S promoter present in the pBI221 vector.
Transient expression experiments and dual luciferase reporter assay
The methods were majorly following description of Chuang et al.46 with some modification. The PaMLS promoter plasmids, PaHB5 plasmid, and RL2-pJD301 (relina luciferase, internal control) were isolated by using the High-Speed Plasmid Mini Kit (Geneaid) coated on gold particles 1.6 μm in diameter by co-precipitation. Before particle bombardment, each floral organ was separated from the full-opening flower, and then placed on a central core 2 cm in diameter on solid agar medium. These sections were bombarded by using a Modle Biolistic PDS-1000/He system (BioRad) at 1100 psi helium gas pressure, 28.5 inch-of-Hg vacuum and 9 cm target distance. After bombardment, floral buds were grown at room temperature for 18 to 20 hr to allow for expression of the luciferase protein. Luciferase activity in the transfected floral buds was measured by using the dual-luciferase reporter assay system (Promega). To prepare cell lysates, transfected floral buds were ground into fine powder by the addition of liquid nitrogen, and 1X phosphate buffered saline was added. The firefly (Photinus pyralis) luciferase reporter assay involved adding cell lysates to Luciferase Assay Reagent II. Then, firefly luciferase luminescence was quenched and renilla luciferase (internal control) was activated by adding Stop & GloR Reagent. The luciferase activity was measured with the TD-20/20 Luminometer system (BD Monolight 3010 C) with a 2-sec pre-measurement delay followed by a 10-sec measurement period for each assay. The relative luciferase activity was calculated as the ratio of firefly to renilla luciferase activity.
Supplementary information
Acknowledgements
This work was supported by the Ministry of Science and Technology, Taiwan (MOST 105-2321-B-006-026- and MOST 106-2321-B-006-014-); the Teamwork Projects Funded by Guangdong Natural Science Foundation, China (no. 2017A030312004); the National Natural Science Foundation of China (no. 31870199); the National Key Research and Development Program of China (no. 2018YFD1000401); the Fundamental Research Project of Shenzhen, China (nos. JCYJ20170307170746099 and JCYJ20160531110513702), and the Development Funds for Strategic Emerging Industries of Shenzhen, China (no. XDNYCYFC20160509020004).
Author contributions
W.-C.T. and Z.-J.L. planned and coordinated the project and wrote the manuscript. W.-L.W. and Y.-Y.H. conducted all experimental works. H.-C.L. prepared mRNA extracted from the collected samples. C.-K.L. and L.-J.C. collected and grew the plant material. C.-H.F. and T.-H.H. conducted transcriptome sequencing and analysis. M.-H.C. conducted histology and TEM observation.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
These authors contributed equally: Wan-Lin Wu and Yu-Yun Hsiao.
Contributor Information
Zhong-Jian Liu, Email: zjliu@fafu.edu.cn.
Wen-Chieh Tsai, Email: tsaiwc@mail.ncku.edu.tw.
Supplementary information
is available for this paper at 10.1038/s41598-020-66932-8.
References
- 1.Eastmond PJ, Graham IA. Re-examining the role of the glyoxylate cycle in oilseeds. Trends Plant. Sci. 2001;6:72–78. doi: 10.1016/S1360-1385(00)01835-5. [DOI] [PubMed] [Google Scholar]
- 2.Graham IA. Seed storage oil mobilization. Annu. Rev. Plant. Biol. 2008;59:115–142. doi: 10.1146/annurev.arplant.59.032607.092938. [DOI] [PubMed] [Google Scholar]
- 3.Graham IA, Baker CJ, Leaver CJ. Analysis of the cucumber malate synthase gene promoter by transient expression and gel retardation assays. Plant. J. 1994;6:893–902. doi: 10.1046/j.1365-313X.1994.6060893.x. [DOI] [PubMed] [Google Scholar]
- 4.Graham IA, Denby KJ, Leaver CJ. Carbon Catabolite Repression Regulates Glyoxylate Cycle Gene Expression in Cucumber. Plant. Cell. 1994;6:761–772. doi: 10.1105/tpc.6.5.761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Graham IA, Leaver CJ, Smith SM. Induction of Malate Synthase Gene Expression in Senescent and Detached Organs of Cucumber. Plant. Cell. 1992;4:349. doi: 10.1105/tpc.4.3.349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Finkelstein RR, Lynch TJ. Abscisic Acid Inhibition of Radicle Emergence But Not Seedling Growth Is Suppressed by Sugars. Plant. Physiol. 2000;122:1179. doi: 10.1104/pp.122.4.1179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lee, Y.-I., Yeung, E. C. & Chung, M.-C. In Orchid Biotechnology 23-44 (World Scientific (2007).
- 8.Smith, S. E. & Read, D. In Mycorrhizal Symbiosis (Third Edition) (eds Sally E. Smith & David Read) 419-457 (Academic Press 2008).
- 9.Lee Y-I, Yeung E, Lee N, Chung M-C. Embryology of Phalaenopsis amabilis var. formosa: Embryo development. Botanical Stud. 2008;49:139–146. [Google Scholar]
- 10.Leake JR. Myco-heterotroph/epiparasitic plant interactions with ectomycorrhizal and arbuscular mycorrhizal fungi. Curr. Opin. Plant. Biol. 2004;7:422–428. doi: 10.1016/j.pbi.2004.04.004. [DOI] [PubMed] [Google Scholar]
- 11.Smith ES. Asymbiotic germination of orchid seeds on carbohydrates of fungal origin. N. Phytologist. 1973;72:497–499. doi: 10.1111/j.1469-8137.1973.tb04400.x. [DOI] [Google Scholar]
- 12.Ernst R, Arditti J. Carbohydrate physiology of orchid seedlings. III. Hydrolysis of maltooligosaccharides by phalaenopsis (orchidaceae) seedlings. Am. J. Botany. 1990;77:188–195. doi: 10.1002/j.1537-2197.1990.tb13545.x. [DOI] [PubMed] [Google Scholar]
- 13.Stewart SL, Kane ME. Effects of Carbohydrate Source on Thein Vitroasymbiotic Seed Germination of the Terrestrial Orchidhabenaria Macroceratitis. J. Plant. Nutr. 2010;33:1155–1165. doi: 10.1080/01904161003763757. [DOI] [Google Scholar]
- 14.Kanehisa M, et al. Data, information, knowledge and principle: back to metabolism in KEGG. Nucleic Acids Res. 2014;42:D199–205. doi: 10.1093/nar/gkt1076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhang GQ, et al. The Apostasia genome and the evolution of orchids. Nature. 2017;549:379–383. doi: 10.1038/nature23897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhang GQ, et al. The Dendrobium catenatum Lindl. genome sequence provides insights into polysaccharide synthase, floral development and adaptive evolution. Sci. Rep. 2016;6:19029. doi: 10.1038/srep19029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Swarbreck D, et al. The Arabidopsis Information Resource (TAIR): gene structure and function annotation. Nucleic Acids Res. 2008;36:D1009–1014. doi: 10.1093/nar/gkm965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cai J, et al. The genome sequence of the orchid Phalaenopsis equestris. Nat. Genet. 2015;47:65–72. doi: 10.1038/ng.3149. [DOI] [PubMed] [Google Scholar]
- 19.Holdsworth M, Kurup S, Mkibbin R. Molecular and genetic mechanisms regulating the transition from embryo development to germination. Trends Plant. Sci. 1999;4:275–280. doi: 10.1016/S1360-1385(99)01429-6. [DOI] [Google Scholar]
- 20.Eastmond PJ, et al. Postgerminative growth and lipid catabolism in oilseeds lacking the glyoxylate cycle. Proc. Natl Acad. Sci. USA. 2000;97:5669–5674. doi: 10.1073/pnas.97.10.5669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Harrison CR. Ultrastructural and Histochemical Changes during the Germination of Cattleya aurantiaca (Orchidaceae) Botanical Gaz. 1977;138:41–45. doi: 10.1086/336896. [DOI] [Google Scholar]
- 22.Manning J, Van SJ. The Development and Mobilisation of Seed Reserves in Some African Orchids. Australian J. Botany. 1987;35:343–353. doi: 10.1071/BT9870343. [DOI] [Google Scholar]
- 23.Cooper TG, Beevers H. Mitochondria and Glyoxysomes from Castor Bean Endosperm: ENZYME CONSTITUENTS AND CATALYTIC CAPACITY. J. Biol. Chem. 1969;244:3507–3513. [PubMed] [Google Scholar]
- 24.Knudson L. Nonsymbiotic Germination of Orchid Seeds. Botanical Gaz. 1922;73:1–25. doi: 10.1086/332956. [DOI] [Google Scholar]
- 25.Kudielka RA, Theimer RR. Derepression of glyoxylate cycle enzyme activities in anise suspension culture cells. Plant. Sci. Lett. 1983;31:237–244. doi: 10.1016/0304-4211(83)90061-5. [DOI] [Google Scholar]
- 26.Penfield S, Graham S, Graham IA. Storage reserve mobilization in germinating oilseeds: Arabidopsis as a model system. Biochem. Soc. Trans. 2005;33:380–383. doi: 10.1042/BST0330380. [DOI] [PubMed] [Google Scholar]
- 27.Cornah JE, Germain V, Ward JL, Beale MH, Smith SM. Lipid utilization, gluconeogenesis, and seedling growth in Arabidopsis mutants lacking the glyoxylate cycle enzyme malate synthase. J. Biol. Chem. 2004;279:42916–42923. doi: 10.1074/jbc.M407380200. [DOI] [PubMed] [Google Scholar]
- 28.Valadares RB, Perotto S, Santos EC, Lambais MR. Proteome changes in Oncidium sphacelatum (Orchidaceae) at different trophic stages of symbiotic germination. Mycorrhiza. 2014;24:349–360. doi: 10.1007/s00572-013-0547-2. [DOI] [PubMed] [Google Scholar]
- 29.Tsai W-C, Kuoh C-S, Chuang M-H, Chen W-H, Chen H-H. Four DEF-Like MADS Box Genes Displayed Distinct Floral Morphogenetic Roles in Phalaenopsis Orchid. Plant. Cell Physiol. 2004;45:831–844. doi: 10.1093/pcp/pch095. [DOI] [PubMed] [Google Scholar]
- 30.Tsai, W.-C., Pan, Z.-J., Su, Y.-Y. & Liu, Z.-J. In International Review of Cell and Molecular Biology Vol. 311 (ed Kwang W. Jeon) 157–182 (Academic Press 2014). [DOI] [PubMed]
- 31.Hong YF, et al. Convergent starvation signals and hormone crosstalk in regulating nutrient mobilization upon germination in cereals. Plant. Cell. 2012;24:2857–2873. doi: 10.1105/tpc.112.097741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rook F, Hadingham SA, Li Y, Bevan MW. Sugar and ABA response pathways and the control of gene expression. Plant, Cell Environ. 2006;29:426–434. doi: 10.1111/j.1365-3040.2005.01477.x. [DOI] [PubMed] [Google Scholar]
- 33.Buscaill P, Rivas S. Transcriptional control of plant defence responses. Curr. Opin. Plant. Biol. 2014;20:35–46. doi: 10.1016/j.pbi.2014.04.004. [DOI] [PubMed] [Google Scholar]
- 34.Wang HJ, et al. Transcriptomic adaptations in rice suspension cells under sucrose starvation. Plant. Mol. Biol. 2007;63:441–463. doi: 10.1007/s11103-006-9100-4. [DOI] [PubMed] [Google Scholar]
- 35.Sun C, et al. A novel WRKY transcription factor, SUSIBA2, participates in sugar signaling in barley by binding to the sugar-responsive elements of the iso1 promoter. Plant. Cell. 2003;15:2076–2092. doi: 10.1105/tpc.014597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Leivar P, Quail PH. PIFs: pivotal components in a cellular signaling hub. Trends Plant. Sci. 2011;16:19–28. doi: 10.1016/j.tplants.2010.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Stewart JL, Maloof JN, Nemhauser JL. PIF genes mediate the effect of sucrose on seedling growth dynamics. PLoS One. 2011;6:e19894. doi: 10.1371/journal.pone.0019894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Balilashaki, K., Gantait, S., Naderi, R. & Vahedi, M. Capsule formation and asymbiotic seed germination in some hybrids of Phalaenopsis, influenced by pollination season and capsule maturity. Physiology and Molecular Biology of Plants 21(3), 341–347 (2015). [DOI] [PMC free article] [PubMed]
- 39.Chen Y-Y, et al. C- and D-class MADS-Box Genes from Phalaenopsis equestris (Orchidaceae) Display Functions in Gynostemium and Ovule Development. Plant. Cell Physiol. 2012;53:1053–1067. doi: 10.1093/pcp/pcs048. [DOI] [PubMed] [Google Scholar]
- 40.Tamura K, et al. MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol. Biol. Evol. 2011;28:2731–2739. doi: 10.1093/molbev/msr121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Bolger AM, Lohse M, Usadel B. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics. 2014;30:2114–2120. doi: 10.1093/bioinformatics/btu170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Niu SC, et al. De novo transcriptome assembly databases for the butterfly orchid Phalaenopsis equestris. Sci. Data. 2016;3:160083. doi: 10.1038/sdata.2016.83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Mortazavi A, Williams BA, McCue K, Schaeffer L, Wold B. Mapping and quantifying mammalian transcriptomes by RNA-Seq. Nat. Methods. 2008;5:621–628. doi: 10.1038/nmeth.1226. [DOI] [PubMed] [Google Scholar]
- 44.Chang WC, Lee TY, Huang HD, Huang HY, Pan RL. PlantPAN: Plant promoter analysis navigator, for identifying combinatorial cis-regulatory elements with distance constraint in plant gene groups. BMC Genomics. 2008;9:561. doi: 10.1186/1471-2164-9-561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Jin J, Zhang H, Kong L, Gao G, Luo J. PlantTFDB 3.0: a portal for the functional and evolutionary study of plant transcription factors. Nucleic Acids Res. 2014;42:D1182–1187. doi: 10.1093/nar/gkt1016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Chuang Y-C, et al. A Dual Repeat Cis-Element Determines Expression of GERANYL DIPHOSPHATE SYNTHASE for Monoterpene Production in Phalaenopsis Orchids. Front. Plant. Sci. 2018;9:765–765. doi: 10.3389/fpls.2018.00765. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.