Abstract
An influenza A (H3N2) virus epidemic occurred in China in 2017 and the causative strain failed to bind red blood cells (RBCs), which may affect receptor binding and antibody recognition. The objective of this study was to analyze the genetic characteristics and glycosylation changes of this novel H3N2 strain. We directly sequenced the hemagglutinin (HA) genes of H3N2 clinical specimens collected from patients with acute respiratory tract infection during 2017 in Guangdong, China. We aligned these sequences with those of A/Hong Kong/1/1968 (H3N2) and A/Brisbane/10/2007 (H3N2). Glycosylation changes were analyzed by C18 Chip-Q-TOF-MS. A/China/LZP/2017 (H3N2) was negative by HA assay, but was positive by quantitative real-time Polymerase Chain Reaction (qPCR) and direct immunofluorescence assay (DFA). We found that the HA1 residue 160T of A/China/LZP/2017 (H3N2) could block virus binding to receptors on RBCs. Furthermore, the ASN (N)-X-Thr (T) motif at HA1 residues 158–160, encoding a glycosylation site as shown by C18 Chip-Q-TOF-MS, predominated worldwide and played a critical role in RBC receptor binding. Ten glycoforms at HA1 residue 158 were identified [4_3_1_0, 5_6_0_1, 3_3_0_1, 4_4_3_0, 6_7_0_0 (SO3), 3_6_2_0, 4_3_1_2 (SO3), 7_5_2_0 (SO3), 3_6_2_1 (SO3), and 3_7_0_2]. Glycosylation changes at HA1 residues 158–160 of a circulating influenza A (H3N2) virus in Guangdong, China, in 2017 blocked binding to RBC receptors. Changes to these HA1 residues may have reduced protective antibody responses as well. Understanding these critical epitopes is important for selecting vaccine strains.
Keywords: A(H3N2), receptor binding, antibody recognition, glycosylation possessing, glycoforms
Introduction
Influenza A virus H3N2 first emerged in 1968, and frequently leads to annual outbreaks within the human population (Nair et al., 2011). Important contributions to the pandemic of influenza have come from hemagglutinin (HA), which is a major surface antigen of the virus. Mutations in the HA may cause affecting receptor binding and immune response. Thus, understanding the novel mutation is critical for developing strategies to control the pandemic of influenza A (H3N2).
The hemagglutinin (HA) protein plays an important role in binding host cell receptors and releasing the viral RNA into the cell (Beer et al., 2018). HA is composed of two polypeptides, HA1 and HA2. HA1 contains the receptor binding site in a globular head domain and targets sialic acid residues on host cells. HA2 forms a stem-like structure containing a fusion domain (N-terminal) and a transmembrane anchor domain (C-terminal), and mediates fusion of the virus envelope and endosomal membrane (Skehel and Wiley, 2000). Both regions contain N-linked oligosaccharides (Keil et al., 1985), which may interfere with the function of HA. The majority of oligosaccharides are conserved and promote fusion activity. But glycans near the antigenic epitopes may influence the antibody recognition and those near the proteolytic activation site may affect the infectivity of the influenza virus (Skehel et al., 1984; Deshpande et al., 1987).
HA mutations of influenza A (H3N2) have been emerging with changes of glycosylation since 1968. Influenza A (H3N2) has re-assorted with an H2N2 virus, the former virus is capable of transmission between humans (Westgeest et al., 2014; Poucke et al., 2015). The H3N2 virus mutated so quickly that the World Health Organization (WHO) has recommended 28 vaccine strains (Lin et al., 2017). The rapid mutation rate of the H3N2 virus has been proposed to lead to generation of a novel virus in 2–5 years (Park et al., 2009). In the 2014/15 season, two new clades started to emerge. Clade 3c.2a, characterized by the F159Y and K160T mutations, bore a novel potential glycosylation site (Chambers et al., 2015; Skowronski et al., 2016). Another clade in circulation was 3c.3a (including strain A/Switzerland/9715293/2013) (Chambers et al., 2015). During the 2016/17 season, clade 3c.2a separated into two new clades. The 3c.2a1, characterized by amino acid changes such as T135K, N171K, and D122N, may have resulted in the loss of a glycosylation site (Melidou et al., 2017). By the 2017/18 season, clade 3c.2a2 began to split into cluster I, bearing the I58V and S219K mutations, and cluster II, bearing the N122D and S262N mutations. The N122D mutation resulted in the loss of a glycosylation site (Harvala et al., 2017). Generally, the HA1 genes of H3N2 viruses undergo continuous mutations result in losing or possessing of glycosylation to reduce host immune recognition and to enhance transmissibility (Hensley et al., 2009; Popova et al., 2012). The majority of neutralizing antibodies induced by vaccination or infection do not target the receptor-binding site (RBS), which is surrounded by highly variable regions of HA1, and prevent virus binding to cell receptors (Gerhard et al., 1981; Knossow et al., 1984; Whittle et al., 2011).
Besides, the HA protein is critical for the virulence and host preference as it binds to sialic acid receptors on epithelial cells (Shinya et al., 2006; de Wit et al., 2010). It was reported that mutation of HA glycosylation sites can affect the specificity and affinity for receptor binding (Mair et al., 2014). As the amount of glycans becomes more, the binding ability for the receptor is weakened (Abe et al., 2004). However, influenza will undergo multiple mutations to make a successful adaptation to humans.
Overall, the number and position of N-linked glycans in HA have influenced the evolution of H3N2 viruses and can contribute to immune evasion (Wanzeck et al., 2011; Tate et al., 2014). In our study, we identified H3N2 clinical strains from Guangdong, China, in 2017 that were not able to bind RBCs. The surface of RBCs contains many receptors for binding by HA. RBCs clump together because of interactions between the HA of virus particles and RBCs, leading to a lattice formation, which called the hemagglutination assay (Grimes, 2002). This suggested that H3N2 virus epitopes may change and influence binding to host cell receptors as well as antibody responses. Therefore, it is necessary to provide further insights into the genetic characteristics and glycosylation changes of H3N2 viruses.
Materials and Methods
Cells and Viruses
Madin-Darby canine kidney (MDCK) cells were purchased from the American Tissue Culture Collection (ATCC). The cells were cultured in minimum Eagle’s medium containing 10% heat-inactivated fetal bovine serum. Influenza A viruses A/Brisbane/10/2007 (H3N2) (B10) and A/Hong Kong/4801/2014 (H3N2) (HK4801) were also purchased from the ATCC. Influenza A viruses A/China/LZP/2017 (H3N2) LZP) and A/China/GMU-SKLRD03/2017 (H3N2) (GMU03) were detected and isolated from clinical specimens. Collection of clinical specimens was approved by the Ethics Committee of The First Affiliated Hospital of Guangzhou Medical University (2014 No. 50; Guangzhou, China; File number: 2014-50, date: June 2, 2015). Written informed consent was obtained from study participants.
Viral Genome Characterization and Reverse Genetics
RNA of the four H3N2 strains was extracted and reverse transcribed. The sequences of LZP and GMU03 were deposited in GenBank (LZP: Accession numbers MK053902 to MK053909; GMU03: Accession numbers MN658587). The eight viral segments of different strains were amplified and cloned into a pHW2000 plasmid system as previously described (Wang et al., 2019). The seven segments of LZP (NA, PB1, PB2, PA, NP, M, NS) were used as a backbone to rescue recombinant viruses (LZP-B10, LZP-HK4801, LZP-GMU03) bearing the HA genes of B10, HK4801, and GMU03, respectively.
Hemagglutination Assay
The hemagglutination assay was performed in a V-bottom 96-well microtiter plate. The most concentrated sample was in the first well, and subsequent wells were serially diluted twofold in phosphate-buffered saline (PBS). The first row served as a negative control and contained only PBS. Different strains of virus were detected by 0.5% solution of turkey RBCs.
Quantitative Real-Time PCR (qPCR)
RNA from wild type and recombinant H3N2 viruses was extracted using the QIAamp Viral RNA Mini Kit (Qiagen, Germany) and analyzed by qPCR using the ABI 7500 Real-time PCR System (Wang et al., 2019).
Direct Immunofluorescence Assay (DFA)
The nucleoproteins (NPs) of H3N2 viruses were detected using the D3 Ultra DFA Respiratory Virus Screen and ID Kit (Healthline Media UK Ltd., Brighton, United Kingdom). H3N2 strains (LZP, GMU03) and B10- or HK4801-infected cells were fixed in 4% paraformaldehyde at 37°C for 30 min. Fluorescein isothiocyanate (FITC)-labeled mouse anti-NP antibody was used to detect nucleoprotein. The cells were observed using a fluorescence microscope (Zeiss Axiovert 135, Zeiss, Oberkochen, Germany).
Bioinformatic Analysis
The HA genes of B10 and HK4801 were obtained from the National Center for Biotechnology Information (NCBI) Influenza Virus Resource Database and the HA genes of LZP and GMU03 were sequenced by Sangon Biotech (Shanghai, China). Both sequences were analyzed using Vector NTI (Invitrogen, United States). The prevalence of the T160K, N158K and the substitutions at residues 158–160 were analyzed through the Global Initiative on Sharing All Influenza Data (GISAID) database1.
Mutation of H3N2 HA1 N158K and T160K
Mutations were introduced at HA1 residues 158 and 160 using the QuikChange site-directed mutagenesis kit (Agilent Technologies, Santa Clara, United States). HA1-N158K and HA1-T160K mutant viruses were rescued as described previously (Hoffmann et al., 2000).
Enzymatic Digestion of Influenza Proteins
Influenza proteins were purified by filtration through a 10-kDa cutoff centrifugal filter, and sequentially treated with 10 mM dithiothreitol and 2 M hydroxylamine. The samples were alkylated with 55 mM iodoacetamide. Next, the samples were concentrated and buffer-exchanged buffer into H2O using a 3-kDa cutoff centrifugal filter. The protein concentration was determined using a Bio-Rad protein assay.
Fifty micrograms of protein were diluted with 100 mM Tris-HCl containing 10 mM CaCl2 (pH 8.0) to a final concentration of 1 μg/μL. Two micrograms of chymotrypsin were added and incubated for 16 h incubation at 25°C. The digested samples were loaded onto the preconditioned porous graphitic carbon cartridge and targeted glycopeptides were eluted with 50% acetonitrile (ACN) containing 0.1% trifluoroacetic acid. The collected fraction was dried using a speed vacuum. Finally, the dried residue was reconstituted in 50 μL of MilliQ H2O prior to analysis.
Glycopeptide Profiling by C18 Chip-Q-TOF-MS
Chromatographic separation of glycopeptides was performed on an Agilent 1260 Infinity HPLC Chip LC system (Agilent, Santa Clara, CA, United States) equipped with a Polaris-HR C18 chip (G4240-62030, Agilent). Three microliters of each sample were loaded onto the enrichment column in a solution of 0.1% formic acid (FA) and 3% ACN in water at 3 μL/min. The mobile phase used in the nano-pump consisted of 0.1% FA in water (A) and 0.1% FA in ACN (B). The gradient was as follows: 3–40% B for 15 min, 40–70% B for 1 min, 70% B for 1 min, 70–3% B for 0.5 min, and 3% B for 10 min. The flow rate was 0.45 μL/min.
Glycopeptide profiling was carried out on an Agilent 6550 iFunnel Q-TOF MS. The dry gas (N2) was flowed at 11 L/min at 250°C. MS spectra were acquired in positive mode with the mass range m/z 450–3000. Mass correction was enabled using reference masses of m/z 922.0098.
Ethics Approval and Consent to Participate
All clinical specimens have been approved from the Ethics Committee of The First Affiliated Hospital of Guangzhou Medical University (2014 no. 50; Guangzhou, China) (File number: 2014-50 date: June 2, 2015). Written informed consent was obtained from the participant.
Results
Detection of H3N2 Clinical Specimens
LZP and GMU03 were negative by hemagglutination assay. The positive controls, B10 and HK4801 were positive and their hemagglutination titers were 1:64 and 1:512, respectively (Table 1 and Supplementary Figure S1). To determine whether the two clinical isolates were truly negative for hemagglutination activity, we used qPCR and DFA. The CT values of LZP and GMU03 were 21.85 and 23.14, respectively, by qPCR (Table 1). The viral NPs of the H3N2 viruses were detected by DFA using a FITC-labeled mouse anti-NP antibody (Figure 1).
TABLE 1.
qPCR for detecting HA in H3N2 clinical specimens.
| Species of virus | Hemagglutination assay | qPCR (CT values) |
| A/China/LZP/2017 (H3N2) | (–) | 21.85 |
| A/China/GMU-SKLRD03/2017 (H3N2) | (–) | 23.14 |
| A/Brisbane/10/2007 | (+) 1:64 | 15.08 |
| A/Hong Kong/4801/2014 (H3N2) | (+) 1:512 | 14.24 |
(+), positive; (–), negative. Throat swabs (LZP, GMU03) were collected from patients with acute respiratory tract infection (ARTI) in Guangdong, China during 2017. The viral RNA/DNA was extracted and reverse transcribed. The HA of A (H3N2) was determined by qPCR kit. Meanwhile, the clinical specimens were tested using the HA assay.
FIGURE 1.
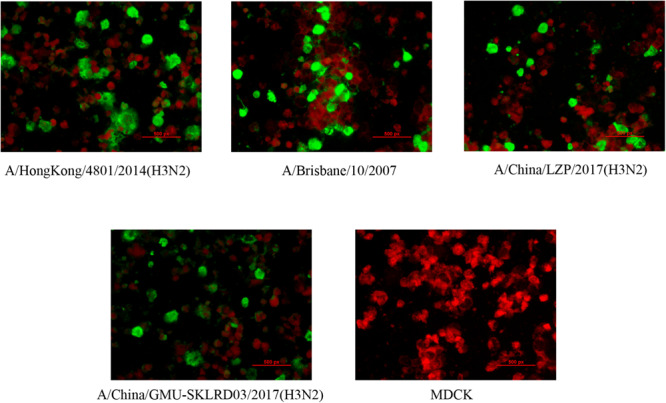
Immunofluorescence assay for detecting H3N2 clinical specimens. The nucleoproteins (NPs) of H3N2 viruses were determined by the D3 Ultra DFA Respiratory Virus Screen and ID Kit. H3N2 strains (LZP, GMU03) and B10- or HK4801-infected cells were fixed in 4% paraformaldehyde at 37°C for 30 min. FITC-labeled mouse anti-NP antibody was used to detect nucleoprotein. The cells were observed using a fluorescence microscope.
Determination of Critical Viral Proteins
To confirm whether the HA protein affected the binding ability of viruses to RBCs, LZP-03, LZP-B10, and LZP-4801 H3N2 viruses were rescued as recombinant viruses. LZP and LZP-GMU03 recombinant viruses were both negative by hemagglutination assay (Figure 2). However, the LZP-B10 and LZP-HK4801 recombinant viruses showed hemagglutination activity at titers of 1:64 and 1:16, respectively. Thus, negative results in hemagglutination assays were caused by the HA proteins of H3N2 viruses.
FIGURE 2.
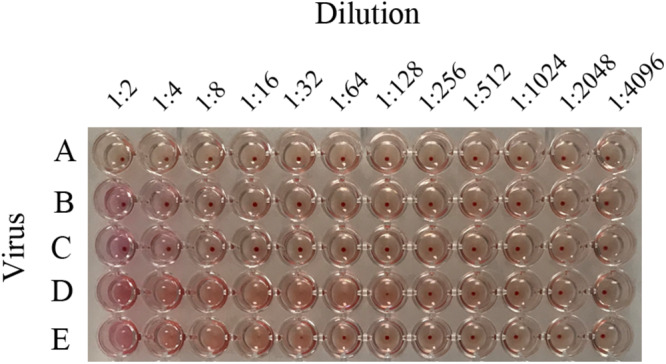
Hemagglutination titers of H3N2 recombinant viruses. To figure out whether HA protein of LZP led to unable to bind the RBCs, the seven segments of LZP (NA, PB1, PB2, PA, NP, M, NS) were used as a backbone to rescue recombinant viruses (LZP-B10, LZP-HK4801, LZP-GMU03) bearing the HA genes of B10, HK4801, and GMU03, respectively. The recombinant viruses and LZP were determined by HA assay. A: PBS; B: LZP; C: LZP-GMU03; D: LZP-B10; E: LZP-HK4801.
Identification of Mutated Sites in A/China/LZP/2017 (H3N2)
To identify important mutations, the HA sequences of LZP and GMU03 were compared with those of B10 and HK4801. Two mutations in the HA protein were found: HA1-K121N and HA1-T160K.
Critical Nature of the N158K and T160K Mutations in A/China/LZP/2017 (H3N2)
According to the GISAID database, a Thr substitution at HA1 residue 160 H3N2 viruses rose in frequency from 2013 to the present day. By contrast, the peak frequency of K160 occurred in 2012 (2848/2850 isolates, 99.93%) and then gradually declined (Figure 3). After incorporating a T160K mutation in LZP, the hemagglutination assay was positive with a titer of 1:64. However, the frequency of K160 started to rise in 2018. In addition, another critical substitution of LZP was identified at HA1 residue 158. When Asn was substituted with Lys at this position, a positive result was also obtained in the hemagglutination assay (Figure 4). Both N158K and T160K of LZP could change the binding ability to RBCs. From 2013 to present, the frequency of the N-X-T motif at residues 158–160 increased worldwide and became dominant in 2015 (5198/6805 isolates, 76.4%), where X is any amino acid except Pro. In Guangdong, China, the N-X-T motif began to dominate in 2016 (6/8 isolates, 75%) (Figures 5, 6).
FIGURE 3.
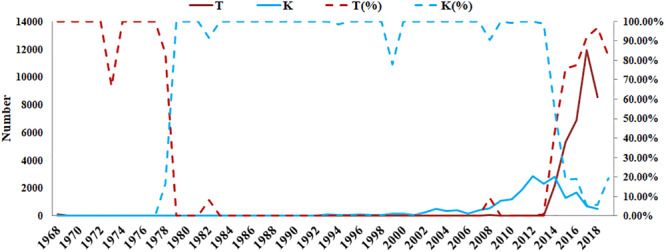
Prevalence of T and K residues at HA1 position 160 in the world since 1968. The sequences of 62330 clinical strains worldwide from 1968 to 2019 were collected from GISAID. The T and K residues at HA1 position 160 were analyzed. The y-axis on the left: the number of HA1-160T and HA1-160K; The y-axis on the right: the percentage of HA1-160T and HA1-160K.
FIGURE 4.
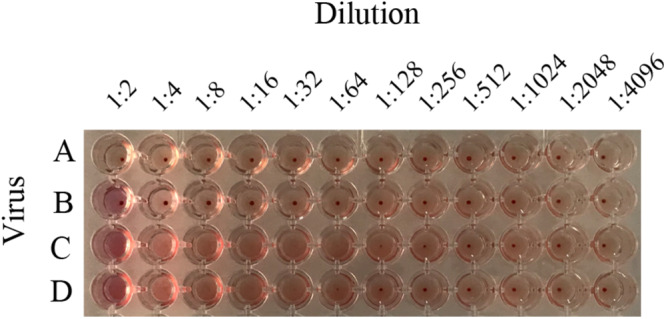
Hemagglutination titers of H3N2 mutant strains. To distinguish whether the possessing of glycosylation motif site at position 158 failed to bind RBCs, recombinant virus containing HA1-160K, HA1-158K, and LZP were determined by HA assay. A: PBS; B: LZP; C: T160K; D: N158K.
FIGURE 5.
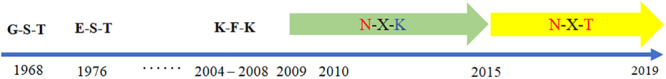
Worldwide prevalence of the glycosylation motif site at position 158 from 1968 to 2019. The sequences of 62,330 clinical strains worldwide from 1968 to 2019 were collected from GISAID. The substitutes at position 158–160 were analyzed.
FIGURE 6.
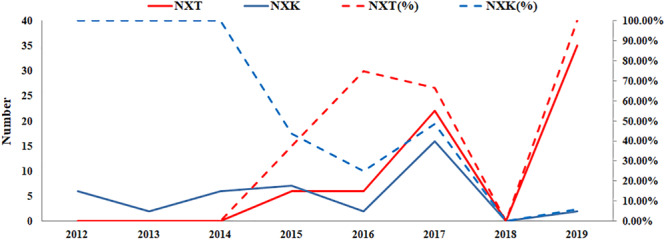
Prevalence of the N-X-T/K motif in Guangdong, China from 2012 to 2019. The sequences of 106 clinical strains in Guangdong, China from 2012 to 2019 were collected from GISAID. The substitutes N-X-T/K at position 158–160 were analyzed. The y-axis on the left: the number of the substitutes N-X-T and N-X-K at HA1 residues 158–160; The y-axis on the right: the percentage of the substitutes N-X-T and N-X-K at HA1 residues 158–160.
Glycopeptide Profiling of HA1 Residue 158
N-linked glycans can only be attached to the nitrogen atom of an N residue that forms part of the consensus sequence N-X-Ser(S)/T. Therefore, one chymotryptic glycopeptide at residues 154–159 (peptide sequence: LTHLNY) of HA, which possessed a glycosylation site at HA1 residue 158, was identified in LZP. Based on the high-resolution MS data and matching with the database, 10 glycoforms were identified at residue HA1 position 158 including three neutral glycans (3_6_2_0, 4_3_1_0, and 4_4_3_0), three acidic glycans (5_6_0_1, 3_3_0_1, and 3_7_0_2), and four sulfated glycans [6_7_0_0 (SO3), 4_3_1_2 (SO3), 7_5_2_0 (SO3), and 3_6_2_1 (SO3)] (Figure 7). Both the N158K and T160K mutations resulted in loss of this glycosylation site.
FIGURE 7.
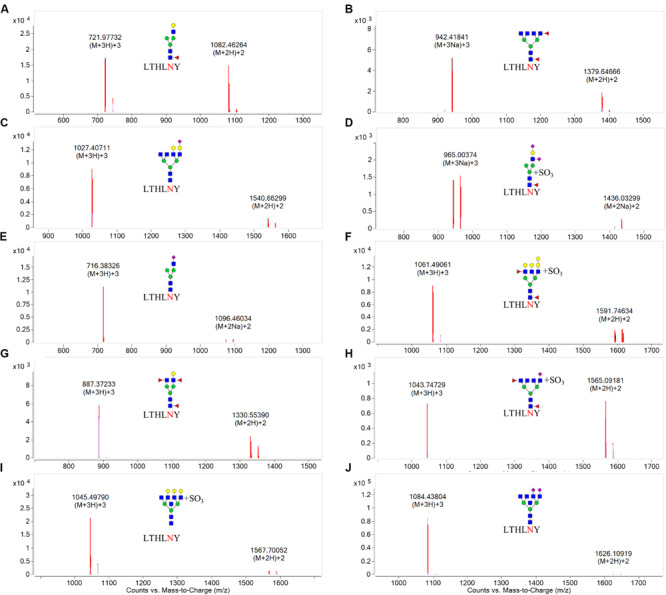
Overlaid extracted compound chromatograms (ECCs) of identified HA1 glycopeptides containing the glycosylation site at position 158. HA1 proteins extracted from influenza were digested with chymotrypsin at 25°C for 16 h, followed by profiling the glycopeptides by C18 chip-Q-TOF-MS. The glycoforms at position 158 were then identified based on high-resolution MS data. (A): 4_3_1_0, (B): 3_6_2_0, (C): 5_6_0_1, (D): 4_3_1_2 (SO3), (E): 3_3_0_1, (F): 7_5_2_0 (SO3), (G): 4_4_3_0, (H): 3_6_2_1 (SO3), (I): 6_7_0_0 (SO3), (J): 3_7_0_2.
Discussion
Influenza is a global threat and represents a pressing public health challenge worldwide (Schoenbaum, 2004). In China, an outbreak of influenza A H3N2 occurred during 2017 (Supplementary Figure S2). We found that some H3N2 strains in Guangdong did not bind RBCs and tested negative by hemagglutination assay. To confirm this result, we collected several clinical H3N2 strains, along with B10 and HK4801 as positive controls, and analyzed their genetic characteristics. As HA protein is present on enveloped viruses such as influenza viruses, it is able to bind receptors on the membranes of RBCs to cause agglutination (Grimes, 2002). By inserting the HA genes of GMU03, B10, HK4801 into an LZP backbone, we were able to use a hemagglutination assay to deeply analyze the HA proteins of LZP and GMU03. We found that HA1 121K and 160T may play an important role in binding to RBCs. According to the GISAID database, the frequency of HA1 K160 has risen since 2017 worldwide, but T160 was still dominant in these years. Another residue, HA1 158, was also investigated. We mutated N158K and T160K and found that both strains were positive by HA assay, which suggested that mutations at both residues were responsible for failure to bind RBCs. In Guangdong Province, more isolates bore N-X-T substitutions at HA1 residues 158–160 during 2017. Further, we found that HA1 residue 158 of LZP bore an N-linked glycosylation site by enzymatic digestion of influenza proteins and C18 Chip-Q-TOF-MS analysis. In summary, our study confirmed that glycosylation at HA1 residues 158–160 can block receptor binding to RBCs.
It was reported that the loss and possessing of the glycosylation sites in the head region could affect recognition by antibodies. During the 2016/2017 season, Melidou et al. (2017) demonstrated that the T135K, N171K, and D122N substitutions were located within antigenic sites and potentially affected viral antigenicity, as they resulted in losses of N-linked glycosylation sites. Furthermore, the I58V and S219K mutations of clade 3c.2a2, cluster I and the N122D and S262N mutations of cluster II were found to result in the loss of a glycosylation site during the 2017/18 season (Harvala et al., 2017). Nevertheless, since transmitting between the human population, HK/68 have gained up to seven glycosylation sites on the HA globular head and five on the stem from the only original two sites (Yang et al., 2015; Alymova et al., 2016). In our study, we found that strains encoding 160T and 158N began to dominate in Guangdong Province, China during the 2015/16 season, leading to a new potential glycosylation site. Similar to previous studies, they concluded that the glycosylation site was a crucial factor affecting the antibody response. Cueno et al. (2019) demonstrated that residues 158–160 in the H3N2 hemagglutinin globular head had structural significance for antigenic drift by modeling. Moreover, Beer et al. (2018) screened the epitopes of anti-A/Victoria/361/2011 neutralizing mouse monoclonal antibodies (mAbs) and found that HA1 residue 158 was glycosylated leading to reduce binding by mAbs. Zost et al. (2017) also found that the generation of protective antibody responses against a H3N2 vaccine strain was affected during the 2016–2017 H3N2 influenza season due to the substitutions within HA1 residue 160 resulting in a new glycosylation site in the HA protein, which affected antigenicity. Generally, the addition or deletion of glycosylation sites on the globular head is vital for escaping the immune response of human. We supposed that the possessing of the glycan at HA1 residue 158 may be a important contribution for antigenic drift in Guangdong, China in recent years.
Not only the glycosylation site, the length and the type of the glycans on the HA head region may be other reasons underlying our observations. In our study, we discovered 10 different glycoforms at residue 158 [4_3_1_0, 5_6_0_1, 3_3_0_1, 4_4_3_0, 6_7_0_0 (SO3), 3_6_2_0, 4_3_1_2 (SO3), 7_5_2_0 (SO3), 3_6_2_1 (SO3), and 3_7_0_2]. As the length of the glycans, Cheng-Chi et al. (2009) reported that antibodies raised against the fully glycosylated HA were less binding affinity to antibody than bearing a single N-linked GlcNAc at each glycosylation site and implicated that removal of structural and non-essential glycans on viral glycoproteins might be an effective approach for vaccine design. Melissa Boeijen reviewed that long oligosaccharide side chains can block the binding of neutralizing antibody due to coverage of the antibody binding site and mentioned that galactose and fucose were long glycans (Boeijen, 2013). In the study, the length of glyforms were ranked as followed: 7_5_2_0 (SO3) > 4_3_1_2 (SO3), 5_6_0_1 > 4_3_1_0, 4_4_3_0, 6_7_0_0 (SO3) > 3_3_0_1, 3_6_2_1 (SO3), 3_7_0_2 > 3_6_2_0. Therefore, we suggested that 7_5_2_0 (SO3) should be highly concerned. Additionally, the type of oligosaccharides side chains is critical as well to result in antigenic drift. A recent study by An et al. (2019) supposed that the globular head region with high-mannose and complex glycans would promote the virus well shielding under the immune system. As we found, 3_6_2_0, 5_6_0_1, 7_5_2_0 (SO3), 3_6_2_1 (SO3), 6_7_0_0 (SO3), 3_7_0_2 are large, complex glycans on the N158 glycosite. Thus, not only the length but also the types of glycoform are crucial factors to affect immune responses, disturbing antibody recognition by steric hindrance of the binding site.
Since the N-X-T motif at residue 158–160 started to increase in prevalence worldwide in 2015, the WHO still recommended the vaccine strain (A/Hong Kong/4801/2014) until September 2017 (Allen and Ross, 2018). In Guangdong, China, the N-X-T motif became dominant in 2016. Before September 2017, the potential glycosylation site at residue 158 resulted in disruption of antibody recognition. Despite the WHO updating the vaccine strain to A/Singapore/INFIMH-16-0019/2016 bearing a glycosylation site at residue 158, an H3N2 outbreak still occurred in Guangdong, China in 2019. This may affect the vaccine strategy of local government or represent new drift mutations in H3N2 viruses. However, the N-X-T motif at residues 158–160 and the resulting potential glycosylation site will be a focus in Guangdong, China in the future.
A limitation of our study was that we lacked a large number of clinical specimens to be analyzed. In addition, we will continue to test protective antibody responses against the circulating H3N2 virus by generating mAbs and human serum.
Conclusion
Glycosylation at HA1 residues 158–160 of circulating H3N2 viruses in Guangdong, China in 2017 blocked binding to RBC receptors. Introduction of glycosylation sites and glycoform variation can affect protective antibody responses, and thus are critical for guiding vaccine strain selection in specific areas.
Data Availability Statement
The datasets generated for this study are available on request to the corresponding author.
Ethics Statement
The studies involving human participants were reviewed and approved by Ethics Committee of The First Affiliated Hospital of Guangzhou Medical University. The patients/participants provided their written informed consent to participate in this study.
Author Contributions
XW, WS, and J-RW contributed to conceive and design the experiments. ZZ and L-FY contributed to perform the experiments and wrote the original draft. ZL, XX, and ZY contributed to analyze the data and provide the data interpretation. All authors contributed to the study design, data interpretation and revisions to the text, and approved the final text and agreed to be accountable for the work.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thank the volunteers of this study for their generous participation.
Funding. This work was supported by the National Natural Science Foundation of China (Grant No. U1502226) and the General Project of Guangzhou Medical University (Grant No. SKLRD-MS-201908), Guangzhou Medical University High-level University Clinical Research and Cultivation Program (Guangzhou Medical University released [2017] No. 160).
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2020.01318/full#supplementary-material
References
- Abe Y., Takashita E., Sugawara K., Matsuzaki Y., Muraki Y., Hongo S. (2004). Effect of the addition of oligosaccharides on the biological activities and antigenicity of influenza A/H3N2 virus hemagglutinin. J. Virol. 78 9605–9611. 10.1128/jvi.78.18.9605-9611.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allen J. D., Ross T. M. (2018). H3N2 influenza viruses in humans: viral mechanisms, evolution, and evaluation. Hum. Vaccines 14 1840–1847. 10.1007/s40266-018-0537-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alymova I. V., York I. A., Air G. M., Cipollo J. F., Gulati S., Baranovich T., et al. (2016). Glycosylation changes in the globular head of H3N2 influenza hemagglutinin modulate\\r receptor binding without affecting virus virulence. Sci. Rep. 6:36216. 10.1038/srep36216 [DOI] [PMC free article] [PubMed] [Google Scholar]
- An Y., Parsons L. M., Jankowska E., Melnyk D., Joshi M., Cipollo J. F. (2019). N-Glycosylation of Seasonal influenza vaccine hemagglutinins: implication for potency testing and immune processing. J. Virol. 93:e01693-18. 10.1128/jvi.01693-18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beer K., Dai M., Howell S., Rijal P., Townsend A. R., Lin Y., et al. (2018). Characterization of neutralizing epitopes in antigenic site B of recently circulating influenza A(H3N2) viruses. J. Gen. Virol. 99 1001–1011. 10.1099/jgv.0.001101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boeijen M. (2013). Glycosylation of the influenza A virus hemagglutinin protein. Int. J. Nuclear Med. Biol. 11 103–105. [Google Scholar]
- Chambers B. S., Parkhouse K., Ross T. M., Alby K., Hensley S. E. (2015). Identification of Hemagglutinin Residues Responsible for H3N2 Antigenic Drift during the 2014-2015 influenza season. Cell Rep. 12 1–6. 10.1016/j.celrep.2015.06.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng-Chi W., Juine-Ruey C., Yung-Chieh T., Che-Hsiung H., Yu-Fu H., Shih-Wei C., et al. (2009). Glycans on influenza hemagglutinin affect receptor binding and immune response. Proc. Natl. Acad. Sci. U.S.A. 106 18137–18142. 10.1073/pnas.0909696106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cueno M. E., Shiotsu H., Nakano K., Sugiyama E., Kikuta M., Usui R., et al. (2019). Structural significance of residues 158-160 in the H3N2 hemagglutnin globular head: a computational study with implications in viral evolution and infection. J. Mol. Graph. Model. 89 33–40. 10.1016/j.jmgm.2019.02.007 [DOI] [PubMed] [Google Scholar]
- de Wit E., Munster V. J., van Riel D., Beyer W. E., Rimmelzwaan G. F., Kuiken T., et al. (2010). Molecular determinants of adaptation of highly pathogenic avian influenza H7N7 viruses to efficient replication in the human host. J. Virol. 84 1597–1606. 10.1128/jvi.01783-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deshpande K. L., Fried V. A., Ando M., Webster R. G. (1987). Glycosylation affects cleavage of an H5N2 influenza virus hemagglutinin and regulates virulence. Proc. Natl. Acad. Sci. U.S.A. 84 36–40. 10.1073/pnas.84.1.36 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerhard W., Yewdell J., Frankel M. E., Webster R. (1981). Antigenic structure of influenza virus haemagglutinin defined by hybridoma antibodies. Nature 290 713–717. 10.1038/290713a0 [DOI] [PubMed] [Google Scholar]
- Grimes S. E. (2002). “A Basic Laboratory Manual for the Small-Scale Production and Testing of I-2 Newcastle Disease Vaccine,” in 10. Haemagglutination test (Rockville, MD: RAP publication; ). Available online at: http://www.fao.org/3/ac802e/ac802e00.htm#Contents [Google Scholar]
- Harvala H., Frampton D., Grant P., Raffle J., Nastouli E. (2017). Emergence of a novel subclade of influenza A(H3N2) virus in London, December 2016 to January 2017. Eur. Commun. Dis. Bull. 22:30466. 10.2807/1560-7917.ES.2017.22.8.30466 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hensley S. E., Das S. R., Bailey A. L., Schmidt L. M., Hickman H. D., Jayaraman A., et al. (2009). Hemagglutinin receptor binding avidity drives influenza A virus antigenic drift. Science 326 734–736. 10.1126/science.1178258 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffmann E., Neumann G., Kawaoka Y., Hobom G., Webster R. G. (2000). A DNA transfection system for generation of influenza A virus from eight plasmids. Proc. Natl. Acad. Sci. U.S.A. 97 6108–6113. 10.1073/pnas.100133697 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keil W., Geyer R., Dabrowski J., Dabrowski U., Niemann H., Stirm S., et al. (1985). Carbohydrates of influenza virus. Structural elucidation of the individual glycans of the FPV hemagglutinin by two-dimensional 1H n.m.r. and methylation analysis. EMBO J 4 2711–2720. 10.1002/j.1460-2075.1985.tb03991.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knossow M., Daniels R. S., Douglas A. R., Skehel J. J., Wiley D. C. (1984). Three-dimensional structure of an antigenic mutant of the influenza virus haemagglutinin. Nature 311 678–680. 10.1038/311678a0 [DOI] [PubMed] [Google Scholar]
- Lin Y., Wharton S. A., Whittaker L., Dai M., Ermetal B., Lo J., et al. (2017). The characteristics and antigenic properties of recently emerged subclade 3C.3a and 3C.2a human influenza A(H3N2) viruses passaged in MDCK cells. Influenza Other Respir. Viruses 11 263–274. 10.1111/irv.12447 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mair C. M., Ludwig K., Herrmann A., Sieben C. (2014). Receptor binding and pH stability - how influenza A virus hemagglutinin affects host-specific virus infection. Biochim. Biophys. Acta 1838 1153–1168. 10.1016/j.bbamem.2013.10.004 [DOI] [PubMed] [Google Scholar]
- Melidou A., Gioula G., Exindari M., Ioannou E., Gkolfinopoulou K., Georgakopoulou T., et al. (2017). Influenza A(H3N2) genetic variants in vaccinated patients in northern Greece. J. Clin. Virol. 94 29–32. 10.1016/j.jcv.2017.07.003 [DOI] [PubMed] [Google Scholar]
- Nair H., Brooks W. A., Katz M., Roca A., Berkley J. A., Madhi S. A., et al. (2011). Global burden of respiratory infections due to seasonal influenza in young children: a systematic review and meta-analysis. Lancet 378 1917–1930. 10.1016/s0140-6736(11)61051-9 [DOI] [PubMed] [Google Scholar]
- Park A. W., Daly J. M., Lewis N. S., Smith D. J., Wood J. L., Grenfell B. T. (2009). Quantifying the impact of immune escape on transmission dynamics of influenza. Science 326 726–728. 10.1126/science.1175980 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Popova L., Smith K., West A. H., Wilson P. C., James J. A., Thompson L. F., et al. (2012). Immunodominance of antigenic site B over site A of hemagglutinin of recent H3N2 influenza viruses. PLoS One 7:e41895. 10.1371/journal.pone.0041895 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poucke S. V., Doedt J., Baumann J., Qiu Y., Matrosovich M. (2015). Role of Substitutions in the Hemagglutinin in the Emergence of the 1968 Pandemic influenza virus. J. Virol. 89 12211–12216. 10.1128/JVI.01292-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schoenbaum S. C. (2004). The great influenza: the epic story of the deadliest plague in history. J. Public Health Policy 25 435–443. 10.1080/01947648.2015.1262197 [DOI] [PubMed] [Google Scholar]
- Shinya K., Ebina M., Yamada S., Ono M., Kasai N., Kawaoka Y. (2006). Avian flu: influenza virus receptors in the human airway. Nature 440 435–436. 10.1038/440435a [DOI] [PubMed] [Google Scholar]
- Skehel J. J., Stevens D. J., Daniels R. S., Douglas A. R., Knossow M., Wilson I. A., et al. (1984). A carbohydrate side chain on hemagglutinins of Hong Kong influenza viruses inhibits recognition by a monoclonal antibody. Proc. Natl. Acad. Sci. U.S.A. 81 1779–1783. 10.1073/pnas.81.6.1779 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skehel J. J., Wiley D. C. (2000). Receptor binding and membrane fusion in virus entry: the influenza hemagglutinin. Annu. Rev. Biochem. 69 531–569. 10.1146/annurev.biochem.69.1.531 [DOI] [PubMed] [Google Scholar]
- Skowronski D. M., Sabaiduc S., Chambers C., Eshaghi A., Li Y. (2016). Mutations acquired during cell culture isolation may affect antigenic characterisation of influenza A(H3N2) clade 3C.2a viruses. Eur. Commun. Dis. Bull. 21:30112. 10.2807/1560-7917.ES.2016.21.3.30112 [DOI] [PubMed] [Google Scholar]
- Tate M. D., Job E. R., Deng Y. M., Gunalan V., Maurer-Stroh S., Reading P. C. (2014). Playing hide and seek: how glycosylation of the influenza virus hemagglutinin can modulate the immune response to infection. Viruses 6 1294–1316. 10.3390/v6031294 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y., Zeng Z., Chen Q., Yan W., Chen Y., Xia X., et al. (2019). Pterodontic acid isolated from Laggera pterodonta suppressed RIG-I/NF-KB/STAT1/Type I interferon and programmed death-ligand 1/2 activation induced by influenza A virus in vitro. Inflammopharmacology 27 1255–1263. 10.1007/s10787-019-00571-y [DOI] [PubMed] [Google Scholar]
- Wanzeck K., Boyd K. L., McCullers J. A. (2011). Glycan shielding of the influenza virus hemagglutinin contributes to immunopathology in mice. Am. J. Respir. Crit. Care Med. 183 767–773. 10.1164/rccm.201007-1184OC [DOI] [PMC free article] [PubMed] [Google Scholar]
- Westgeest K. B., Russell C. A., Lin X., Spronken M. I., Bestebroer T. M., Bahl J., et al. (2014). Genomewide analysis of reassortment and evolution of human influenza A(H3N2) viruses circulating between 1968 and 2011. J. Virol. 88 2844–2857. 10.1128/jvi.02163-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whittle J. R., Zhang R., Khurana S., King L. R., Manischewitz J., Golding H., et al. (2011). Broadly neutralizing human antibody that recognizes the receptor-binding pocket of influenza virus hemagglutinin. Proc. Natl. Acad. Sci. U.S.A. 108 14216–14221. 10.1073/pnas.1111497108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang H., Carney P. J., Chang J. C., Guo Z., Villanueva J. M., Stevens J. (2015). Structure and receptor binding preferences of recombinant human A(H3N2) virus hemagglutinins. Virology 477 18–31. 10.1016/j.virol.2014.12.024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zost S. J., Parkhouse K., Gumina M. E., Kim K., Diaz Perez S., Wilson P. C., et al. (2017). Contemporary H3N2 influenza viruses have a glycosylation site that alters binding of antibodies elicited by egg-adapted vaccine strains. Proc. Natl. Acad. Sci. U.S.A. 114 12578–12583. 10.1073/pnas.1712377114 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets generated for this study are available on request to the corresponding author.


