Abstract
Objective
Incomplete and static reaction picklists in the allergy module led to free-text and missing entries that inhibit the clinical decision support intended to prevent adverse drug reactions. We developed a novel, data-driven, “dynamic” reaction picklist to improve allergy documentation in the electronic health record (EHR).
Materials and Methods
We split 3 decades of allergy entries in the EHR of a large Massachusetts healthcare system into development and validation datasets. We consolidated duplicate allergens and those with the same ingredients or allergen groups. We created a reaction value set via expert review of a previously developed value set and then applied natural language processing to reconcile reactions from structured and free-text entries. Three association rule-mining measures were used to develop a comprehensive reaction picklist dynamically ranked by allergen. The dynamic picklist was assessed using recall at top k suggested reactions, comparing performance to the static picklist.
Results
The modified reaction value set contained 490 reaction concepts. Among 4 234 327 allergy entries collected, 7463 unique consolidated allergens and 469 unique reactions were identified. Of the 3 dynamic reaction picklists developed, the 1 with the optimal ranking achieved recalls of 0.632, 0.763, and 0.822 at the top 5, 10, and 15, respectively, significantly outperforming the static reaction picklist ranked by reaction frequency.
Conclusion
The dynamic reaction picklist developed using EHR data and a statistical measure was superior to the static picklist and suggested proper reactions for allergy documentation. Further studies might evaluate the usability and impact on allergy documentation in the EHR.
INTRODUCTION
Documentation of adverse reactions to drugs, foods, and healthcare product ingredients, such as adhesives and latex, in the electronic health record (EHR) is critical to patient safety. Most EHR systems have an allergy module—or allergy list—to support this documentation. Despite being called an “allergy list,” diverse adverse reactions beyond allergies (eg, intolerances, contraindications) are documented in this list. Allergy list documentation is a routine component of ambulatory and hospitalized care1; more than 35% of patients have at least 1 EHR-documented allergy, and 4% have 3 or more EHR-documented allergies.2 While several allergy data elements can be captured in the allergy list (eg, reaction, severity, and type), there are only 2 routinely reported data elements: the culprit agent (also called allergen, such as a drug or food) and the resulting adverse reaction(s), such as rash, hives, or anaphylaxis. Complete and accurate reaction documentation is important to support clinical decision-making and inform safe prescribing in healthcare settings.
The current EHR allergy module has notable limitations that reduce its usefulness for clinical decision support.3–10 Most current EHR allergy modules rely on commercial or local dictionaries shown to the user as a single, static picklist and ordered alphabetically. Even among hospitals with the same commercial EHR vendor, reaction picklists vary by site; coded reaction options range from just 10 to more than 100 reactions. For example, a large Massachusetts healthcare system’s reaction picklist contains 46 reactions (see Supplementary MaterialTable 1), while a Colorado healthcare system’s picklist contains a different set of 156 reactions. Without a coded reaction lexicon comprehensive enough to encode the diverse reactions encountered in clinical practice, reaction entries are often incomplete and/or entered as free text.11 Indeed, up to half of reaction fields are left blank, and at least one-sixth of reactions are entered as free text.12
Table 1.
Characteristics of the development and validation datasets from the EHR allergy database
| Characteristics | Allergy Data |
||
|---|---|---|---|
| Development | Validation | Total | |
| Patients, n | 1 683 245 | 272 108 | 1 850 444 |
| Allergy entries, n | 3 743 553 | 490 774 | 4 234 327 |
| Active allergy entries, n (%) | 3 280 743 (87.6) | 475 583 (96.9) | 3 756 326 (88.7) |
| Allergy entries with free-text comments, n (%) | 1 535 657 (41.0) | 156 193 (31.8) | 1 691 850 (40.0) |
| Allergy entries with at least 1 reaction, n (%) | 2 171 548 (58.0) | 276 348 (56.3) | 2 448 538 (57.8) |
| Allergens before consolidation, n | 10 265 | 5968 | 11 181 |
| Allergens after consolidation, n | 6720 | 4818 | 7463 |
| Allergens after consolidation and have at least 1 associated reaction, n | 5553 | 3419 | 6127 |
| Unique reactions used after the refinement and reaction list reconciliation, n | 468 | 459 | 469 |
The high prevalence of free-text and missing entries may be due to the inadequacy of reaction picklists and/or the inconvenience of searching for the most appropriate reaction using a static alphabetized picklist. To address this, we used natural language processing (NLP) to analyze a large volume of EHR allergen-reaction entries to develop a comprehensive reaction value set of 787 reactions.12 Although the value set is a comprehensive reaction list, it would be too cumbersome for providers to search the entire list to enter the appropriate reaction(s). As such, in the present study, we developed a novel, data-driven, “dynamic” reaction picklist through a combination of manual review efforts and data-mining techniques. With the dynamic picklist, once an allergen is selected, the reaction picklist is populated with the most probable reactions, thereby allowing providers to easily select coded reaction(s).
MATERIALS AND METHODS
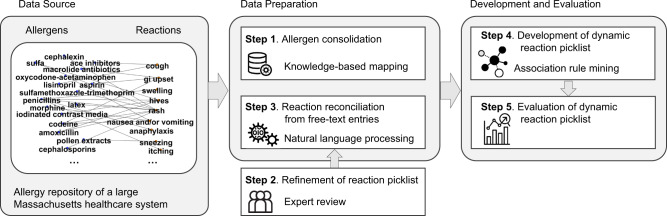
The study method was comprised of 5 steps: (1) allergen consolidation, (2) refinement of reaction value set, (3) reaction reconciliation from free-text entries, (4) development of dynamic reaction picklist, and (5) evaluation of the dynamic reaction picklist (see Figure 1).
Figure 1.
An overview of the workflow for the development and evaluation of the proposed dynamic reaction picklist. The source data were the allergy entries in an allergy repository of a large Massachusetts healthcare system containing allergens and reactions. The data preparation processes include (1) allergen consolidation conducted through knowledge-based mapping, (2) refinement of reaction picklist conducted through exclusive manual expert review, and (3) reaction reconciliation from free-text entries using natural language processing. The dynamic reaction picklist was then developed using association rule-mining measures and evaluated.
Clinical setting and data sources
This study was conducted at a large Massachusetts healthcare system that includes Massachusetts General Hospital and Brigham and Women’s Hospital (Boston). The healthcare system initially used an in-house developed EHR system but switched to a commercial EHR vendor (ie, Epic) beginning in 2015. We used EHR allergy data from all patients within our healthcare system between around March 28, 1980 and October 8, 2019. Data were entered by healthcare team members, and each patient may have 1 or multiple allergy entries, with each allergy entry corresponding to 1 allergen and any number of reactions. The allergy data were separated into development and validation datasets, with the development dataset including allergy entries from 1980 to October 8, 2018, and the validation dataset including entries from October 9, 2018 to October 8, 2019. Inactive allergy entries and entries consisting of “No Known Allergies” were excluded.
Allergen consolidation
The reference allergen table contained 42 027 allergens among which are many synonyms, duplicates, brand names, or other variations. We consolidated allergens by developing a mapping among allergens when they had (1) a versioning history in the database, (2) the same name, or (3) the same ingredients and allergen groups. First, for allergens that were indicated as expired or hidden but still associated with active allergy entries in our database, we mapped them to active allergens using the recorded mapping history. Second, we consolidated allergens with the same names by mapping the less frequently entered allergen(s) to the 1 entered most frequently. Furthermore, we consolidated allergens that had the same ingredients and allergen groups. To identify those allergens, we used the mappings between allergens and allergen groups, between allergens and First Databank (FDB) medications, and between FDB medications and RxNorm ingredients.13 We excluded allergens at the drug class level (eg, penicillins) from the consolidation process, as we tried to avoid merging drug class-level allergens with specific drug allergens under that class. Among allergens with the same ingredients and allergen groups, we chose the allergen with the most entries in the database as the preferred allergen term and considered the rest synonyms. After the allergen consolidation, we updated the allergen entries in our database using the preferred allergen names and generated an allergen and reaction co-occurrence table.
Refinement of reaction value set
We refined the reaction value set through expert panel review, using the following 3-part process: (1) merging 2 reaction lists, (2) identifying clinician preferred terms, and (3) consolidating reaction terms that were overly granular, similar, or less-frequently used.
First, we merged the reaction picklist used in our EHR allergy module with our reaction value set. There were notable differences between the reaction terms included in the 2 datasets. The current EHR reaction picklist contained 46 reactions, including commonly reported reactions (eg, rash, hives, itching) and serious reactions (eg, anaphylaxis, Stevens-Johnson syndrome). It did not include many other common reactions, such as rhinitis, syncope, or bleeding. In a prior study, we identified a comprehensive reaction value set of 787 reactions,12 but the concepts in this value set were much more granular compared to the EHR picklist. To address this, we mapped the terms in the reaction value set to those in the picklist, using the latter as preferred terms. For example, we mapped “dyspnea” in the reaction value set to “shortness of breath” in the picklist, and similarly mapped “nausea” and “nausea and vomiting” to “nausea and/or vomiting.”
Second, as our goal was to develop a reaction picklist that could be used by clinicians for reaction documentation in the EHR, we updated the preferred terms of the reaction value set based on clinicians’ usage preference. This was accomplished by calculating the occurrence of the SNOMED CT14 preferred terms and their synonyms in the allergy database. We then identified a subset of reaction concepts whose synonyms were more frequently entered by clinicians in the allergy database than the SNOMED CT preferred terms. These reaction concepts and the most frequent synonyms were manually reviewed by at least 2 clinicians (FG, SNS, ZTK, KGB) to determine preferred reaction terms for data entry. For example, “blood in urine” was the preferred term for a SNOMED CT concept (ID: 34436003), but its synonym “hematuria” was much more frequently mentioned in the free-text comments in our allergy database and was therefore chosen as the preferred term in the refined reaction value set. Similarly, while “dyssomnia” was the preferred term in SNOMED CT, “sleep disturbance” occurred more frequently in our database and, as such, was chosen as the preferred term in the picklist.
Third, an expert panel with allergy subject matter expertise (FG, KGB, SY, LZ, YL, LW) manually reviewed the merged reaction list to further evaluate and consolidate the included reactions. Specifically for the final reaction picklist, we consolidated terms with similar meanings (eg, mapping “feeling nervous” to “anxiety”) and terms with parent-child relationships (eg, mapping “finger swelling” and “hand swelling” to “swelling of extremities”). We also identified and removed general or vague terms (eg, “toxicity,” “addiction,” “allergic reaction”) and condition terms (eg, “diabetes,” “bronchitis”) from the list. All decisions to merge or remove reactions were agreed upon by at least 2 of the experts.
Reaction reconciliation from free-text entries
We used an NLP tool suite called Medical Text Extraction, Reasoning, and Mapping System (MTERMS)15,16 to process the free-text comments field in our database to extract any reaction terms mentioned. For this task, we used a knowledge-based approach within the MTERMS allergy module. Using a lexicon,12 it extracted the reaction terms used by our clinicians and mapped them to the corresponding preferred terms in the refined reaction value set. It also handled contextual information, such as negations. For each allergy entry, we excluded any reactions found in free text that were also among the coded reactions for that entry. This resulted in an updated table of allergy entries where each allergy entry contained all unique reactions in the refined reaction value set.
Development of the dynamic reaction picklist
Based on the updated allergen-reaction entries, we developed the dynamic reaction picklist. We explored 3 approaches to ranking reaction concepts with respect to a given allergen substance: support, modified lift (lift’), and derived term frequency inverse document frequency (ie, tf-idf’).17,18 The formulations of the 3 measures that we used to develop the dynamic picklist are defined in the following formulas., , and are the proportions of allergy entries containing allergen x and reaction y and co-occurring allergen x and reaction y, respectively. is the frequency of allergy entries in the database in which x and y co-occurred. is the frequency of allergy entries containing allergen x, N is the total number of allergens, and is the number of allergens that co-occurred with reaction y. Note that the original formula for lift tends to raise the association scores for extremely rare reactions, such as those with only 1 occurrence. We added the square root function to the to reduce the effect of the lift of rare reactions, thus the modified is denoted as .
| (1) |
| (2) |
| (3) |
The above formulas were applied only to those reactions that co-occurred with an allergen. If an allergen did not co-occur with all of the reactions in the reaction value set, it was impossible to form a dynamic reaction picklist for that allergen that includes the full value set. Thus, for each allergen, we ranked the reactions that never co-occurred with that allergen by the frequency of each reaction’s occurrence in the development dataset and concatenated that list to the list of reactions that did co-occur with that allergen. This ensures that each allergen will have a fully ranked reaction picklist.
Evaluation of the dynamic reaction picklist
We used retrospective allergy entries documented between October 09, 2018 and October 08, 2019 to evaluate the relevance, effectiveness, and usefulness of the proposed dynamic picklist. Allergy entries with new allergens that never occurred in the development dataset or without any associated reactions were excluded from the validation dataset.
To choose proper evaluation metrics, we considered this ideal scenario: When a clinician documents an allergy in the EHR, after choosing the allergen, the dynamic reaction list should suggest the relevant reactions among the top-k returned list. The smaller the k, the more efficiently the clinician can select the correct reaction. If the user cannot find the desired reaction(s) among the top k, they might search among the entire list, in which case the dynamic reaction list becomes less useful. Therefore, in our evaluation, we investigated the performance of the dynamic reaction picklist with the top-k suggested reactions at k ≤ 15 under the assumption that a list of more than 15 terms on a single screen might be less acceptable to clinicians. Precision and recall at top k are 2 classic evaluation metrics for recommendation systems, where precision at k is the proportion of recommended items in the top-k set that are relevant, while recall at top k is the proportion of relevant items found in the top-k recommendations. We argue that, in our application scenarios, it is more important to have all true positive reaction terms among the suggested top k (k ≤ 15) reactions (recall), while it is less important to measure the proportion of positive reactions to the entire suggested list (precision) as the number of true positive reactions was often small, ranging from 1 to a few. Therefore, we measured the performance of the dynamic reaction list in terms of the recall at top k (k ≤ 15), which is the proportion of true positive reactions among the top k suggested reactions by the dynamic reaction picklist to the total number of true reactions in the validation dataset. In addition, we compared the performance of 3 dynamic reaction picklists ranked by support, lift’, and tfidf’ and the static reaction picklist, where the static reaction picklist was a list of reactions ranked by their total occurrence frequency in the development dataset, irrespective of allergen chosen. We used the Friedman test to verify the significance of the difference, followed by pairwise comparisons using the Nemenyi posthoc test.19
RESULTS
As of October 8, 2019, the allergy database contained 4 234 327 allergy entries of 1 850 444 unique patients among which 3 756 326 were active allergy reaction entries (excluding “No Known Allergies” entries and allergy entries marked as inactive) with 2 289 511 active allergy entries having 1 or more reactions. The development dataset contained 3 743 628 allergy entries of 1 683 678 patients (see Table 1). Among these, 3 280 743 (87.6%) were active allergy entries, 1 535 657 (41.0%) were free-text reaction comments, and 2 171 548 (58.0%) allergy entries had at least 1 reaction in the refined reaction value set. The validation dataset contained a total of 490 774 allergy entries corresponding to 272 108 patients. Among these, 475 583 (96.9%) were active allergy entries, 156 193 (31.8%) were free-text reaction comments, and 276 348 (56.3%) allergy entries had at least 1 reaction.
The allergen reference table in our EHR database contained a total of 42 639 allergens, but only 26.2% (n = 11 181) of these allergens had allergy entries in patient records, with 24.1% (n = 10 265) of allergens in the development dataset, and 14.0% (n = 5968) of allergens in the validation dataset. Through the allergen consolidation process, a total of 13 177 (30.9%) allergens were mapped to other allergens in the allergen reference table. First, a total of 5668 allergens which were indicated as “deleted” or “hidden” were simply mapped to their current allergen versions. Second, 3438 allergens were mapped to other allergens by name. Furthermore, a total of 4071 allergens were mapped to other allergens with which they shared allergen groups and RxNorm ingredients. After consolidation, the number of allergens was reduced from 42 639 to 29 462. Among the consolidated allergen list, 7463 allergens were present in the entire allergy dataset, with 6720 (90.0%) in the development dataset and 4818 (64.5%) in the validation dataset. However, counting only those allergens that had at least 1 recorded resulting reaction, 6127 allergens were present in the entire allergy dataset, with 5553 in the development dataset, 3419 in the validation dataset, and 3265 in both datasets.
During the manual refinement of the reaction lexicon, the original reaction value set of 787 reaction terms was consolidated to 490 reactions. The refined reaction value set was submitted to the National Library of Medicine (NLM) Value Set Authority Center (VSAC). Supplementary MaterialTable 2 shows the full refined reaction value set with mappings to SNOMED CT concepts. The 50 most frequent reactions are listed in Table 2.
Table 2.
Top 50 most prevalent adverse reactions in the reaction picklist
| Ranking | Reaction | Allergy entries (N = 3 743 553) n (%) | Ranking | Reaction | Allergy entries (N = 3 743 553) n (%) |
|---|---|---|---|---|---|
| 1 | Rash | 557 188 (14.9) | 26 | Hypotension | 12 090 (0.3) |
| 2 | Hives | 339 952 (9.1) | 27 | Rhinitis | 11 199 (0.3) |
| 3 | GI upset | 197 981 (5.3) | 28 | Musculoskeletal pain | 11 108 (0.3) |
| 4 | Nausea and/or vomiting | 171 474 (4.6) | 29 | Pain | 9891 (0.3) |
| 5 | Itching | 139 414 (3.7) | 30 | Dystonia | 9758 (0.3) |
| 6 | Anaphylaxis | 118 274 (3.2) | 31 | Syncope | 9163 (0.2) |
| 7 | Swelling | 88 620 (2.4) | 32 | Facial swelling | 8612 (0.2) |
| 8 | Cough | 59 154 (1.6) | 33 | Throat tightness | 7878 (0.2) |
| 9 | Mental status change | 53 961 (1.4) | 34 | Edema | 7870 (0.2) |
| 10 | Shortness of breath | 46 713 (1.2) | 35 | Throat swelling | 7691 (0.2) |
| 11 | Angioedema | 46 651 (1.2) | 36 | Tongue swelling | 7573 (0.2) |
| 12 | Diarrhea | 37 997 (1.0) | 37 | Seizures | 7467 (0.2) |
| 13 | Headaches | 35 765 (0.1) | 38 | Swelling of extremities | 6996 (0.2) |
| 14 | Dizziness | 31 974 (0.9) | 39 | Lip swelling | 6893 (0.2) |
| 15 | Myalgia | 27 511 (0.7) | 40 | Agitation | 6555 (0.2) |
| 16 | Palpitations | 24 948 (0.7) | 41 | Bleeding | 6521 (0.2) |
| 17 | Bronchospasm | 24 041 (0.6) | 42 | Dermatitis | 6481 (0.2) |
| 18 | Sneezing | 23 911 (0.6) | 43 | Nasal congestion | 6378 (0.2) |
| 19 | Wheezing | 18 364 (0.5) | 44 | Confusion | 6221 (0.2) |
| 20 | Anxiety | 17 075 (0.5) | 45 | Abdominal pain | 6090 (0.2) |
| 21 | Erythema | 16 128 (0.4) | 46 | Chest pain | 5887 (0.2) |
| 22 | Flushing | 15 105 (0.4) | 47 | Renal toxicity | 5753 (0.2) |
| 23 | Fever | 14 751 (0.4) | 48 | Burning sensation | 5601 (0.1) |
| 24 | Fatigue | 13 135 (0.4) | 49 | Blister | 5587 (0.1) |
| 25 | Hallucinations | 12 732 (0.3) | 50 | Gastrointestinal hemorrhage | 5584 (0.1) |
We used the development dataset, or the 2 171 548 allergy entries, to create the dynamic reaction picklists for the allergens occurring in the development dataset. Table 3 shows the 15 most common reactions for amoxicillin and ibuprofen ranked by 3 different association measures. We validated the performance of 3 dynamic reaction picklists and the static reaction picklist using the recall of the top-k suggested reactions. Figure 2 shows the recalls at top k (k ≤ 15) of the dynamic reaction picklists ranked by support, lift’, and tf-idf’ and the static picklist ranked by the reaction frequency in the database. For example, the recall at top 15 for support is 0.822 indicating that, if we suggest the top 15 reactions ranked by support, 82.2% of reactions in the validation dataset were among the top 15 suggested reactions. The dynamic reaction picklist ranked by support achieved recalls of 0.632, 0.763, and 0.822 at top 5, 10, 15, respectively, while tfidf’ achieved recalls of 0.593, 0.738, and 0.804, and lift’ achieved recalls of 0.533, 0.674, and 0.749 at top 5, 10, 15, respectively. The static reaction picklist ranked by frequency achieved recalls of 0.504, 0.655, and 0.734 at top 5, 10, and 15, respectively. Using the Friedman test, the 4 picklists showed significant differences in their recalls at top k (k ≤ 15), with P value <.001. With the posthoc test, the best dynamic picklist ranked by support significantly outperformed the static picklist in terms of the recall at top k (k ≤ 15), P value <.001. Supplementary MaterialTable 3 shows the top 15 relevant reactions associated with certain common drug allergens including penicillins, sulfonamide antibiotics, amoxicillin, and codeine, ranked by support.
Table 3.
Top 15 most common reactions for Amoxicillin and Ibuprofen using 3 association measures
| Rank | Top 15 most common reactions for Amoxicillin |
Top 15 most common reactions for Ibuprofen |
||||
|---|---|---|---|---|---|---|
| Support | Lift’ | Tfidf’ | Support | Lift’ | Tfidf’ | |
| 1 | Rash | Rash | Rash | GI upset | GI upset | GI upset |
| 2 | Hives | Hives | Hives | Hives | Hives | Hives |
| 3 | GI upset | Diarrhea | GI upset | Rash | Swelling | Swelling |
| 4 | N/V | GI Upset | Diarrhea | Swelling | GI Hemorrhage | Rash |
| 5 | Itching | Itching | N/V | N/V | Bleeding | N/V |
| 6 | Diarrhea | N/V | Itching | Anaphylaxis | Facial swelling | GI Hemorrhage |
| 7 | Swelling | Maculopapular rash | Swelling | Itching | Rash | Angioedema |
| 8 | Anaphylaxis | Vaginitis | Anaphylaxis | Angioedema | Angioedema | Anaphylaxis |
| 9 | SOB | Swelling | Vaginitis | SOB | Gastritis | Bleeding |
| 10 | Angioedema | Erythema multiforme | Maculopapular rash | GI Hemorrhage | Ulcer | Itching |
| 11 | Vaginitis | Anaphylaxis | Angioedema | Bronchospasm | Peptic ulcer | Facial swelling |
| 12 | Fever | Rash w/ joint painsa | SOB | Facial swelling | Eye swelling | SOB |
| 13 | Maculopapular rash | SOB | Fever | Bleeding | Renal toxicity | Renal toxicity |
| 14 | Erythema | Morbilliform rash | Rash w/ joint painsa | Wheezing | N/V | Bronchospasm |
| 15 | Facial swelling | Angioedema | Erythema multiforme | Lip swelling | Lip swelling | Wheezing |
Abbreviations: GI hemorrhage, gastrointestinal hemorrhage; N/V, nausea and/or vomiting; SOB, shortness of breath.
Rash w/joint pains (serum sickness).
Figure 2.
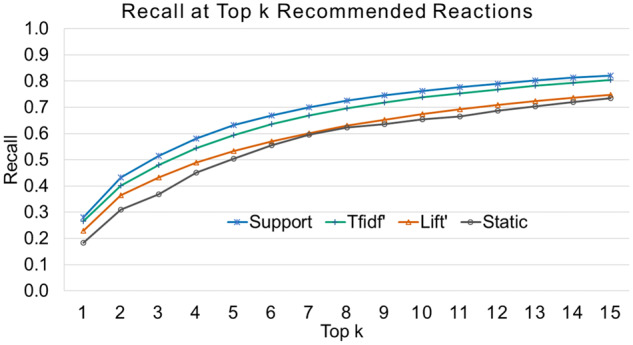
The recalls at top k of the dynamic reaction picklists ranked by support, tfidf’, and lift’ and the static reaction picklist ranked by reaction frequency.
DISCUSSION
In this study, we identified an allergy reaction value set consisting of 490 reaction concepts for EHR allergy documentation and disseminated this refined value set to the NLM VSAC. We created a dynamic reaction picklist that ranked reactions dynamically based on specific allergen by leveraging association rule-mining algorithms and more than 3 decades of allergy entries of a large Massachusetts healthcare system. In validating the dynamic reaction picklist using 1 year of retrospective allergy entries, we found that among the 3 association-mining measures (ie, support, lift’, and tfidf’) for generating the dynamic reaction picklist, support had the best recall for the top-ranked reactions outperforming the other 2 ranking algorithms. The dynamic reaction picklist ranked by support significantly outperformed the static reaction picklist ranked by frequency, indicating that it was able to suggest more probable reactions for allergy documentation. This study contributes to the standardization of a reaction value set for EHR allergy documentation as the value set was developed based on a standard terminology (ie, SNOMED CT) and can be easily adopted or adapted across healthcare systems. The study also contributes to a novel conceptual implementation of dynamic reaction picklists that will support the development of a prototype allergy entry module that has the potential to vastly improve EHR allergy reaction documentation. The novel idea of a dynamic picklist is generalizable and can be applied to other domains to facilitate efficient and accurate EHR documentation.
During the course of the reaction value set development, we identified multiple challenges related to using proprietary knowledge bases or standard terminologies for such tasks. Most current commercial EHR systems allow users to configure the clinical content of different modules. Users often purchase proprietary knowledge bases which they supplement with a certain level of customization. However, for adverse reactions, no standard or commercial products are available; therefore, the number and type of reactions included in the reaction picklist often varies across EHR implementations. In a prior study, we attempted to develop a reaction value set using SNOMED CT, a large and comprehensive standard terminology, and free-text allergy reaction entries.12,14 While SNOMED CT might be used to generate a value set for the reaction picklist, certain challenges remain. First, there are some synonymous concepts not listed as synonyms, such as “throat tightness” and “tightness in throat.” Second, the concepts in SNOMED CT are highly granular, and clinicians may not find it clinically meaningful to differentiate between similar but distinct concepts (eg, “itchy rash” and “red rash” versus “rash”). In addition, the free-text entries in the allergy database contained concepts related to patient diagnoses (eg, “pneumonia”) and unspecific language (“changes in skin texture”, “toxic effect”), introducing noise to the reaction value set. These issues limited our ability to directly use SNOMED CT to develop the reaction value set and dynamic picklists. Therefore, extensive manual curation combined with data-mining was necessary in order to develop a high-quality reaction value set.
The refinement of the reaction value set was conducted by a team of diverse backgrounds using a systematic approach. The advising expert panel included an allergist, an emergency physician/clinical informatician, a nephrologist/clinical informatician, a pharmacist, and 2 additional clinical informaticians. When updating the clinician-preferred terms, we used a data-driven approach so that the majority of selected terms were based on the frequency with which they were entered in the database, although some preferred terms were based on the experts’ choices and agreed upon by at least 2 experts. When reducing granular terms, we not only consolidated terms of similar meanings but also merged some child-level (ie, more specific) concepts to their parent terms based on the experts’ opinions. For example, while there are a total of 38 SNOMED CT concepts related to pain, we maintained only those concepts describing pain in a major body location with distinct and meaningful clinical interpretations (eg, “back pain,” “chest pain,” “abdominal pain,” “joint pain,” “neck pain”), and merged pain in other locations (eg, “pain in face,” “foot pain,” “pain in upper limb,” “aching pain”) with the concept of “pain.” Applying this rule, the number of pain concepts was reduced from 38 to 11. Although we believe these changes resulted in an improved value set, further investigation is required to understand how these reaction changes impact EHR allergy documentation.
The allergen consolidation process, although conducted to facilitate development of dynamic reaction picklists, revealed problems with our EHR’s existing knowledge base, including duplicate entries and similar entries. About one-third of allergens were able to be mapped to other allergens. The benefit of allergen consolidation is that by combining the frequencies of reactions that co-occurred with multiple individual allergens that all refer to the same substance, the dynamic picklist can be more accurately ranked for that substance.
It was not surprising that support was the best association rule-mining measure for suggesting relevant reactions for allergy reaction documentation. Support worked well because it suggested reactions based on allergen-reaction co-occurrence in the development dataset, and the distribution of allergen-reaction pairs in the validation dataset may follow similar patterns. In addition, lift’ and tf-idf’, which tended to raise the associations between allergens and rare reactions, yielded lower recall at top k, perhaps because our validation dataset has an uneven distribution of allergen-reaction pairs with a high frequency of common reactions. However, regardless of the measure used, all 3 dynamic reaction picklists outperformed the static reaction picklist. Based on these results, we recommend the healthcare systems that employ EHR systems that use static picklists consider the adoption of dynamic reaction picklists to facilitate and improve allergy documentation.
It is worth mentioning that in our current EHR allergy module, 3 reaction types (ie, allergy, intolerance, and contraindication) are available for clinicians to choose from when entering a reaction. This field was rarely used by clinicians, however, resulting in most reactions being recorded as the “allergy” reaction type. Contraindications are conditions such as glucose-6-phosphate dehydrogenase (G6PD) deficiency or renal failure in which case certain medications should be avoided. While these contraindications should perhaps be entered elsewhere in the EHR—such as the problem list—in this work, we did not remove contraindications but used the original data for our analysis. Our ongoing effort is to better classify these reaction entries to facilitate further analyses. Once adverse reaction types and contraindications are well-defined and differentiated, we can build additional functions to inform clinicians dynamically of where to document these items.
This study has several limitations worth noting. First, although we used a diverse team of experts and a data-driven approach, some changes (eg, choosing preferred terms, mapping, removal) were subjective and may be biased by the experts’ training, background, and experiences. Second, during the allergen consolidation, we used the mappings between FDB medications and RxNorm concepts as a source to identify the allergens’ ingredients. However, due to differences in the coverage of these medication terminology systems, many allergens could not be identified by their ingredients in RxNorm and thus were left unconsolidated. Allergens with complicated ingredients (eg, vitamins, food) may be less likely to be consolidated due to the challenge of listing all their ingredients. Third, some newer allergens (eg, recently approved drugs) listed in the EHR reference list did not have allergy entries in the allergy database, and such allergens would temporarily need to have static reaction lists. However, dynamic picklists for those allergens could be developed in the future; as we noticed a growth in the number of allergens with allergy entries in the database, for example, there were 743 new allergens with allergy entries in the dataset’s final year (October 9, 2018 through October 9, 2019). Finally, this study was conducted in a single healthcare system, and the dynamic reaction picklist was generated in the context of an Epic EHR system and a terminology system for allergens and medications provided by FDB. Although both Epic and FDB are among the top EHR vendors in the US, the portability of the dynamic reaction picklist to other institutions may be impacted due to differences in allergy-reaction frequency distribution across different patient populations as well as in EHR vendors. We therefore suggest that the dynamic picklist be modified by considering these factors prior to adoption.
CONCLUSION
In summary, we identified an enhanced reaction value set of 490 reactions and found that dynamic reaction picklists developed using data-mining association rules and retrospective allergy reaction entries can suggest relevant reactions for improved allergy module reaction entries. Future work includes enhancing reaction type and severity definition and evaluating whether these enhancements improve reaction documentation and/or reduce free-text entries using a prototype user interface that implements the dynamic reaction picklist.
FUNDING
This research was supported with funding from the Agency for HealthCare Research and Quality (AHRQ) grant R01HS025375 and the National Institute of Allergy and Infectious Diseases (NIAID) of National Institute of Health (NIH) grant 1R01AI150295.
AUTHOR CONTRIBUTIONS
LZ obtained funding. LW, FG, and LZ led the conceptualization and design of the work. LW, SY, KGB, FG, YL, SNS, ZTK, and LZ contributed to the refinement of the reaction picklist. SB performed the reaction consolidation. LW conducted the data collection, statistical analysis, and interpretation of the results and drafted the manuscript. SB, SY, KGB, and LZ critically revised the manuscript. All authors provided feedback and take responsibility for the final approval of the version to be published and are accountable for all aspects of the work.
SUPPLEMENTARY MATERIAL
Supplementary material is available at Journal of the American Medical Informatics Association online.
Supplementary Material
ACKNOWLEDGMENTS
The authors thank Adam Wright, PhD for valuable advice with this research.
CONFLICT OF INTEREST STATEMENT
None declared.
REFERENCES
- 1. Gandhi TK, Weingart SN, Borus J, et al. Adverse drug events in ambulatory care. N Engl J Med 2003; 348 (16): 1556–64. [DOI] [PubMed] [Google Scholar]
- 2. Zhou L, Dhopeshwarkar N, Blumenthal KG, et al. Drug allergies documented in electronic health records of a large healthcare system. Allergy 2016; 71 (9): 1305–13. [DOI] [PubMed] [Google Scholar]
- 3. Ariosto D. Factors contributing to CPOE opiate allergy alert overrides. AMIA Annu Symp Proc 2014; 2014: 256–65. [PMC free article] [PubMed] [Google Scholar]
- 4. Bryant AD, Fletcher GS, Payne TH.. Drug interaction alert override rates in the Meaningful Use era: no evidence of progress. Appl Clin Inform 2014; 05 (03): 802–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Isaac T, Weissman JS, Davis RB, et al. Overrides of medication alerts in ambulatory care. Arch Intern Med 2009; 169 (3): 305–11. [DOI] [PubMed] [Google Scholar]
- 6. Jani YH, Barber N, Wong IC.. Characteristics of clinical decision support alert overrides in an electronic prescribing system at a tertiary care paediatric hospital. Int J Pharm Pract 2011; 19 (5): 363–6. [DOI] [PubMed] [Google Scholar]
- 7. McCoy AB, Thomas EJ, Krousel-Wood M, Sittig DF.. Clinical decision support alert appropriateness: a review and proposal for improvement. Ochsner J 2014; 14 (2): 195–202. [PMC free article] [PubMed] [Google Scholar]
- 8. Slight SP, Beeler PE, Seger DL, et al. A cross-sectional observational study of high override rates of drug allergy alerts in inpatient and outpatient settings, and opportunities for improvement. BMJ Qual Saf 2017; 26 (3): 217–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Topaz M, Seger DL, Lai K, et al. High override rate for opioid drug-allergy interaction alerts: current trends and recommendations for future. Stud Health Technol Inform 2015; 216: 242–6. [PMC free article] [PubMed] [Google Scholar]
- 10. Topaz M, Seger DL, Slight SP, Goss F, et al. Rising drug allergy alert overrides in electronic health records: an observational retrospective study of a decade of experience. J Am Med Inform Assoc 2016; 23 (3): 601–8. [DOI] [PubMed] [Google Scholar]
- 11. Blumenthal KG, Wickner PG, Lau JJ, Zhou L.. Stevens-Johnson syndrome and toxic epidermal necrolysis: a cross-sectional analysis of patients in an integrated allergy repository of a large health care system. J Allergy Clin Immunol Pract 2015; 3 (2): 277–80 e1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Goss FR, Lai KH, Topaz M, et al. A value set for documenting adverse reactions in electronic health records. J Am Med Informatics Association 2018; 25 (6): 661–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Liu S, Ma W, Moore R, Ganesan V, Nelson S.. RxNorm: prescription for electronic drug information exchange. IT Prof 2005; 7 (5): 17–23. [Google Scholar]
- 14. Donnelly K. SNOMED-CT: the advanced terminology and coding system for eHealth. Stud Health Technol Inform 2006; 121: 279–90. [PubMed] [Google Scholar]
- 15. Zhou L, Plasek JM, Mahoney LM, et al. Using medical text extraction, reasoning and mapping system (MTERMS) to process medication information in outpatient clinical notes. AMIA Annu Symp Proc 2011; 2011: 1639–48. [PMC free article] [PubMed] [Google Scholar]
- 16. Goss FR, Plasek JM, Lau JJ, Seger DL, Chang FY, Zhou L.. An evaluation of a natural language processing tool for identifying and encoding allergy information in emergency department clinical notes. AMIA Annu Symp Proc 2014; 2014: 580–8. [PMC free article] [PubMed] [Google Scholar]
- 17. Wang L, Del Fiol G, Bray BE, Haug PJ.. Generating disease-pertinent treatment vocabularies from MEDLINE citations. J Biomed Inform 2017; 65: 46–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Wright A, Chen ES, Maloney FL.. An automated technique for identifying associations between medications, laboratory results and problems. J Biomed Inform 2010; 43 (6): 891–901. [DOI] [PubMed] [Google Scholar]
- 19. Demšar J. Statistical comparisons of classifiers over multiple data sets. J Mach Learn Res 2006; 7: 1–30. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.