Abstract
Objective
Accurate electronic phenotyping is essential to support collaborative observational research. Supervised machine learning methods can be used to train phenotype classifiers in a high-throughput manner using imperfectly labeled data. We developed 10 phenotype classifiers using this approach and evaluated performance across multiple sites within the Observational Health Data Sciences and Informatics (OHDSI) network.
Materials and Methods
We constructed classifiers using the Automated PHenotype Routine for Observational Definition, Identification, Training and Evaluation (APHRODITE) R-package, an open-source framework for learning phenotype classifiers using datasets in the Observational Medical Outcomes Partnership Common Data Model. We labeled training data based on the presence of multiple mentions of disease-specific codes. Performance was evaluated on cohorts derived using rule-based definitions and real-world disease prevalence. Classifiers were developed and evaluated across 3 medical centers, including 1 international site.
Results
Compared to the multiple mentions labeling heuristic, classifiers showed a mean recall boost of 0.43 with a mean precision loss of 0.17. Performance decreased slightly when classifiers were shared across medical centers, with mean recall and precision decreasing by 0.08 and 0.01, respectively, at a site within the USA, and by 0.18 and 0.10, respectively, at an international site.
Discussion and Conclusion
We demonstrate a high-throughput pipeline for constructing and sharing phenotype classifiers across sites within the OHDSI network using APHRODITE. Classifiers exhibit good portability between sites within the USA, however limited portability internationally, indicating that classifier generalizability may have geographic limitations, and, consequently, sharing the classifier-building recipe, rather than the pretrained classifiers, may be more useful for facilitating collaborative observational research.
Keywords: electronic phenotyping, electronic health records, cohort identification, phenotype, machine learning
INTRODUCTION
Electronic phenotyping refers to the task of identifying patients within an electronic health record (EHR) who match a defined clinical profile.1 Accurate phenotyping is critical to support observational research, pragmatic clinical trials, quality improvement evaluations, and clinical decision support systems.2,3 However, issues such as missingness, accuracy, and heterogeneity in EHR data present major challenges to effective phenotyping.4
The traditional approach to phenotyping has been rule-based, where a cohort is manually defined with inclusion and exclusion criteria based on structured data, such as diagnosis codes, laboratory results, and medications.5 Although several collaborative networks exist for generating and sharing rule-based definitions, including the Phenotype Knowledge Base (PheKB), Phenotype Execution and Modeling Architecture (PheMA), and CALIBER,6–8 these phenotypes are typically too labor-intensive to create and require multiple rounds of review by domain experts.9
Recent efforts to establish common data models for EHRs, including the Observational Health Data Sciences and Informatics (OHDSI)10 and the Informatics for Integrating Biology and the Bedside initiatives,11 are enabling large-scale observational research and algorithm deployment across sites. To make use of this infrastructure, we need the ability to generate complex, generalizable phenotypes rapidly.3,12
Supervised machine learning has emerged as a way to generate phenotypes in a high-throughput manner.1 By incorporating a wide range of EHR features, statistical methods have shown robust performance for complex phenotypes including chronic pain and rheumatoid arthritis13,14 with some evidence to indicate portability (preserved classification accuracy) across sites.15 The major bottleneck for supervised machine learning is access to labeled training data, which traditionally requires manual chart review by clinicians.
To address the scarcity of labeled training data, Chen et al used active learning to intelligently select training samples for labeling, demonstrating that classifier performance could be preserved with fewer samples.16 Another trend is the use of “silver standard training sets,” a semisupervised approach where training samples are labeled using an imperfect heuristic rather than by manual review.17–22 The intuition is that noise-tolerant classifiers trained on imperfectly labeled data will abstract higher order properties of the phenotype beyond the original labeling heuristic (so-called “noise-tolerant learning”23). Halpern et al have described the anchor learning framework where the presence of “anchor” references, which are highly predictive of a phenotype and are conditionally independent of other features (ie, best predicted by the phenotype itself), are used to define an imperfect training cohort for phenotype classifiers.19 Similarly, Agarwal et al developed the XPRESS (eXtraction of Phenotypes from Records using Silver Standards) pipeline, where noisy training samples are defined based on highly specific keyword mentions in a patient’s EHR.17 This led to the development of the APHRODITE R-package, an open-source implementation of the XPRESS framework with dynamic anchor learning built on the OHDSI common data model, which has shown comparable performance to rule-based definitions for 2 phenotypes (type 2 diabetes and myocardial infarction).24
The current work addresses 2 questions resulting from the use of APHRODITE. The first is about the labeling function used to generate imperfectly labeled training data. While APHRODITE uses the mention of a single phrase, we hypothesize that a high-precision labeling heuristic based on multiple keyword (or phrase) mentions may improve classifier performance in situations where phenotyping precision is critical. In addition, APHRODITE was evaluated using balanced cohorts of cases and controls; in real-world situations where the number of controls far outnumbers cases, a higher-precision labeling function may perform better. We investigate the improvement obtained via complex labeling functions across a spectrum of 10 different phenotypes, and using real-world disease prevalence in the test data.
The second question is about APHRODITE’s ability to port both the final classifiers and the underlying training “recipes” between OHDSI sites. A recent study demonstrated the translation of PheKB definitions into executable EHR queries that ported across 6 different health systems7; however, the portability of classifier-based approaches such as APHRODITE has yet to be rigorously assessed. We conduct reciprocal experiments where we evaluate the performance of phenotype classifiers trained at our academic medical center on the EHRs of 2 other health systems and, conversely, evaluate the performance of classifiers trained externally on our data. We find that phenotype classifiers perform well across OHDSI sites, though portability may be limited by underlying differences in EHR data at various sites.
MATERIALS AND METHODS
Data sources
We used longitudinal EHR data from Stanford Hospital & Clinics and Lucile Packard Children’s Hospital, Columbia University Medical Center, and Seoul National University Bundang Hospital (SNUBH) to construct and evaluate phenotype classifiers. At Stanford, patient data was extracted from the Stanford Medicine Research Data Repository clinical data warehouse and included nearly 1.8 million patients and 53 million unique visits. The dataset used at Columbia comprised 5.7 million patients. At SNUBH, the dataset included over 1.8 million patients. Patient data at each institution were composed of coded diagnoses, laboratory tests, medication orders, and procedures. All data at the 3 institutions were mapped to the Observational Medical Outcomes Partnership (OMOP) Common Data Model (CDM), which serves as a shared standard representation of clinical data across multiple data sources and institutions. We summarize site-specific differences in each dataset via the number of concepts (eg, diagnosis, medication, procedure codes) recorded per person for each concept type, which provides insight into both the extent of data captured per patient and the availability of certain feature types in each dataset, in Supplementary Figure 1.
This study was reviewed and approved by Institutional Review Boards at Stanford University, Columbia University, and SNUBH.
Phenotype selection and classifier development
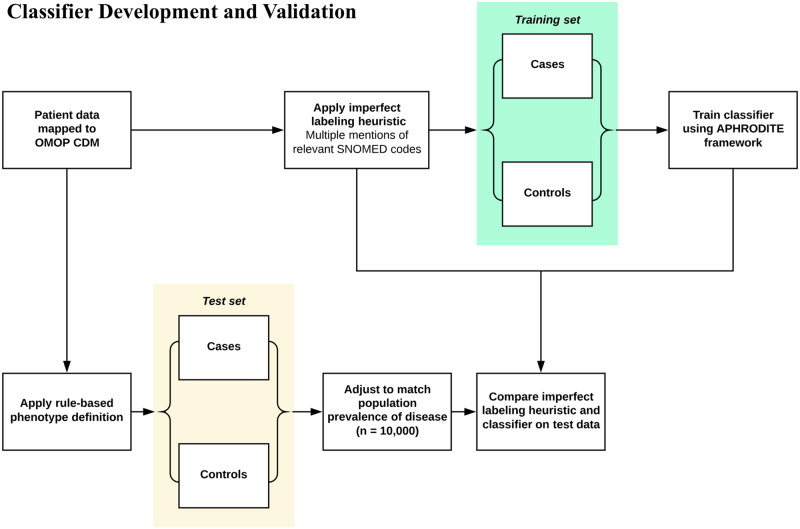
We selected 10 phenotypes (appendicitis, type 2 diabetes mellitus, cataracts, heart failure, abdominal aortic aneurysm, epileptic seizure, peripheral arterial disease, adult onset obesity, glaucoma, and venous thromboembolism) for which rule-based definitions have been created by either the Electronic Medical Records and Genomics (eMERGE) or OHDSI networks. We developed classifiers for each phenotype using the APHRODITE framework, an R-package built for the OMOP CDM that can be used to construct phenotype classifiers using imperfectly labeled training data. In previous work, the labeling heuristic used with APHRODITE was based on single mentions of relevant terms in textual data. In this study, we used multiple mentions of disease-specific codes as our labeling function. In particular, we identified cases by searching patients’ clinical data for at least 4 mentions of any relevant SNOMED code associated with the phenotype of interest (Figure 1). We identified all relevant codes by using vocabulary tables and existing relationships between concepts within the OMOP CDM. Patients who did not meet this multiple mention criteria were considered controls for training purposes, and the ratio of training cases to controls was set to 1:1. We required 4 mentions to balance ensuring that our labeling function was precise with finding a sufficient number of training cases for classifier development. Increasing the mentions required to greater than 4, though likely more precise, resulted in small (less than 500 cases) training set sizes, which we anticipated would result in poor performance.24 When sufficient cases were not identified, we incrementally lowered the number of mentions to identify more cases.
Figure 1.
Development and validation of phenotype classifiers. Training sets were constructed by applying multiple mentions-based imperfect labeling functions to our patient data extract. Patients with multiple mentions of any SNOMED codes relevant to the phenotype of interest were considered training cases. Patients who did not meet this criterion were labeled as training controls. Random forest classifiers were built for each phenotype using 5-fold cross validation. The test set was constructed using OMOP implementations of rule-based phenotype definitions. Test cases were randomly sampled from the cohort of patients selected by the rule-based definitions. Test controls were sampled from the remaining patients. For each phenotype, the imperfect labeling function used to generate the training set and the corresponding classifier were evaluated using the rule-based phenotype-derived test sets.
Once the training cohort was identified, we represented patient data with the following feature types: visits, observations, lab results, procedures and drug exposures. Frequency counts were calculated for each feature capturing the entire course of patients’ EHR records. We chose not to exclude a single mention of relevant disease-specific codes as potential features used by classifiers, since our labeling function was based on multiple mentions. Random forest classifiers were trained for each phenotype using 5-fold cross validation.
Classifier validation with cohorts derived from rule-based definitions
Development of evaluation sets
Rule-based definitions were used to identify the cohort of patients comprising the test set for each phenotype (Figure 1). Two of the definitions, appendicitis and cataracts, were OMOP implementations of definitions that were publicly available on PheKB, a repository of phenotype algorithms developed by the eMERGE network. The other 8 definitions were developed and evaluated collaboratively by several members of the OHDSI network with clinician oversight. Although PheKB definitions have been shown to favor precision and have low recall relative to manual chart review, these rule-based definitions were the best available ground truth label for this experiment.
Rule-based definitions were implemented using ATLAS, an open source software tool for building patient cohorts with OMOP CDM-mapped data. Test cases were identified by randomly sampling the cohort of patients selected by the rule-based definitions. Test controls were identified by randomly sampling from the remaining patients. All test sets were composed of 10 000 patients, with the proportion of cases set equal to the population prevalence of the corresponding phenotype. Any patients used to train classifiers were excluded from test sets.
Local validation of phenotype classifiers
We evaluated the performance of our classifiers by running them on the test sets derived from our rule-based definitions. Classifiers were evaluated locally by using our patient data extract. For reference, we also assessed the performance of the “multiple mentions” labeling heuristic described previously. Performance was reported in terms of recall and precision.
Performance of classifiers across multiple sites
To evaluate the portability of our phenotype classifiers, we shared the 10 classifiers developed on our patient data extract with 2 other institutions within the OHDSI network, Columbia University and SNUBH. Since both these institutions have mapped their patient datasets to meet OMOP CDM specifications, we were able to share our classifiers without any modification. Classifier performance was evaluated in a process identical to the one used locally at Stanford; at both sites, rule-based definitions were used to derive the cohort of patients comprising the test set for each phenotype.
We further assessed the portability of phenotype classifiers by performing a reciprocal experiment in which models were built at Columbia and SNUBH and then evaluated at both the development site and at Stanford. Classifiers were constructed for all 10 phenotypes using the same method that was used locally. Specifically, we employed the same labeling approach to generate training sets for each phenotype; patients with multiple mentions of relevant disease-specific codes were considered training cases, while others were considered training controls. Once classifiers were developed at both sites, we evaluated their performance using test sets that were constructed from rule-based definitions.
Comparing demographics of cases across sites
While rule-based definitions offer an alternative to manual chart review for the generation of test sets, development of these definitions is ultimately labor-intensive and limits the speed with which classifiers can be evaluated. To circumvent the need for rule-based definitions and chart review, we propose comparing the demographics of patients identified as cases by our classifiers across different sites as a proxy for model validation. For this experiment, we randomly selected 150 000 patients at each site and used classifiers developed at our institution to identify cases for each phenotype. We then compared the cohorts of patients labeled as cases across different sites with respect to key demographics, such as age and sex. The purpose of this was to evaluate whether the classifiers not only showed comparable performance across sites, but also identified comparable cohorts in terms of basic demographic features.
RESULTS
Local performance of classifiers
We first compared the performance of our phenotype classifiers with the “multiple mentions” labeling heuristic used to identify training cases for each phenotype. Table 1 shows the recall and precision of both of these phenotyping approaches. Requiring multiple disease-specific code mentions to classify patients as cases yields a mean precision of 0.99, as it is likely that patients with several mentions of a relevant code have the associated phenotype. Achieving high precision, however, results in noticeably low recall. The mean recall for requiring multiple mentions was 0.17.
Table 1.
Test set performance of labeling heuristic requiring multiple disease-specific code mentions compared to phenotype classifiers trained with data labeled using this multiple mentions approach
| Phenotype | Prevalence of cases in test set | Multiple mentions of SNOMED code |
APHRODITE classifier |
Recall boost using classifier | Precision loss using classifier | |||
|---|---|---|---|---|---|---|---|---|
| No. of mentions | Recall | Precision | Recall | Precision | ||||
| Appendicitis | 0.05 | 2 | 0.31 | 1.00 | 0.97 | 0.99 | 0.66 | 0.01 |
| T2DM | 0.14 | 4 | 0.24 | 0.99 | 0.60 | 0.91 | 0.36 | 0.08 |
| Cataracts | 0.17 | 4 | 0.07 | 0.97 | 0.63 | 0.93 | 0.56 | 0.04 |
| HF | 0.02 | 4 | 0.33 | 0.94 | 0.99 | 0.56 | 0.66 | 0.38 |
| AAA | 0.04 | 4 | 0.22 | 0.99 | 0.53 | 0.97 | 0.31 | 0.02 |
| Epileptic seizure | 0.02 | 4 | 0.06 | 1.00 | 0.22 | 0.94 | 0.17 | 0.06 |
| PAD | 0.05 | 4 | 0.18 | 0.98 | 0.91 | 0.91 | 0.72 | 0.07 |
| Adult onset obesity | 0.36 | 4 | 0.20 | 1.00 | 0.29 | 0.91 | 0.09 | 0.09 |
| Glaucoma | 0.01 | 4 | 0.08 | 1.00 | 0.22 | 0.88 | 0.14 | 0.12 |
| VTE | 0.01 | 4 | 0.03 | 1.00 | 0.69 | 0.22 | 0.66 | 0.78 |
Abbreviations: AAA, abdominal aortic aneurysm; HF, heart failure; T2DM, type 2 diabetes mellitus; PAD, peripheral arterial disease; VTE, venous thromboembolism.
Classifiers built with training data labeled using the multiple code mentions heuristic showed markedly improved recall with relatively small losses in precision. The mean recall boost observed was 0.43 while the mean precision loss was 0.17. Seven classifiers showed precision losses that were less than 0.10. Classifiers for 2 phenotypes, heart failure and venous thromboembolism, had more considerable losses in precision (−0.38 and −0.78, respectively).
Performance of classifiers across sites
We evaluated the portability of our classifiers by assessing their performance at Columbia and SNUBH. Table 2 summarizes performance at these 2 sites. When classifiers were tested at Columbia, mean recall and precision decreased marginally by 0.08 and 0.01, respectively, compared to local performance. Classifiers tested at SNUBH had more significant losses in performance. Mean recall and precision decreased by 0.18 and 0.10, respectively.
Table 2.
Classifier performance at 3 sites within OHDSI network. Phenotype classifiers constructed at Stanford were shared with Columbia and SNUBH, and evaluated using test sets derived locally at each site using rule-based definitions. Furthermore, classifiers built at Columbia and SNUBH were shared with Stanford and evaluated using similarly constructed test sets. Blue denotes values equal to 1, white denotes values equal to 0
| Development site |
Stanford |
Columbia |
SNUBH |
Stanford |
Columbia |
SNUBH |
||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Validation site |
Stanford |
Columbia |
SNUBH |
Stanford |
Columbia |
Stanford |
SNUBH |
Stanford |
Columbia |
SNUBH |
Stanford |
Columbia |
Stanford |
SNUBH |
| Phenotype | Recall | Precision | ||||||||||||
| Appendicitis | 0.97 | 0.9 | 0.09 | 0.82 | 0.75 | 0.52 | 0.1 | 0.99 | 0.9 | 0.56 | 0.83 | 0.48 | 0.13 | 0.98 |
| T2DM | 0.6 | 0.63 | 0.77 | 0.58 | 0.69 | 0.67 | 0.75 | 0.91 | 0.86 | 0.75 | 0.75 | 0.83 | 0.51 | 0.89 |
| Cataracts | 0.63 | 0.45 | 0.84 | 0.8 | 0.6 | 0.35 | 0.84 | 0.93 | 0.79 | 0.85 | 0.74 | 0.74 | 0.42 | 0.74 |
| HF | 0.99 | 0.97 | 0.8 | 0.99 | 0.97 | 0.71 | 0.82 | 0.56 | 0.67 | 0.75 | 0.47 | 0.4 | 0.11 | 0.66 |
| AAA | 0.53 | 0.24 | 0.54 | 0.59 | 0.78 | 0.33 | 0.57 | 0.97 | 0.75 | 0.87 | 0.96 | 0.97 | 0.13 | 0.47 |
| Epileptic seizure | 0.22 | 0.3 | 0.28 | 0.41 | 0.79 | 0.46 | 0.11 | 0.94 | 0.87 | 0.55 | 0.79 | 0.57 | 0.08 | 0.68 |
| PAD | 0.91 | 0.89 | 0.57 | 0.48 | 0.85 | 0.46 | 0.55 | 0.91 | 0.87 | 0.68 | 0.69 | 0.57 | 0.24 | 0.59 |
| Adult onset obesity | 0.29 | 0.33 | 0.07 | 0.14 | 0.59 | 0.39 | 0.07 | 0.91 | 0.93 | 0.73 | 0.85 | 0.89 | 0.68 | 0.8 |
| Glaucoma | 0.22 | 0.18 | 0.11 | 0.34 | 0.4 | 0.22 | 0.12 | 0.88 | 0.78 | 0.65 | 0.69 | 0.8 | 0.06 | 0.75 |
| VTE | 0.69 | 0.34 | 0.2 | 0.21 | 0.71 | 0.46 | 0.19 | 0.2 | 0.71 | 0.83 | 0.51 | 0.21 | 0.05 | 0.78 |
Abbreviations: AAA, abdominal aortic aneurysm; HF, heart failure; T2DM, type 2 diabetes mellitus; PAD, peripheral arterial disease; VTE, venous thromboembolism.
We also assessed classifier portability by constructing models at Columbia and SNUBH, and evaluating their performance at Stanford. Classifiers built at Columbia performed comparably to those developed at Stanford, with these classifiers having mean recall and precision values of 0.54 and 0.73, respectively. In contrast, classifiers developed at SNUBH did not perform well at Stanford. For these classifiers, we observed mean recall and precision values of 0.46 and 0.24, respectively. Notably, these classifiers performed better when tested locally at SNUBH (mean recall and precision values of 0.41 and 0.73, respectively), indicating that performance dropped when classifiers were ported to Stanford.
Classifier evaluation by comparing demographic features of cases across sites
We further examined classifier performance by evaluating the demographics of patients classified as cases for each phenotype at all 3 sites. We used the classifiers built at Stanford to select cases at all sites. The aim of this is to assess both the overall performance and portability of classifiers by determining whether classifiers identify comparable cohorts of patients across sites. Overall, age and proportion of each sex were similar among all patients at the 3 sites; mean age was 39.3, 39.9, and 40.4, and the proportion of males was 0.45, 0.45, and 0.48 at Stanford, Columbia, and SNUBH, respectively. There was considerable variability in demographics of patients selected as cases by classifiers at each site. For instance, for 7 of the 10 phenotypes, there was a statistically significant difference in the proportion of males identified as cases. Similar variation existed with regards to mean age of cases at the 3 sites (Supplementary Table 1).
DISCUSSION
This study outlines a method for generating high-precision phenotyping classifiers in a semisupervised manner. We demonstrate that classifiers trained using a high-precision labeling heuristic (ie, multiple mentions of disease-specific codes) are able to preserve precision while boosting recall relative to the original labeling function. While it may be difficult to predict a priori which phenotypes would benefit most from classifier generation, we believe that conditions likely to benefit most are those like peripheral arterial disease, appendicitis, and cataracts that have common medications, procedures, and lab values that can help identify additional patients who might be missed by simply requiring multiple mentions of disease-specific SNOMED codes. This recall boost may be particularly important in situations of low disease prevalence, in which identifying additional cases is necessary for building sufficiently large cohorts for observational studies. Furthermore, these classifiers are significantly faster to generate than rule-based phenotype definitions and do not rely on expert clinical input. This may be a template for high-throughput creation of phenotyping classifiers in a way that optimizes precision and recall which would greatly facilitate observational research.3
An important advantage of building phenotype classifiers with the APHRODITE framework is the ability to easily exchange models across sites,24 however this has not been previously assessed. We evaluated model portability in this study by sharing phenotype classifiers developed at Stanford with Columbia and SNUBH. Performance at both of these sites was generally good, with minimal losses in recall and precision at Columbia (0.08 and 0.01, respectively) and a larger performance drop at SNUBH (0.18 and 0.10, respectively). We suspect that the larger drop in performance at SNUBH, which is an international site, is likely related to regional differences in EHR data and how clinical concepts are coded across sites even within the same common data model. We observed larger differences in the availability of concept types (eg, diagnosis, medication, procedure codes) between Stanford and SNUBH than Stanford and Columbia which likely impacts classifier portability between these sites.
In a reciprocal experiment, Columbia and SNUBH both constructed classifiers and shared them for evaluation at Stanford. Classifiers built at Columbia performed well when tested at Stanford, with these classifiers showing similar performance to those developed natively at Stanford; both mean recall and precision were within 0.1 points of Stanford classifiers. In comparison, we observed a considerable performance drop when classifiers were built at SNUBH and ported to Stanford. Although SNUBH classifiers demonstrated a mean precision value of 0.73 when evaluated at SNUBH, this dropped to 0.24 when tested at Stanford.
The poor portability of classifiers developed at SNUBH suggests that sharing the classifier-building recipe may prove more useful than sharing the pretrained classifiers themselves since site-specific differences in EHR data appear to substantially impact performance. Notably, in the majority of comparisons, classifiers trained locally at the site of evaluation, using the labeling function to generate training data, performed at least as well or better than classifiers trained elsewhere. The APHRODITE framework specifically offers the ability to exchange classifier-building recipes. Unlike traditional supervised learning approaches for phenotyping which require manually searching for patients to construct the training set, sharing a high-precision labeling function for developing a large imperfectly labeled training set and rebuilding classifiers at any given site is efficient and feasible. Given that constructing a phenotype classifier with APHRODITE requires minimal work and that differences in EHR data (ie, the availability of concept types) across sites affect performance, we propose that sharing the classifier-building recipe is favorable to sharing the classifiers themselves in the majority of situations.
This study was limited by the use of PheKB definitions as the gold standard for classifier evaluation. Although these definitions have been reviewed by clinical experts, they are still rule-based definitions with imperfect classification accuracy. While the use of these definitions provided a standardized way to assign “ground-truth” labels across multiple international sites, in future our classifier pipeline could be assessed using clinician-labeled test sets or phenotype algorithm evaluation tools that have been recently developed, such as PheValuator.25 Additionally, the feature engineering scheme used in training classifiers is relatively rudimentary—simply a frequency count of all structured data elements. The performance of our classifiers may therefore be seen as a conservative estimate of such semisupervised learning. More sophisticated feature engineering regimens such as EHR embeddings, incorporating temporal trends in lab values, or some extracts from the unstructured data, would likely improve performance. Finally, use of this APHRODITE-based pipeline relies on sites mapping their EHR data to the OMOP CDM.
CONCLUSION
We demonstrate a high-throughput pipeline for developing and sharing phenotype classifiers using APHRODITE with a high-precision labeling heuristic. These classifiers are easier to create than rule-based definitions and can be shared across sites within the OHDSI network. We establish good portability between Stanford and Columbia in both directions but limited performance when sharing classifiers with SNUBH, indicating that the generalizability of phenotype classifiers may have geographic limitations due to differences in EHR data. In this situation, sharing the classifier training recipe (ie, providing the labeling function for generating a large imperfectly labeled training set and training a local classifier) rather than the pretrained models may be more useful for generating classifiers and thus facilitating collaborative observational research.
FUNDING
This study was supported by NLM R01LM011369-06, NLM R01LM006910, and a research grant by Janssen Research and Development LLC.
AUTHOR CONTRIBUTIONS
NHS, MK, MS, and JMB envisioned the study and designed the experiments. MK, MS, TF, GH, BR, and SY conducted the experiments at each of their respective institutions. MK, MS, and NHS performed the analysis and wrote the paper. All authors participated in editing the manuscript and approve of its final version.
SUPPLEMENTARY MATERIAL
Supplementary material is available at Journal of the American Medical Informatics Association online.
Supplementary Material
ACKNOWLEDGMENTS
We would like to thank the members of the Stanford Center for Biomedical Informatics Research and the Observational Health Data Sciences and Informatics community for their support throughout this study.
CONFLICT OF INTEREST STATEMENT
MS currently works at Google DeepMind. This work was completed prior to his employment and represents personal views only.
REFERENCES
- 1. Banda JM, Seneviratne M, Hernandez-Boussard T, et al. Advances in electronic phenotyping: from rule-based definitions to machine learning models. Annu Rev Biomed Data Sci. 2018; 1 (1): 53–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Richesson RL, Hammond WE, Nahm M, et al. Electronic health records based phenotyping in next-generation clinical trials: a perspective from the NIH Health Care Systems Collaboratory. J Am Med Inform Assoc. 2013; 20 (e2): e226–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Hripcsak G, Albers DJ.. Next-generation phenotyping of electronic health records. J Am Med Inform Assoc. 2013; 20 (1): 117–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Pathak J, Kho AN, Denny JC.. Electronic health records-driven phenotyping: challenges, recent advances, and perspectives. J Am Med Inform Assoc. 2013; 20 (e2): e206–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Shivade C, Raghavan P, Fosler-Lussier E, et al. A review of approaches to identifying patient phenotype cohorts using electronic health records. J Am Med Inform Assoc. 2014; 21 (2): 221–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Kirby JC, Speltz P, Rasmussen LV, et al. PheKB: a catalog and workflow for creating electronic phenotype algorithms for transportability. J Am Med Inform Assoc. 2016; 23 (6): 1046–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Pacheco JA, Rasmussen LV, Kiefer RC, et al. A case study evaluating the portability of an executable computable phenotype algorithm across multiple institutions and electronic health record environments. J Am Med Inform Assoc. 2018; 25 (11): 1540–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Denaxas SC, George J, Herrett E, et al. Data resource profile: cardiovascular disease research using linked bespoke studies and electronic health records (CALIBER). Int J Epidemiol. 2012; 41 (6): 1625–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Newton KM, Peissig PL, Kho AN, et al. Validation of electronic medical record-based phenotyping algorithms: results and lessons learned from the eMERGE network. J Am Med Inform Assoc. 2013; 20 (e1): e147–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Hripcsak G, Duke JD, Shah NH, et al. Observational Health Data Sciences and Informatics (OHDSI): opportunities for observational researchers. Stud Heal Technol Inform. 2015; 216: 574–8. [PMC free article] [PubMed] [Google Scholar]
- 11. Kohane IS, Churchill SE, Murphy SN.. A translational engine at the national scale: informatics for integrating biology and the bedside. J Am Med Inform Assoc. 2012; 19 (2): 181–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Hripcsak G, Shang N, Peissig PL, et al. Facilitating phenotype transfer using a common data model. J Biomed Inform. 2019; 96: 103253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Tian TY, Zlateva I, Anderson DR.. Using electronic health records data to identify patients with chronic pain in a primary care setting. J Am Med Inform Assoc. 2013; 20 (e2): e275–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Carroll RJ, Eyler AE, Denny JC.. Naïve Electronic Health Record phenotype identification for rheumatoid arthritis. AMIA Annu Symp Proc. 2011; 2011: 189–96. [PMC free article] [PubMed] [Google Scholar]
- 15. Carroll RJ, Thompson WK, Eyler AE, et al. Portability of an algorithm to identify rheumatoid arthritis in electronic health records. J Am Med Inform Assoc. 2012; 19 (e1): e162–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Chen Y, Carroll RJ, Hinz ERM, et al. Applying active learning to high-throughput phenotyping algorithms for electronic health records data. J Am Med Inform Assoc. 2013; 20 (e2): e253–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Agarwal V, Podchiyska T, Banda JM, et al. Learning statistical models of phenotypes using noisy labeled training data. J Am Med Inform Assoc. 2016; 23 (6): 1166–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Halpern Y, Choi Y, Horng S, et al. Using anchors to estimate clinical state without labeled data. AMIA Annu Symp Proc. 2014; 2014: 606–15. [PMC free article] [PubMed] [Google Scholar]
- 19. Halpern Y, Horng S, Choi Y, et al. Electronic medical record phenotyping using the anchor and learn framework. J Am Med Inform Assoc. 2016; 23 (4): 731–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Beaulieu-Jones BK, Greene CS.. Consortium PRO-AALSCT. Semi-supervised learning of the electronic health record for phenotype stratification. J Biomed Inform. 2016; 64: 168–78. [DOI] [PubMed] [Google Scholar]
- 21. Yu S, Chakrabortty A, Liao KP, et al. Surrogate-assisted feature extraction for high-throughput phenotyping. J Am Med Inform Assoc. 2017; 24 (e1): e143–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Murray SG, Avati A, Schmajuk G, et al. Automated and flexible identification of complex disease: building a model for systemic lupus erythematosus using noisy labeling. J Am Med Informatics Assoc. 2019; 26 (1): 61–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Simon HU. General bounds on the number of examples needed for learning probabilistic concepts. J Comput Syst Sci. 1996; 52 (2): 239–54. [Google Scholar]
- 24. Banda JM, Halpern Y, Sontag D, et al. Electronic phenotyping with APHRODITE and the Observational Health Sciences and Informatics (OHDSI) data network. AMIA Jt Summits Transl Sci Proc. 2017; 2017: 48–57. [PMC free article] [PubMed] [Google Scholar]
- 25. Swerdel JN, Hripcsak G, Ryan PB.. PheValuator: development and evaluation of a phenotype algorithm evaluator. J Biomed Inform. 2019; 97: 103258. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.