Abstract
Interspecies interactions among oral microorganisms in the pathogenic biofilms causing dental caries have not yet been elucidated in detail. We herein demonstrated that indole and its derivatives induced biofilm formation by Streptococcus mutans. Indole is an intercellular signaling molecule that is produced by oral bacteria other than S. mutans. The amounts of biofilm and extracellular DNA were significantly increased by the addition of indole and 4-hydroxyindole (4-HI). An examination with quorum sensing mutants showed that the induction of biofilm formation by indole and 4-HI required a quorum sensing system. These results suggest that this intercellular signaling molecule plays a role in pathogenic biofilm formation.
Keywords: Biofilm, Quorum sensing, Streptococcus mutans, indole
A wide variety of microorganisms inhabit the oral cavity (Griffen et al., 2012), and the dynamics of the oral microbial community influence the onset of oral infectious disease (Lamont et al., 2018). Biofilms are the most important habitat for oral microorganisms, and consist of multiple species of oral bacteria in nature. Intra- and interspecies interactions in the oral microbiota influence the composition and development of oral diseases in dynamic communities; however, the underlying mechanisms remain elusive. Dental caries, which is a typical oral infectious disease, is caused by pathogenic biofilms that form on the tooth surface (Bowen et al., 2018). In biofilms, cells and self-produced extracellular polymeric substances tightly packed in the community restrict the diffusion of microbial metabolites, such as lactic acid, resulting in the creation of an acidic microenvironment that causes dental decay. Streptococcus mutans plays a pivotal role in the formation of cariogenic biofilms by producing insoluble extracellular polysaccharides and lactic acid from carbohydrates, such as starch and sucrose (Hamada and Slade, 1980; Loesche, 1986). Due to the importance of S. mutans in dental caries, the mechanisms underlying biofilm formation have been investigated in detail. Moreover, the actual oral biofilm consists of a number of species of oral bacteria, and previous studies demonstrated that interspecies coaggregation constructed polymicrobial biofilms (Kolenbrander et al., 2010). Based on the paradigm of interspecies communication among oral bacteria, interspecies signaling to S. mutans appears to facilitate cariogenic biofilm formation and microbial ecological interactions in the oral cavity (Nakanishi et al., 2018). Signaling molecules are some of the most important factors in multispecies biofilm development, and, for example, autoinducer-2 has been identified as a universal interspecies signaling molecule that enhances multispecies biofilm formation by the oral commensal bacteria Actinomyces naeslundii and Streptococcus oralis (Rickard et al., 2006). In the oral cavity, indole and its derivatives are candidates for interspecies signaling molecules. Indole is an aromatic heterocyclic molecule that is synthesized from a tryptophan by a number of oral bacteria and functions as an intercellular signaling molecule in microbial communities (Lee and Lee, 2010). Indole and its derivatives act as metabolic signals and play a role in biofilm formation (Hu et al., 2010). The periodontal bacterium Fusobacterium nucleatum produces indole, which promotes its own biofilm formation (Sasaki-Imamura et al., 2010). Moreover, indole has been detected in human saliva (Tonzetich et al., 1967; Cooke et al., 2003). These findings indicate that indole functions as a biofilm-related interspecies signaling molecule in the oral cavity.
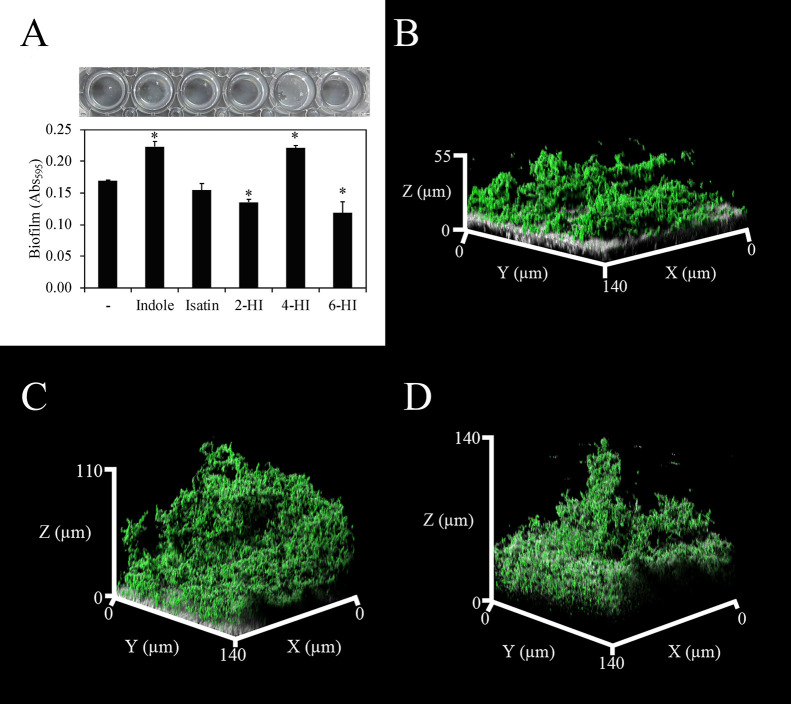
In the present study, we investigated the influence of indole and its derivatives on biofilm formation by cariogenic S. mutans. S. mutans does not produce indoles as its own signal molecules and does not possess orthologous genes encoding receptors for indole, which have been detected in other bacteria (Lee and Lee, 2010). The materials and methods used are shown in Supplementary materials. Three strains of S. mutans were used in the biofilm formation assay with or without 500 μM indole. The addition of indole increased biofilm formation by the three strains more than that dimethyl sulfoxide (DMSO), which was used as the solvent (Supplementary Fig. S1A). Thus, we used the most commonly used laboratory strain UA159 in subsequent experiments. Some indole derivatives, such as hydroxyindoles and isatin, are produced by many microbial species as metabolites of indoles through the actions of monooxygenase and dioxygenase (Lee and Lee, 2010). Moreover, oxidized indole derivatives have been shown to influence the bacterial behaviors of non-indole-producing bacteria similar to indole (Melander et al., 2014). Therefore, the effects of four types of indole derivatives, namely, 2-hydroxyindole (2-HI), 4-HI, 6-HI, and isatin, on biofilm formation by S. mutans were examined (Fig. 1A). The results obtained showed that the addition of 4-HI and indole increased biofilm formation, whereas that of isatin, 2-HI, and 6-HI did not (Fig. 1A). Therefore, modifications to indole appeared to mostly abolish the biofilm induction effect; however, specific modifications, for example, 4-hydroxylation, maintained this activity. Approximately 0.3–50 μM indole has been detected in human saliva (Tonzetich et al., 1967; Cooke et al., 2003). To verify that the concentrations of indole and 4-HI were sufficient to influence biofilm formation, concentration series of indole and 4-HI (0.5, 5, 50, and 500 μM) were added to media, and the influence of these concentrations on biofilm formation was investigated. The results obtained showed that the biofilm-inducing effects of indole and 4-HI were detectable at the lowest concentration (0.5 μM, Supplementary Fig. S1B). Therefore, the concentration of indole present in the human oral cavity appears to be sufficient to stimulate biofilm formation by S. mutans. Based on the appearance of biofilms in the presence of indole or 4-HI, the effects of 4-HI on biofilm formation appeared to differ from those of indole (Fig. 1A, upper panel). Confocal microscopic observations showed that indole and 4-HI increased biofilm thickness more than DMSO (Fig. 1B, C, and D). However, the bottom of the biofilm, i.e., the surface of hydroxyapatite, in the presence of 4-HI was not observed, although the bottoms of the biofilms with DMSO and indole were clearly visualized by reflection microscopy (shown as gray colors in Fig. 1B and C). This signal decay was attributed to the impenetrability of the excitation laser at the deeper position of the biofilm. A highly dense biofilm structure caused reflection signal extinction. The area occupancy of biofilms in a cross-section, i.e., the density of biofilm cells at a certain plane, was increased more by the addition of 4-HI (68.4±6.8%) than the DMSO control (56.5±1.7%) and indole (58.0±4.6%). These results suggest that the biofilm structure was denser under the 4-HI added condition. Thus, 4-HI appears to increase the density of biofilms and potentially reinforce their structure. Extracellular insoluble polysaccharides and extracellular DNA (eDNA) are the main components in biofilms formed by S. mutans, and promote their structural integrity and adhesion to substratum surfaces, respectively (Leme et al., 2006; Bowen and Koo, 2011; Das et al., 2011; Klein et al., 2015). However, sucrose, which is necessary for extracellular insoluble polysaccharide production (Wood and Critchley, 1966), was not present in the medium used in the present study. Hence, we extracted and quantified the amounts of eDNA in biofilms to clarify whether indole and 4-HI increased eDNA release. We found significant increases in eDNA levels in the presence of indole and 4-HI (Fig. 2). These results suggested that increases in biofilm formation by indole and 4-HI were associated with eDNA release. Moreover, a large amount of eDNA may contribute to a dense biofilm architecture under 4-HI added conditions. eDNA plays a role in biological functions other than biofilm formation. The presence of eDNA in biofilms confers increased resistance against aminoglycoside (Wilton et al., 2016). Oral bacteria may utilize eDNA as a nutrient source to survive nutrient-limited conditions and chelating metal ions (Jakubovics and Grant Burgess, 2015). Thus, increases in eDNA following the addition of indoles may be advantageous for the growth of S. mutans. Further investigations are needed to elucidate the impact of eDNA release induced by indoles on the ecology of S. mutans.
Fig. 1.
Effects of indole and its derivatives on biofilm formation by Streptococcus mutans. Effects of indole and its derivatives on biofilm formation by Streptococcus mutans strain UA159 (A). Values represent the means and standard deviations of three biological replicates. Asterisks indicate a significant difference from the DMSO control (shown as “–”) (P<0.05), as evaluated by Dunnett’s test (A). A representative of at least three independent experiments is shown. Confocal laser scanning microscopic images of the biofilm grown with DMSO as a control (B), 500 μM indole (C), and 4-HI (D). Regarding microscopic visualization, biofilms were formed on the surfaces of hydroxyapatite disks. Green indicates the cells of S. mutans labeled by SYTO9, and gray the reflection signals representing non-cell parts, particularly arrows. The gray color at the bottom of the images indicates the surfaces of the hydroxyapatite disks. Visualization of biofilms formed on a non-transparent substratum was performed following a previously described procedure (Inaba et al., 2013), and a detailed procedure is shown in Supplementary materials. Each projection shows fields of 140×140 μM (x-y) as indicated. Microscopic images were taken of at least three random positions, and representative images are shown.
Fig. 2.
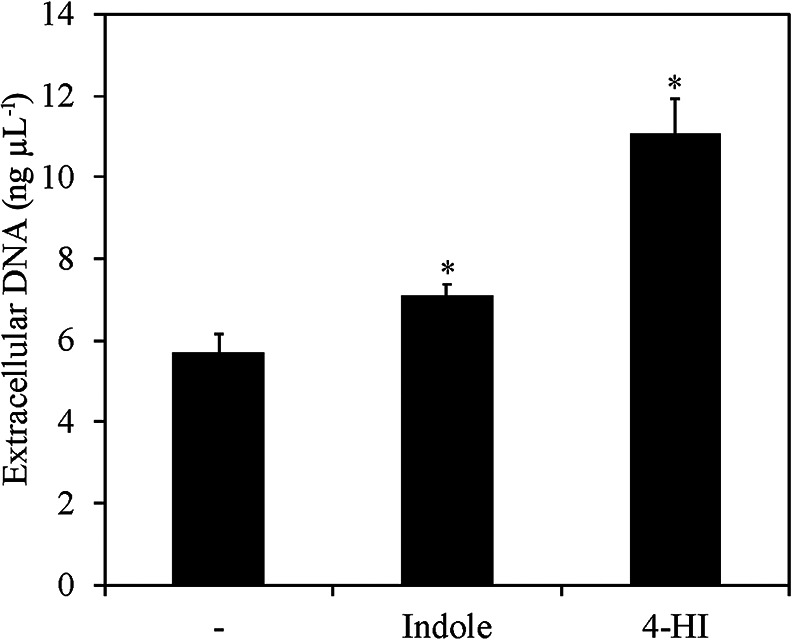
Quantification of eDNA. The amounts of eDNA in biofilms. Cells were grown in TSB medium with or without 500 μM indole and 4-HI at 37°C for 12 h in a 24-well microtiter plate. A detailed procedure for eDNA quantification is shown in Supplementary materials. Representative data from two independent experiments are shown in the figure. Values represent the means and standard deviations of at least three biological replicates. Asterisks indicate a significant difference (P<0.05), as evaluated by Dunnett’s test.
In S. mutans, eDNA is released as a consequence of cell death via the intercellular signaling pathway, generally called quorum sensing. Two quorum sensing signals induce cell death and eDNA release in S. mutans in a cell density-dependent manner and consist of competence-stimulating peptide (CSP) (Perry et al., 2009) and sigX-inducing peptide (XIP) (Wenderska et al., 2012). The comC and sigX genes encode CSP and XIP, respectively. Therefore, we quantified the biofilm amounts of ΔcomC and ΔsigX, which are strains with deficient CSP production and sigma factor stimulated by XIP, respectively (Fig. 3). The results obtained showed that biofilm formation was not increased by the addition of indole to ΔcomC and ΔsigX. Moreover, since LytF autolysin, the expression of which is induced via quorum sensing pathways, was shown to cause the autolysis of cells (Dufour and Lévesque, 2013), we also examined ΔlytF in the biofilm formation assay. The biofilm amounts of ΔlytF were significantly increased by the addition of indole. These results indicate that indole induced biofilm formation via CSP and XIP quorum sensing pathways; however, LytF autolysin was not necessary for the stimulation of biofilm formation by indole. Although the interconnection between CSP and the XIP signaling pathway currently remains unclear, the activation of the XIP signaling pathway requires CSP in complex medium (Reck et al., 2015). Therefore, the present results suggest that indole acts through the XIP quorum sensing pathway, but is mediated by an unknown autolysis pathway. Moreover, the addition of 4-HI significantly decreased biofilm formation by quorum sensing mutants, suggesting that 4-HI exhibited growth inhibitory activity independent of quorum sensing pathways. In the wild type, 4-HI inhibited cell growth and decreased maximum cell density at the stationary phase more than DMSO and indole (Supplementary Fig. S2). This inhibitory effect was not restored in the quorum sensing mutants; furthermore, the growth rates of these mutants were suppressed more than the wild type (Supplementary Fig. S2). These results suggest that 4-HI exhibits antibiotic-like activity, resulting in eDNA release independent of quorum sensing. A subminimal inhibitory concentration of antibiotics has been shown to induce eDNA release and eDNA-dependent biofilm formation by Staphylococci (Kaplan et al., 2011; 2012). Similarly, the antibiotic-like activity of 4-HI may induce eDNA release, resulting in the induction of biofilm formation in S. mutans. ΔcomC is more sensitive to some antimicrobial agents, such as ampicillin and chlorhexidine (Wang et al., 2013), suggesting that the quorum sensing cascade confers tolerance to 4-HI. Thus, the growth inhibitory effects of 4-HI may significantly decrease biofilm volumes in quorum sensing mutants.
Fig. 3.
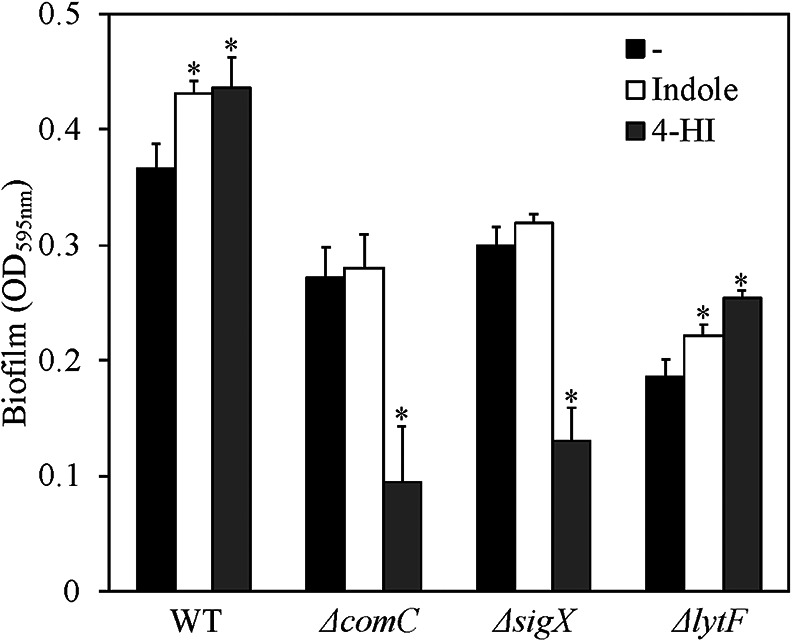
Biofilm assay for quorum sensing mutants. Biofilm amounts of quorum sensing mutants. Biofilms were formed at the bottom of a 24-well microtiter plate. A biofilm assay was performed according with the procedure shown in Supplementary materials. Asterisks indicate a significant difference based on three biological replicates from the DMSO control (P<0.05), as evaluated by Dunnett’s test. At least three independent experiments were performed, and representative experiments are shown.
Although the different effects of indole and its derivatives imply the complexity of interspecies interactions in the oral microbiome, the present study revealed for the first time a potential interaction between cariogenic S. mutans and indole-producing bacteria during biofilm formation. Indole is converted to oxidized indole derivatives, such as isatin, 2-HI, 3-HI, and 4-HI, by the enzymatic activity of bacteria (Rui et al., 2005). Our results demonstrated that indole modifications and metabolism were crucially important for the biofilm-stimulating activity of indole, and further studies on the metabolic dynamics of indole in oral microbiota are awaited. Interspecies interactions via signaling molecules may shape mixed oral biofilm formation and the composition of the community, and indole produced by the oral microbiota may be a candidate signaling molecule to control the oral microbiota. To elucidate interspecies interactions among S. mutans and oral commensal bacteria, we need to investigate the molecular mechanisms underlying the responses to indole and its derivatives in S. mutans during biofilm formation. Furthermore, comprehensive metagenomic, metatranscriptomic, and metabolome analyses of the oral microbiome producing or responding to indole will contribute to a more detailed understanding of oral microbial ecology based on intercellular communications. The saliva of patients with periodontal disease contains high amounts of indole and some periodontopathogenic bacteria, such as Porphyromonas gingivalis, Prevotella sp., and F. nucleatum, which produce indole (Berg et al., 1946; Sasaki-Imamura et al., 2010; Sasaki-Imamura et al., 2011). Further investigations on intercellular interactions in the oral cavity via indole will also provide critical insights into the onset and treatment of oral infectious diseases, dental caries, and periodontitis.
Citation
Inaba, T., Obana, N., Habe, H., and Nomura, N. (2020) Biofilm Formation by Streptococcus mutans is Enhanced by Indole via the Quorum Sensing Pathway. Microbes Environ 35: ME19164.
https://doi.org/10.1264/jsme2.ME19164
Supplementary Material
Acknowledgements
This research was financially supported by JSPS Grants-in-Aid for Challenging Research (Exploratory) and Encouragement of Young Scientist (Grant Numbers 19K22297 and 18K15143) and JST ERATO Grant Number JPMJER1502, Japan.
References
- Berg M., Burrill D.Y., and Fosdick L.S. (1946) Chemical studies in periodontal disease III: putrefaction of salivary proteins. J Dent Res 25: 231–246. [DOI] [PubMed] [Google Scholar]
- Bowen W.H., and Koo H. (2011) Biology of Streptococcus mutans-derived glucosyltransferases: Role in extracellular matrix formation of cariogenic biofilms. Caries Res 45: 69–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowen W.H., Burne R.A., Wu H., and Koo H. (2018) Oral biofilms: Pathogens, matrix, and polymicrobial interactions in microenvironments. Trends Microbiol 26: 229–242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooke M., Leeves N., and White C. (2003) Time profile of putrescine, cadaverine, indole and skatole in human saliva. Arch Oral Biol 48: 323–327. [DOI] [PubMed] [Google Scholar]
- Das T., Sharma P.K., Krom B.P., van der Mei H.C., and Busscher H.J. (2011) Role of eDNA on the adhesion forces between Streptococcus mutans and substratum surfaces: Influence of ionic strength and substratum hydrophobicity. Langmuir 27: 10113–10118. [DOI] [PubMed] [Google Scholar]
- Dufour D., and Lévesque C.M. (2013) Cell death of Streptococcus mutans induced by a quorum-sensing peptide occurs via a conserved streptococcal autolysin. J Bacteriol 195: 105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griffen A.L., Beall C.J., Campbell J.H., Firestone N.D., Kumar P.S., Yang Z.K., et al. (2012) Distinct and complex bacterial profiles in human periodontitis and health revealed by 16S pyrosequencing. ISME J 6: 1176–1185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamada S., and Slade H.D. (1980) Biology, immunology, and cariogenicity of Streptococcusmutans. Microbiol Rev 44: 331–384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu M., Zhang C., Mu Y., Shen Q., and Feng Y. (2010) Indole affects biofilm formation in bacteria. Indian J Microbiol 50: 362–368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inaba T., Ichihara T., Yawata Y., Toyofuku M., Uchiyama H., and Nomura N. (2013) Three-dimensional visualization of mixed species biofilm formation together with its substratum. Microbiol Immunol 57: 589–593. [DOI] [PubMed] [Google Scholar]
- Jakubovics N.S., and Grant Burgess J. (2015) Extracellular DNA in oral microbial biofilms. Microbes Infect 17: 531–537. [DOI] [PubMed] [Google Scholar]
- Kaplan J.B., Jabbouri S., and Sadovskaya I. (2011) Extracellular DNA-dependent biofilm formation by Staphylococcus epidermidis RP62A in response to subminimal inhibitory concentrations of antibiotics. Res Microbiol 162: 535–541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaplan J.B., Izano E.A., Gopal P., Karwacki M.T., Kim S., Bose J.L., et al. (2012) Low levels of β-lactam antibiotics induce extracellular DNA release and biofilm formation in Staphylococcus aureus. mBio 3: e00198–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klein M.I., Hwang G., Santos P.H., Campanella O.H., and Koo H. (2015) Streptococcus mutans-derived extracellular matrix in cariogenic oral biofilms. Front Cell Infect Microbiol 5: 10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolenbrander P.E., Palmer Jr. R.J., Periasamy S., and Jakubovics N.S. (2010) Oral multispecies biofilm development and the key role of cell-cell distance. Nat Rev Microbiol 8: 471–480. [DOI] [PubMed] [Google Scholar]
- Lamont R.J., Koo H., and Hajishengallis G. (2018) The oral microbiota: dynamic communities and host interactions. Nat Rev Microbiol 16: 745–759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee J.-H., and Lee J. (2010) Indole as an intercellular signal in microbial communities. FEMS Microbiol Rev 34: 426–444. [DOI] [PubMed] [Google Scholar]
- Leme A.F.P., Koo H., Bellato C.M., Bedi G., and Cury J.A. (2006) The role of sucrose in cariogenic dental biofilm formation—New insight. J Dent Res 85: 878–887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loesche W.J. (1986) Role of Streptococcus mutans in human dental decay. Microbiol Rev 50: 353–380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melander R.J., Minvielle M.J., and Melander C. (2014) Controlling bacterial behavior with indole-containing natural products and derivatives. Tetrahedron 70: 6363–6372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakanishi Y., Yamamoto T., Obana N., Toyofuku M., Nomura N., and Kaneko A. (2018) Spatial distribution and chemical tolerance of Streptococcus mutans within dual-species cariogenic biofilms. Microbes Environ 33: 455–458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perry J.A., Cvitkovitch D.G., and Levesque C.M. (2009) Cell death in Streptococcus mutans biofilms: a link between CSP and extracellular DNA. FEMS Microbiol Lett 299: 261–266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reck M., Tomasch J., and Wagner-Döbler I. (2015) The alternative sigma factor sigX controls bacteriocin synthesis and competence, the two quorum sensing regulated traits in Streptococcus mutans. PLoS Genet 11: e1005353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rickard A.H., Palmer Jr R.J., Blehert D.S., Campagna S.R., Semmelhack M.F., Egland P.G., et al. (2006) Autoinducer 2: a concentration-dependent signal for mutualistic bacterial biofilm growth. Mol Microbiol 60: 1446–1456. [DOI] [PubMed] [Google Scholar]
- Rui L., Reardon K.F., and Wood T.K. (2005) Protein engineering of toluene ortho-monooxygenase of Burkholderia cepacia G4 for regiospecific hydroxylation of indole to form various indigoid compounds. Appl Microbiol Biotechnol 66: 422–429. [DOI] [PubMed] [Google Scholar]
- Sasaki-Imamura T., Yano A., and Yoshida Y. (2010) Production of indole from L-tryptophan and effects of these compounds on biofilm formation by Fusobacterium nucleatum ATCC 25586. Appl Environ Microbiol 76: 4260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sasaki-Imamura T., Yoshida Y., Suwabe K., Yoshimura F., and Kato H. (2011) Molecular basis of indole production catalyzed by tryptophanase in the genus Prevotella. FEMS Microbiol Lett 322: 51–59. [DOI] [PubMed] [Google Scholar]
- Tonzetich J., Eigen E., King W.J., and Weiss S. (1967) Volatility as a factor in the inability of certain amines and indole to increase the odour of saliva. Arch Oral Biol 12: 1167–1175. [DOI] [PubMed] [Google Scholar]
- Wang W.-L., Liu J., Huo Y.-B., and Ling J.-Q. (2013) Bacteriocin immunity proteins play a role in quorum-sensing system regulated antimicrobial sensitivity of Streptococcus mutans UA159. Arch Oral Biol 58: 384–390. [DOI] [PubMed] [Google Scholar]
- Wenderska I.B., Lukenda N., Cordova M., Magarvey N., Cvitkovitch D.G., and Senadheera D.B. (2012) A novel function for the competence inducing peptide, XIP, as a cell death effector of Streptococcus mutans. FEMS Microbiol Lett 336: 104–112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilton M., Charron-Mazenod L., Moore R., and Lewenza S. (2016) Extracellular DNA acidifies biofilms and induces aminoglycoside resistance in Pseudomonas aeruginosa. Antimicrob Agents Chemother 60: 544–553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wood J.M., and Critchley P. (1966) The extracellular polysaccharide produced from sucrose by a cariogenic streptococcus. Arch Oral Biol 11: 1039–1042. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.