Abstract
Background:
There is strong evidence for a role of type 2 cytokines in the pathogenesis of eosinophilic esophagitis (EoE); however, heterogeneity in type 2 gene expression has not been examined.
Objective:
We examined type 2 immunity-associated gene expression in esophageal biopsies aiming to determine the degree of cytokine heterogeneity and its potential clinical significance.
Methods:
Patients (n=312) were recruited from 10 sites associated with the Consortium of Eosinophilic Gastrointestinal Disease Researchers (CEGIR). In addition to histologic and endoscopic assessment, esophageal biopsies were examined for expression of 96 genes within the EoE Diagnostic Panel (EDP).
Results:
Five subgroups of patients with active EoE were identified by unsupervised clustering based on expression of IL4, IL5, IL13, CCL26, TSLP, CLC, CCR3, and CPA3. These groups differed in age (P < 0.02) and EDP score (P <1.08E-30) but not eosinophil levels. Group V patients had the highest expression of IL5, TSLP, CCL26, and genes associated with tissue remodeling such as COL8A1, ACTG2 and TSPAN12. IL5 and IL13 were highly expressed in group IV; however, groups IV and V differed in age (34 vs. 14 years, P < 0.001). Groups II and III, which exhibited intermediate expression of IL5 and CPA3, were differentiated by high TSLP and IL13 in group III.
Conclusion:
We observed heterogeneous type 2 gene expression among patients with active EoE. Type 2 gene overexpression was not directly proportional to disease features; this was especially true for tissue remodeling events. These findings highlight a clinical opportunity for leveraging molecular endotypes to implement personalized medicine in EoE.
Keywords: Allergy, Cytokine, Eosinophil, Fibrosis, IL-5, IL-13, Inflammation, Precision Medicine, Resolution, T-helper type 2
Graphical Abstract
Capsule Summary
This study demonstrates heterogeneity of type 2 immunity in eosinophilic esophagitis independent of esophageal eosinophil levels. These data substantiate the presence of clinical endotypes and suggest heterogeneity of responses to anti-type 2 therapy.
INTRODUCTION
Eosinophilic esophagitis (EoE) is a chronic inflammatory disease triggered by delayed immune hypersensitivity to foods and aeroallergens resulting in the accumulation of excessive eosinophils in the esophagus.1-3 Children and adult patients can present with vomiting, dysphagia and food impaction.4
Substantial evidence is accumulating supporting that EoE is driven by a type 2 immune response, associated with IL-4, IL-5 and IL-13.5-15 In particular, mRNA expression of IL5 and IL13 (and IL4 to a lesser extent) are increased in the esophagus of EoE patients compared with controls.6, 16, 17 IL-5 is required for induction of experimental EoE in mice, and overexpression leads to esophageal inflammation and remodeling.10 IL-5 induces integrin ligation, activation, and survival of eosinophils, all of which can be inhibited in vivo in humans with a neutralizing antibody against IL-5, which is now approved for the treatment of eosinophilic asthma.18, 19 IL-13 appears to be particularly important in EoE as overexpression in mice induces cardinal features of the disease, including induction of esophageal eotaxin expression, eosinophil accumulation, epithelial hyperplasia, angiogenesis and fibrosis, processes that occur in the human disease.15 In addition, IL-13 induces impaired barrier function of esophageal epithelial cells partially via upregulation of calpain-14 (CAPN14), which is encoded by a gene located at the site of one of the chief EoE susceptibility loci, 2p23.12, 15, 20-22 Genetic studies have also identified genome wide susceptibility at 5q22, the thymic stromal lymphopoietin (TSLP) locus. TSLP-activated dendritic cells cause T cell polarization to Th2 cells that serve as sources of IL-5 and IL-13 in EoE.7, 23, 24 Antibodies against IL-5 and IL-13, including its receptor IL4Rα (also a receptor for IL-4) reduce esophageal eosinophils and improve endoscopic, histologic and/or esophageal transcriptomic abnormalities in early clinical studies, but the effects are variable.5, 11, 13, 14, 25, 26 Despite strong evidence for a role of type 2 cytokines in EoE pathogenesis, implementation of targeted therapeutics is hampered, in part, by the inability to predict responders and non-responders and the heterogeneity of treatment response.
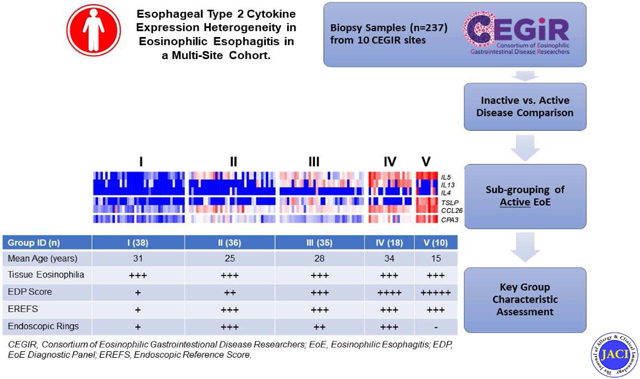
Levels of esophageal eosinophils correlate with the severity of endoscopic findings.27 Yet, disease severity associates with multiple histological parameters better than esophageal eosinophil counts alone.28 Transcript expression profiling of esophageal biopsies has identified unique disease specific transcripts, notably rich in genes that encode type 2 cytokines or products induced by these cytokines (e.g. IL-13 induced eotaxin-3).6, 17, 29-31 An EoE diagnostic panel (EDP), composed of 96 genes involved in diverse functions including inflammation, remodeling, and ion transport, effectively differentiates patients with active EoE from control individuals including EoE patients in remission.17 Molecular profiling with the EDP has identified substantial molecular heterogeneity associated with distinct histologic and endoscopic features which lead to the identification of three distinct disease endotypes.16 Herein, we examine 237 adult and pediatric EoE patient biopsies and clinical features from 10 sites associated with the Consortium of Eosinophilic Gastrointestinal Disease Researchers (CEGIR).32, 33 Esophageal biopsies were examined for the expression of type 2 immunity associated gene products, aiming to test for the presence of patient heterogeneity, based on type 2 immune responses, and its significance.
METHODS
Patient Recruitment
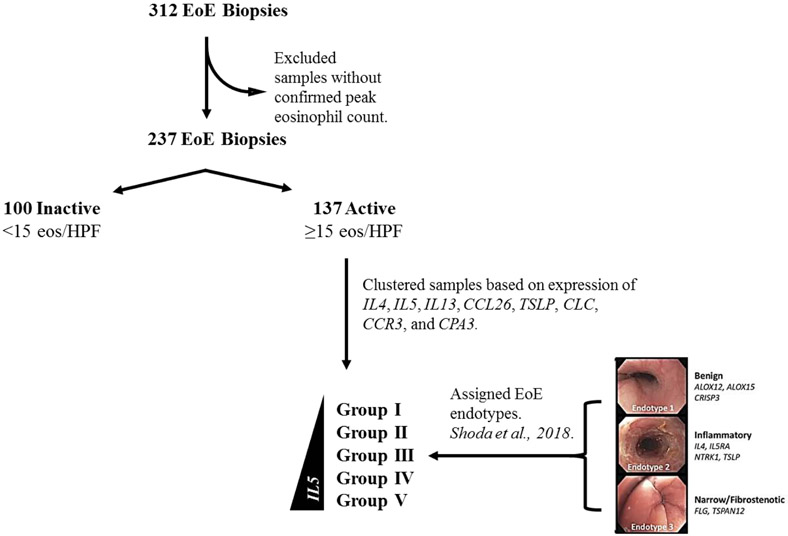
This study was conducted within the context of CEGIR, a national collaborative network of 16 academic centers caring for adults and children with eosinophilic gastrointestinal disorders (Supplemental Table 1). The CEGIR clinical trial, Outcomes Measures in Eosinophilic Gastrointestinal disorders across the Ages (OMEGA), is a longitudinal cohort study aimed at understanding the natural history of EoE, eosinophilic gastritis, and eosinophilic colitis during routine clinical care.16, 32-34 With this aim, demographic, clinical, endoscopic and histologic data, as well as gastrointestinal and blood samples have been prospectively collected from 2015 through 2018. Ten sites provided the EoE patient and sample data for this study; samples without peak eosinophil counts were excluded from the current analysis (Fig. 1; Supplemental Table 2 ). Clinical features of subjects were determined during standard of care evaluation with intake and follow up forms. All subject data were stored at the Data Management and Coordinating Center (DMCC) at the University of South Florida in Tampa, FL and were systematically extracted from the database. We defined patients with active EoE as having symptomatic esophageal dysfunction and a peak count of 15 or more eosinophils per high-power field (HPF); samples with no documented eosinophil count at the time of biopsy were excluded from the current study. Specimens obtained from patients with confirmed EoE with fewer than 15 peak eosinophils per HPF were classified as inactive. Pediatric subjects were defined as those less than 18 years of age. Atopy was defined based on self-report of allergic rhinitis, dermatitis, asthma, or food allergy. The definition of “response to steroid” was determined using a positive/negative response to whether swallowed topical steroids had been effective on the basis of symptom and pathology. These data were captured across 10 sites by the CEGIR questionnaire, as reported.16, 35 This study was approved by the institutional review boards of the participating institutions via a central institutional review board at Cincinnati Children’s Hospital Medical Center (CCHMC). Participants provided written informed consent.
FIG 1:
Workflow diagram depicting sample inclusion and categorization criteria. Biopsies with confirmed active EoE were sorted into 5 groups by K-means clustering using inflammatory genes IL4, IL5, IL13, CCL26, TSLP, CLC, CCR3, and CPA3. Using a published algorithm16, patients with active EoE were assigned to one of three EoE endotypes: benign (EoEe1), inflammatory (EoEe2), or fibrostenotic (EoEe3).
Biopsy acquisition
We obtained distal esophageal biopsy specimens during endoscopy; 2-3 specimens were taken from the area 2-4 cm above the lower esophageal sphincter.30 Gene expression in unfixed biopsy specimens was analyzed by qPCR with the EDP as previously described.16, 17 The EoE transcriptome as measured by the EDP is enriched for epithelial genes as reported although it also has embedded markers of T cells, mast cells and eosinophils.17 Changes in cycle thresholds (Δ Ct) for all genes of interest was calculated by subtracting the Ct of GAPDH. In order to control for the potential effect of variable GAPDH detection on ΔCt values for samples with Ct = 40 (the lower limit of detection), ΔCt values associated with Ct=40 were removed, the minimum expression of each gene was identified, and ΔCt values associated with Ct = 40 were replaced with (minimum-1) values. The EDP score (previously described as EoE score) was calculated by summing ΔCT values of the 76 highly dysregulated genes (ΣΔCT) in the panel, as described previously.16, 17 In order to reflect the disease-specific expression signature and disease severity, the CT sums were separately derived for the upregulated and downregulated genes, and then their two sums were combined. Of note, the EDE score is inversely correlated with disease severity. The assignment of the subject’s EoE endotype based on the expression of 8 genes in the panel of genes was established by a method comprising linear discriminant analysis determining a probability distance using Mahalanobis distance.16 By using the same algorithm, probabilities and predicted EoE endotypes were calculated based on the highest predicted probability.
Data analysis
Unsupervised clustering was used to identify groups of patients with active EoE based on expression of Th2-associated genes previously shown to be upregulated in EoE; IL4, IL5, IL13, CXCL8, CCR3, CCL26, TSLP, as well as markers of eosinophils (CLC), and mast cells (CPA3). The selection of K=5 for patient clustering was based on obtaining a sufficient fit, the results of a sensitivity analysis, and also to limit depletion of cluster size. Gene expression, EDP score, and age were compared between these groups using one-way ANOVA with Bonferroni post-test, and significance with respect to inactive patients was assessed with Dunnett’s post-test. Values from the histologic scoring system (HSS),28 endoscopic reference scoring system (EREFS),27, 36 and peak eosinophil count were compared with the non-parametric Kruskal-Wallis test and Dunn’s post-test. Features of the HSS were scored for grade (0-3) and stage (0-3), which were combined and the sums normalized to a range of 0-1 for the total scores. Lamina propria fibrosis could not be assessed in over 30% of samples, therefore these measurements were excluded from the current analysis. Features of the EREFS were scored (0-2) and features were added to obtain total scores in the distal and proximal esophagus; proximal and distal scores were, in turn, added to obtain a final score.
To characterize groups of patients with active EoE, we examined the distribution of previously described EoE endotypes. To assign endotypes, an algorithm with discriminant analysis was performed based on expression of 8 genes expression previously described.16 Data compilation, filtration and K-means clustering were performed in R Studio (version 3.5.1) and statistical analyses were completed in GraphPad Prism (version 8.0.1); statistical significance was assigned at p-values less than 0.01. Morpheus (https://software.broadinstitute.org/morpheus/) was used to generate heatmaps.
RESULTS
EoE Patient Characteristics
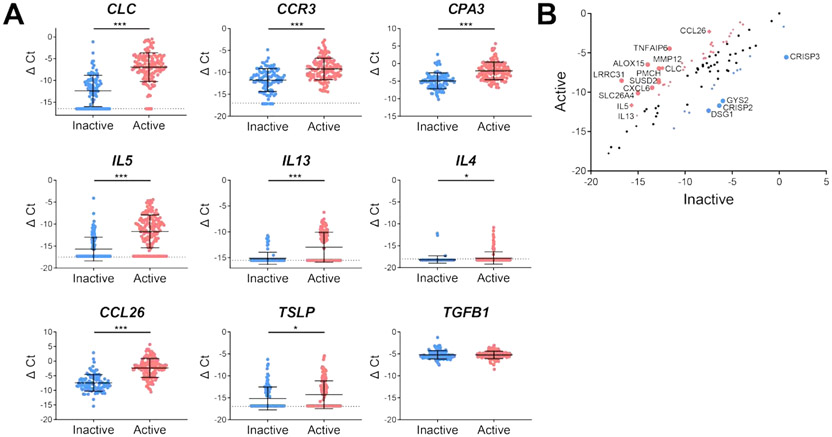
Patient age ranged from 4 to 71 years with mean age of 26 years (Table 1). A majority of the patients were male (67%) (Table 1). A subset of patients were on proton-pump inhibitor, dietary elimination and topical steroid therapy (Table 1). Patients with active EoE had mean value of 50 eosinophils/HPF, whereas patients with inactive EoE had 3 eosinophils/HPF (Table 1). The EDP total score differed in active and inactive group (123 vs 332, P < 0.0001; Table 1). Consistent with the increase in esophageal eosinophil levels, expression of esophageal CLC mRNA was increased in active EoE compared with inactive EoE (ΔΔCt = 5.45, P < 0.0001; Fig. 2 ). Levels of CPA3, a mast cell specific transcript, were increased in biopsies from active EoE patients compared with patients with inactive EoE (ΔΔCt = 2.84, P < 0.0001; Fig. 2).
TABLE 1:
Patient Demographics
| Inactive n = 100 |
Active n = 137 |
P value | |
|---|---|---|---|
| Age (years ± SD; range) | 23 ± 17 (4-67) | 28 ± 17 (5-71) | 0.07 |
| Male (%) | 64.0% | 69.3% | |
| White (%) | 95.0% | 92.7% | |
| EDP Score * | 332 ± 83 | 123 ± 136 | < 0.0001 |
| Peak Eosinophil Count (eosinophils/HPF) | 3 ± 4 | 50 ± 34 | < 0.0001 |
| Atopic Status | |||
| Allergic Rhinitis (%) | 44.0% | 38.7% | |
| Asthma (%) | 24.0% | 25.5% | |
| Eczema (%) | 26.0% | 24.1% | |
| Food Allergy (%) | 42.0% | 40.9% | |
| Ongoing Diet Therapy (%) | 50.0% | 51.1% | |
| Current PPI Usage (%) | 34.0% | 39.4% | |
| Current Oral Systemic Steroids (%) | 0% | 0% | |
| Current Swallowed Topical Steroids (%) | 59.0% | 47.4% | |
| History of Steroid Usage (%) | 68.0% | 71.5% | |
| Response to Steroid – Symptom (% Effective) | 73.5% | 63.3% | |
| Response to Steroid – Pathology (% Effective) | 64.7% | 51.0% |
EDP score calculated as the sum of delta Ct values.
FIG 2:
Th2-associated gene expression in patients with active and inactive EoE (A) Expression of genes associated with eosinophils (CLC and CCR3) or mast cells (CPA3), Th2 activation (IL5, IL13, IL4, and TSLP), and eosinophil chemotaxis (CCL26) was measured and normalized by subtracting the Ct of GAPDH (ΔCt; active EoE n = 137, inactive EoE n = 100). Significance was assessed by Mann-Witney test. Dotted lines indicate limit of detection (B) Expression of genes in the EDP was averaged across active and inactive patients and differentially expressed genes were identified and annotated. Genes with ∣ΔΔCt∣ > 2 are indicated in blue if their expression was higher in inactive patients or in red if their expression was higher in active patients. * P < 0.05, *** P < 0.001. The data shown includes the cumulative results of all samples performed over 78 Custom TaqMan Gene Expression Array Cards. Experiments each supported the trends shown.
Type 2 cytokine profiles in EoE
Of the upregulated genes in active vs. inactive disease, we noted numerous type 2 immunity associated genes, such as IL5, TSLP, and IL13 (Fig. 2). These included increases in several IL-13-inducible genes including CCL26 (ΔΔCt = 5.12, P < 0.0001), SLC26A4 (ΔΔCt = 4.88, P < 0.0001), MMP12 (ΔΔCt = 4.32, P < 0.0001), LRRC31 (ΔΔCt = 8.21, P < 0.0001) and ALOX15 (ΔΔCt = 7.47, P < 0.0001). Focusing on type 2 associated cytokines, we observed increased IL5 (ΔΔCt = 4.02, P < 0.0001) and IL13 (ΔΔCt = 2.15, P < 0.0001). By contrast we observed only a modest increase in TSLP (ΔΔCt = 0.09, P < 0.05) and no significant change in TGFB1.
Expression of type 2 inflammatory genes distinguishes subgroups of active EoE
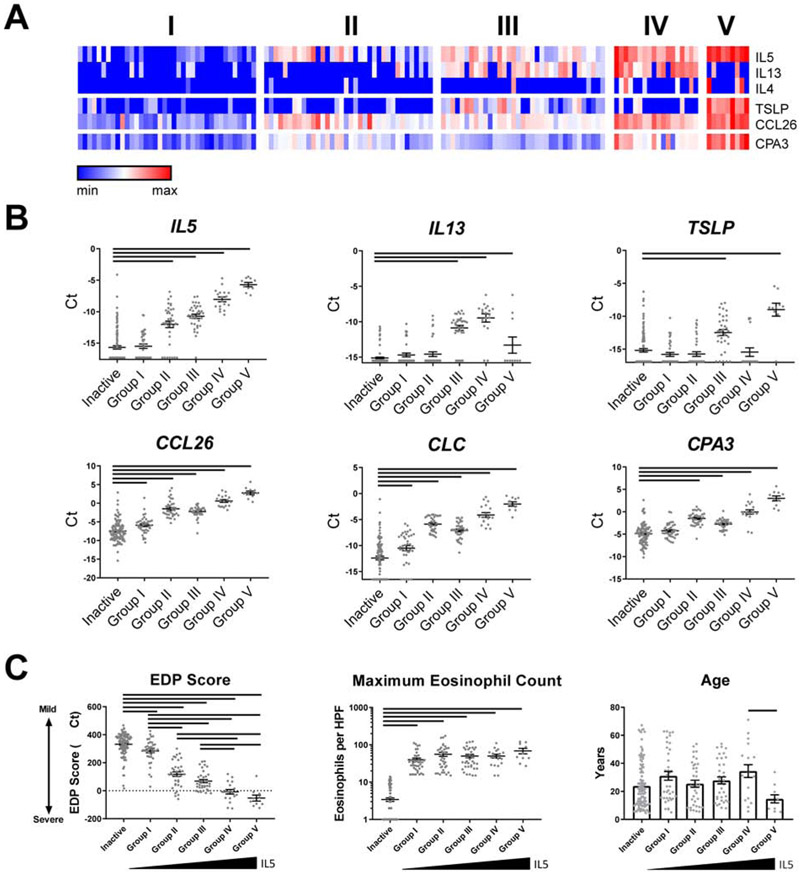
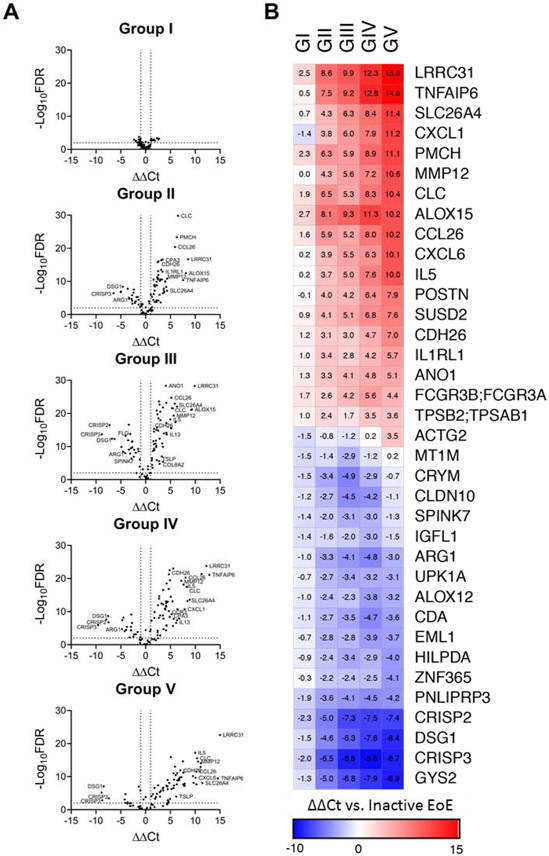
We set out to identify unique subgroups of patients with active EoE based on differential expression of type 2 cytokines (IL4, IL5, TSLP and IL13) and the allergic inflammatory cell specific markers CLC and CPA3. Using unsupervised clustering, we identified five groups of patients with low (Group I), intermediate (Groups II and III), and high (Groups IV and V) expression of type 2 cytokine genes, which were ordered according to expression of IL5 (Fig. 3, A). IL5 and CPA3 were not significantly increased in Group I with respect to patients with inactive EoE (Fig. 3, B ). Eosinophil levels were comparable among all 5 subgroups (Fig. 3, C). Whereas IL13 was significantly increased in Groups III-V, the increase in Groups III and IV was most dramatic. TSLP was increased specifically in Groups III and V. Of note, IL4 was universally low; it was undetectable in 95 of 97 patients with inactive EoE, and in all Group I patients; conversely, IL4 expression was detectable in approximately 20% of IL5-high patients with active EoE (Groups IV and V; Fig. 3, A).
FIG 3:
Expression of Th2 genes in active EoE identifies distinct patient groups. (A) Five subgroups of active EoE were identified by K-means clustering based on expression of IL4, IL5, IL13, TSLP, CCR3, CCL26, CLC, and CPA3. (B) Normalized expression of Th2 genes was compared in inactive patients and 5 groups of active patients. Bars indicate statistical differences (P < 0.05) between inactive patients and specified groups, assessed by one-way ANOVA with Dunnett’s post-test. (C) EDP score and patient age were compared between inactive patients and active groups I-V by one-way ANOVA with Tukey post-test. Maximum eosinophil count was compared via Kruskal-Wallis test with Dunn’s post-test. Bars indicate P < 0.05. The data shown includes the cumulative results of all samples performed over 78 Custom TaqMan Gene Expression Array Cards.
Groups I-V differed significantly in total EDP score, both compared to one-another and compared to patients with inactive EoE (Fig. 3, C). Groups that did not significantly differ in EDP score were Groups II and III, the IL5-intermediate groups, as well as Groups IV and V, the IL5-high groups (Fig. 3, C). While there was no difference in age between patients with active or inactive EoE (Table 1), patients in Group IV were significantly older than patients with inactive EoE (P < 0.05; Fig. 3, C). Moreover, treatment status did not account for patient grouping, as chi-square analysis did not reveal significant enrichment of PPI, steroid usage, or ongoing diet therapy in any of the five patient groups (Supplemental Table 4).
Cytokine heterogeneity associates with histologic and endoscopic phenotypes as well as molecular endotypes
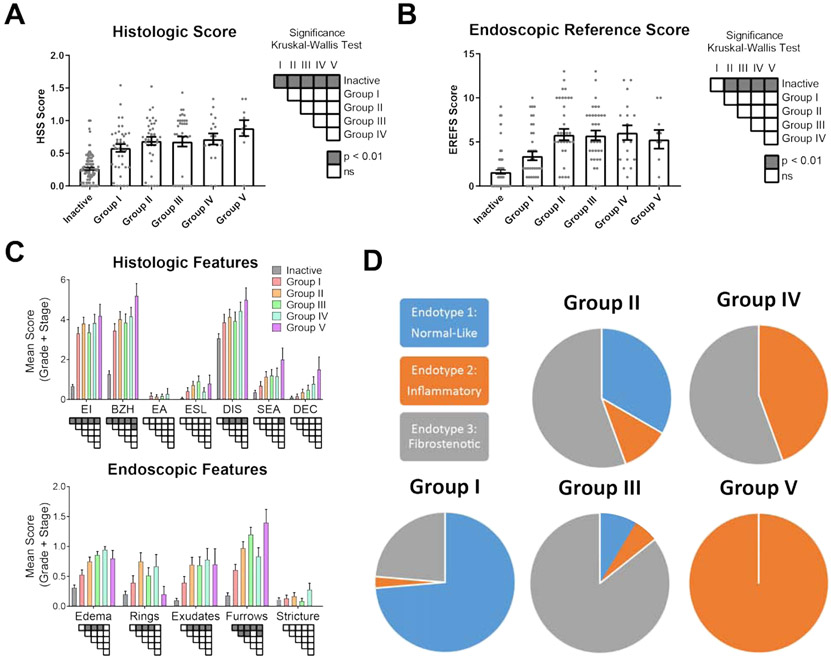
We aimed to determine if the identified patient subgroups associated with histological and endoscopic features. In patients in Group I with low IL5 expression, we observed that HSS grade and stage scores were higher than in the inactive group corroborating their assignment as active EoE; however, we observed no significant increase in total endoscopic scores (EREFS) with respect to inactive EoE (Fig. 4, A-B). Peak eosinophil counts did not differ among the active groups; however, significant differences between active and inactive disease patients and significant intergroup differences occurred among the individual features of the HSS (Fig. 4, C; Supplemental Table 3). The most frequent abnormalities were eosinophilic inflammation (EI), basal zone hyperplasia (BZH) and dilated intercellular spaces (DIS); EI and BZH were significantly increased in all groups compared to inactive EoE, BZH was significantly increased in Group V compared to Group I, and DIS was significantly increased in Groups II-V compared to inactive EoE. Surface epithelial alteration (SEA) was significantly increased in Group V compared to inactive patients. Eosinophil abscesses (EA), eosinophil surface layering (ESL) and dyskeratotic epithelial cells (DEC) were less common, and significant increases were not found. ESL was most common in Group III and DEC in Group V. Group V had the highest scores for EI, BZH, DIS, SEA and DEC.
FIG 4:
Inflammatory and fibrostenotic phenotypes among IL5-high and intermediate groups. (A) Histologic and (B) endoscopic scoring of patients with inactive EoE or in Groups I-V. Statistical significance determined by Kruskal-Wallis post-test between individual groups is indicated as shading. (C) Features of the EoE histology scoring system (HSS) and EoE endoscopic reference score (EREFS) were scored for grade (0-3) and stage (0-3) across Groups I-V. Significance between active patient subgroups and inactive patients was calculated with two-way ANOVA and Tukey post-test (shading;P < 0.01). (D) EoE endotype assignment in groups I-V. Groups were found to be dependent on endotypes according to Chi Square Exact Test (p < 0.0001). Eosinophilic infiltrates (EI); basal zone hyperplasia (BZH); eosinophilic abscess (EA); eosinophilic surface layering (ESL); dilated intercellular spaces (DIS); surface epithelial alteration (SEA); dyskeratotic epithelial cells (DEC).
Focusing on endoscopic features, the identified patient subgroups had unique associations. Group I did not have a significant increase in rings or edema, whereas edema was elevated across Groups II-IV (Fig. 4, B). Exudates were elevated across Groups II-V. Although furrowing was observed in all patient groups with respect to inactive disease, the most severe furrowing was observed in Groups II, III and V, which were significantly greater than Group I. Of note, rings (or trachealization) were highest in Groups II, III and IV and were absent in Group V.
We aimed to understand the relationship between the identified cytokine heterogeneity and previously described EoE endotypes. Absence of rings in Group V patients led us to question whether these biopsies shared characteristics with the inflammatory EoE endotype (EoEe2), which also exhibited a notable lack of rings.16 Whereas Group V overlapped entirely with EoEe2, Group IV represented a combination of EoEe2 and the fibrostenotic endotype, EoEe3 (Fig. 4, D). Groups II and III were primarily represented by EoEe3, while the remaining patients conssited of the normal-like endotype, EoEe1, as well as EoEe2. Finally, Group I consisted primarily of patients assigned to EoEe1, while the remainder of patients consisted of EoEe3 and only 1 individual from EoEe2. Collectively, our data substantiate distinct EoE endotypes, independent of esophageal eosinophil levels in active biopsies, based on cytokine expression.
Group-specific gene dysregulation corroborates endotype membership
We aimed to determine which genes and/or pathways associated with the unique IL5-related endotypes. By enumerating the 10 genes that were the most highly upregulated in each group, we identified several genes that were upregulated in all five groups including ALOX15, CCL26, and LRRC31, which are induced by IL-13, as well as CLC and PMCH (Fig. 5, B). MMP12, SLC26A4, and TNFAIP were among the most induced genes of Groups II-V. In Group III, there was notable upregulation of structural remodeling genes MMP12 and COL8A2 paired with a decrease in genes associated with epithelial integrity and differentiation such as FLG, DSG1, and SPINK7 –this result corroborates the observation that Group III is largely congruent with the fibrostenotic endotype ( EoEe3; Fig. 5, A ).16 Whereas IL13 was preferentially expressed in Group IV, several IL-13-dependent transcripts were preferentially expressed in Group V, including LRRC31 and SLC26A4. Similarly, we observed that both groups preferentially expressed specific genes associated with structural remodeling, specifically COL8A2 in Group IV and ACTG2 as well as MMP12 in Group V.
FIG 5:
Differential gene expression in Th2 subgroups. (A) The ΔΔ Ct values (ΔCtGROUP - ΔCtINACTIVE) and accompanying statistical significance (-log10FDR) were calculated for the entire EDP in each group with respect to patients with inactive EoE. Dotted lines indicate significance thresholds at – log10(FDR)>2 and ∣ ΔΔ Ct∣ > 1. Genes of particular interest are annotated. (B) ΔΔ Ct values of the highest up- and downregulated genes in each group with respect to inactive EoE.
DISCUSSION
Herein, we have examined the relationship between type 2 gene expression and EoE related disease features. In this study, we examined heterogeneous expression of the type 2 cytokines IL5 and IL13 because of their known roles in promoting eosinophil survival and activation through both direct and indirect mechanisms and the emerging set of therapies that target these cytokines and/or their receptors.5, 11, 13, 14, 18 We have identified subgroups of patients with active EoE that have low, intermediate, and high expression of IL5, and furthermore demonstrated concomitant upregulation of genes associated with fibrotic tissue remodeling in IL5-intermediate patient groups (Fig. 6, A). Treatment status did not account for differential gene expression that determines patient grouping, as chi-square analysis did not reveal significant enrichment of PPI, steroid usage, or ongoing diet therapy in any of the five patient groups. Importantly, of the two groups with high IL5 expression, IL13 expression was elevated in only one of them, suggesting the existence of a mechanism by which regulation of IL5 and IL13 gene expression is discordant either spatially or temporally, as group IV patients were markedly older in age than group V patients We have demonstrated a positive relationship between high IL5 expression and increased disease severity as assessed by the total EDP score and endoscopic parameters. Specifically, we found that patients with active EoE but low expression of IL5 did not exhibit a significant global increase in endoscopic disease severity scoring, although they did exhibit higher scores compared to inactive patients via histologic scoring. These findings support the practice of obtaining biopsies from patients suspected of having EoE even if the endoscopic appearance is normal.
FIG 6:
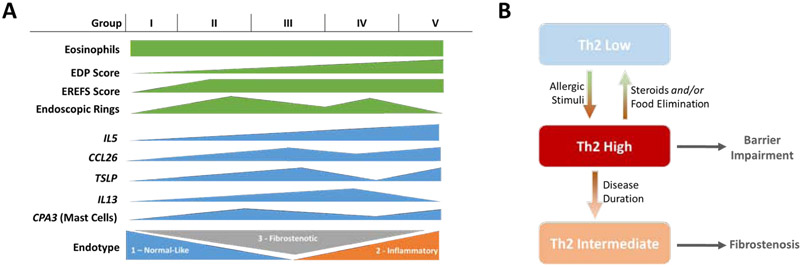
Summary of study results and proposed disease mechanism. (A) Groups I-V as defined in this study were not statistically different on the basis of eosinophil count but exhibited increased disease severity as assessed by cumulative EDP scoring. Expression of cytokines and chemokines varied among the 5 groups, which also differed in terms of membership in EoE endotypes 1-3. (B) Model depicting patient progression from Th2-low phenotype (congruent with EoEe1) to a Th2-high phenotype (congruent with EoEe2) following allergic or inflammatory insult. Upon steroid treatment, food elimination, or biologic therapy the Th2-gene expression decreases and patients either resolve inflammation by reverting to a Th2-low phenotype or develop a fibrostenotic (EoEe3) signature.
Previous studies have reported a correlation between eosinophil counts by histology and expression of inflammatory genes including IL5, IL13, and CCL26 6, 17. In contrast, by focusing specifically on patients with >15 eosinophils/HPF, we describe differential gene expression in groups of patients with no difference in esophageal eosinophil counts. However, we observed discordance between CLC transcript and eosinophil levels. This result may be due to the presence of extracellular vesicles containing CLC mRNA, which may be indicative of eosinophil activation. Further investigation will be needed to establish a relationship between CLC transcript, intact eosinophils, and extracellular eosinophil-derived vesicles in biopsy tissue.
A recent single cell RNA sequencing study of esophageal residing CD3+ T cells23 has identified the cellular source of IL-5 and IL-13 to an enriched population of activated effector memory T cells (designated the T8 cell population), consistent with prior reports.37, 38 The single cell RNA sequencing paper was limited to 17 subjects; whereas, the current study examines over 200 subjects, thus allowing a deeper analysis of patient heterogeneity, as well as phenotypic associations. On a single cell level, only a minority of pathogenic effector Th2 cells produce only IL-5, whereas most produce IL-13. Collectively, our data substantiate patient specific differences in the presence of CD3+ T cells based on IL-5 and IL-13 expression, and that this heterogeneity associates and is likely causal, at least in part, with distinct disease features.
Heterogeneity of disease presentation may be indicative of chronological stages of disease or distinct disease mechanisms. Recently, we have reported the identification of three molecular endotypes of active EoE with distinctive histologic and endoscopic features and presented evidence that patients transition from a normal-like endotype (EoEe1) to an inflammatory (EoEe2) and finally a fibrostenotic (EoEe3) endotype.16 Although samples in this report represent a single timepoint, the data substantiate previous reports that EoE progression is not a simple linear pathway from low to high Th2 cytokine expression.39 By comparing the current findings with endotype assignments, these collective results support a disease mechanism that transitions from IL5-low/normal to IL5-high/inflammatory and finally to IL5-intermediate/fibrostenotic.
Whereas expression of type 2 cytokines IL13 and TSLP were detected at low levels in patients with low IL5 expression or with inactive EoE, IL4 transcript was undetectable in these patients and was only expressed at a low level in patients with active EoE and high IL5 expression. It is possible that active protein production and signaling is not adequately accounted for by mRNA quantification. We have recently reported differential cytokine expression in T-cells from patients with EoE, and observed that IL-4 was readily detected by flow cytometry but not in RNA-sequencing data, indicating that its regulation may occur post-translationally and that biologically significant changes in IL-4 signaling may not be detected at the level of mRNA quantification.23 Moreover, significant evidence exists that although IL-4 and IL-13 share receptor components, due to differential receptor subunit usage, IL-4 exerts biological activity at substantially lower concentrations than IL-13;40-42 it is therefore possible that IL-4 signaling is a critical component of the signaling events in patients from Groups IV and V, since low expression may be sufficient to skew the inflammatory environment. Furthermore, as cytokines largely exert their activity in local tissue microenvironments and immunological synapses, mRNA quantification in bulk tissue may fail to detect bioactive quantities of IL-4 expression.
High-throughput EoE genetic studies used to identify common genetic susceptibility elements (single nucleotide polymorphisms; SNPs) and altered gene expression have implicated several genes associated with type 2 inflammation. Thymic stromal lymphopoietin (TSLP), which acts on DCs to promote Th2 polarization, was identified by GWAS as the site of multiple SNPs that are associated with EoE.7, 43, 44 We have shown that upregulation of TSLP expression in EoE patients is affected by the presence of a risk allele in the 5q22 locus.43 Whether patient genotype at the TSLP locus accounts for patients with high IL5 expression but low TSLP expression, as in Group IV, will be the subject of future study.
Improved understanding of the relationship between type 2 gene expression and clinical progression may enable selection of therapeutic interventions that target IL-13, IL-4Rα and/or IL-5. Anti-IL-13 leads to improvement in clinical outcomes in EoE and suggests the presence of responders and non-responders.9, 13 Trials using anti-IL-5 in EoE have shown significant improvement in some patients evidenced by decreased eosinophilia and tissue remodeling.5, 11, 14, 25, 26 Anti-IL-5 (mepolizumab and reslizumab) has utility in asthma but only in the eosinophilic endotype8; as such, it is reasonable to expect that anti-IL-5 may be useful for EoE in patients with specific endotypes. Our data supports the importance of studying anti-IL-5 therapeutics in patients from groups IV and V. Moreover, a recent report suggests that factors aside from IL-5 signaling may drive eosinophil survival in allergic disease45, therefore cytokine expression alone may not predict eosinophil survival and it would be beneficial to develop functional assays to assess eosinophil responsiveness to cytokines in inflammatory patients. It is interesting to speculate that anti-IL-4Ra and anti-IL-13 based therapeutics may be particularly effectively against subgroup III and IV, which harbor the highest levels of IL-13 mRNA.
Due to the nature of the CEGIR patient cohort, in this analysis we were able to retrospectively examine the relationship between inflammatory gene expression and patient characteristics in EoE; however, we are unable to make conclusive statements regarding the patient population as a whole or over time. We found that K-means clustering using 5 clusters resulted in reproducible groupings with biologically meaningful distinctions; however, the current study is not sufficiently powered to claim that these 5 groups are intrinsic to the EoE population or to propose diagnostic or prognostic value of these clusters. Future study will be necessary to evaluate patient outcomes in relation to type 2 gene expression. Moreover, this is one of several studies that suggest a progression from normal-like to inflammatory and finally to fibrostenotic phenotypes. A longitudinal analysis to examine changes in patient gene expression and phenotype over time will be an important future step for the field. Future studies should also be designed to account for atopic status. Finally, whereas atopic status is reported for some patients in the CEGIR cohort, this information is not available for all patients and it therefore not feasible to observe associations between atopic status and inflammatory gene expression. It will be important for future studies to standardize the definition of atopic status and to ensure thorough collection of this information.
In conclusion, we have identified substantial heterogeneity of type 2 cytokine associated gene expression in EoE, resulting in the identification of five groups of active EoE patients, all of which do not differ significantly in esophageal eosinophil counts. Our findings support a non-linear model to explain EoE pathogenesis, where cytokine expression is heterogeneous and associated with distinct endoscopic and histological features, and that elevated IL5 is not directly proportional to the degree of histological pathology and endoscopic abnormalities, esophageal eosinophilia, and tissue remodeling. Our data supports a disease model wherein a Th2 low response transitions to a Th2 high response associated with decreased barrier function, likely in response to allergic stimuli or food antigen exposure. Effective treatment reduces eosinophil numbers and is associated with decreased inflammatory gene expression, and we propose that under certain conditions this decrease in inflammatory gene expression results in patients reverting to a relatively normal-like gene expression signature; however, over time patients, gene expression adopts a fibrotic profile with intermediate inflammatory expression. Intermediate Th2 responses are observed in older patients and are associated with fibrostenotic remodeling; interestingly, eosinophil levels remain relatively stable throughout these phases. While this hypothesis needs to be validated by longitudinal studies, we present this pathogenic model for consideration (as summarized in Fig. 6, B). Our model is consistent with the finding that the development of fibrostenosis is proportional to the length of time that EoE is untreated46; however, we are not proposing that this is the only pathway to the fibrostenotic endotype. These findings provide a potential molecular basis for heterogeneity of patient responsiveness to therapeutic intervention and an emerging opportunity for personalized medicine approaches in the EoE field.
Supplementary Material
Clinical Implications.
In addition to illuminating distinct disease mechanisms, the heterogeneity of IL5, IL13 and TSLP expression described herein is likely to impact patient response to biological agents that target these cytokines.
Acknowledgements
The authors are grateful to Shawna Hottinger for editorial assistance. CEGIR investigators are grateful to their colleagues and clinical support staff for procuring biopsies and clinical data.
Funding
CEGIR (U54 AI117804) is part of the Rare Disease Clinical Research Network (RDCRN), an initiative of the Office of Rare Diseases Research (ORDR), National Center for Advancing Translational Sciences (NCATS), and is co-funded by National Institute of Allergy and Infectious Diseases (NIAID), National Institute of Diabetes and Digestive and Kidney Diseases (NIDDK), and NCATS. CEGIR is also supported by patient advocacy groups including the American Partnership for Eosinophilic Disorders (APFED), and Campaign Urging Research for Eosinophilic Disease (CURED).
Abbreviations
- ACTG2
Actin Gamma 2
- ALOX15
Arachidonate 15-Lipoxygenase
- ANOVA
Analysis of Variance
- BZH
Basal Zone Hyperplasia
- CCL26
C-C Motif Chemokine Ligand 26
- CCR3
C-C Motif Chemokine Receptor 3
- CEGIR
Consortium of Eosinophilic Gastrointestional Disease Researchers
- CLC
Charcot Leyden Crystal
- COL8A2
Collagen Type 8 Alpha 2-Chain
- DEC
Dyskeratotic Epithelial Cells
- DIS
Dilated Intercellular Spaces
- DSG1
Desmoglein1
- EA
Eosinophil Abscesses
- EDP
Eosinophilic Esophagitis Diagnostic Panel
- EI
Eosinophil Infiltration
- EoE
Eosinophilic Esophagitis
- EoEe
Eosinophilic Esophagitis endotype
- EREFS
Endoscopic Reference Score
- FLG
Filaggrin
- GWAS
Genome Wide Association Study
- HPF
High Power Field
- HSS
Histology Scoring System
- IL
Interleukin
- LRRC31
Leucine Rich Repeat Containing 31
- MMP12
Matrix Metalloprotease 12
- SEA
Surface Epithelial Alteration
- SLC26A4
Solute carrier Family 26 Member 4
- SPINK7
Serine Peptidase Inhibitor, Kazal Type 7
- TGFB1
Transforming Growth Factor Beta −1
- TNFAIP
Tumor Necrosis Factor Alpha Induced Protein
- TSLP
Thymic Stromal Lymphopoietin
- TSPAN12
Tetraspanin 12
Footnotes
Disclosures
M.E.R. is a consultant for Pulm One, Spoon Guru, Celgene, Astra Zeneca, Allakos, and ClostraBio, and has an equity interest in Pulm One, Spoon Guru, and ClostraBio and royalties from reslizumab (Teva Pharmaceuticals). M.E.R. is an inventor of patents, owned by Cincinnati Children’s. G.W.F. has received research support from Celgene/Receptos, Regeneron, Shire and Adare. M.H.C. is a consultant for Allakos, AstraZeneca, Esocap, Shire/Takeda, Regeneron, and Receptos/Celgene and has received research funding from Shire/Takeda, Regeneron and Receptos/Celgene. S.K.G. is a consultant for Abbott, Allakos, QOL, Meritage, and Receptos and receives research support from Shire. E.S.D. is a consultant for Adare, Aimmune, Alivio, Allakos, AstraZeneca, Banner, Calypso, Enumeral, Esocap, Gossamer Bio, GSK, Receptos/Celegene, Regeneron, Robarts, Salix, and Shire/Takeda, has received research funding from Adare, Allakos, Meritage, Miraca, Nutricia, Receptos/Celgene, Regeneron, and Shire/Takeda, and has received educational grants from Allakos, Banner, and Holoclara. S.S.A. is a consultant for Regeneron and AImmune, is an inventor of oral viscous budesonide, patented by UCSD and licensed by Shire, and has research funding from Ferring Research Institute. J.M.S. is a consultant for Regeneron and DBV Technology, and his research is supported by NIH, EATS foundation, AImmune Therapeutics, FARE and DBV Technology. I.H. is a consultant for Regeneron, Receptos, Shire, Allakos and Adare and has received research funding from Regeneron, Receptos, Shire and Adare. G.T.F. is a consultant for Shire and a co-founder of EnteroTrack. All other authors declare they have no competing interests.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Simon D, Cianferoni A, Spergel JM, Aceves S, Holbreich M, Venter C, et al. Eosinophilic esophagitis is characterized by a non-IgE-mediated food hypersensitivity. Allergy 2016; 71:611–20. [DOI] [PubMed] [Google Scholar]
- 2.Lucendo AJ, Molina-Infante J, Arias A, von Arnim U, Bredenoord AJ, Bussmann C, et al. Guidelines on eosinophilic esophagitis: evidence-based statements and recommendations for diagnosis and management in children and adults. United European Gastroenterol J 2017; 5:335–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Dellon ES, Liacouras CA, Molina-Infante J, Furuta GT, Spergel JM, Zevit N, et al. Updated International Consensus Diagnostic Criteria for Eosinophilic Esophagitis: Proceedings of the AGREE Conference. Gastroenterology 2018; 155:1022–33 e10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Walsh SV, Antonioli DA, Goldman H, Fox VL, Bousvaros A, Leichtner AM, et al. Allergic esophagitis in children: a clinicopathological entity. Am J Surg Pathol 1999; 23:390–6. [DOI] [PubMed] [Google Scholar]
- 5.Assa'ad AH, Gupta SK, Collins MH, Thomson M, Heath AT, Smith DA, et al. An antibody against IL-5 reduces numbers of esophageal intraepithelial eosinophils in children with eosinophilic esophagitis. Gastroenterology 2011; 141:1593–604. [DOI] [PubMed] [Google Scholar]
- 6.Blanchard C, Stucke EM, Rodriguez-Jimenez B, Burwinkel K, Collins MH, Ahrens A, et al. A striking local esophageal cytokine expression profile in eosinophilic esophagitis. J Allergy Clin Immunol 2011; 127:208–17, 17 e1-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kottyan LC, Davis BP, Sherrill JD, Liu K, Rochman M, Kaufman K, et al. Genome-wide association analysis of eosinophilic esophagitis provides insight into the tissue specificity of this allergic disease. Nat Genet 2014; 46:895–900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Haldar P, Brightling CE, Hargadon B, Gupta S, Monteiro W, Sousa A, et al. Mepolizumab and exacerbations of refractory eosinophilic asthma. N Engl J Med 2009; 360:973–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hirano I, Collins MH, Assouline-Dayan Y, Evans L, Gupta S, Schoepfer AM, et al. RPC4046, a Monoclonal Antibody Against IL13, Reduces Histologic and Endoscopic Activity in Patients With Eosinophilic Esophagitis. Gastroenterology 2019; 156:592–603 e10. [DOI] [PubMed] [Google Scholar]
- 10.Mishra A, Wang M, Pemmaraju VR, Collins MH, Fulkerson PC, Abonia JP, et al. Esophageal remodeling develops as a consequence of tissue specific IL-5-induced eosinophilia. Gastroenterology 2008; 134:204–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Otani IM, Anilkumar AA, Newbury RO, Bhagat M, Beppu LY, Dohil R, et al. Anti-IL-5 therapy reduces mast cell and IL-9 cell numbers in pediatric patients with eosinophilic esophagitis. J Allergy Clin Immunol 2013; 131:1576–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rochman M, Travers J, Abonia JP, Caldwell JM, Rothenberg ME. Synaptopodin is upregulated by IL-13 in eosinophilic esophagitis and regulates esophageal epithelial cell motility and barrier integrity. JCI Insight 2017; 2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rothenberg ME, Wen T, Greenberg A, Alpan O, Enav B, Hirano I, et al. Intravenous anti-IL-13 mAb QAX576 for the treatment of eosinophilic esophagitis. J Allergy Clin Immunol 2015; 135:500–7. [DOI] [PubMed] [Google Scholar]
- 14.Straumann A, Conus S, Grzonka P, Kita H, Kephart G, Bussmann C, et al. Anti-interleukin-5 antibody treatment (mepolizumab) in active eosinophilic oesophagitis: a randomised, placebo-controlled, double-blind trial. Gut 2010; 59:21–30. [DOI] [PubMed] [Google Scholar]
- 15.Zuo L, Fulkerson PC, Finkelman FD, Mingler M, Fischetti CA, Blanchard C, et al. IL-13 induces esophageal remodeling and gene expression by an eosinophil-independent, IL-13R alpha 2-inhibited pathway. J Immunol 2010; 185:660–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Shoda T, Wen T, Aceves SS, Abonia JP, Atkins D, Bonis PA, et al. Eosinophilic oesophagitis endotype classification by molecular, clinical, and histopathological analyses: a cross-sectional study. The Lancet Gastroenterology & Hepatology 2018; 3:477–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wen T, Stucke EM, Grotjan TM, Kemme KA, Abonia JP, Putnam PE, et al. Molecular diagnosis of eosinophilic esophagitis by gene expression profiling. Gastroenterology 2013; 145:1289–99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Johansson MW, Gunderson KA, Kelly EA, Denlinger LC, Jarjour NN, Mosher DF. Anti-IL-5 attenuates activation and surface density of beta(2) -integrins on circulating eosinophils after segmental antigen challenge. Clin Exp Allergy 2013; 43:292–303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fulkerson PC, Schollaert KL, Bouffi C, Rothenberg ME. IL-5 triggers a cooperative cytokine network that promotes eosinophil precursor maturation. J Immunol 2014; 193:4043–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.D'Mello RJ, Caldwell JM, Azouz NP, Wen T, Sherrill JD, Hogan SP, et al. LRRC31 is induced by IL-13 and regulates kallikrein expression and barrier function in the esophageal epithelium. Mucosal Immunol 2016; 9:744–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Davis BP, Stucke EM, Khorki ME, Litosh VA, Rymer JK, Rochman M, et al. Eosinophilic esophagitis-linked calpain 14 is an IL-13-induced protease that mediates esophageal epithelial barrier impairment. JCI Insight 2016; 1:e86355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wheeler JC, Vanoni S, Zeng C, Waggoner L, Yang Y, Wu D, et al. 17beta-Estradiol protects the esophageal epithelium from IL-13-induced barrier dysfunction and remodeling. J Allergy Clin Immunol 2019; 143:2131–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wen T, Aronow BJ, Rochman Y, Rochman M, Kc K, Dexheimer PJ, et al. Single-cell RNA sequencing identifies inflammatory tissue T cells in eosinophilic esophagitis. J Clin Invest 2019; 130:2014–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kottyan LC, Rothenberg ME. Genetics of eosinophilic esophagitis. Mucosal Immunol 2017; 10:580–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Straumann A Treatment of eosinophilic esophagitis: diet, drugs, or dilation? Gastroenterology 2012; 142:1409–11. [DOI] [PubMed] [Google Scholar]
- 26.Spergel JM, Rothenberg ME, Collins MH, Furuta GT, Markowitz JE, Fuchs G, 3rd, et al. Reslizumab in children and adolescents with eosinophilic esophagitis: results of a double-blind, randomized, placebo-controlled trial. J Allergy Clin Immunol 2012; 129:456–63, 63 e1-3. [DOI] [PubMed] [Google Scholar]
- 27.Hirano I, Moy N, Heckman MG, Thomas CS, Gonsalves N, Achem SR. Endoscopic assessment of the oesophageal features of eosinophilic oesophagitis: validation of a novel classification and grading system. Gut 2013; 62:489–95. [DOI] [PubMed] [Google Scholar]
- 28.Collins MH, Martin LJ, Alexander ES, Boyd JT, Sheridan R, He H, et al. Newly developed and validated eosinophilic esophagitis histology scoring system and evidence that it outperforms peak eosinophil count for disease diagnosis and monitoring. Dis Esophagus 2017; 30:1–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wen T, Rothenberg ME. Clinical Applications of the Eosinophilic Esophagitis Diagnostic Panel. Front Med (Lausanne) 2017; 4:108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Dellon ES, Yellore V, Andreatta M, Stover J. A single biopsy is valid for genetic diagnosis of eosinophilic esophagitis regardless of tissue preservation or location in the esophagus. J Gastrointestin Liver Dis 2015; 24:151–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Dellon ES, Veerappan R, Selitsky SR, Parker JS, Higgins LL, Beitia R, et al. A Gene Expression Panel is Accurate for Diagnosis and Monitoring Treatment of Eosinophilic Esophagitis in Adults. Clin Transl Gastroenterol 2017; 8:e74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Cheng K, Gupta SK, Kantor S, Kuhl JT, Aceves SS, Bonis PA, et al. Creating a multi-center rare disease consortium - the Consortium of Eosinophilic Gastrointestinal Disease Researchers (CEGIR). Transl Sci Rare Dis 2017; 2:141–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gupta SK, Falk GW, Aceves SS, Chehade M, Collins MH, Dellon ES, et al. Consortium of Eosinophilic Gastrointestinal Disease Researchers: Advancing the Field of Eosinophilic GI Disorders Through Collaboration. Gastroenterology 2019; 156:838–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Aceves SS, King E, Collins MH, Yang GY, Capocelli KE, Abonia JP, et al. Alignment of parent- and child-reported outcomes and histology in eosinophilic esophagitis across multiple CEGIR sites. J Allergy Clin Immunol 2018; 142:130–8 e1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Shoda T, Wen T, Caldwell JM, Collins MH, Besse JA, Osswald GA, et al. Molecular, Endoscopic, Histologic and Circulating Biomarker-Based Diagnosis of Eosinophilic Gastritis: Multi-Site Study. J Allergy Clin Immunol 2019, 145(1): 255–269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wechsler JB, Bolton SM, Amsden K, Wershil BK, Hirano I, Kagalwalla AF. Eosinophilic Esophagitis Reference Score Accurately Identifies Disease Activity and Treatment Effects in Children. Clin Gastroenterol Hepatol 2018; 16:1056–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wambre E, Bajzik V, DeLong JH, O'Brien K, Nguyen QA, Speake C, et al. A phenotypically and functionally distinct human TH2 cell subpopulation is associated with allergic disorders. Sci Transl Med 2017; 9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Upadhyaya B, Yin Y, Hill BJ, Douek DC, Prussin C. Hierarchical IL-5 expression defines a subpopulation of highly differentiated human Th2 cells. J Immunol 2011; 187:3111–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Dellon ES, Kim HP, Sperry SL, Rybnicek DA, Woosley JT, Shaheen NJ. A phenotypic analysis shows that eosinophilic esophagitis is a progressive fibrostenotic disease. Gastrointest Endosc 2014; 79:577–85 e4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Gour N, Wills-Karp M. IL-4 and IL-13 signaling in allergic airway disease. Cytokine 2015; 75:68–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Perkins C, Wills-Karp M, Finkelman FD. IL-4 induces IL-13-independent allergic airway inflammation. J Allergy Clin Immunol 2006; 118:410–9. [DOI] [PubMed] [Google Scholar]
- 42.Finkelman FD, Yang M, Perkins C, Schleifer K, Sproles A, Santeliz J, et al. Suppressive effect of IL-4 on IL-13-induced genes in mouse lung. J Immunol 2005; 174:4630–8. [DOI] [PubMed] [Google Scholar]
- 43.Rothenberg ME, Spergel JM, Sherrill JD, Annaiah K, Martin LJ, Cianferoni A, et al. Common variants at 5q22 associate with pediatric eosinophilic esophagitis. Nat Genet 2010; 42:289–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sleiman PM, Wang ML, Cianferoni A, Aceves S, Gonsalves N, Nadeau K, et al. GWAS identifies four novel eosinophilic esophagitis loci. Nat Commun 2014; 5:5593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Geslewitz WE, Percopo CM, Rosenberg HF. Eosinophil persistence in vivo and sustained viability ex vivo in response to respiratory challenge with fungal allergens. Clin Exp Allergy 2018; 48:29–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Schoepfer AM, Safroneeva E, Bussmann C, Kuchen T, Portmann S, Simon HU, et al. Delay in diagnosis of eosinophilic esophagitis increases risk for stricture formation in a time-dependent manner. Gastroenterology 2013; 145:1230–6 e1-2. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.