Abstract
Background
The genetic predisposition to elite athletic performance has been a controversial subject due to the underpowered studies and the small effect size of identified genetic variants. The aims of this study were to investigate the association of common single-nucleotide polymorphisms (SNPs) with endurance athlete status in a large cohort of elite European athletes using GWAS approach, followed by replication studies in Russian and Japanese elite athletes and functional validation using metabolomics analysis.
Results
The association of 476,728 SNPs of Illumina DrugCore Gene chip and endurance athlete status was investigated in 796 European international-level athletes (645 males, 151 females) by comparing allelic frequencies between athletes specialized in sports with high (n = 662) and low/moderate (n = 134) aerobic component. Replication of results was performed by comparing the frequencies of the most significant SNPs between 242 and 168 elite Russian high and low/moderate aerobic athletes, respectively, and between 60 elite Japanese endurance athletes and 406 controls. A meta-analysis has identified rs1052373 (GG homozygotes) in Myosin Binding Protein (MYBPC3; implicated in cardiac hypertrophic myopathy) gene to be associated with endurance athlete status (P = 1.43 × 10−8, odd ratio 2.2). Homozygotes carriers of rs1052373 G allele in Russian athletes had significantly greater VO2max than carriers of the AA + AG (P = 0.005). Subsequent metabolomics analysis revealed several amino acids and lipids associated with rs1052373 G allele (1.82 × 10–05) including the testosterone precursor androstenediol (3beta,17beta) disulfate.
Conclusions
This is the first report of genome-wide significant SNP and related metabolites associated with elite athlete status. Further investigations of the functional relevance of the identified SNPs and metabolites in relation to enhanced athletic performance are warranted.
Keywords: GWAS, SNP, metabolomics, metabolites, elite athletes, endurance
Background
Elite athletic performance is a multi-factorial trait with input from both genetic and environmental factors. The superior performance of elite athletes has been historically considered an outcome of a special talent shaped by intensive training. The talent is now believed to be a product of additive genetic components predisposing the athlete to endurance, speed, strength, flexibility and coordination trainability under the control of strong environmental cues including exercise and nutrition. In this model, the genetic predisposition together with ability to respond to training are the keys to the superior physical performance of elite athletes (Georgiades et al., 2017).
Sports can be classified according to the type and intensity of the exercise required to perform during competition. The percentage of maximal oxygen uptake (VO2max) is a detrimental factor in the categorization of endurance sports, as it reflects the maximal cardiac output, the oxygen transport capacity, and the blood volume (Bergh et al., 2000). Accordingly, sports can be divided into sport events with low, moderate and high aerobic (dynamic) component (Mitchell et al., 2005). Similarly, the percent of maximal voluntary contraction (MVC), which reflects the greatest amount of tension a muscle can generate and hold, is used to classify sports into sporting disciplines with low, moderate and high power component (Mitchell et al., 2005).
Classical twin and family genetic studies have suggested that VO2max is up to 94% inherited (Bouchard et al., 1998; Peeters et al., 2009). Genome-wide association studies (GWAS) in athletes versus non-athletes have uncovered many new loci in association with VO2max (Rankinen et al., 2010; Bouchard et al., 2011) and elite endurance performance (Ahmetov et al., 2015). A more recent review of genetic predisposition to elite athletic endurance has highlighted 100 endurance variants (Semenova et al., 2019). However, despite some initial evidence suggesting identification of genetic variants in GWAS studies, further studies did not replicate/validate these findings hindered by a small sample size and complex phenotype (Pitsiladis et al., 2016). One of the first GWAS in athletes using 143 K single-nucleotide polymorphisms (SNPs) and subsequent meta-analysis of 45 promising genetic markers in 1,520 endurance athletes and 2,760 controls has revealed only one statistically significant marker (rs558129 at GALNTL6) associated with endurance status in world class athletes, but not at genome wide level of significance (Rankinen et al., 2016). Therefore, the genetic predisposition to endurance traits remains unclear, largely due to the relatively underpowered elite athletes’ cohorts. Recently, a polymorphism in human homeostatic iron regulator protein was found to be associated with elite endurance athlete status and aerobic capacity in Russian athletes (Semenova et al., 2020).
Metabolomics analysis has presented a novel tool to validate genomics data by providing an intermediate phenotype (metabolites) in association with the identified genetic variants (Kastenmuller et al., 2015; Tanaka et al., 2016). Pilot metabolomics studies have revealed differences in the metabolic signature of moderate and high endurance elite athletes, such as steroid biosynthesis, fatty acid metabolism, oxidative stress and energy-related molecular pathways (Al-Khelaifi et al., 2018, 2019a). Recently, a study investigating metabolic GWAS of elite athletes showed novel genetically influenced metabolites associated with athletic performance. These included two novel genetic loci in FOLH1 and VNN1 in association with N-acetyl-aspartyl-glutamate and linoleoyl ethanolamide, respectively, and one novel locus linking genetic variant in SULT2A1 and androstenediol (3alpha, 17alpha) monosulfate in endurance athletes (Al-Khelaifi et al., 2019b).
In this study, we aimed to investigate the association of multiple SNPs and endurance athlete status in a relatively large cohort of European elite athletes specialized in sports with high and low/moderate aerobic component using GWAS approach and replicate our findings in elite Russian and Japanese athletes. We also aimed to perform functional validation using VO2max testing and metabolomics analysis by identifying metabolites that are associated with significant endurance-related SNPs.
Results
Genome-Wide Association Study
Athletes from the discovery cohort were classified into different groups of sports following previously published sports classification criteria (Mitchell et al., 2005), as shown in Table 1.
TABLE 1.
Classification of GWAS participants according to sports classes.
| Low/moderate (<70% VO2max) | High (>70% VO2max) | Total | ||||
| High (>50% MVC) | Wrestling and Judo (8M) | Skate boarding (2M) | Modern Pentathlon (1F) | 287 | ||
| Kayaking (1F) | Rowing (9M/8F) | Biathlon (2M/1F) | ||||
| Weightlifting (14M/7F) | Boxing (4M/7F) | Cycling (157M/49F) | Triathlon (8M/9F) | |||
| Moderate | Jumping (athletics) (1F) | Handball (19M/3F) | Skiing Cross Country (3M/1F) | Basketball (3M) | 165 | |
| (20–50% MVC) | Rugby (15M) | Aquatics (3M/2F) | ||||
| Athletics other (41M/26F) | Sprint (2M) | Hockey (4M/1F) | Swimming (25M/16F) | |||
| Low (<20% MVC) | Baseball (2M) | Long-Distance running and marathon (37M/12F) | Tennis (3M/3F) | 344 | ||
| Volleyball (2M) | ||||||
| Table tennis (9M) | Soccer (256M/1F) | Ultra-running (1F) | Football (17M/1F) | |||
| 134 | 662 | 796 | ||||
Distribution of elite athletes in various categories based on sport type-associated peak dynamic (maximal oxygen uptake percentage; VO2max) and peak static (maximal voluntary muscle contraction percentage; MVC) components achieved during competition as described previously (Mitchell et al., 2005).
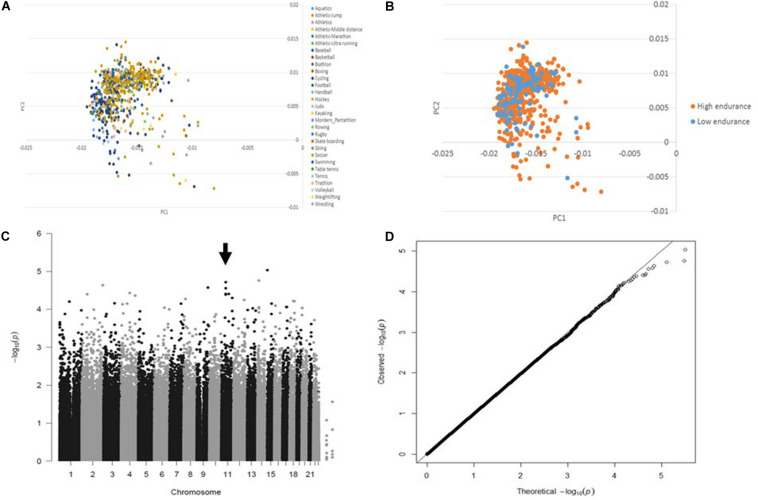
The principle component analysis (PCA) of the genotyping data revealed no influence of sport disciplines (Figure 1A) or training modality (i.e., sports with low/moderate versus high aerobic component) (Figure 1B) on genotype distribution. Following quality control data processing, genotyping of 341385 SNPs in 796 European elite athletes revealed several variants associated with endurance athlete status, but none reached GWAS level of significance. Table 2 shows top SNPs (P < 5 × 10−5) with their odd ratios (OR) in relation to elite athletic endurance, location according to function genome variation server (GVS), gene name and minor allele frequency (MAF) in sports with high and low/moderate aerobic component. MAF in non-elite athletes from 1,000 genome project were used as a reference. Figure 1 shows Manhattan (C) and quartile-quartile (QQ) plots (D) of GWAS hits associated with endurance.
FIGURE 1.
GWAS data quality control. PCA shows no difference in the genotype distribution among sport disciplines (A) or between groups (sports with low/moderate versus high aerobic component) (B) Manhattan (arrow indicates significant SNPs) (C) and Quantile-quantile (no evidence of genomic inflation, lambda GC = 1.006) (D) plots illustrating GWAS results in association with endurance.
TABLE 2.
Top GWAS SNPs associated with endurance athlete status from the discovery study.
| rsID | Chromosome | Position | Reference base | N | OR | Standard error | P value | Function GVS | Gene list | MAF-high aerobic N = 662 | MAF- moderate/low aerobic N = 134 | MAF-non-athletes |
| rs8029108 | 15 | 22945314 | C | 795 | 0.5293 | 0.1435 | 9.23 × 10–6 | intron | CYFIP1 | 0.4448 | 0.403 | G = 0.36 |
| kgp5680198 | 14 | 34627202 | C | 792 | 0.5151 | 0.1545 | 1.75 × 10–5 | intergenic | LOC102724945 | 0.2135 | 0.3246 | C = 0.27 |
| rs10838681 | 11 | 47275064 | A | 794 | 0.5208 | 0.1526 | 1.92 × 10–5 | intron | NR1H3 | 0.233 | 0.3496 | A = 0.35 |
| kgp2861067 | 2 | 234653039 | T | 795 | 0.2227 | 0.3551 | 2.34 × 10–5 | intron | UGT1A10 | 0.01815 | 0.0597 | T = 0.013 |
| kgp11512684 | 9 | 123798492 | A | 793 | 0.202 | 0.3808 | 2.66 × 10–5 | intron | C5 | 0.01364 | 0.04887 | A = 0.016 |
| rs1052373 | 11 | 47354787 | A | 796 | 0.5393 | 0.1475 | 2.81 × 10–5 | missense | MYBPC3 | 0.2764 | 0.3955 | T = 0.39 |
| rs17020631 | 4 | 94380515 | G | 795 | 0.3064 | 0.2866 | 3.68 × 10–5 | intron | GRID2 | 0.0287 | 0.09398 | G = 0.09 |
| rs1949886 | 11 | 80311066 | A | 796 | 4.346 | 0.3573 | 3.92 × 10–5 | intergenic | none | 0.1329 | 0.03731 | A = 0.15 |
| rs7120118 | 11 | 47286290 | C | 796 | 0.5455 | 0.1475 | 3.97 × 10–5 | intron | NR1H3 | 0.2696 | 0.3881 | C = 0.38 |
Replication of Endurance SNPs in Russian and Japanese Elite Athlete Cohorts
Replication of results was performed by comparing the frequencies of the most significant SNPs (P < 10−5) in 242 elite Russian high and 168 low/moderate aerobic athletes, and in 60 elite Japanese endurance athletes and 406 controls. Out of the 9 top SNPs identified form the GWAS discovery stage, the rs1052373 (MYBPC3) and rs7120118 (NR1H3) showed significant association with endurance in Russian and Japanese (P < 0.05). However, the association was driven by a dominant model since results of this analysis showed over representation for rs1052373 GG and rs7120118 TT genotypes in the high endurance group. A subsequent meta-analysis has confirmed the over representation of the rs1052373 GG and rs7120118 TT genotypes in high endurance sports at genome-wide and Bonferroni levels of significance (1.43 × 10–8 and 1.66 × 10–7, respectively) (Table 3). The combined analysis showed no evidence of heterogeneity and direction of association was similar in all three cohorts.
TABLE 3.
SNPs associated with Endurance athlete status from the discovery, replication and meta-analysis.
| Chr | SNP | RG | GWAS |
Russian |
Japanese |
Combined |
||||||
| P | OR (95% CI) | P | OR (95% CI) | P | OR (95% CI) | P | OR (95% CI) | I2 | Phet | |||
| 11 | rs1052373 | GG | 5.48 × 10–6 | 2.61 (1.72–3.94) | 0.012 | 1.67 (1.12–2.49) | 0.0027 | 2.92 (1.41–6.05) | 1.43 × 10–8 | 2.17 (1.67–2.84) | 35 | 0.2 |
| 11 | rs7120118 | TT | 1.26 × 10–5 | 2.49 (1.65–3.75) | 0.016 | 1.64 (1.10–2.45) | 0.0352 | 2.48 (1.10–5.56) | 1.66 × 10–7 | 2.07 (1.59–2.70) | 12 | 0.3 |
OR, odds ratio for the risk genotype; CI, confidence interval; I2, heterogeneity statistics; Phet, P value for heterogeneity.
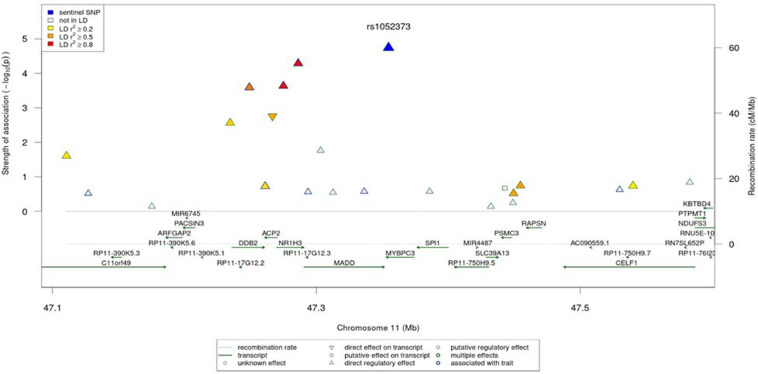
The regional association plot for the rs1052373 G allele in MYBPC3 gene revealed a number of SNPs in the same LD block in association with high endurance including the rs7120118 T allele in NR1H3 gene (Figure 2).
FIGURE 2.
Regional association plot for the region around rs1052373. The colors correspond to different LD thresholds, where LD is computed between the sentinel SNP (lowest P-value, colored in blue) and all SNPs. Shapes of markers correspond to their functionality as described in the legend.
To validate the potential functionality of the identified GWAS SNPs, association of the identified two SNPs (rs1052373 G and rs7120118 T alleles) with VO2max was investigated in a subgroup of the Russian replication cohort in which VO2max data was available. This included 32 elite Russian long-distance athletes [19 biathletes, 13 cross-country skiers; 17 females, age 23.5 (3.5) years; 15 males, age 21.3 (4.1) years]. The rs1052373 GG carriers had significantly greater VO2max than carriers of the AA + AG (P = 0.005 adjusted for sex). Similarly, rs7120118 TT carriers showed a trend of higher VO2max than carriers of the CC + CT (P = 0.053 adjusted for sex).
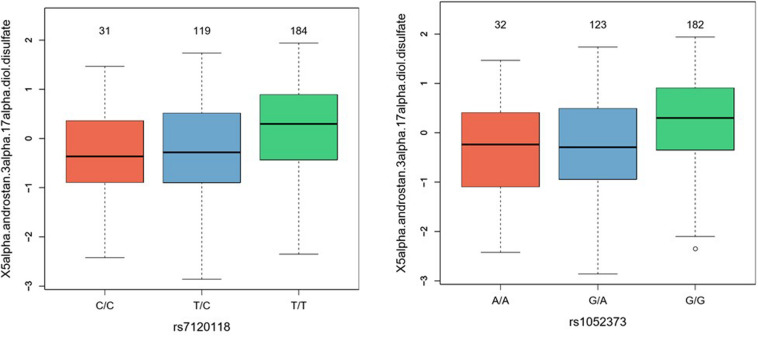
For further validation of the potential functionality of the identified GWAS SNPs, metabolomics of 750 metabolites was carried out in a subset of the discovery cohort (n = 490) and enriched metabolic pathways associated with the rs1052373 G allele and rs7120118 T alleles were determined (Table 4). Among the metabolic pathways associated with rs56330321 and rs7120118, various lipids and amino acids were significantly altered by their genotypes. However, only 5alpha-androstan-3alpha,17alpha-diol disulfate reached Bonferroni level of significance (Table 4), exhibiting higher levels in rs1052373 GG and rs7120118 TT carriers compared to AA + AG and CC + TC carriers, respectively (Figure 3).
TABLE 4.
Metabolites that belong to the significantly enriched phospholipids pathway Top metabolites associated with significant SNPs.
| SNP | Beta | SE.Beta | P | Metabolites | SUPER_PATHWAY | SUB_PATHWAY |
| rs1052373 | –0.36 | 0.08 | 1.82 × 10–5 | 5alpha-androstan-3alpha,17alpha-diol disulfate | Lipid | Androgenic steroids |
| –0.25 | 0.07 | 0.000248 | 2-hydroxy-3-methylvalerate | Amino Acid | Leucine, Isoleucine and Valine Metabolism | |
| –0.23 | 0.07 | 0.000879 | alpha-hydroxyisovalerate | Amino Acid | Leucine, Isoleucine and Valine Metabolism | |
| 0.31 | 0.09 | 0.000928 | xylose | Carbohydrate | Pentose Metabolism | |
| –0.23 | 0.07 | 0.001226 | N1-methylinosine | Nucleotide | Purine Metabolism, (Hypo)Xanthine/Inosine containing | |
| –0.23 | 0.07 | 0.001315 | palmitoleoylcarnitine (C16:1)* | Lipid | Fatty Acid Metabolism(Acyl Carnitine) | |
| –0.23 | 0.07 | 0.001509 | 2-hydroxyadipate | Lipid | Fatty Acid, Dicarboxylate | |
| –0.22 | 0.07 | 0.001516 | 2-methylcitrate/homocitrate | Energy | TCA Cycle | |
| –0.21 | 0.07 | 0.001933 | myristoleoylcarnitine (C14:1)* | Lipid | Fatty Acid Metabolism(Acyl Carnitine) | |
| rs7120118 | –0.33 | 0.08 | 5.17 × 10–5 | 5alpha-androstan-3alpha,17alpha-diol disulfate | Lipid | Androgenic Steroids |
| –0.27 | 0.07 | 0.000136 | 2-hydroxy-3-methylvalerate | Amino Acid | Leucine, Isoleucine and Valine Metabolism | |
| –0.24 | 0.07 | 0.000582 | alpha-hydroxyisovalerate | Amino Acid | Leucine, Isoleucine and Valine Metabolism | |
| –0.24 | 0.07 | 0.000715 | N1-methylinosine | Nucleotide | Purine Metabolism, (Hypo)Xanthine/Inosine containing | |
| 0.31 | 0.09 | 0.001004 | xylose | Carbohydrate | Pentose Metabolism | |
| –0.23 | 0.07 | 0.001527 | 2-hydroxyadipate | Lipid | Fatty Acid, Dicarboxylate | |
| 0.28 | 0.09 | 0.001966 | 5-acetylamino-6-formylamino-3-methyluracil | Xenobiotics | Xanthine Metabolism | |
| –0.22 | 0.07 | 0.002116 | alpha-hydroxyisocaproate | Amino Acid | Leucine, Isoleucine and Valine Metabolism | |
| –0.22 | 0.07 | 0.002216 | 2-methylcitrate/homocitrate | Energy | TCA Cycle | |
| –0.22 | 0.07 | 0.002266 | glycerol | Lipid | Glycerolipid Metabolism |
FIGURE 3.
Boxplots representing levels of 5alpha-androstan-3alpha,17alpha-diol disulfate in rs7120118 and rs1052373 genotype groups.
Discussion
Genetic predisposition into cardiorespiratory fitness and response to exercise training has been previously described (Lortie et al., 1982; Prud’homme et al., 1984; Hamel et al., 1986; Bouchard et al., 1994, 1998, 1999). Since endurance performance sports are characterized by increased cardiorespiratory capacity, genetic predisposition into elite endurance performance is also expected to be genetically influenced (Guth and Roth, 2013). However, genetic studies of elite athletic endurance showed inconsistent results (Guth and Roth, 2013; Ahmetov and Fedotovskaya, 2015; Pitsiladis et al., 2016; Wang et al., 2016). The aims of this study were to carry out the largest GWAS study of elite European athletes to date using a unique SNP microarray that is enriched with genes involved in different metabolic pathways with direct influence on various physiological pathways characteristic of elite athletes. GWAS results have revealed a number of novel SNPs associated with endurance but none reached the GWAS level of significance. Replication of the top identified SNP associations in two independent cohorts of elite athletes from Russia and Japan has confirmed the association of rs7120118 and rs1052373 with endurance athlete status. Subsequent meta-analysis of the three cohorts has revealed for the first time that both SNPs were associated with endurance athlete status at genome-wide and Bonferroni level of significance, respectively. Functional validation has revealed the association of the two SNPs with increased Vo2max and levels of the testosterone precursor 5alpha-androstan-3alpha,17alpha-diol disulfate.
The top identified GWAS significant SNP (rs1052373) is located within MYBPC3 gene. MYBPC3 codes for a myosin-associated protein expressed in the cross-bridge-bearing zone (C region) of A bands in striated muscle. The phosphorylation of MYBPC3 protein modulates cardiac contraction (Moss et al., 2015). Mutations in MYBPC3 were previously associated with a lower super-relaxed state in patients with hypertrophic cardiomyopathy (HCM) (McNamara et al., 2017). Intense exercise can trigger heart remodeling to compensate for the elevations in blood pressure or volume by increasing muscle mass. Hence, hearts of the endurance athletes typically exhibit an eccentric cardiac hypertrophy with increased cavity dimension and wall thickness (Pelliccia et al., 1991; Hedman et al., 2015), which is influenced by the type of sport performed (Pelliccia, 1996; Pelliccia et al., 1999; Maron and Pelliccia, 2006). As a result, the endurance-trained heart can deliver a large maximal systolic volume (35% larger than untrained heart) in order to produce a large cardiac output (Ogawa et al., 1992; Pelliccia et al., 1999). Since carriers of the GG allele exhibit a benign phenotype of HCM according to NIH’s ClinVar database (Landrum et al., 2018), the mild phenotype may be enhancing exercise-triggered physiological adaptations. The seemingly dominant effect of rs1052373 GG on increased VO2max and endurance may support this added advantage although more studies are needed to confirm this finding. These adaptations, however, might be associated with a greater risk of cardiovascular disease. Indeed, we have recently shown that endurance athletes with high cardiovascular demand (higher blood pressure and stroke volume) show metabolic signature consistent with higher risk of cardiovascular disease (Al-Khelaifi et al., 2019a). When investigating the expression quantitative trait loci (eQTLs) associated with rs1052373, a number of genes was identified including SPI1, MYBPC3, MADD, ACP2 and NR1H3 (Ray et al., 1990; Tang and Chu, 2002; Mannan et al., 2004; Wu et al., 2012; Carrier et al., 2015; Theofilopoulos and Arenas, 2015). Interestingly, eQTL (GTEx) showed that rs1052373 polymorphism is associated with expression level of MADD and ACP2 in heart, but not MYBPC3. Since MAP kinase plays an important role of cardiac hypertrophy (Zhang et al., 2003), the association between rs1052373 polymorphism and VO2max and endurance may also be explained by MADD expression, although this needs further validatoin. Information related to function and associated diseases with these genes are summarized in Supplementary Table S1.
The other significant association was between rs7120118 TT carriers and high endurance. Rs7120118 is located in NR1H3 gene that codes for a nuclear receptor regulating macrophage function, lipid homeostasis and inflammation. NR1H3, also known as liver X Receptor Alpha (LXRA), plays an important role in the regulation of cholesterol homeostasis including adrenal steroidogenesis (Repa et al., 2002; Cummins et al., 2006). The association of rs7120118 with high endurance could be reflecting the high linkage disequilibrium (r2 = 0.89, P < 0.0001) between rs7120118 TT and the potentially functional rs1052373 GG. It could, however, be related to increased synthesis of the testosterone precursor 5alpha-androstan-3alpha,17alpha-diol disulfate since NR1H3 regulates hypothalamo-pituitary–adrenal steroidogenesis (Handa et al., 2011). Indeed, we have previously shown that high-endurance athletes exhibit elevated levels of several sex hormone steroids involved in testosterone synthesis including 5alpha-androstan-3alpha,17alpha-diol disulfate (Al-Khelaifi et al., 2018) with implication on improving performance due to enhanced glucose metabolism and protein synthesis in the muscle (Sato et al., 2008). The functional relevance of these associations remains to be further validated.
Study limitations: The lack of information about participants and the heterogeneity of their sport groups were major limitations of this study. To overcome these limitations and to increase the power of the study, genotyping was compared between athletes who belong to high endurance versus moderate endurance performance sports instead of power versus endurance due to the overlap between the two classes as per Mitchell’s categorization (Mitchell et al., 2005). Other limitations included using add-on replication studies (Russian and Japanese cohorts) rather than using a carefully designed replication. However, differences were confirmed in each study separately and the subsequent meta-analysis confirmed the significance of the association of the two SNPs with endurance.
Conclusion
This study reports the first GWAS significant SNP (rs1052373) in MYBPC3 in association with endurance athlete status with a direct relevance to cardiac hypertrophy and contraction. The SNP is associated with increased VO2max and elevated levels of the testosterone precursor androstenediol (3beta,17beta) disulfate, both phenotypes that potentially contribute to the superior performance of endurance athletes. This study also identifies a second SNP (rs7120118) associated with endurance at Bonferroni level of significance in NR1H3. This SNP could be either working independently of rs1052373 through influencing steroidogenesis or could be acting as a marker of rs1052373. Further investigations of the functional relevance of the identified SNPs and associated metabolites in relation to enhanced athletic performance are warranted.
Methods
The aim of this study is to investigate the genetic predisposition to elite athletic endurance through conducting the largest GWAS in elite athletes to date, followed by functional validation through aerobic capacity testing and metabolomics analysis to shed light on the underlying mechanisms of genetic associations.
Participants
Discovery Study
Seven hundred and ninety six consented European international-level athletes (645 males, 151 females) from different sports disciplines who participated in national or international sports events and tested negative for doping substances at anti-doping laboratories in Qatar (ADLQ) and Italy (FMSI) were included in this study. No other information of participants was available due to the strict anonymization process undertaken by the anti-doping laboratories. This study was performed in line with the World Medical Association Declaration of Helsinki – Ethical Principles for Medical Research Involving Human Subjects. All protocols were approved by the Institutional Research Board of ADLQ (F2014000009). Athletes were dichotomized into groups with different aerobic (dynamic) and power (static) components (Table 1) based on their sport types as described previously (Mitchell et al., 2005). Table 1 further lists the number of participants based on various analyses as per sport type in each class/group and their genders.
Replication Studies
The first replication study involved 410 Russian athletes [187 females, age 25.3 (4.1) years, 223 males, age 25.7 (4.3) years]. Athletes were dichotomized into two groups with different aerobic (dynamic) and power (static) components based on their sport types. Group 1 (242 athletes with high aerobic component) included biathletes (n = 19), cross-country skiers (n = 16), 800–10,000 m runners (n = 9), rowers (n = 9), kayakers (n = 30), canoers (n = 8), speed skaters (n = 12), short-trackers (n = 3), swimmers (n = 38), cyclists (n = 5), race walkers (n = 6), boxers (n = 43), badminton players (n = 11), basketball players (n = 6), water polo players (n = 12), football players (n = 9), and ice hockey players (n = 6). Group 2 (168 athletes with low aerobic component) included 100–400 m runners (n = 8), wrestlers (n = 44), alpine skiers (n = 2), sailors (n = 2), synchronized swimmer (n = 1), taekwondo athletes (n = 5), baseball players (n = 10), volleyball players (n = 19), table tennis players (n = 5), softball players (n = 5), rhythmic gymnasts (n = 7), chess players (n = 5), throwers (n = 6), athletics jumpers (n = 16), ski jumpers (n = 2), weightlifters (n = 25), ure skaters (n = 6). All athletes were Olympic team members (International level; all Caucasians of Eastern European descent) who have tested negative for doping substances. The Russian study was approved by the Ethics Committee of the Federal Research and Clinical Center of Physical-chemical Medicine of the Federal Medical and Biological Agency of Russia. Written informed consent was obtained from each participant. The study complied with the guidelines set out in the Declaration of Helsinki and ethical standards in sport and exercise science research. The experimental procedures were conducted in accordance with the set of guiding principles for reporting the results of genetic association studies defined by the STrengthening the REporting of Genetic Association studies (STREGA) Statement.
The second replication study involved endurance athletes (n = 60) and controls (n = 406) from Japan. All endurance athletes were track and field competitors who participated in endurance events from 800 m to marathon. In addition, all athletes were international athletes who had competed at major international competitions. All controls were healthy Japanese individuals. All subjects gave written informed consent before their inclusion in the study. The study protocols were approved by the ethics committee of the Juntendo University and was conducted according to the Declaration of Helsinki.
Aerobic Capacity Testing
VO2max in biathletes and cross-country skiers was determined using an incremental test to exhaustion on a treadmill HP Cosmos (Germany). The initial speed was 7 km/h, the increment was 0.1 km/h every 10 s. O2max was determined breath by breath using a MetaMax 3B-R2 gas analysis system. O2max was recorded as the highest mean value observed over a 30 s period.
Genotyping
Discovery Study
DNA was extracted from leukocytes (venous blood) samples from all participants using DNeasy Blood & Tissue kit (Qiagen) following manufacturer’s instructions. The concentration and the quality of DNA were assessed using the Nanodrop (Thermo Fisher) and Qubit Fluorometer (Invitrogen) to ensure sufficient amount and quality of DNA were obtained for genotyping. Illumina Drug Core array-24 BeadChips was chosen for the genotyping of 476,728 SNPs in the 796 European elite athletes collected for Anti-Doping analysis (discovery cohort). This array contains over 240,000 highly-informative genome-wide tag SNPs and a novel ∼200,000 custom marker set designed to support studies of drug target validation and treatment response. The assay required 200 ng of DNA sample as input with a concentration of at least 50 ng/μl. All further procedures were performed according to the instructions of Infinium HD Assay according to manufacturer’s instructions. Briefly, 4 μl of obtained DNA was mixed with Illumina amplification reagents and incubated overnight at 37oC in hybridization oven. On the second day, enzymatic reagents were used to fragment the amplified DNA then precipitated by centrifugation. Subsequently, re-suspended pellet was loaded in the beadchip then incubated overnight at 48oC in hybridization oven. On third day, beadchips underwent enzymatic base extension and fluorescent staining. Lastly, after coating, the beadchips were imaged using iScan.
Replication Studies
Molecular genetic analysis in Russian cohorts was performed with DNA samples obtained from leukocytes (venous blood). Four ml of venous blood were collected in tubes containing EDTA (Vacuette EDTA tubes, Greiner Bio-One, Austria). Blood samples were transported to the laboratory at 4°C and DNA was extracted on the same day. DNA extraction and purification were performed using a commercial kit according to the manufacturer’s instructions (Technoclon, Russia) and included chemical lysis, selective DNA binding on silica spin columns and ethanol washing. Extracted DNA quality was assessed by agarose gel electrophoresis at this step. HumanOmni1-Quad BeadChips (Illumina Inc, United States) were used for genotyping of 1,140,419 SNPs in athletes and controls. The assay required 200 ng of DNA sample as input with a concentration of at least 50 ng/μl. Exact concentrations of DNA in each sample were measured using a Qubit Fluorometer (Invitrogen, United States). All further procedures were performed according to the instructions of Infinium HD Assay. For the second replication study, total DNA was isolated from saliva or venous blood using Oragene⋅DNA Collection Kits (DNA genotek, Ontario, Canada) or QIAamp DNA blood Maxi Kit (QIAGEN, Hilden, Germany), respectively. The total DNA content was measured using a NanoDrop 8000 spectrophotometer (Thermo Fisher Scientific, MA, United States). Subsequently, DNA samples were adjusted to a concentration of 50 ng/μL with TE buffer and were stored at 4°C. Total DNA samples were genotyped for more than 700,000 markers using the Illumina® HumanOmniExpress Beadchip.
Data Extraction and SNP Identification
Raw data was extracted, peak-identified and QC processed using Illumina iScan hardware and software. These systems are built on a web-service platform utilizing Microsoft’s NET technologies, which run on high-performance application servers and fiber-channel storage arrays in clusters to provide active failover and load-balancing.
Metabolomics
Screening of serum metabolites was performed in 490 elite athletes (Supplementary Table S2) using protocols established at Metabolon, Durham, NC, United States. The platform utilizes Waters ACQUITY ultra-performance liquid chromatography (UPLC) and a Thermo Scientific Q-Exactive high resolution/accurate mass spectrometer interfaced with a heated electrospray ionization (HESI-II) source and Orbitrap mass analyzer operated at 35,000 mass resolution. Detailed protocol and QC measures were previously published (Evans et al., 2009; Al-Khelaifi et al., 2018).
Statistical Analysis
Following genotyping using Illumina’s Drug Core SNP array, analysis was performed using Plink v1.9. Quality control measures were applied to the genotype data set to exclude samples with low genotype call rate or excess heterozygosity. Accordingly, SNPs with a genotype call rate <98%, minor allele frequency <1%, or deviating from Hardy-Weinberg equilibrium (P < 10–6) were excluded. After filtering the data with the above criteria, 341,385 SNPs were used in analysis. Population background was determined using principal component analysis (PCA) in comparision to samples from HapMap project and only samples with European ancestry were included in the analysis. The analysis in European and Russian cohorts was performed using linear or logistic regression models. A model incorporating sports grouped by training modalities (i.e., sports with high versus low/moderate aerobic component) was used for the discovery cohort after incorporating gender and PCA components 1, 2, 3 & 4 as covariates in the model. A stringent Bonferroni level of significance of P ≤ 0.05/341385 = 1.46 × 10–7 was used to define significant associations. To perform the meta-analysis, the Cochrane Review Manager version 5.3 was used. Random and fixed effect models were applied. The heterogeneity degree between the studies was assessed with the I2 statistics. Associations between SNPs and metabolite levels were computed using lm function in R (version 3.3.1) while correcting for gender, hemolysis and PCA. An additive inheritance model was used (SNPs were coded as 0,1,2 according to their genotype group. Pathway enrichment analyses were carried out using Chi square tests to identify pathways with enriched metabolites ranked by P-value from the linear model since Bonferroni level of significance was not observed.
Data Availability Statement
The SNP data supporting this study is available at: https://figshare.com/articles/GWAS_elite_endurance_athletes/12199760. Summary statistics will be made available through the NHGRI-EBI GWAS Catalog: https://www.ebi.ac.uk/gwas/downloads/summary-statistics.
Ethics Statement
This study was performed in accordance with the World Medical Association Declaration of Helsinki. All protocols were approved by the Institutional Research Board of anti-doping lab Qatar (F2014000009). The patients/participants provided their written informed consent to participate in this study.
Author Contributions
All authors contributed to sample collection, analysis, manuscript writing, and manuscript review and acceptance of final version. ME is responsible for the integrity of the work as a whole.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
Authors would like to thank Qatar National Research Fund (QNRF) for funding this project. Grant number NPRP7-272-1-041 (ME, KS, CG, and FB). An earlier version of this manuscript has been released as a pre-print at [ResearchSqure], (Fatima et al., 2019).
Abbreviations
- ACP2, acid phosphatase 2
Lysosomal
- ADLQ
anti-doping laboratories in Qatar
- FDR
false discovery rate
- FMSI, Laboratorio Antidoping
Federazione Medico Sportiva Italiana
- GVS
genome variation server
- GWAS
genome-wide association studies
- HESI-II
high resolution/accurate mass spectrometer interfaced with a heated electrospray ionization
- MADD
MAP kinase activating death domain
- MAF
minor allele frequency
- MVC
maximal voluntary contraction
- MYBPC3, myosin binding protein C
cardiac
- NR1H3
nuclear receptor subfamily 1 group H member 3
- OR
odds ratio
- Spi-1
Spi-1 proto-oncogene
- UPLC
ultra-performance liquid chromatography
- VO2max
maximal oxygen uptake.
Footnotes
Funding. This study was funded by Qatar National Research Fund (QNRF), Grant number NPRP7-272-1-041 (ME, KS, CG, and FB). The funding body had no role in the design of the study and collection, analysis, and interpretation of data and in writing the manuscript.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fgene.2020.00595/full#supplementary-material
References
- Al-Khelaifi F., Diboun I., Donati F., Botre F., Abraham D., Hingorani A., et al. (2019b). Metabolic GWAS of elite athletes reveals novel genetically-influenced metabolites associated with athletic performance. Sci. Rep. 9:19889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Al-Khelaifi F., Diboun I., Donati F., Botre F., Alsayrafi M., Georgakopoulos C., et al. (2018). A pilot study comparing the metabolic profiles of elite-level athletes from different sporting disciplines. Sports Med. Open 4:2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Al-Khelaifi F., Donati F., Botre F., Latiff A., Abraham D., Hingorani A., et al. (2019a). Metabolic profiling of elite athletes with different cardiovascular demand. Scand. J. Med. Sci. Sports 29 933–943. [DOI] [PubMed] [Google Scholar]
- Ahmetov I. I., Fedotovskaya O. N. (2015). Current progress in sports genomics. Adv. Clin. Chem. 70 247–314. 10.1016/bs.acc.2015.03.003 [DOI] [PubMed] [Google Scholar]
- Ahmetov I., Kulemin N., Popov D., Naumov V., Akimov E., Bravy Y., et al. (2015). Genome-wide association study identifies three novel genetic markers associated with elite endurance performance. Biol. Sport 32 3–9. 10.5604/20831862.1124568 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bergh U., Ekblom B., Astrand P. O. (2000). Maximal oxygen uptake “classical” versus “contemporary” viewpoints. Med. Sci. Sports Exerc. 32 85–88. [DOI] [PubMed] [Google Scholar]
- Bouchard C., Tremblay A., Despres J. P., Theriault G., Nadeau A., Lupien P. J., et al. (1994). The response to exercise with constant energy intake in identical twins. Obes Res. 2 400–410. 10.1002/j.1550-8528.1994.tb00087.x [DOI] [PubMed] [Google Scholar]
- Bouchard C., Daw E. W., Rice T., Perusse L., Gagnon J., Province M. A., et al. (1998). Familial resemblance for VO2max in the sedentary state: the HERITAGE family study. Med. Sci. Sports Exerc. 30 252–258. 10.1097/00005768-199802000-00013 [DOI] [PubMed] [Google Scholar]
- Bouchard C., An P., Rice T., Skinner J. S., Wilmore J. H., Gagnon J., et al. (1999). Familial aggregation of VO(2max) response to exercise training: results from the HERITAGE Family Study. J. Appl. Physiol. 87 1003–1008. 10.1152/jappl.1999.87.3.1003 [DOI] [PubMed] [Google Scholar]
- Bouchard C., Sarzynski M. A., Rice T. K., Kraus W. E., Church T. S., Sung Y. J., et al. (2011). Genomic predictors of the maximal O(2) uptake response to standardized exercise training programs. J. Appl. Physiol. 110 1160–1170. 10.1152/japplphysiol.00973.2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carrier L., Mearini G., Stathopoulou K., Cuello F. (2015). Cardiac myosin-binding protein C (MYBPC3) in cardiac pathophysiology. Gene. 573 188–197. 10.1016/j.gene.2015.09.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cummins C. L., Volle D. H., Zhang Y., McDonald J. G., Sion B., Lefrancois-Martinez A. M., et al. (2006). Liver X receptors regulate adrenal cholesterol balance. J. Clin. Investigat. 116 1902–1912. 10.1172/jci28400 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evans A. M., DeHaven C. D., Barrett T., Mitchell M., Milgram E. (2009). Integrated, nontargeted ultrahigh performance liquid chromatography/electrospray ionization tandem mass spectrometry platform for the identification and relative quantification of the small-molecule complement of biological systems. Anal. Chem. 81 6656–6667. 10.1021/ac901536h [DOI] [PubMed] [Google Scholar]
- Fatima A.-K., Yousri N. A., Albagha O., Semenova E. A., Kostryukova E. S., Kulemin N. A., et al. (2019). Genome-wide association study reveals novel genetic markers associated with endurance athlete status. Res. Squre. 10.21203/rs.2.14107/v1 [DOI] [Google Scholar]
- Georgiades E., Klissouras V., Baulch J., Wang G., Pitsiladis Y. (2017). Why nature prevails over nurture in the making of the elite athlete. BMC Genomics 18(Suppl. 8):835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guth L. M., Roth S. M. (2013). Genetic influence on athletic performance. Curr. Opin. Pediatr. 25 653–658. 10.1097/mop.0b013e3283659087 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamel P., Simoneau J. A., Lortie G., Boulay M. R., Bouchard C. (1986). Heredity and muscle adaptation to endurance training. Med. Sci. Sports Exerc. 18 690–696. [PubMed] [Google Scholar]
- Handa R. J., Sharma D., Uht R. A. (2011). role for the androgen metabolite, 5alpha androstane 3beta, 17beta diol (3beta-diol) in the regulation of the hypothalamo-pituitary-adrenal axis. Front. Endocrinol. 2:65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hedman K., Tamas E., Bjarnegard N., Brudin L., Nylander E. (2015). Cardiac systolic regional function and synchrony in endurance trained and untrained females. BMJ Open Sport Exerc. Med. 1:e000015. 10.1136/bmjsem-2015-000015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kastenmuller G., Raffler J., Gieger C., Suhre K. (2015). Genetics of human metabolism: an update. Hum. Mol. Genet. 24 R93–R101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Landrum M. J., Lee J. M., Benson M., Brown G. R., Chao C., Chitipiralla S., et al. (2018). ClinVar: improving access to variant interpretations and supporting evidence. Nucleic Acids Res. 46 D1062–D1067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lortie G., Bouchard C., Leblanc C., Tremblay A., Simoneau J. A., Theriault G., et al. (1982). Familial similarity in aerobic power. Hum. Biol. 54 801–812. [PubMed] [Google Scholar]
- Mannan A. U., Roussa E., Kraus C., Rickmann M., Maenner J., Nayernia K., et al. (2004). Mutation in the gene encoding lysosomal acid phosphatase (Acp2) causes cerebellum and skin malformation in mouse. Neurogenetics 5 229–238. 10.1007/s10048-004-0197-9 [DOI] [PubMed] [Google Scholar]
- Maron B. J., Pelliccia A. (2006). The heart of trained athletes: cardiac remodeling and the risks of sports, including sudden death. Circulation 114 1633–1644. 10.1161/circulationaha.106.613562 [DOI] [PubMed] [Google Scholar]
- McNamara J. W., Li A., Lal S., Bos J. M., Harris S. P., van der Velden J., et al. (2017). MYBPC3 mutations are associated with a reduced super-relaxed state in patients with hypertrophic cardiomyopathy. PLoS One 12:e0180064. 10.1371/journal.pone.0180064 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitchell J. H., Haskell W., Snell P., Van Camp S. P. (2005). Task force 8: classification of sports. J. Am. Coll. Cardiol. 45 1364–1367. 10.1016/j.jacc.2005.02.015 [DOI] [PubMed] [Google Scholar]
- Moss R. L., Fitzsimons D. P., Ralphe J. C. (2015). Cardiac MyBP-C regulates the rate and force of contraction in mammalian myocardium. Circ. Res. 116 183–192. 10.1161/circresaha.116.300561 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogawa T., Spina R. J., Martin W. H., III, Kohrt W. M., Schechtman K. B., Holloszy J. O., et al. (1992). Effects of aging, sex, and physical training on cardiovascular responses to exercise. Circulation 86 494–503. 10.1161/01.cir.86.2.494 [DOI] [PubMed] [Google Scholar]
- Pelliccia A., Maron B. J., Spataro A., Proschan M. A., Spirito P. (1991). The upper limit of physiologic cardiac hypertrophy in highly trained elite athletes. N. Engl. J. Med. 324 295–301. 10.1056/nejm199101313240504 [DOI] [PubMed] [Google Scholar]
- Pelliccia A., Culasso F., Di Paolo F. M., Maron B. J. (1999). Physiologic left ventricular cavity dilatation in elite athletes. Ann. Intern. Med. 130 23–31. [DOI] [PubMed] [Google Scholar]
- Pelliccia A. (1996). Determinants of morphologic cardiac adaptation in elite athletes: the role of athletic training and constitutional factors. Int. J. Sports Med. 17(Suppl. 3) S157–S163. [DOI] [PubMed] [Google Scholar]
- Peeters M. W., Thomis M. A., Beunen G. P., Malina R. M. (2009). Genetics and sports: an overview of the pre-molecular biology era. Med. Sport Sci. 54 28–42. 10.1159/000235695 [DOI] [PubMed] [Google Scholar]
- Pitsiladis Y. P., Tanaka M., Eynon N., Bouchard C., North K. N., Williams A. G., et al. (2016). Athlome Project Consortium: a concerted effort to discover genomic and other “omic” markers of athletic performance. Physiol. Genomics 48 183–190. 10.1152/physiolgenomics.00105.2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prud’homme D., Bouchard C., Leblanc C., Landry F., Fontaine E. (1984). Sensitivity of maximal aerobic power to training is genotype-dependent. Med. Sci. Sports Exerc. 16 489–493. 10.1249/00005768-198410000-00012 [DOI] [PubMed] [Google Scholar]
- Rankinen T., Roth S. M., Bray M. S., Loos R., Perusse L., Wolfarth B., et al. (2010). Advances in exercise, fitness, and performance genomics. Med. Sci. Sports Exerc. 42 835–846. 10.1249/mss.0b013e3181d86cec [DOI] [PubMed] [Google Scholar]
- Rankinen T., Fuku N., Wolfarth B., Wang G., Sarzynski M. A., Alexeev D. G., et al. (2016). No evidence of a common DNA variant profile specific to world class endurance athletes. PLoS One 11:e0147330. 10.1371/journal.pone.0147330 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ray D., Culine S., Tavitain A., Moreau-Gachelin F. (1990). The human homologue of the putative proto-oncogene Spi-1: characterization and expression in tumors. Oncogene 5 663–668. [PubMed] [Google Scholar]
- Repa J. J., Berge K. E., Pomajzl C., Richardson J. A., Hobbs H., Mangelsdorf D. J. (2002). Regulation of ATP-binding cassette sterol transporters ABCG5 and ABCG8 by the liver X receptors alpha and beta. J. Biol. Chem. 277 18793–18800. 10.1074/jbc.m109927200 [DOI] [PubMed] [Google Scholar]
- Sato K., Iemitsu M., Aizawa K., Ajisaka R. (2008). Testosterone and DHEA activate the glucose metabolism-related signaling pathway in skeletal muscle. Am. J. Physiol. Endocrinol. Metab. 294 E961–E968. [DOI] [PubMed] [Google Scholar]
- Semenova E., Fuku N., Ahmetov I. (2019). “Genetic profile of elite endurance athletes,” in Sports, Exercise, and Nutritional Genomics: Current Status and Future Directions, eds Barh D., Ahmetov I. (Cambridge, MA: Academic Press; ), 73–104. 10.1016/b978-0-12-816193-7.00004-x [DOI] [Google Scholar]
- Semenova E. A., Miyamoto-Mikami E., Akimov E. B., Al-Khelaifi F., Murakami H., Zempo H., et al. (2020). The association of HFE gene H63D polymorphism with endurance athlete status and aerobic capacity: novel findings and a meta-analysis. Eur. J. Appl. Physiol. 120 665–673. 10.1007/s00421-020-04306-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka M., Wang G., Pitsiladis Y. P. (2016). Advancing sports and exercise genomics: moving from hypothesis-driven single study approaches to large multi-omics collaborative science. Physiol. Genom. 48 173–174. 10.1152/physiolgenomics.00009.2016 [DOI] [PubMed] [Google Scholar]
- Theofilopoulos S., Arenas E. (2015). Liver X receptors and cholesterol metabolism: role in ventral midbrain development and neurodegeneration. F1000Prime Rep. 7:37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang J., Chu G. (2002). Xeroderma pigmentosum complementation group E and UV-damaged DNA-binding protein. DNA Repair (Amst). 1 601–616. 10.1016/s1568-7864(02)00052-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang G., Tanaka M., Eynon N., North K. N., Williams A. G., Collins M., et al. (2016). The future of genomic research in athletic performance and adaptation to training. Med. Sport Sci. 61 55–67. [DOI] [PubMed] [Google Scholar]
- Wu C. K., Huang Y. T., Lee J. K., Chiang L. T., Chiang F. T., Huang S. W., et al. (2012). Cardiac myosin binding protein C and MAP-kinase activating death domain-containing gene polymorphisms and diastolic heart failure. PLoS One 7:e35242. 10.1371/journal.pone.0035242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang W., Elimban V., Nijjar M. S., Gupta S. K., Dhalla N. S. (2003). Role of mitogen-activated protein kinase in cardiac hypertrophy and heart failure. Exp. Clin. Cardiol. 8 173–183. [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The SNP data supporting this study is available at: https://figshare.com/articles/GWAS_elite_endurance_athletes/12199760. Summary statistics will be made available through the NHGRI-EBI GWAS Catalog: https://www.ebi.ac.uk/gwas/downloads/summary-statistics.