Abstract
Stroke is the second leading cause of death and a significant cause of disability worldwide. Recent advances in DNA sequencing, proteomics, metabolomics, and computational tools are dramatically increasing access to the identification of host–microbiota interactions in systemic diseases. In this review, we describe the accumulating evidence showing how human microbiota plays an essential role in cerebrovascular diseases. We introduce the symbiotic relationships between microbiota and the mucosal immune system, focusing on differences by anatomical sites. Microbiota directly or indirectly contributes to the pathogenesis of traditional vascular risk factors including age, obesity, diabetes mellitus, dyslipidemia, and hypertension. Moreover, recent studies proposed independent effects of the microbiome on the progression of various subtypes of stroke through direct microbial invasion, exotoxins, functional amyloids, inflammation, and microbe-derived metabolites. We propose the critical concept of gene-microbial interaction to elucidate the heterogeneity of stroke and provide possible therapeutic avenues. We suggest ways to resolve the vast inter-individual diversity of cerebrovascular disease and mechanisms for personalized prevention and treatment.
Keywords: Cerebrovascular disease, immune system, stroke, microbiota–oral–brain axis, microbiota–gut–brain axis, gene–microbial interactions, functional bacterial amyloid
Background
Stroke is one of the leading causes of death and a primary cause of disability worldwide. Traditional risk factors contributing to the development of stroke include age, obesity, diabetes mellitus, dyslipidemia, hypertension, smoking, physical inactivity, diet, and alcohol consumption.1 However, in most cases of stroke, the trigger of the event remains unknown. In fact, not all elderly patients with hypertension, diabetes mellitus, or smoking have strokes. Furthermore, the contribution of these traditional risk factors to stroke differs among ethnicities,1 suggesting the need for personalized prevention of stroke. Recently, rapid advances in metagenomics, meta-transcriptomics, and meta-proteogenomics have identified novel host–microbiota interactions.2 Moreover, emerging evidence has demonstrated the role of both infectious diseases and dybiosis (imbalance in the microbiota) in the development of stroke.3 Increased consideration of the effects of the microbiota will provide us with crucial benefits to deepen our understanding of the pathophysiology of stroke. Here, we comprehensively review the influence of microbiota on risk factors, atherosclerosis, the subtypes of stroke, and vascular cognitive impairment (VCI), while also examining the underlying molecular mechanisms.
Tissue-specific immunity and microbiota
The difference in microbiota between the oral cavity and gut
The human gut is home to several trillions of microbes that form a bidirectional relationship with the human host.4 Time-series metagenomic analyses using next-generation sequencing5 and phylogeny-based metric6 revealed that the human microbiota begins to colonize in early life.7 Exposure to microbes starts before birth as some bacteria in the mother’s oral cavity can be transmitted through the placenta.8 The route of delivery, breastfeeding, infant care-associated behavior, diet, and gender altogether contribute to the establishment of microbiota in different body sites, including the skin, oral cavity, and gastrointestinal (GI) tract during infancy.9 As a consequence of ecological processes, in adults, the microbiota of the oral cavity is distinct from that of the GI tract.10 A statistical approach using Dirichlet multinomial mixture models, which assigned samples to community subtypes, demonstrated that changes of oral bacterial populations may subsequently give rise to a bacterial community shift in the gut, resulting in dysbiosis.11
The mucosal immune system of the oral cavity and the gastrointestinal tract
The GI mucosal immune system consists of three significant compartments, the epithelium, lamina propria, and gut-associated lymphoid tissue. The mucosal immune system in the oral cavity, as the gateway to the GI tract, shares functions with the GI tract in the following four points12:
Enterocytes and Paneth cells produce antimicrobial peptides (immunoglobulins, defensins, lysozyme, and cathelicidins), and goblet cells produce mucin in the GI tract, which contributes to innate host defense against gut microbiota.13 In the oral cavity, oral epithelial cells, neutrophils, and salivary glands secrete these antimicrobial peptides.14
Intraepithelial lymphocytes, especially regulatory T cells, respond to antigens presented with major histocompatibility complex molecules and express toll-like receptors (TLRs); they also eliminate foreign antigens and exhibit surveillance functions in both the GI tract and oral cavity.12
Dendritic cells (DCs) in the GI tract and oral cavity acquire luminal antigens via extension processes. After taking up the antigen, DCs migrate into the mucosa-associated lymphoid tissue and lymph nodes, initiating the adaptive immune response by inducing T cell proliferation and differentiation.15,16
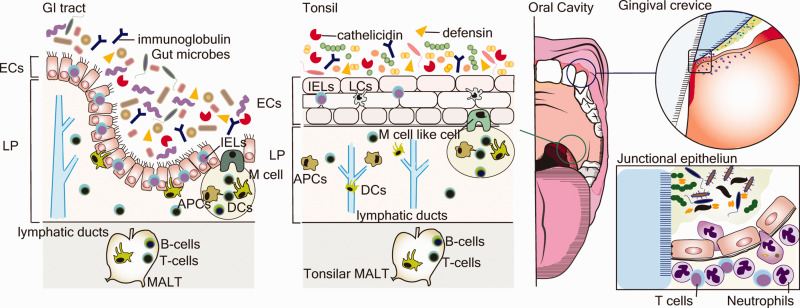
In the GI tract, microfold cells (M cells) transport luminal antigens to antigen presenting cells, stimulating B cells in follicle to differentiate into plasma cells in Peyer’s patches. Corresponding to M cells in the GI tract, a group of M cell-like cells has been identified in the epithelium of the palatine tonsil crypt in the oral cavity.17
On the other hand, the gingival crevice in the mouth has unique anatomical and immunological features. The junctional epithelium, where the mucosa meets the tooth at the base of gingival tissue, has few layers of thickness attached to the tooth via hemidesmosomes. The junctional epithelium connection to the tooth is highly permeable, allowing the bidirectional passage of oral microbiota, host-protective factors, and inflammatory cells (Figure 1).18 Both histological studies and flow cytometry analyses showed that neutrophils, resident terminally differentiated memory subset of CD4+ and CD8+ cells, and Foxp3+ regulatory T cells are more abundant in the gingival crevice than in the GI tract.19,20
The role of microbiota in the host immune system
The Human Microbiome Project showed that the oral and gut microbiota play essential roles in the maturation of the host immune systems (Figure 2)21:
The regulation of IgA secretion involves crosstalk between innate immunity and commensal bacteria. Recent approaches assessing the coating of gut microbes with host-derived IgA suggested that microbiota-reactive and polyreactive IgA antibodies arise naturally in all naïve B cell populations, significantly in IgA-secreting plasma cells.22 Nevertheless, evidence from germ-free mice and fecal microbiota transplantation (FMT) revealed that commensal bacteria increased secretion of IgA and antimicrobial peptides into the gut lumen,23 which indicates that the orchestra of innate immunity and commensal bacteria is necessary to maintain IgA synthesis in the local mucosal environment. Commensal bacteria induce IgA production through living bacteria and bacterial products,24 as well as intestinal epithelial cell-derived B-cell stimulating and activating factors.25
Hematopoiesis and innate immunity. The microbiota can impact stem-cell-derived myeloid cell development, and the size of the bone marrow myeloid cell pool correlates with the complexity of the gut microbiota.26 Circulating TLR ligands and short-chain fatty acids (SCFAs), which are gut microbial metabolites derived from carbohydrates, promote DC and monocyte trafficking into the bloodstream (reviewed in Thaiss et al.27). Moreover, SCFAs play essential roles in the central nervous system, critically mediating the maturation and function of brain microglia and the blood–brain barrier (BBB).28
Bridge between innate and adaptive immunity. Interleukin (IL)-17-producing effector T helper (Th17) cells are crucial for maintaining tissue physiology. Th17 acts on epithelial cells to enhance anti-microbial defense and epithelial barrier integrity via IL-17 and IL-22.29 Th17 cells initially have a central role in preventing infection by several species of pathogenic bacteria; however, stimulation with IL-2330 or high salt diet31 makes Th17 cells proinflammatory. Furthermore, it is now becoming clear that specific microbes, including segmented filamentous bacteria (SFB), Bifidobacterium adolescentis, Escherichia coli, Citrobacter rodentium, and strains of Clostridia, can induce Th17 through independent mechanisms.32 While these microbes mediate host defense, they can also cause pathological systemic inflammation. Moreover, Benakis et al.33 proposed that microbiota influence IL17-positive γδ T cells, which translocate form gut to meninges in experimental stroke, and also effect the balance between regulatory T cells and γδ T cells, which modulate neuroinflammation in ischemic stroke.
Traditional risk factors and microbiota
Aging
Aging is one of the most critical risk factors for all subtypes of stroke.34 The proinflammatory status accompanied by aging, which was termed “inflamm-aging” in 2000 by Claudio Franceschi,35 is a risk factor for cerebrovascular and neurodegenerative diseases.36 Among the underlying mechanisms for inflamm-aging, dysbiosis is central due to its crosstalk with dysregulation of metabolism and immune system,35 along with genomic instability and mitochondrial dysfunctions.37 In the elderly, the composition of gut microbiota is different from that of younger adults, with a higher proportion of Bacteroides spp, and a reduction of Bifidobacteria.38 Moreover, loss of gut microbiota diversity is associated with increased frailty.39 Notably, reduced production of SCFAs accelerates not only nuclear factor-κB-mediated inflammation but also increases intestinal permeability, leading to bacterial translocation and higher levels of IL-6 and tumor necrosis factor-α.40 FMT promotes the recovery from inflamm-aging, which was accompanied by an improved SCFA production and decreased inflammatory cytokines.41 Using elderly germ-free and conventional mice, Thevaranjan et al.42 demonstrated that age-related dysbiosis increases intestinal permeability and inflammatory biomarkers.
Metabolic diseases
In randomized controlled trials, lean-donor FMT in subjects with metabolic syndrome improved obesity and insulin sensitivity, suggesting that the microbiota can regulate host metabolism as an “organ.”43 Accumulating evidence illustrates more detailed mechanisms linking either the presence of specific bacteria or the production of bacteria-derived metabolites with metabolic disorders.44 Akkermansia muciniphila is gaining attention as a possible “next-generation” probiotic.45 A. muciniphila improves several metabolic parameters in both model mice46 and even in a proof-of-concept exploratory human study.47 Various studies demonstrated the effect of microbiota-derived metabolites of carbohydrates and proteins on host metabolism. Reduced production of SCFAs, including acetate and propionate, has inconsistent effects on energy homeostasis and glucose and lipid metabolism.48 Acetate, the most abundant SCFA, induces glucagon-like peptide (GLP)-1 production in enteroendocrine L cells through G-protein-coupled receptor (GPR)-43, improving insulin sensitivity.49 Additionally, rodent studies demonstrated that acetate reaches the hypothalamus through the BBB, which induces the production of gamma-aminobutyric acid, resulting in suppression of central appetite.50 On the other hand, propionate causes peptide YY production through GPR41, which inhibits intestinal motility and increases the absorption rate of nutrients through the intestinal epithelium.51 The advance of metabolomic analyses enabled discovery of the molecular mechanisms between bacteria-derived metabolites of amino-acids and host glucose metabolism. Important examples are the beneficial role of indole and its derivatives and the harmful effect of imidazole propionate, a microbial metabolites of histidine. Indole and its derivatives increase the production of the incretin hormone GLP-1 from intestinal enteroendocrine cells and modulate innate and adaptive immune response via the aryl hydrocarbon receptor.52 Koh et al.53 identified that imidazole propionate impairs insulin receptor activation via the p38γ-p62-mammalian target of rapamycin complex1 (mTORC1) pathway in peripheral tissues. Furthermore, microbial-derived metabolites control bile acid homeostasis via farnesoid X receptors, which can also influence glucose metabolism.54
Hypertension
Hypertension is the leading cause of stroke and heart diseases. Genetic and environmental factors influence the individual variation of increased blood pressure (BP). Extensive populational genome-wide association studies (GWAS) identified several genetic factors associated with blood pressure control, including the renin-angiotensin-aldosterone system, sympathetic nervous system, atrial natriuretic peptide signaling, and dopaminergic system; however, these genetic factors contribute to only 3% of hypertension in humans.55 Now, growing evidence supports different roles of the GI tract in BP regulation, not only as an organ for salt absorption, but also controlling autonomic and endocrine functions. The gut microbiome in patients with hypertension is comprised by a higher percentage of bacteria from the genus Prevotella, which includes periodontal pathogens, such as Klebsiella, Porphyromonas, and Actinomyces.56 Increasing evidence reveals that microbial-derived metabolites also have key roles in BP regulation, by regulating the gut–cardiorenal axis (reviewed in Marques et al.57). Acetate and propionate reduce BP via GPRs and transcriptome changes. Although propionate can partially increase BP through the release of renin in renal juxtaglomerular cells via olfactory receptor 78, acetate and propionate have stronger depressive effects through vasodilation via GPR41.57 Hydrogen sulfide, which is produced from cysteine by both epithelial cells and gut microbiota, exerts hypotensive effects due to peripheral vasodilation and decreases heart rate in experimental studies.58
Stroke and acute or chronic infectious burden
As mentioned above, microbiota–host interactions are associated with various vascular risk factors of stroke, including aging, metabolic diseases, and hypertension. Recent case-crossover studies suggested acute infection as a possible trigger of stroke.59 Cumulative exposure to chronic infectious diseases, or “infectious burden,” has been measured using serological profiles against several pathogens and inflammatory biomarkers. Using an analyzed serological profile for Chlamydia pneumoniae, Helicobacter pylori, cytomegalovirus, and herpes simplex virus, a prospective cohort study revealed that infectious burden is associated with onset of ischemic stroke.60 Another prospective study showed that higher value of serum procalcitonin, a surrogate biomarker for chronic bacterial infection, is linked to small vessel stroke.61 Inflammatory and coagulation cascades are shared signaling pathways underlying both infectious diseases and stroke; however, there are additional way in which the microbiota can influence stroke (Figure 3).
Large artery atherosclerosis and microbiota
Intra- and extracranial large artery atherosclerosis occurs concomitantly with systemic atherosclerosis. Atherosclerosis occurs at sites in the arterial tree where the laminar flow is disrupted and presents multiple pathological processes, including accumulation of lipid and extracellular matrix, migration of inflammatory cells, plaque rupture, and luminal thrombosis.62 A meta-analysis assessing bacteria in atherosclerotic plaque samples confirmed the abundance of oral commensal bacteria, including Streptococcus mutans, Porphyromonas gingivalis, and Aggregatibacter actinomycetemcomitans, which suggests that oral bacteria can translocate into the bloodstream and subsequently enter atherosclerotic plaques.63 Bacteremia and endotoxemia are mainly sensed by host cells via microorganism-associated molecular patterns (MAMPs)-host pattern reorganization receptors (PRRs) signaling, including peptidoglycan-nucleotide binding oligomerization domain-containing protein 1, lipopeptide-heterodimeric TLR2 and TLR6, or lipopolysaccharide (LPS)-TLR4.64 These MAMP-PRR pathways increase the production of inflammatory cytokines and accelerate atherosclerosis.64 However, another study showed no discernible difference in bacterial DNA amount and community between symptomatic and asymptomatic carotid artery stenosis, suggesting that other factors are critical in determining plaque vulnerability.65 Jie et al.66 reported that the abundance of Streptococcus spp. in gut microbiota was associated with atherosclerosis, which introduces another mechanism, whereby ectopic translocation of oral microbe may cause gut dysbiosis and impact on bacterial metabolisms. Several lines of research assessing gut microbial-derived metabolites proposed other underlying mechanisms of atherosclerosis. Trimethylamine N-oxide (TMAO), a choline-derived metabolite produced by gut microbiota and host hepatic flavin monooxygenase, is associated with increased risk of coronary heart diseases and large artery ischemic stroke in humans.67,68 In animal studies, TMAO was shown to promote cholesterol accumulation and subsequent foam cell formation,69 inflammation,70 and enhanced platelet hyperactivity in the vessel wall.71 Among SCFAs, butyrate may have a protective role against atherosclerosis. In animal models, oral administration of butyrate and butyrate-producing bacterial genus was inversely correlated with atherosclerotic lesions.72 In humans, butyrate-producing bacteria and butyrate concentration in the gut were lower among subjects with multiple vascular risk factors.73 The precise etiologies are unproven, but increase of intestinal epithelium integrity, regulatory T cell induction, and histone deacetylase (HDAC) inhibition, which results in decreased the amount of bacterial translocation, inflammation, and reduced epigenetic modulation, are possible mechanisms.74
Cardioembolic stroke
Atrial fibrillation (Af) is one of the most important causes of cardioembolic stroke. Cardiac autonomic ganglionated plexuses facilitate electrophysiological modulation of the atrium and arrhythmogenesis.75 A few studies have revealed the association between Af and microbiota in humans.76 In a recent cohort study, a higher level of plasma TMAO was a significant independent risk factor of Af.77 In a canine model of Af, TMAO increased the instability of atrial electrophysiology and aggravated acute atrial remodeling.78
Cerebral small vessel disease
Cerebral small vessel disease (CSVD), including lacunar infarction, cerebral microbleeds (CMBs), enlarged perivascular spaces, leukoaraiosis, and cortical atrophy, is commonly observed in neuroimaging among elderly persons. CSVD contains two major pathological classifications, hypertensive vasculopathy and cerebral amyloid angiopathy (CAA), and is clinically relevant to stroke, including recent subcortical small infarction and intracerebral hemorrhage. Furthermore, the CSVD burden is highly relevant to cognitive decline.79,80 In a multiethnic cohort study, procalcitonin was associated with subclinical CSVD.81 Additionally, LPS administration induces CMBs in wild-type mice.82 Hereafter, we review the specific role of microbiota in the development of each subtype of CSVD.
Hypertensive vasculopathy
Pathological features of hypertensive vasculopathy include lipohyalinosis and fibrinoid necrosis. BBB disruption and inflammation are two crucial etiologies of hypertensive vasculopathy. Nakano et al.,83 using vessel occlusion model mice, reported that S. mutans expressing the collagen-binding protein Cnm (Cnm-positive S. mutans) is associated with intracerebral hemorrhage (ICH).83 Another in vivo study showed that Cnm as the major virulence factor for adherence and invasion of human coronary artery endothelial cells.84 Approximately 15% of S. mutans have cnm gene among Japanease population.85 We have reported that Cnm-positive S. mutans in the oral cavity is associated with both hypertensive ICH and deep CMBs in a hospital cohort study,86 and cognitive decline accompanied by increased burden of CMBs in a populational cohort study.87 We hypothesized that Cnm-positive S. mutans translocates from the oral cavity into the bloodstream, adheres to and invades cerebral small vessels, and evokes local inflammation.79,86 However, to elucidate this brain-oral-microbial axis, a more detailed molecular assessment in vitro and in animal models, and more extensive cohort studies in multiethnic populations are necessary.
Cerebral amyloid angiopathy
Even though there is little human data investigating the role of the microbiome in patients with cerebral amyloid angiopathy (CAA), there is accumulating evidence about Alzheimer’s disease (AD), which has shared pathophysiology with CAA. The amyloid cascade hypothesis suggests that vascular amyloid-β (Aβ) accumulation plays a central role in elucidating the pathogenesis of AD.88 However, repeated failures of anti-amyloidogenic trials prompted us to explore the brain–microbiota axis as an alternative to the amyloid cascade hypothesis, from initiation to aggravation of AD and CAA. We have proposed that functional bacterial amyloid proteins in the gut may cause cross-seeding of Aβ via a prion-like mechanism. In studies of aged rats, we showed that exposure to bacterial amyloid initiates cross-seeding of cerebral alpha synuclein aggregation and neuroinflammation.89 For instance, curli, a functional amyloid protein expressed by E. coli and other bacteria, can accelerate cross-seeding of amyloid formation in vivo and in vitro.90 Of particular interest are the findings of Venegas et al.,91 who noted cross-seeding of aggregation of Aβ in the brain caused by neuroinflammation through NLR family pyrin domain containing 3 (NLPR3)-apoptosis-associated speck-like protein containing a caspase recruitment domain (ASC-SPECK)-inflammasome in microglia. Another possible hypothesis supported by mounting evidence is the role of Aβ as a broad-spectrum antibiotic against bacteria, fungi, and enveloped viruses.92 Eimer et al.93 showed that Aβ oligomers inhibit microbe adhesion to host cells, and Aβ fibrils entrap microbes using 3D human neural cell cultures. Metabolomics on post-mortem brain showed that human herpesvirus 7 was associated with increased Aβ metabolism in late-onset AD.94 Collectively, these studies suggest that the microbiota, which is intimately connected with innate immune system activation, is a critical initiating factor and novel therapeutic target for AD and CAA.95
Future directions
Gene–microbial interaction
The single nucleotide polymorphisms (SNPs) of microbial-derived metabolite receptors and PRRs can contribute to the susceptibility of metabolic diseases and atherosclerosis. Among microbial-derived metabolites, SNPs of GPR modulate the effects of SCFAs on hypertension and type 2 diabetes mellitus,96 and SNPs of farnesoid X receptors affect lipid and glucose metabolism.97 Among PRRs, TLR-4 SNPs are associated not only with the differences in LPS responsiveness but also with progression of atherosclerosis or ischemic stroke.98 Some studies report the association between oligomerization domain-containing protein 1 genotypes and susceptibility to cerebrovascular diseases.99 However, these links between SNPs and cerebrovascular diseases are controversial for different ethnicities, implying that combination of microbiome and genome-wide association studies, called “mGWAS,” is essential to unravel the complexity of cerebrovascular diseases.100
Figure 1.
The mucosal immune system of the oral cavity and GI tract. The oral and GI immune systems share similar anatomical compartments, epithelium, LP, and MALT. IELs reside within the epithelial layer. LP-resident DCs directly uptake luminal antigens via extension process and migrate to MALTs, tonsillar MALTs in the oral cavity, and GALTs in the GI tract. M-cells in the epithelium of Peyer’s patches and M-cell-like cells in the epithelium of tonsil crypts pass the antigens to DCs or macrophages. Instead, the immunological interactions in the gingiva are specific to the oral cavity. The gingival crevice, the inner base of the gingiva, is lined with a few layers of non-keratinized highly permeable thin epithelium. In the gingival crevice, neutrophils and oral microbiome constantly transmigrate through JE and mediate inflammation even in steady state. DC: dendritic cell; GALT: gut-associated lymphoid tissue; GI: gastrointestinal; IEL: intraepithelial lymphocyte; JE: junctional epithelium; LP: lamina propria; MALT: mucosa-associated lymphoid tissue; M-cell: microfold cell.
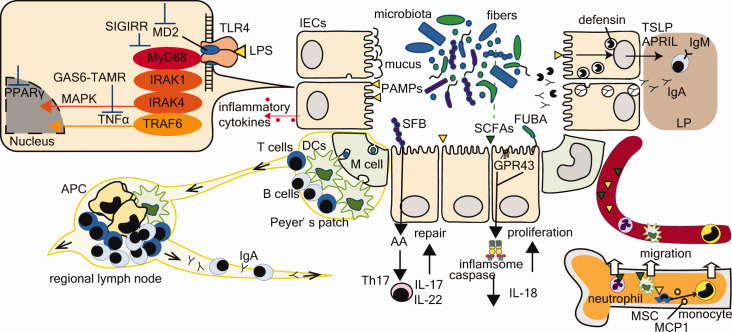
Figure 2.
The interactions of the microbiota with the host immune system. Bidirectional interactions between the mucosal immune system and the microbiota. These include (1) the mucus layer, (2) epithelial cells (intercellular tight junction, antimicrobial peptide secretion, recognitions of MAMPs), (3) innate immunity (hematopoiesis, IgA secretion in LP, Inflammasome), and (4) acquired immunity (Th17 cell activation, DCs in LP, M-cells and APC cells in Peyer’s patches, regional lymph nodes). As a recognition of MAMPs in epithelial cells, for instance, TLR4-MD2 complex binding to LPS activates NF-κB and MAPK via the MyD88-dependent signaling pathway. Microbes control inflammatory reaction in epithelial cells activating or inhibiting these pathways (GAS6-TAMR axis, MD2, MyD88, SIGIRR, PPARγ). Metabolites or MAMPs of microbiota have a role in educating the host innate system. In bone marrow, SCFAs promote DC maturation and PAMPs induce MCP1 production in MSC, resulting in monocyte migration. Epithelial cell-derived cytokines TSLP and APRIL promote class-switch recombination and the production of IgA by B cells in intestinal LP, transmitted into mucosal layer. Not only microbiota, but also FUBA or SCFAs influence the inflammasome involving caspase 1 and proinflammatory cytokines. Th17 cell differentiation induced by commensal microbiota, including SBF, produce IL-17 and IL-22, which play a homeostatic role in microbiota–host interactions. The M-cell of Peyer’s patches transfers the luminal antigen to DCs and APCs, which is drained and induces T cell-dependent B cell maturation in MLNs. APC: antigen-presenting cell; APRIL: aproliferation-inducing ligand; DCs: dendritic cells; FUBA: functional bacterial amyloid; GAS6: growth-arrest specific gene 6; G-CSF: granulocyte-colony stimulating factor; GPR43: G-protein coupled receptor 43; IgA: immunoglobulin A; IL: interleukin; LP: lamina propria; LPS: lipopolysaccharide; M cell: microfold cell; MAMPs: microorganism-associated molecular patterns; MAPK: mitogen-activated protein kinase; MCP1: monocyte chemoattractant protein 1; MD2: myeloid differentiation factor 2; MLN: mesenteric lymph node; MSC: mesenchymal stem cell; MyD88: myeloid differentiation factor 88; NF-κB: nuclear factor-κB; PPARγ: peroxisome proliferator activated receptor γ; SBF: segmented filamentous bacterium; SCFAs: short-chain fatty acids; SIGIRR: single immunoglobulin interleukin-1 receptor-related molecule; TAMR: tyro3 axl mer receptor; TLR: Toll-like receptor; TSLP: thymic stromal lymphopoietin.
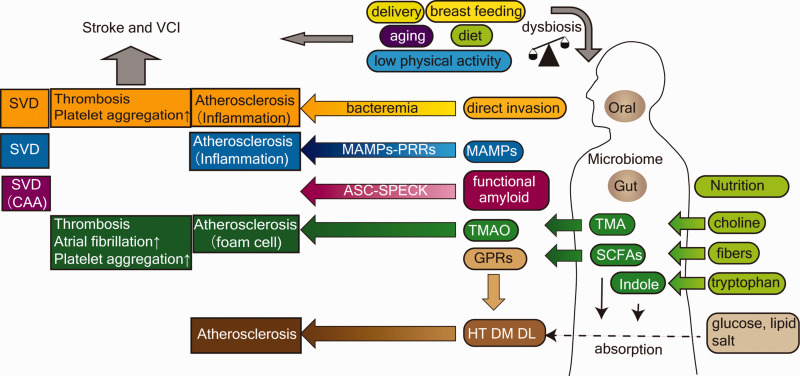
Figure 3.
Overview of the interplay between microbiota and stroke or vascular cognitive impairment. Beside the traditional risk factors, including HT, DM, DL, physical activity, and aging, mounting evidence has shown the role of gut microbiota in the development of stroke. Delivery, breastfeeding, aging, diet, and physical activity are bidirectionally related to dysbiosis. There are several possible mechanisms between oral and gut microbiota and the development of stroke and VCI. Microbes in the oral cavity can act directly on the cerebrovascular system via bacteremia. PRRs recognize MAMPs, for example TLRs and LPS, lipoteichoic acid, or peptidoglycan, which subsequently stimulate immune cells and induce the production of inflammatory cytokines. Among bacterial metabolites, TMA and TMAO interact with coagulation and inflammatory cascades in atherosclerotic plaque. SCFAs and indoles affect the absorption rate of nutrients and have hormonal activity in metabolic homeostasis via various types of GPRs. Bacteria-derived functional amyloid induces Aβ aggregation in CAA through inflammasome secretion or the ASC-SPECK system. Aβ: Amyloid β; ASC-SPECK: adaptor protein apoptosis-associated speck-like protein containing a caspase recruitment domain; CAA: cerebral amyloid angiopathy; GPRs: G-protein-coupled receptors; DL dyslipidemia; DM: diabetes mellitus; HT: hypertension; LPS: lipopolysaccharide; MAMPs: microorganism-associated molecular patterns; SCFAs: short-chain fatty acids; SVD: small vessel disease; TLRs: toll-like receptors; TMA: trimethylamine; TMAO: Trimethylamine N-oxide; VCI: vascular cognitive impairment; PRRs: pattern recognition receptors
Gene–microbial interaction also gives us the novel insight that human genetics and microbiota have shared mechanisms in the development of each subtype of stroke, including modulation of HDAC in large artery atherosclerosis, modulation of type IV collagen in hypertensive vasculopathy, and modulation of Aβ-NLPR3-ASC-SPECK signaling in CAA via apolipoprotein E (APOE) and triggering receptor expressed on myeloid cells (TREM2). HDAC is a pivotal epigenetic factor modulating chromatin topology and regulating gene expression. HDAC9 polymorphisms are associated with large artery atherosclerosis and stroke, which is inhibited by butyrate, as previously mentioned.101 Additionally, COL4A1 and COL4A2 polymorphisms are associated with hemorrhagic stroke, disrupting type IV collagen of the vascular basement protein,102 where Cnm-positive S. mutans adhere.87,103 APOE and TREM2 are the most influential genetic risk factors for AD and CAA. Recent studies found that both APOE and TREM2 are included in the Aβ-NLPR3-ASC-SPECK signaling pathway in microglia.104 These findings indicate that gene–microbial interactions will be valuable in revealing the unknown etiologies of stroke. These studies will also be critical in finding intervention strategies depending on genotype.
Prevention perspectives
The antibiotic eradication of Helicobacter pylori has been useful as a treatment of peptic ulceration, fulfilling Koch’s postulates. As described in this review, the microbiota–oral–brain and microbiota–gut–brain axis suggest therapeutic avenues to prevent or treat stroke. However, the clinical efficacy of prophylactic antibiotics for stroke prevention is conflicting,105 which suggests that unfavorable effects of antibiotics might outweigh the potential benefits. Accumulating evidence from basic and translational researches provide deeper insights into the applicable strategies beyond antibiotics: bacterial cell-to-cell communication, microbiome, microbial-derived metabolites, and immune modulation.
Bacterial cell-to-cell communication
The quorum-sensing (QS) system of bacterial cell-to-cell communication is a novel therapeutic target. Microbial QS systems regulate the expression of many virulence factors, biofilm synthases, and even clustered regularly interspaced short palindromic repeat (CRISPR)-associated defense system.106 QS-interfering agents that potentially reduce bacterial virulence and prevent biofilm formation of infectious pathogens are expected to influence metabolic disorders affecting the microbiome.107 However, most QS-interfering agents are still in preclinical trials. Further research is necessary to assess the side effects toward non-target bacteria and the possibility of resistance development in humans.
Microbiome
Prebiotics are compounds that are digested by bacteria which may be beneficial to the host, depending on the nature of the host’s microbiome. Probiotics, the direct administration of beneficial microbes, and FMT are attractive therapies to shape a healthy microbiome and modify immune and metabolic balance. However, these microbiome-modifying therapies bear many unsolved questions.108,109 Personalized nutrition in combination with beneficial microbes might be an integrated approach to microbiome-modifying therapies.110 Furthermore, as the proof-of-concept study with A. muciniphila implicated, increasing translational studies on next-generation probiotics demonstrated that core species are possible new therapeutic tools for metabolic diseases, atherosclerosis, and cerebrovascular diseases.111
Microbial-derived metabolites
Roberts et al.112 proposed that small molecules inhibiting microbial production of TMA in the gut lumen suppress thrombosis formation and prevent atherosclerosis. Chambers et al.113 showed that oral administration of inulin-propionate ester can deliver propionate to colon and ameliorate obesity and glucose homeostasis. This indicates the beneficial role of SCFAs and GPR modulators as novel preventive interventions for metabolic disorders and atherosclerosis. This work illustrates that the new strategies of leveraging the host–microbiota interactions as druggable molecular targets show promise
Immunomodulation
The microbiota interacts with the host immune system through immune cell polarization, stimulating MAMP-PRR signals, Aβ-NLPR3-ASC-SPECK signals, and inducing anti- or pro-inflammatory cytokines. Several studies attempted to investigate new therapeutics modulating these chemical mediators.114,115 Fingolimod, a sphingosine-1-phosphate receptor modulator, salvaged penumbra tissues in patients with acute internal carotid artery or middle cerebral artery occlusion in a clinical pilot trial.116 Subcutaneous administration of anakinra, an IL-1 receptor antagonist, significantly decreased IL-6 and C-reactive protein, which are associated with worse clinical outcomes after stroke, in a randomized phase 2 clinical trial.117 mTORC1 is another possible therapeutic target associated with the host immune–microbiota interaction. Recent evidence suggests that rapamycin, mTORC1 inhibitor, induces neuroprotective autophagy and pleiotropic effects on BBB and inflammation.118 Microbiota and microbial metabolites control mTORC1 expression.119 Finally, the collagen-binding microbial surface components recognize adhesive matrix molecules, which are common invasive and adhesive factors for various types of virulent Gram-positive cocci,120 might become possible targets for vaccine design to prevent infectious endocarditis and specific subtypes of CSVDs.
Conclusions
Firstly, it must be emphasized that the microbiota is our most substantial environmental exposure. They represent an enormous potential threat to survival, a threat to which the evolution of both the host and the microbes has adapted over the past several million years. Thus, it is critical that the host tolerates beneficial organisms, and that pathogens do not acquire immune tolerance. It is similarly vital for the microbes that we tolerate them. These inter-relationships are the foundation for the influences of the microbiota on immune systems. We propose several interactions by which the microbiota may impact stroke and VCI in humans. Mechanisms involved in these interactions include innate and adaptive immunity, epigenetic alteration of protein expression, proteostasis, hematopoiesis, coagulation, hormone secretion, autonomic nerve system, and metabolism.
Secondly, in particular, we have shown the differences in the microbiota and host defense systems between the GI tract and oral cavity, suggesting the concept of “Microbiota–Oral–Brain Axis,” which is distinct from the “Microbiota–Gut–Brain Axis.” The role of the “Microbiota–Gut–Brain Axis” in human health is receiving increasing scientific and public attention. However, the influence of “Microbiota–Oral–Brain Axis” has not yet been properly recognized for its importance. According to the U.S. National Library of Medicine, PubMed database, there were 22,349 citations for “microbiota” and “gut,” while there were only 4743 citations for “microbiota” and “oral” (4 February 2020). Bidirectional gut–brain interactions involve several signaling molecules, immune mediators, and gut hormones by the microbiota. Conversely, considering that the nasal and oral cavities are anatomically close to the brain, microbiota and their metabolites can have direct influences through the blood, the cranial nerves and the central nervous system.
Despite promising data in both animal and human studies, it is challenging to integrate these findings into personalized prevention and treatment for stroke. As described above, the microbiota is influenced by genetics, nutrition, and other lifestyle factors. Thus, epidemiological research requires careful consideration of ethnicity, genetics, and lifestyle factors.
Funding
The author(s) disclosed receipt of the following financial support for the research, authorship, and/or publication of this article: This work was supported by the Grant-in-Aid for Japan Society for Promotion of Science Fellows to S.T (20 J), Grant-in-Aid for Challenging Exploratory Research to M.I. (16K14573, 19K22610), Japan Heart Foundation Research Grant, Smoking Research Foundation, Novartis Research Grants, the University of Louisville and the Jewish Heritage Fund for Excellence, W. Cowan and the family of E. A Ford II.
Declaration of conflicting interests
The author(s) declared no potential conflicts of interest with respect to the research, authorship, and/or publication of this article.
ORCID iD
Shuichi Tonomura https://orcid.org/0000-0001-7092-2121
References
- 1.Feigin VL, Roth GA, Naghavi M, et al. Global burden of stroke and risk factors in 188 countries, during 1990-2013: a systematic analysis for the Global Burden of Disease Study 2013. Lancet Neurol 2016; 15: 913–924. [DOI] [PubMed] [Google Scholar]
- 2.Clemente JC, Ursell LK, Parfrey LW, et al. The impact of the gut microbiota on human health: an integrative view. Cell 2012; 148: 1258–1270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Grau AJ, Urbanek C, Palm F.Common infections and the risk of stroke. Nat Rev Neurol 2010; 6: 681–694. [DOI] [PubMed] [Google Scholar]
- 4.Li JH, Jia HJ, Cai XH, et al. An integrated catalog of reference genes in the human gut microbiome. Nat Biotechnol 2014; 32: 834–841. [DOI] [PubMed] [Google Scholar]
- 5.Metzker ML.Sequencing technologies – the next generation. Nat Rev Genet 2010; 11: 31–46. [DOI] [PubMed] [Google Scholar]
- 6.Paliy O, Agans R.Application of phylogenetic microarrays to interrogation of human microbiota. FEMS Microbiol Ecol 2012; 79: 2–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Milani C, Duranti S, Bottacini F, et al. The first microbial colonizers of the human gut: composition, activities, and health implications of the infant gut microbiota. Microbiol Mol Biol Rev 2017; 81: e00036–17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Aagaard K, Ma J, Antony KM, et al. The placenta harbors a unique microbiome. Sci Transl Med 2014; 6: 237ra265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Backhed F, Roswall J, Peng Y, et al. Dynamics and stabilization of the human gut microbiome during the first year of life. Cell Host Microbe 2015; 17: 690–703. [DOI] [PubMed] [Google Scholar]
- 10.Costello EK, Lauber CL, Hamady M, et al. Bacterial community variation in human body habitats across space and time. Science 2009; 326: 1694–1697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ding T, Schloss PD.Dynamics and associations of microbial community types across the human body. Nature 2014; 509: 357–360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wu RQ, Zhang DF, Tu E, et al. The mucosal immune system in the oral cavity – an orchestra of T cell diversity. Int J Oral Sci 2014; 6: 125–132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chung LK, Raffatellu M.GI pros: antimicrobial defense in the gastrointestinal tract. Semin Cell Dev Biol 2019; 88: 129–137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gorr SU, Abdolhosseini M.Antimicrobial peptides and periodontal disease. J Clin Periodontol 2011; 38: 126–141. [DOI] [PubMed] [Google Scholar]
- 15.Cerovic V, Bain CC, Mowat AM, et al. Intestinal macrophages and dendritic cells: what’s the difference? Trends Immunol 2014; 35: 270–277. [DOI] [PubMed] [Google Scholar]
- 16.Cutler CW, Jotwani R.Dendritic cells at the oral mucosal interface. J Dent Res 2006; 85: 678–689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Novak N, Haberstok J, Bieber T, et al. The immune privilege of the oral mucosa. Trends Mol Med 2008; 14: 191–198. [DOI] [PubMed] [Google Scholar]
- 18.Lamster IB.Evaluation of components of gingival crevicular fluid as diagnostic tests. Ann Periodontol 1997; 2: 123–137. [DOI] [PubMed] [Google Scholar]
- 19.Dutzan N, Konkel JE, Greenwell-Wild T, et al. Characterization of the human immune cell network at the gingival barrier. Mucosal Immunol 2016; 9: 1163–1172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Park JY, Chung H, DiPalma DT, et al. Immune quiescence in the oral mucosa is maintained by a uniquely large population of highly activated Foxp3(+) regulatory T cells. Mucosal Immunol 2018; 11: 1092–1102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Fulde M, Hornef MW.Maturation of the enteric mucosal innate immune system during the postnatal period. Immunol Rev 2014; 260: 21–34. [DOI] [PubMed] [Google Scholar]
- 22.Bunker JJ, Erickson SA, Flynn TM, et al. Natural polyreactive IgA antibodies coat the intestinal microbiota. Science 2017; 358: eaan6619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Macpherson AJ, Harris NL.Interactions between commensal intestinal bacteria and the immune system. Nat Rev Immunol 2004; 4: 478–485. [DOI] [PubMed] [Google Scholar]
- 24.Tezuka H, Ohteki T.Regulation of IgA production by intestinal dendritic cells and related cells. Front Immunol 2019; 10: 1891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.He B, Xu W, Santini PA, et al. Intestinal bacteria trigger T cell-independent immunoglobulin A(2) class switching by inducing epithelial-cell secretion of the cytokine APRIL. Immunity 2007; 26: 812–826. [DOI] [PubMed] [Google Scholar]
- 26.Khosravi A, Yanez A, Price JG, et al. Gut microbiota promote hematopoiesis to control bacterial infection. Cell Host Microbe 2014; 15: 374–381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Thaiss CA, Zmora N, Levy M, et al. The microbiome and innate immunity. Nature 2016; 535: 65–74. [DOI] [PubMed] [Google Scholar]
- 28.Erny D, Hrabe de Angelis AL, Jaitin D, et al. Host microbiota constantly control maturation and function of microglia in the CNS. Nat Neurosci 2015; 18: 965–977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Weaver CT, Elson CO, Fouser LA, et al. The Th17 pathway and inflammatory diseases of the intestines, lungs, and skin. Annu Rev Pathol 2013; 8: 477–512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.McGeachy MJ, Chen Y, Tato CM, et al. The interleukin 23 receptor is essential for the terminal differentiation of interleukin 17-producing effector T helper cells in vivo. Nat Immunol 2009; 10: 314–324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Faraco G, Brea D, Garcia-Bonilla L, et al. Dietary salt promotes neurovascular and cognitive dysfunction through a gut-initiated TH17 response. Nat Neurosci 2018; 21: 240–249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Atarashi K, Tanoue T, Ando M, et al. Th17 cell induction by adhesion of microbes to intestinal epithelial cells. Cell 2015; 163: 367–380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Benakis C, Brea D, Caballero S, et al. Commensal microbiota affects ischemic stroke outcome by regulating intestinal γδ T cells. Nat Med 2016; 22: 516–523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.O’Donnell MJ, Xavier D, Liu L, et al. Risk factors for ischaemic and intracerebral haemorrhagic stroke in 22 countries (the INTERSTROKE study): a case-control study. Lancet 2010; 376: 112–123. [DOI] [PubMed] [Google Scholar]
- 35.Franceschi C, Garagnani P, Parini P, et al. Inflammaging: a new immune-metabolic viewpoint for age-related diseases. Nat Rev Endocrinol 2018; 14: 576–590. [DOI] [PubMed] [Google Scholar]
- 36.Stephenson J, Nutma E, van der Valk P, et al. Inflammation in CNS neurodegenerative diseases. Immunology 2018; 154: 204–219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ferrucci L, Fabbri E.Inflammageing: chronic inflammation in ageing, cardiovascular disease, and frailty. Nat Rev Cardiol 2018; 15: 505–522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Buford TW. ( Dis)Trust your gut: the gut microbiome in age-related inflammation, health, and disease. Microbiome 2017; 5: 80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.O’Toole PW, Jeffery IB.Gut microbiota and aging. Science 2015; 350: 1214–1215. [DOI] [PubMed] [Google Scholar]
- 40.Thorburn AN, Macia L, Mackay CR.Diet, metabolites, and “western-lifestyle” inflammatory diseases. Immunity 2014; 40: 833–842. [DOI] [PubMed] [Google Scholar]
- 41.Spychala MS, Venna VR, Jandzinski M, et al. Age-related changes in the gut microbiota influence systemic inflammation and stroke outcome. Ann Neurol 2018; 84: 23–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Thevaranjan N, Puchta A, Schulz C, et al. Age-associated microbial dysbiosis promotes intestinal permeability, systemic inflammation, and macrophage dysfunction. Cell Host Microbe 2017; 21: 455–466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kootte RS, Levin E, Salojarvi J, et al. Improvement of insulin sensitivity after lean donor feces in metabolic syndrome is driven by baseline intestinal microbiota composition. Cell Metab 2017; 26: 611–619. [DOI] [PubMed] [Google Scholar]
- 44.Cani PD.Microbiota and metabolites in metabolic diseases. Nat Rev Endocrinol 2019; 15: 69–70. [DOI] [PubMed] [Google Scholar]
- 45.Anhe FF, Schertzer JD, Marette A.Bacteria to alleviate metabolic syndrome. Nat Med 2019; 25: 1031–1033. [DOI] [PubMed] [Google Scholar]
- 46.Plovier H, Everard A, Druart C, et al. A purified membrane protein from Akkermansia muciniphila or the pasteurized bacterium improves metabolism in obese and diabetic mice. Nat Med 2017; 23: 107–113. [DOI] [PubMed] [Google Scholar]
- 47.Depommier C, Everard A, Druart C, et al. Supplementation with Akkermansia muciniphila in overweight and obese human volunteers: a proof-of-concept exploratory study. Nat Med 2019; 25: 1096–1103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Canfora EE, Meex RCR, Venema K, et al. Gut microbial metabolites in obesity, NAFLD and T2DM. Nat Rev Endocrinol 2019; 15: 261–273. [DOI] [PubMed] [Google Scholar]
- 49.Kimura I, Inoue D, Hirano K, et al. The SCFA receptor GPR43 and energy metabolism. Front Endocrinol 2014; 5: 85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Frost G, Sleeth ML, Sahuri-Arisoylu M, et al. The short-chain fatty acid acetate reduces appetite via a central homeostatic mechanism. Nat Commun 2014; 5: 11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Samuel BS, Shaito A, Motoike T, et al. Effects of the gut microbiota on host adiposity are modulated by the short-chain fatty-acid binding G protein-coupled receptor, Gpr41. Proc Natl Acad Sci U S A 2008; 105: 16767–16772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Roager HM, Licht TR.Microbial tryptophan catabolites in health and disease. Nat Commun 2018; 9: 3294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Koh A, Molinaro A, Stahlman M, et al. Microbially produced imidazole propionate impairs insulin signaling through mTORC1. Cell 2018; 175: 947–961. [DOI] [PubMed] [Google Scholar]
- 54.Ahmad TR, Haeusler RA.Bile acids in glucose metabolism and insulin signalling – mechanisms and research needs. Nat Rev Endocrinol 2019; 15: 701–712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Ehret GB, Ferreira T, Chasman DI, et al. The genetics of blood pressure regulation and its target organs from association studies in 342,415 individuals. Nat Genet 2016; 48: 1171–1184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Li J, Zhao F, Wang Y, et al. Gut microbiota dysbiosis contributes to the development of hypertension. Microbiome 2017; 5: 14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Marques FZ, Mackay CR, Kaye DM.Beyond gut feelings: how the gut microbiota regulates blood pressure. Nat Rev Cardiol 2018; 15: 20–32. [DOI] [PubMed] [Google Scholar]
- 58.Yang G, Wu L, Jiang B, et al. H2S as a physiologic vasorelaxant: hypertension in mice with deletion of cystathionine gamma-lyase. Science 2008; 322: 587–590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Sebastian S, Stein LK, Dhamoon MS.Infection as a stroke trigger. Stroke 2019; 50: 2216–2218. [DOI] [PubMed] [Google Scholar]
- 60.Elkind MS, Ramakrishnan P, Moon YP, et al. Infectious burden and risk of stroke: the northern Manhattan study. Arch Neurol 2010; 67: 33–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Katan M, Moon YP, Paik MC, et al. Procalcitonin and midregional proatrial natriuretic peptide as markers of ischemic stroke the northern Manhattan Study. Stroke 2016; 47: 1714–1719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Stary HC.Natural history and histological classification of atherosclerotic lesions – an update. Arterioscl Throm Vasc Biol 2000; 20: 1177–1178. [DOI] [PubMed] [Google Scholar]
- 63.Chhibber-Goel J, Singhal V, Bhowmik D, et al. Linkages between oral commensal bacteria and atherosclerotic plaques in coronary artery disease patients. NPJ Biofilms Microbiomes 2016; 2: 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Brown JM, Hazen SL.Microbial modulation of cardiovascular disease. Nat Rev Microbiol 2018; 16: 171–181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Jonsson AL, Hallenius FF, Akrami R, et al. Bacterial profile in human atherosclerotic plaques. Atherosclerosis 2017; 263: 177–183. [DOI] [PubMed] [Google Scholar]
- 66.Jie ZY, Xia HH, Zhong SL, et al. The gut microbiome in atherosclerotic cardiovascular disease. Nat Commun 2017; 8: 845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Tang WH, Wang Z, Levison BS, et al. Intestinal microbial metabolism of phosphatidylcholine and cardiovascular risk. N Engl J Med 2013; 368: 1575–1584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Yin J, Liao SX, He Y, et al. Dysbiosis of gut microbiota with reduced trimethylamine-N-oxide level in patients with large-artery atherosclerotic stroke or transient ischemic attack. J Am Heart Assoc 2015; 4: e002699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Wang ZN, Klipfell E, Bennett BJ, et al. Gut flora metabolism of phosphatidylcholine promotes cardiovascular disease. Nature 2011; 472: 57–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Haghikia A, Li XS, Liman TG, et al. Gut microbiota-dependent trimethylamine N-oxide predicts risk of cardiovascular events in patients with stroke and is related to proinflammatory monocytes. Arterioscler Thromb Vasc Biol 2018; 38: 2225–2235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Zhu WF, Gregory JC, Org E, et al. Gut microbial metabolite TMAO enhances platelet hyperreactivity and thrombosis risk. Cell 2016; 165: 111–124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Kasahara K, Krautkramer KA, Org E, et al. Interactions between Roseburia intestinalis and diet modulate atherogenesis in a murine model. Nat Microbiol 2018; 3: 1461–1471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Zeng X, Gao X, Peng Y, et al. Higher risk of stroke is correlated with increased opportunistic pathogen load and reduced levels of butyrate-producing bacteria in the gut. Front Cell Infect Microbiol 2019; 9: 4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Bultman SJ.Bacterial butyrate prevents atherosclerosis. Nat Microbiol 2018; 3: 1332–1333. [DOI] [PubMed] [Google Scholar]
- 75.Shen MJ, Zipes DP.Role of the autonomic nervous system in modulating cardiac arrhythmias. Circ Res 2014; 114: 1004–1021. [DOI] [PubMed] [Google Scholar]
- 76.Zuo K, Li J, Li K, et al. Disordered gut microbiota and alterations in metabolic patterns are associated with atrial fibrillation. Gigascience 2019; 8: giz058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Svingen GFT, Zuo H, Ueland PM, et al. Increased plasma trimethylamine-N-oxide is associated with incident atrial fibrillation. Int J Cardiol 2018; 267: 100–106. [DOI] [PubMed] [Google Scholar]
- 78.Yu LL, Meng GN, Huang B, et al. A potential relationship between gut microbes and atrial fibrillation: trimethylamine N-oxide, a gut microbe-derived metabolite, facilitates the progression of atrial fibrillation. Int J Cardiol 2018; 255: 92–98. [DOI] [PubMed] [Google Scholar]
- 79.Ihara M, Yamamoto Y.Emerging evidence for pathogenesis of sporadic cerebral small vessel disease. Stroke 2016; 47: 554–560. [DOI] [PubMed] [Google Scholar]
- 80.Jokinen H, Koikkalainen J, Laakso HM, et al. Global burden of small vessel disease-related brain changes on MRI predicts cognitive and functional decline. Stroke 2020; 51: 170–178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Katan M, Moon Y, von Eckardstein A, et al. Procalcitonin and midregional proatrial natriuretic peptide as biomarkers of subclinical cerebrovascular damage: the northern Manhattan Study. Stroke 2017; 48: 604–610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Sumbria RK, Grigoryan MM, Vasilevko V, et al. A murine model of inflammation-induced cerebral microbleeds. J Neuroinflammation 2016; 13: 218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Nakano K, Hokamura K, Taniguchi N, et al. The collagen-binding protein of Streptococcus mutans is involved in haemorrhagic stroke. Nat Commun 2011; 2: 485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Abranches J, Miller JH, Martinez AR, et al. The collagen-binding protein Cnm is required for Streptococcus mutans adherence to and intracellular invasion of human coronary artery endothelial cells. Infect Immun 2011; 79: 2277–2284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Nakano K, Nomura R, Taniguchi N, et al. Molecular characterization of Streptococcus mutans strains containing the Cnm gene encoding a collagen-binding adhesin. Arch Oral Biol 2010; 55: 34–39. [DOI] [PubMed] [Google Scholar]
- 86.Tonomura S, Ihara M, Kawano T, et al. Intracerebral hemorrhage and deep microbleeds associated with cnm-positive Streptococcus mutans; a hospital cohort study. Sci Rep 2016; 6: 20074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Watanabe I, Kuriyama N, Miyatani F, et al. Oral Cnm-positive Streptococcus mutans expressing collagen binding activity is a risk factor for cerebral microbleeds and cognitive impairment. Sci Rep 2016; 6: 38561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Hardy J, Allsop D.Amyloid deposition as the central event in the aetiology of Alzheimer’s diseases. Trends Pharmacol Sci 1991; 12: 383–388. [DOI] [PubMed] [Google Scholar]
- 89.Friedland RP, Chapman MR.The role of microbial amyloid in neurodegeneration. PLoS Pathog 2017; 13: e1006654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Chen SG, Stribinskis V, Rane MJ, et al. Exposure to the functional bacterial amyloid protein curli enhances alpha-synuclein aggregation in aged Fischer 344 rats and Caenorhabditis elegans. Sci Rep 2016; 6: 34477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Venegas C, Kumar S, Franklin BS, et al. Microglia-derived ASC specks cross-seed amyloid-beta in Alzheimer’s disease. Nature 2017; 552: 355–361. [DOI] [PubMed] [Google Scholar]
- 92.Gosztyla ML, Brothers HM, Robinson SR.Alzheimer’s amyloid-beta is an antimicrobial peptide: a review of the evidence. J Alzheimers Dis 2018; 62: 1495–1506. [DOI] [PubMed] [Google Scholar]
- 93.Eimer WA, Vijaya Kumar DK, Navalpur Shanmugam NK, et al. Alzheimer’s disease-associated beta-amyloid is rapidly seeded by herpesviridae to protect against brain infection. Neuron 2018; 99: 56–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Readhead B, Haure-Mirande JV, Funk CC, et al. Multiscale analysis of independent Alzheimer’s cohorts finds disruption of molecular, genetic, and clinical networks by human herpesvirus. Neuron 2018; 99: 64–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Makin S.The amyloid hypothesis on trial. Nature 2018; 559: S4–S7. [DOI] [PubMed] [Google Scholar]
- 96.Cvijanovic N, Feinle-Bisset C, Young RL, et al. Oral and intestinal sweet and fat tasting: impact of receptor polymorphisms and dietary modulation for metabolic disease. Nutr Rev 2015; 73: 318–334. [DOI] [PubMed] [Google Scholar]
- 97.Koutsounas I, Theocharis S, Delladetsima I, et al. Farnesoid x receptor in human metabolism and disease: the interplay between gene polymorphisms, clinical phenotypes and disease susceptibility. Exp Opin Drug Metab Toxicol 2015; 11: 523–532. [DOI] [PubMed] [Google Scholar]
- 98.Kiechl S, Lorenz E, Reindl M, et al. Toll-like receptor 4 polymorphisms and atherogenesis. New Engl J Med 2002; 347: 185–192. [DOI] [PubMed] [Google Scholar]
- 99.Dueker ND, Beecham A, Wang LY, et al. Rare variants in NOD1 associated with carotid bifurcation intima-media thickness in dominican republic families. PLoS One 2016; 11: 15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Hall AB, Tolonen AC, Xavier RJ.Human genetic variation and the gut microbiome in disease. Nat Rev Genet 2017; 18: 690–699. [DOI] [PubMed] [Google Scholar]
- 101.Prestel M, Prell-Schicker C, Webb T, et al. The atherosclerosis risk variant rs2107595 mediates allele-specific transcriptional regulation of HDAC9 via E2F3 and Rb1. Stroke 2019; 50: 2651–2660. [DOI] [PubMed] [Google Scholar]
- 102.Falcone GJ, Woo D.Genetics of spontaneous intracerebral hemorrhage. Stroke 2017; 48: 3420–3424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Ihara M, Tonomura S, Yamamoto Y, et al. Collagen-binding Streptococcus mutans tied to cerebral microbleeds and intracerebral hemorrhage. Future Neurol 2018; 13: 219–224. [Google Scholar]
- 104.Shi Y, Holtzman DM.Interplay between innate immunity and Alzheimer disease: APOE and TREM2 in the spotlight. Nat Rev Immunol 2018; 18: 759–772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Heianza Y, Zheng Y, Ma W, et al. Duration and life-stage of antibiotic use and risk of cardiovascular events in women. Eur Heart J 2019; 40: 3838–3845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Defoirdt T.Quorum-sensing systems as targets for antivirulence therapy. Trends Microbiol 2018; 26: 313–328. [DOI] [PubMed] [Google Scholar]
- 107.Kalia VC, Patel SKS, Kang YC, et al. Quorum sensing inhibitors as antipathogens: biotechnological applications. Biotechnol Adv 2019; 37: 68–90. [DOI] [PubMed] [Google Scholar]
- 108.Cotillard A, Kennedy SP, Kong LC, et al. Dietary intervention impact on gut microbial gene richness. Nature 2013; 500: 585–588. [DOI] [PubMed] [Google Scholar]
- 109.Suez J, Zmora N, Segal E, et al. The pros, cons, and many unknowns of probiotics. Nat Med 2019; 25: 716–729. [DOI] [PubMed] [Google Scholar]
- 110.Durack J, Lynch SV.The gut microbiome: relationships with disease and opportunities for therapy. J Exp Med 2019; 216: 20–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Satokari R.Modulation of gut microbiota for health by current and next-generation probiotics. Nutrients 2019; 11: E1921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Roberts AB, Gu X, Buffa JA, et al. Development of a gut microbe-targeted nonlethal therapeutic to inhibit thrombosis potential. Nat Med 2018; 24: 1407–1417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Chambers ES, Byrne CS, Morrison DJ, et al. Dietary supplementation with inulin-propionate ester or inulin improves insulin sensitivity in adults with overweight and obesity with distinct effects on the gut microbiota, plasma metabolome and systemic inflammatory responses: a randomised cross-over trial. Gut 2019; 68: 1430–1438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Lambertsen KL, Finsen B, Clausen BH.Post-stroke inflammation-target or tool for therapy? Acta Neuropathol 2019; 137: 693–714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Gordon R, Albornoz EA, Christie DC, et al. Inflammasome inhibition prevents alpha-synuclein pathology and dopaminergic neurodegeneration in mice. Sci Transl Med 2018; 10: eaah4066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Tian DC, Shi K, Zhu Z, et al. Fingolimod enhances the efficacy of delayed alteplase administration in acute ischemic stroke by promoting anterograde reperfusion and retrograde collateral flow. Ann Neurol 2018; 84: 717–728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Smith CJ, Hulme S, Vail A, et al. SCIL-STROKE (subcutaneous interleukin-1 receptor antagonist in ischemic stroke): a randomized controlled phase 2 trial. Stroke 2018; 49: 1210–1216. [DOI] [PubMed] [Google Scholar]
- 118.Hadley G, Beard DJ, Couch Y, et al. Rapamycin in ischemic stroke: old drug, new tricks? J Cereb Blood Flow Metab 2019; 39: 20–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Noureldein MH, Eid AA.Gut microbiota and mTOR signaling: insight on a new pathophysiological interaction. Microbial Pathogen 2018; 118: 98–104. [DOI] [PubMed] [Google Scholar]
- 120.Foster TJ.The MSCRAMM family of cell-wall-anchored surface proteins of Gram-positive Cocci. Trends Microbiol 2019; 27: 927–941. [DOI] [PubMed] [Google Scholar]