Abstract
Introduction
Testing for active SARS-CoV-2 infection is a fundamental tool in the public health measures taken to control the COVID-19 pandemic. Because of the overwhelming use of SARS-CoV-2 reverse transcription (RT)-PCR tests worldwide, the availability of test kits has become a major bottleneck and the need to increase testing throughput is rising. We aim to overcome these challenges by pooling samples together, and performing RNA extraction and RT-PCR in pools.
Methods
We tested the efficiency and sensitivity of pooling strategies for RNA extraction and RT-PCR detection of SARS-CoV-2. We tested 184 samples both individually and in pools to estimate the effects of pooling. We further implemented Dorfman pooling with a pool size of eight samples in large-scale clinical tests.
Results
We demonstrated pooling strategies that increase testing throughput while maintaining high sensitivity. A comparison of 184 samples tested individually and in pools of eight samples showed that test results were not significantly affected. Implementing the eight-sample Dorfman pooling to test 26 576 samples from asymptomatic individuals, we identified 31 (0.12%) SARS-CoV-2 positive samples, achieving a 7.3-fold increase in throughput.
Discussion
Pooling approaches for SARS-CoV-2 testing allow a drastic increase in throughput while maintaining clinical sensitivity. We report the successful large-scale pooled screening of asymptomatic populations.
Keywords: COVID-19, Diagnostics, Group testing, Infectious diseases, RT-PCR
Introduction
An emerging novel severe acute respiratory syndrome-related coronavirus, SARS-CoV-2, is the virus behind the global COVID-19 pandemic. Among the foremost priorities to facilitate efficient public health interventions is a reliable and accessible diagnosis of an active SARS-CoV-2 infection. The standard laboratory diagnosis of COVID-19 involves three main steps, namely viral inactivation and lysis of the nasopharyngeal swab sample, extraction (or purification) of viral RNA and reverse transcription (RT)-PCR. Because of the rapid spread of the virus and the increasing demand for tests, the limited availability of test reagents, mainly RNA extraction kits, has become (and will likely continue to be) a major bottleneck as the pandemic expands [1,2].
Of particular importance is the ability to survey large asymptomatic populations (1) to trace asymptomatic COVID-19 carriers who are otherwise difficult to identify and isolate; (2) to assure key personnel (e.g. healthcare personnel) are not contagious; (3) to screen high-risk populations (such as nursing homes) to help protect them; (4) to accurately estimate the spread of infection and the effectiveness of community measures and social distancing; and (5) to allow and monitor a safe return to work. Efficient and higher throughput diagnostic approaches are needed to support such efforts. While some of these applications (e.g. no. 4) may be achieved with less-sensitive detection approaches, most applications do require adhering to the current high standards of RT-PCR.
Several attempts to address this challenge have been recently reported, and can be categorized into three major approaches. The first approach is to replace PCR-based methods by other direct diagnostic methods such as loop-mediated isothermal amplification [[3], [4], [5], [6]] and clustered regularly interspaced short palindromic repeats (CRISPR)-based diagnostic tools [[7], [8], [9]]. The second approach involves serological surveys [[10], [11], [12], [13]], and the third approach involves the improvement in the capacity of PCR methods by optimization and automation [1,14,15] or by reducing the required number of tests via pooling samples together, known as group testing.
Group testing is a field of research in the intersection of mathematics, computer science and information theory, with applications in biology, communication and more. A group testing algorithm is a testing scheme directed towards minimizing the number of tests conducted on a set of samples by using the ability to test pooled subsets of samples. If a pool of n samples tests negative, all samples must be negative, and therefore their status has been determined in only one test instead of n individual tests. Various group testing algorithms exist, with different assumptions and constraints [16,17]. While many such algorithms, most notably binary splitting, may be very efficient in theory, they might be unsuitable because of practical limitations. Some key constrains are (1) a limit on the number of stages due to the importance of delivering a test result quickly, exemplified by the urgent clinical context of COVID-19 diagnosis; (2) a limit on the ability to dilute samples and still safely identify a single positive sample in a pool; and (3) favourability of simple algorithms which may minimize human error in a laboratory setting.
While several pooling approaches for SARS-CoV-2 detection have been suggested recently [2,[18], [19], [20], [21], [22]], these studies mostly discussed theoretical considerations. Here we describe and demonstrate practical pooling solutions that save time and reagents by performing RNA extraction and RT-PCR on pooled samples. We offer two such pooling approaches, based either on simple (Dorfman) pooling or matrix pooling [23,24], and demonstrate their efficiency and sensitivity in the daily reality of COVID-19.
Methods
Sample collection
At the Hadassah Medical Centre (HMC), two distinct populations are tested for SARS-CoV-2 at present. First, we receive samples from symptomatic patients, from the hospital and from the community. In these samples, about 10% of SARS-CoV-2 tests are positive. Second, we receive samples from prospectively screened asymptomatic populations such as hospital employees and workers in essential industries. According to the Israeli Ministry of Health guidelines all samples were collected using a single swab for combined deep nasal and oropharyngeal collection from the same patient. Nasopharyngeal swab samples were collected in 2 mL of Viral Transport Medium (VTM) or collected directly to 2 mL of Zymo lysis buffer.
RNA extraction and RT-PCR detection for individual tests
For sample lysate preparation 220 μL of sample VTM was added to 280 μL of Zymo lysis buffer. RNA was extracted using MagNA Pure 96 kit (Roche Lifesciences) using the Roche platform and eluted in 60 μL. A 10-μL aliquot of RNA was used in 30 μL of reaction using real-time fluorescent RT-PCR kit (BGI). We followed kit instructions with thermocycler protocol: one cycle at 50°C for 20 min; one cycle at 95°C for 10 min; 40 cycles at 95°C for 15 s; one cycle at 60°C for 30 s. According to the clinical guidelines of the Israeli Ministry of Health at the time experiments were conducted, a sample was defined as positive if the viral genome was detected at threshold cycle (Ct) values ≤35, as indeterminate at Ct values >35 and ≤38, and as negative at Ct values >38.
Pooling approaches
The first pooling strategy is a simple two-stage testing algorithm known as Dorfman pooling [25]. In the first stage, the samples are divided into disjoint pools of n samples each, and each such pool is tested. A negative result implies that all samples in the pool are negative, while a positive result implies that at least one sample in the pool is positive. In the second stage, the samples of each pool that tested positive are individually tested.
To reduce the need to retest positive pools we have also tested a two-stage matrix pooling strategy [23,24], where n 2 samples are ordered in an n × n matrix. Each row and each column are pooled, resulting in 2n tests, times fewer tests than individual testing. If either the number of positive rows or columns is one, the positive samples can be uniquely identified at the intersections of the positive rows and columns. Otherwise, if both the number of positive rows and columns is greater than one, intersections of positive rows and columns will be retested individually.
Testing pooling strategies
Individual barcoded samples were received at the laboratory, inactivated by a lysis buffer and pooled on a Tecan liquid-handling robot. For matrix pool design we pooled equal volumes of sample lysate to a final volume of 450 μL and used MagNA Pure 96 kit (Roche Lifesciences) using the Roche platform. For eight-sample pools, we used QIAsymphony DSP Virus/Pathogen kit on a QIAsymphony platform. We pooled equal volumes of sample lysate to a final volume of 400 μL. Positive pools were validated by individual tests as described above. Both Qiagen kits were used with Zymo lysis buffers, and therefore we skipped the lysis and Proteinase K step. RNA was eluted into 60 μL; 10 μL of RNA was used for a 30-μL reaction using real-time fluorescent RT-PCR kit (BGI). To reduce the risk of contamination, daily ultraviolet irradiation of RNA extraction robots was performed, and different rooms for processing before and after PCR were set up, without mixing personnel or machines between the two compartments.
Note that analysis of pool results requires close attention to indeterminate-result pools, as these may contain individual positive samples. Therefore, all pools detected with Ct ≤ 39 were retested (see Table 2 , batch 3), while maintaining standard criteria for the individual tests when retesting.
Table 2.
Three pooled tests run at the Hadassah Medical Centre
| Batch | No. of samples | No. of pools | No. of positive pools | Ct of positive pools | No. of positive individual samples in the positive pools | Ct of positive individual samples | No. of total tests | No. of tests saved, compared to single sample tests (%) |
|---|---|---|---|---|---|---|---|---|
| 1 | 720 | 90 | 1 | 21.8 | 1 | 19.4 | 98 | 622 (86.4%) |
| 2 | 720 | 90 | 0 | — | — | — | 90 | 630 (87.5%) |
| 3 | 728 | 91 | 3 positives, 1 indeterminate | 25.39 | 2 | 22.05, 28.72 | 123 | 605 (83.1%) |
| 37.44 | 1 | 37.67a | ||||||
| 35.26 | 1 | 34.66 | ||||||
| 38.67 indeterminate | 0 | 38.24 (determined as negative) |
Ct, cycle threshold.
According to Hadassah Medical Centre's protocol for indeterminate values, reverse transcription-PCR was repeated with a different kit, and eventually was determined as positive.
Choosing pooling strategy and parameters
We define the efficiency of a pooling algorithm as the total number of samples divided by the expected number of tests conducted on them. We assume all samples are independent and identically distributed, and denote the probability of a sample to be positive by p (prevalence of detectable COVID-19 patients in the relevant population) and the pool size by n. The efficiency of the algorithms described above depends on both p and n. The best theoretical efficiency is [26]. The efficiency of Dorfman pooling is [25]. We chose a pool size of n = 8 samples as it allows a low false-negative rate (Fig. 1 ) and high efficiency for a wide range of COVID-19 prevalence (Table 1 and Table S1). The prevalence of detectable COVID-19 in an asymptomatic population is estimated to be considerably below 1% [27], and indeed of the 26 576 samples tested in the present study only 0.12% were found positive. Therefore, efficiency is likely to be 5–7.5. For higher prevalence the efficiency of matrix pooling is somewhat higher (Table 1 and supplementary material). We provide a tool (https://github.com/matanseidel/pooling_optimization) to help choose the approach and pool size based on the prevalence.
Fig. 1.
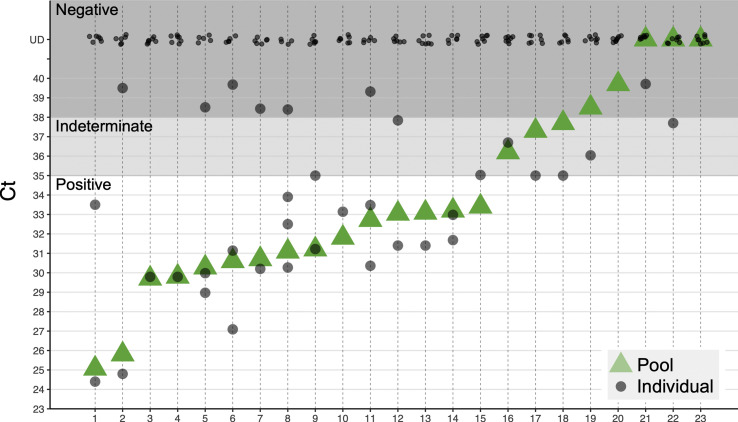
Pooling eight lysates retains clinical sensitivity. Shown are results of 23 pooling experiments, with eight lysates in each pool; 15 pools with positive samples indeed come up positive (pools 1–15), three pools without positive samples come up negative (pools 20, 21, 23) and four out of five pools containing a single indeterminate sample detected as indeterminate (pools 16, 17, 18, 19, 22); Pools containing one or two samples with low amount of SARS-CoV-2 are detected at a similar Ct (pools 9–18), showing clinical sensitivity is retained and the risk of false negatives is minimal. Ct, threshold cycle.
Table 1.
Efficiency of Dorfman and matrix pooling with pool size n = 8 compared with optimal efficiency
| Prevalence (p) | 0.1% | 1% | 2% | 5% | 10% | 20% |
|---|---|---|---|---|---|---|
| Maximal theoretical efficiency | 87.7 | 12.4 | 7.1 | 3.5 | 2.13 | 1.39 |
| Dorfman n = 8 efficiency | 7.5 | 4.9 | 3.6 | 2.2 | 1.44 | 1.04 |
| Matrix n = 8 efficiency | 4.0 | 3.9 | 3.6 | 2.6 | 1.68 | 1.05 |
Study approval
These studies were part of the approved diagnosis optimization and validation procedures at the HMC, and therefore no additional Institutional Review Board approvals were required.
Results
A key requirement of pooled RNA extraction and RT-PCR tests is to retain sufficient sensitivity. Theoretically, pooling eight samples should elevate the Ct of a single positive sample by three cycles. However, reproducibility of RNA extraction and RT-PCR might be affected by other factors. We therefore empirically tested assay sensitivity, when multiple negative samples and one positive sample were mixed at the lysate stage. We tested the pooling of 184 samples into 23 pools of eight samples each, and also tested in parallel each sample individually. This approach yielded highly accurate results, with no loss of diagnostic assay sensitivity: each of the pools that contained one or more positive samples was found to be positive, and all the pools that contained only negative samples were found to be negative (Fig. 1). Of the five pools that contained one individual sample with an ‘indeterminate’ result (in each pool), one was found to be negative per definition of individual tests, but still with Ct < 39, which allows pool retesting.
In addition, we tested matrix pooling (see Methods) by pooling 75 samples into three 5 × 5 matrices, and identified all positive samples accurately (Fig. 2 ). Importantly, the positive samples were detected in both the row and the column pools at a similar cycle in all three tested matrices, suggesting the pooling scheme is robust.
Fig. 2.
Matrix pooling. (a) Scheme for 5 × 5 matrix pooling. Twenty-five samples sorted in a 5 × 5 matrix and each row and each column is pooled into a total of ten pools, on which RNA extraction, reverse transcription and qPCR are performed. In this illustration row B and column 3 are positive (black stars), hence sample B3 is the only positive sample. If more than one row and one column are positive then all samples in intersections need to be retested, as some may be negative. (b) Three 5 × 5 pool matrices were generated (30 pools from 75 lysates). Each matrix (25 lysates that were previously tested individually) included a single lysate positive for SARS-COV-2. As expected, only six pools (one row and one column per matrix) were positive for SARS-COV-2, while 24 pools had threshold cycle (Ct) > 40 (Undetected). Reverse transcription-PCR Ct values of positive pools were nearly identical in the column pool (green) and the row pool (blue), and similar to the values of the individual test of the positive sample (grey).
Given the successful validation of both pooling strategies, and the low prevalence in asymptomatic population, we have adopted a Dorfman pooling protocol of 1:8 and employed it for the routine testing of nasopharyngeal swab samples from screened asymptomatic healthcare personnel, employees of essential industries, and residents and employees of nursing homes.
In the first three batches run at the HMC (Table 2) we tested 2168 samples by pooling, using 311 RNA extraction and RT-PCR reactions (a mere 14% of kits that would have been used in the full individual testing, an increase of sevenfold in throughput). Among these samples, we have identified and individually validated five positive samples, corresponding to a rate of 0.23%. We have then implemented the method in a routine clinical diagnosis setting of asymptomatic populations, testing a total of 26 576 samples with 7.3-fold increase in throughput, identifying a total of 31 positives (0.12%).
Discussion
We demonstrate in a real-life situation the usefulness of pooled sampling starting at the early lysate stage. The simplicity of the method, similarity to currently approved procedures and the fact that we do not require special sample handling or additional information make it easily adoptable on a large scale. This saves time, work and reagents, allowing a considerable throughput increase of clinical diagnostic laboratories and opening the door for efficient screening of large asymptomatic populations for the presence of SARS-CoV-2 infection.
An important consideration before implementing group testing is the expected rate of false-positive and false-negative results. Based on our experience with over 26 500 samples from asymptomatic individuals, we did not encounter any false positives in the pools (see Fig. 1 and Table 2). False negatives are in principle more worrisome when testing in pools, because samples that failed at the RNA extraction step will be missed (while our individual testing includes amplification of a human transcript serving as an internal control for proper RNA extraction and RT-PCR of each sample). To define the magnitude of this potential problem, we examined a set of 13 781 individual tests done at the HMC, which were all expected to show a signal for a human gene serving as internal assay control. Amplification of the human gene failed in 52 samples (0.38%). Thus, we estimate that our current protocol of pooled sampling carries a risk of missing 0.38% of the positive samples. In a population of 1 000 000 individuals tested, of which 1000 are positive (rate of 0.1%), this predicts that four positive individuals will be missed when using pools. We posit that this is a tolerable situation, particularly given the potentially much higher rates of false-negative results due to swab sampling and other errors upstream. Large-scale implementation of the pooling scheme should be carefully done to assure pre-analytical influences (e.g. inadequate sampling, transport time, temperature influences) do not lead to significant loss of signal, which may further increase the risk of false negatives.
Increase in throughput applies to RNA extraction and RT-PCR, but not to viral inactivation and lysis or reporting of results. Reporting at the HMC is automated and was adapted to the pooling scheme and therefore does not require additional work. Viral inactivation and lysis typically take 30 min while RNA extraction and RT-PCR 4-5 hr, and therefore 7.3-fold increase in efficiency translates to ~4.5 increase in efficiency of the entire workflow, or more if efficiency of viral inactivation is increased by other means (e.g. automation).
The increase in efficiency allowed the HMC to survey healthcare personnel and multiple nursing homes, and identify a nursing home with 16 positive individuals, helping to stop the spread at that centre. Specifically, we have demonstrated that pooling lysates from five or eight nasopharyngeal swab samples retains sufficient sensitivity of viral RNA detection, allowing identification of SARS-CoV-2-positive individuals, while increasing throughput fivefold to 7.5-fold.
The prevalence of COVID-19 in the tested population is not always known, which could affect the optimal pool size. This could be addressed either by external estimates, such as a previous run of individual samples, rate of symptomatic patients, or alternative methods such as serological screening or wastewater titres monitoring [28,29]. Alternatively, it is possible to dynamically adapt pooling sizes, when the measured rate of positive samples is different than expected. Finally, some group testing algorithms (reviewed in [17]) estimate the number of positive samples while using a relatively small (logarithmic) number of tests, and may be adapted to clinical constraints and parameters. If samples are not independent, and we have information regarding their dependency, we can further improve efficiency by grouping together dependent samples, that is, samples that are likely all positives or all negatives, such as members of the same family, or samples that are likely to be all negative since they have a low-risk profile. This will increase the number of negative pools, and therefore decrease the overall number of tests conducted. Future improvement of the sensitivity of the test, such as better sets of primers and improved sample collection will allow retaining sensitivity even when pooling a large number of sample lysates together. This will enable further improving efficiency, especially when prevalence is low, by increasing the pool size.
Transparency declaration
All authors disclose no conflicts of interest.
Author contributions
Concept and design: E. Meshorer, Y. Dor, D. Wolf, M. Salton and Y. Drier. Acquisition, analysis, or interpretation of data: R. Ben-Ami, A. Klochendler, M. Seidel, T. Sido, O. Gurel-Gurevich, M. Yassour, E. Meshorer, Y. Dor, D. Wolf, M. Salton and Y. Drier. Software: M. Seidel. Visualization: T. Sido, M. Yassour. Resources: the Hebrew University-Hadassah COVID-19 diagnosis team, G. Benedek, I. Fogel, E. Oiknine-Djian, A. Gertler, Z. Rotstein, B. Lavi, Y. Dor, D. Wolf. Writing original draft, review and editing: Y. Dor, D. Wolf, M. Salton and Y. Drier. Supervision: Y. Dor, D. Wolf, M. Salton and Y. Drier. All authors approved the final manuscript.
Acknowledgements
Y. Drier is supported by an Alon Fellowship and the Concern Foundation/American Friends of the Hebrew University Young Professorship Award. The following people are members of the Hebrew University-Hadassah Medical School COVID-19 diagnosis team and have contributed to the development of the pooled testing pipeline: A. Klochendler, A. Eden, A. Klar, A. Geldman, A. Arbel, A. Peretz, B. Shalom, B.L. Ochana, D. Avrahami-Tzfati, D. Neiman, D. Steinberg, D. Ben Zvi, E. Shpigel, G. Atlan, H. Klein, H. Chekroun, H. Shani, I. Hazan, I. Ansari, I. Magenheim, J. Moss, J. Magenheim, L. Peretz, L. Feigin, M. Saraby, M. Sherman, M. Bentata, M. Avital, M. Kott, M. Peyser, M. Weitz, M. Shacham, M. Grunewald, N. Sasson, N. Wallis, N. Azazmeh, N. Tzarum, O. Fridlich, R. Sher, R. Condiotti, R. Refaeli, R. Ben Ami, R. Zaken-Gallili, R. Helman, S. Ofek, S. Tzaban, S. Piyanzin, S. Anzi, S. Dagan, S. Lilenthal, T. Sido, T. Licht, T. Friehmann, Y. Kaufman, A. Pery, A. Saada, A. Dekel, A. Yeffet, A. Shaag, A. Michael-Gayego, E. Shay, E. Arbib, H. Onallah, K. Ben-Meir, L. Levinzon, L. Cohen-Daniel, L. Natan, M. Hamdan, M. Rivkin, M. Shwieki, O. Vorontsov, R. Barsuk, R. Abramovitch, R. Gutorov, S. Sirhan, S. Abdeen, Y. Yachnin, Y. Daitch.
Editor: F. Allerberger
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.cmi.2020.06.009.
Contributor Information
The Hebrew University-Hadassah COVID-19 Diagnosis Team:
A. Klochendler, A. Eden, A. Klar, A. Geldman, A. Arbel, A. Peretz, B. Shalom, B.L. Ochana, D. Avrahami-Tzfati, D. Neiman, D. Steinberg, D. Ben Zvi, E. Shpigel, G. Atlan, H. Klein, H. Chekroun, H. Shani, I. Hazan, I. Ansari, I. Magenheim, J. Moss, J. Magenheim, L. Peretz, L. Feigin, M. Saraby, M. Sherman, M. Bentata, M. Avital, M. Kott, M. Peyser, M. Weitz, M. Shacham, M. Grunewald, N. Sasson, N. Wallis, N. Azazmeh, N. Tzarum, O. Fridlich, R. Sher, R. Condiotti, R. Refaeli, R. Ben Ami, R. Zaken-Gallili, R. Helman, S. Ofek, S. Tzaban, S. Piyanzin, S. Anzi, S. Dagan, S. Lilenthal, T. Sido, T. Licht, T. Friehmann, Y. Kaufman, A. Pery, A. Saada, A. Dekel, A. Yeffet, A. Shaag, A. Michael-Gayego, E. Shay, E. Arbib, H. Onallah, K. Ben-Meir, L. Levinzon, L. Cohen-Daniel, L. Natan, M. Hamdan, M. Rivkin, M. Shwieki, O. Vorontsov, R. Barsuk, R. Abramovitch, R. Gutorov, S. Sirhan, S. Abdeen, Y. Yachnin, and Y. Daitch
Appendix A. Supplementary data
The following are the Supplementary data to this article:
References
- 1.Fomsgaard A.S., Rosenstierne M.W. An alternative workflow for molecular detection of SARS-CoV-2 - escape from the NA extraction kit-shortage, Copenhagen, Denmark, March 2020. Euro Surveill. 2020;25 doi: 10.2807/1560-7917.ES.2020.25.14.2000398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Yelin I., Aharony N., Shaer-Tamar E., Argoetti A., Messer E., Berenbaum D., et al. Evaluation of COVID-19 RT-qPCR test in multi-sample pools. Clin Infect Dis. 2020 doi: 10.1093/cid/ciaa531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Zhang Y., Odiwuor N., Xiong J., Sun L., Nyaruaba R.O., Wei H., et al. Rapid molecular detection of SARS-CoV-2 (COVID-19) virus RNA using colorimetric LAMP. medRxiv. 2020 doi: 10.1101/2020.02.26.20028373. 02.26.20028373. [DOI] [Google Scholar]
- 4.Yan C., Cui J., Huang L., Du B., Chen L., Xue G., et al. Rapid and visual detection of 2019 novel coronavirus (SARS-CoV-2) by a reverse transcription loop-mediated isothermal amplification assay. Clin Microbiol Infect. 2020;26(6):773–779. doi: 10.1016/j.cmi.2020.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Park G.-S., Ku K., Baek S.-H., Kim S.-J., Kim S.I., Kim B.-T., et al. Development of reverse transcription loop-mediated isothermal amplification (RT-LAMP) assays targeting SARS-CoV-2. J Mol Diagn. 2020;22(6):729–735. doi: 10.1016/j.jmoldx.2020.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ben-Assa N., Naddaf R., Gefen T., Capucha T., Hajjo H., Mandelbaum N., et al. SARS-CoV-2 on-the-spot virus detection directly from patients. medRxiv. 2020 doi: 10.1101/2020.04.22.20072389. 04.22.20072389. [DOI] [Google Scholar]
- 7.Ai J.-W., Zhang Y., Zhang H.-C., Xu T., Zhang W.-H. Era of molecular diagnosis for pathogen identification of unexplained pneumonia, lessons to be learned. Emerg Microbes Infect. 2020;9:597–600. doi: 10.1080/22221751.2020.1738905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lucia C., Federico P.-B., Alejandra G.C. An ultrasensitive, rapid, and portable coronavirus SARS-CoV-2 sequence detection method based on CRISPR-Cas12. bioRxiv. 2020 2020.2.29.971127. [Google Scholar]
- 9.Broughton J.P., Deng X., Yu G., Fasching C.L., Singh J., Streithorst J., et al. Rapid detection of 2019 novel coronavirus SARS-CoV-2 using a CRISPR-based DETECTR lateral flow assay. medRxiv. 2020 doi: 10.1101/2020.03.06.20032334. 03.06.20032334. [DOI] [Google Scholar]
- 10.Guo L., Ren L., Yang S., Xiao M., Chang D., Yang F., et al. Profiling early humoral response to diagnose novel coronavirus disease (COVID-19) Clin Infect Dis. 2020 doi: 10.1093/cid/ciaa310. In press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Xiao S.-Y., Wu Y., Liu H. Evolving status of the 2019 novel coronavirus infection: proposal of conventional serologic assays for disease diagnosis and infection monitoring. J Med Virol. 2020;92:464–467. doi: 10.1002/jmv.25702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhang W., Du R.-H., Li B., Zheng X.-S., Yang X.-L., Hu B., et al. Molecular and serological investigation of 2019-nCoV infected patients: implication of multiple shedding routes. Emerg Microbes Infect. 2020;9:386–389. doi: 10.1080/22221751.2020.1729071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lassaunière R., Frische A., Harboe Z.B., Nielsen A.C.Y., Fomsgaard A., Krogfelt K.A., et al. Evaluation of nine commercial SARS-CoV-2 immunoassays. medRxiv. 2020 doi: 10.1101/2020.04.09.20056325. 04.09.20056325. [DOI] [Google Scholar]
- 14.Barra G.B., Santa Rita T.H., Mesquita P.G., Jacomo R.H., Nery L.F.A. Analytical sensibility and specificity of two RT-qPCR protocols for SARS-CoV-2 detection performed in an automated workflow. medRxiv. 2020 doi: 10.1101/2020.03.07.20032326. 03.07.20032326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kalikiri M.K.R., Hasan M.R., Mirza F., Xaba T., Tang P., Lorenz S. High-throughput extraction of SARS-CoV-2 RNA from nasopharyngeal swabs using solid-phase reverse immobilization beads. medRxiv. 2020 doi: 10.1101/2020.04.08.20055731. 04.08.20055731. [DOI] [Google Scholar]
- 16.Du D., Hwang F.K., Hwang F. World Scientific; 2000. Combinatorial Group Testing and Its Applications. Series on Applied Mathematics vol. 12 (Singapore). ISBN: 9810241070. [DOI] [Google Scholar]
- 17.Aldridge M., Johnson O., Scarlett J. Group testing: an information theory perspective. Found Trends Commun Inf Theory. 2019;15:196–392. [Google Scholar]
- 18.Sinnott-Armstrong N., Klein D., Hickey B. Evaluation of group testing for SARS-CoV-2 RNA. medRxiv. 2020 doi: 10.1101/2020.03.27.20043968. 03.27.20043968. [DOI] [Google Scholar]
- 19.Shani-Narkiss H., Gilday O.D., Yayon N., Landau I.D. Efficient and practical sample pooling high-throughput PCR diagnosis of COVID-19. MedRxiv. 2020 doi: 10.1101/2020.04.06.20052159. [DOI] [Google Scholar]
- 20.Deckert A., Bärnighausen T., Kyei N. Pooled-sample analysis strategies for COVID-19 mass testing: a simulation study. Bull World Health Organ. April 2020;2 doi: 10.2471/BLT.20.257188. E-pub. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Shental N., Levy S., Skorniakov S., Wuvshet V., Shemer-Avni Y., Porgador A., et al. Efficient high throughput SARS-CoV-2 testing to detect asymptomatic carriers. medRxiv. 2020 doi: 10.1101/2020.04.14.20064618. 04.14.20064618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gollier C., Gossner O. Group testing against Covid-19. Covid Econ. April 2020;1(2):32–42. [Google Scholar]
- 23.Kwiatkowski T.J., Jr., Zoghbi H.Y., Ledbetter S.A., Ellison K.A., Chinault A.C. Rapid identification of yeast artificial chromosome clones by matrix pooling and crude lysate PCR. Nucleic Acids Res. 1990;18:7191–7192. doi: 10.1093/nar/18.23.7191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Barillot E., Lacroix B., Cohen D. Theoretical analysis of library screening using a N-dimensional pooling strategy. Nucleic Acids Res. 1991;19:6241–6247. doi: 10.1093/nar/19.22.6241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Dorfman R. The detection of defective members of large populations. Ann Math Stat. 1943;14:436–440. [Google Scholar]
- 26.Li T., Chan C.L., Huang W., Kaced T., Jaggi S. IEEE International Symposium on Information Theory, Honolulu, HI, 2014. 2014. Group testing with prior statistics; pp. 2346–2350. [Google Scholar]
- 27.Lavezzo E., Franchin E., Ciavarella C., Cuomo-Dannenburg G., Barzon L., Del Vecchio C., et al. Suppression of a SARS-CoV-2 outbreak in the Italian municipality of Vo. Nature. 2020 doi: 10.1038/s41586-020-2488-1. 04.17.20053157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Randazzo W., Truchado P., Cuevas-Ferrando E., Simón P., Allende A., Sánchez G. SARS-CoV-2 RNA in wastewater anticipated COVID-19 occurrence in a low prevalence area. Water Res. 2020;181:115942. doi: 10.1016/j.watres.2020.115942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Orive G., Lertxundi U., Barcelo D. Early SARS-CoV-2 outbreak detection by sewage-based epidemiology. Sci Total Environ. 2020;732:139298. doi: 10.1016/j.scitotenv.2020.139298. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.