Abstract
Objective
Predicting patient outcomes using healthcare/genomics data is an increasingly popular/important area. However, some diseases are rare and require data from multiple institutions to construct generalizable models. To address institutional data protection policies, many distributed methods keep the data locally but rely on a central server for coordination, which introduces risks such as a single point of failure. We focus on providing an alternative based on a decentralized approach. We introduce the idea using blockchain technology for this purpose, with a brief description of its own potential advantages/disadvantages.
Materials and Methods
We explain how our proposed EXpectation Propagation LOgistic REgRession on Permissioned blockCHAIN (ExplorerChain) can achieve the same results when compared to a distributed model that uses a central server on 3 healthcare/genomic datasets, and what trade-offs need to be considered when using centralized/decentralized methods. We explain how the use of blockchain technology can help decrease some of the problems encountered in decentralized methods.
Results
We showed that the discrimination power of ExplorerChain can be statistically similar to its counterpart central server-based algorithm. While ExplorerChain inherited some benefits of blockchain, it had a small increased running time.
Discussion
ExplorerChain has the same prerequisites as a distributed model with a centralized server for coordination. In a manner similar to secure multi-party computation strategies, it assumes that participating institutions are honest, but “curious.”
Conclusion
When evaluated on relatively small datasets, results suggest that ExplorerChain, which combines artificial intelligence and blockchain technologies, performs as well as a central server-based method, and may avoid some risks at the cost of efficiency.
Keywords: blockchain distributed ledger technology, privacy-preserving predictive modeling, online machine learning, clinical information systems, decision support systems
INTRODUCTION
Predictive modeling and cross-institutional collaboration
Applying machine learning methods to predict patient outcomes using data sources such as electronic health records and genomics data or healthcare predictive modeling, is an increasingly popular and important area and can facilitate research studies and advance quality improvement initiatives. Specifically, such modeling methods based on artificial intelligence (eg, logistic regression or deep learning algorithms) can help generate scientific evidence for comparative effectiveness research,1 accelerate biomedical discoveries,2 and improve patient care.3 For example, a medical center may use predictive analytics to identify and take measures that can potentially prevent 30-day readmissions, thus improving care while avoiding penalties from regulatory agencies.4,5 Although the concept of predictive modeling is appealing, some diseases and conditions are rare and therefore require data from multiple institutions. To obtain enough data for model construction, sharing healthcare/genomic data across institutions is an intuitive solution to improving models. While a single institution may only have few patient records, taken together, a set of institutions may provide adequate numbers for a generalizable model. Aggregating all cases in a registry or data coordinating center database has been a common strategy to manage these databases. However, this leads to a single point of failure or breach, inaccessibility of data, or improper data disclosure that may place sensitive protected health information at risk, respectively. Once data are passed on to other entities, it is difficult to ascertain the chain of custody, leading to potential misuse.
Distributed predictive modeling and potential blockchain solution
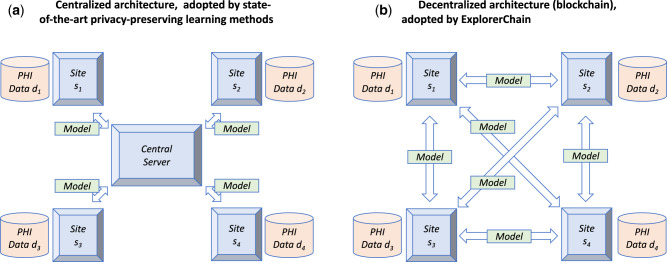
As a result, combining cross-institution data into a single repository for model learning has been challenged as the ideal model to protect privacy of individuals6,7 and/or institutions8,9 while ensuring that no institution has full responsibility for all the data. Multiple algorithms have been proposed to conduct predictive modeling in a distributed way10–18 without the need to aggregate data centrally. That is, by disseminating machine learning models instead of transferring observation-level patient data, the privacy of individuals can be further protected.10–18 However, many of these privacy-preserving modeling algorithms still need a central server to intermediate the process of modeling and combine the global model.10–18 Such an architecture carries its own risks (Figure 1a), such as failure to build, tune, or evaluate a model in case the central coordinating server is down, and the potential for this central coordinating server to try to breach the privacy of individuals or institutions by examining aggregate statistics.
Figure 1.
The network topology for a 4-site (s1, s2, s3 and s4) setting, with each site holding protected health information data (d1, d2, d3 and d4). (a): Centralized network topology adopted by state-of-the-art learning methods. Such an architecture carries several risks: (R1) the sites may not be allowed to transmit data outside specific computer environments due to their institutional policies,19 (R2) the data being disseminated and the transfer records could be altered without a clear way to determine immutability,19–21 (R3) trained models could also be tampered within the central server without being noticed by other participating sites and thus undermines provenance,19,22 (R4) the server represents a single point of failure,23,24 (R5) additional security to protect data is not offered,22 (R6) the client-server architecture may present synchronization problems,19,25–27 and (R7) the sites cannot join/leave the network at any time.28 Furthermore, long-term sustainability of the whole network becomes dependent on the institution that serves as the central coordinating node (R8). Typically, once a coordinating institution is chosen, the network architecture is built around the coordinating center, with nodes serving as data providers that are unable to assume the role when needed. (b): Decentralized network topology (blockchain) adopted by ExplorerChain. Five desirable features22 make blockchain suitable to mitigate the problems faced by centralized architectures: (R1) Blockchain is, by design, decentralized; the verification of transactions is achieved by majority voting.22,29 Each institution can control the use of computational resources. (R2) A blockchain provides an immutable audit trail.19–21 That is, changing the data or records is very difficult.22,29 (R3) The traceable origins certify data provenance.19,22 In our case, each trained model is recorded in a collaborative and distributed ledger, which cannot be updated silently by any of the sites without being noticed. (R4) The peer-to-peer architecture of blockchain ensures that there is no risk of single point of failure,23,24 and thus improves security and robustness. Also, by removing the dependency on a central node, blockchain increases the availability of the models at all sites at all times.30 (R5) The enhanced security/privacy features of blockchain further protect data and models. Additionally, (R6) The blockchain mechanism can remove synchronization conflicts.23,25,31,32 (R7) Each site can join/leave the network freely without imposing overhead on a central server or disrupting the machine learning process.23,24,28 Finally, network long-term sustainability (R8) is increased because its architecture is fully transparent and each participating site can collaborate with low operation/maintenance costs.
To address these challenges, a plausible solution is to adapt distributed databases to build and evaluate predictive models in a peer-to-peer architecture. These peer-to-peer distributed models cannot work with all the data at once. However, partial batch training (ie, batch training at each site) is possible, followed by an online or “transfer learning” strategy in which the next site or peer gets a partially trained model and improves it using its own data. In this sequential peer-to-peer process, it is important that no site constitutes a single point of failure and that all sites can easily verify how the model is evolving.
One of the main problems with online machine learning models (centralized or distributed) is the potential for a limited number of initial inputs force gradient descent methods to be trapped into limited solutions.33 Although the proposed strategy does not eliminate or necessarily mitigate this problem per se, enabling the use of data from multiple sites for model training in a way that does not centralize the data allows more institutions to participate, so the model parameters can evolve in different directions. Also, a major challenge with distributed databases that rely on a central coordinating server is the vulnerability to a single point of failure and potential tampering with logs at the central server side.16,17,22 On the other hand, for distributed databases that rely on a peer-to-peer network, one of the main problems relates to the coordination of access.34 If the path from peer-to-peer is easily traceable and broadcasted to the whole network, this problem is solved. Transparency of usage in the form of an immutable ledger that can be easily verified by the whole network is therefore a desirable feature. When asking queries and building machine learning models using sensitive data from several institutions, maintaining a robust immutable ledger of access is critical. Additionally, resilience to failure at any node and disintermediation (ie, lack of a central authority) are desirable in clinical data research networks. Blockchain technology19–21,23,25,28,29,31,32,35,36 has many of these desirable features, in addition to being developed by a community of developers as opposed to being a custom-tailored software solution that is hard to document and maintain. We propose to treat each institution in a network as a node in a blockchain network in a way that allows batch training at a site, but online training across sites, in a sequential fashion that is resilient to node failure and immutably recorded. We thus take advantage of known characteristics of blockchain technology (resilience to single point of failure, ability to create an immutable ledger, transparency of use, and development by the community) to build a multi-center predictive model.
Blockchain is a decentralized peer-to-peer technology that has evolved significantly since the development of Bitcoin blockchain, which is the underlying technology of Bitcoin crypto currency.29 Blockchain nodes (ie, computers running the blockchain software) compose a distributed peer-to-peer network (Figure 1b) that follow an agreed-upon time stamp and consensus protocol to create a chain of transaction blocks (ie, “blockchain”). Such a blockchain allows nodes to collaboratively generate a consensus ledger without the need of a central intermediary.22,29 The chain can store arbitrary data22 in either space reserved for the “metadata” space of the transactions19,28,37–40 or as part of “smart properties” managed by “smart contracts.”41–45 A blockchain network can be either permissionless (ie, a public network in which any node can participate, such as Bitcoin) or permissioned (ie, a private network in which only authorized nodes can participate, such as most networks that focus on healthcare).22,46
Advantages and disadvantages of blockchain-based modeling
A recent review paper22 summarized the key benefits and potential challenges of blockchain technology and compared blockchain to traditional distributed database management systems. The use of one of the main blockchain platforms ensures that code is maintained, enhanced and documented by the community at large, and not limited to the site that created the infrastructure’s code.46 Among many other decentralized systems, including less-connected systems based on gossip algorithms,47–49 blockchain brings benefits and addresses many current challenges in distributed networks. On the other hand, some challenges related to how certain types of cryptography can be incorporated, scalability, and threat of a 51% attack still exist.16,17,22 Also, there might be potential network security concerns such as leaving particular Internet ports open to allow remote blockchain access, although in permissioned networks this can be protected. A robust method to integrate this distributed ledger technology with the distributed learning algorithms is yet to be investigated in depth. Several methods have been proposed to solve the distributed learning problem with or without blockchain,10–18 as shown in Table 1. GLORE,10 EXPLORER,11 WebGLORE12 and SMAC-GLORE13 are based on a client-server architecture, which possess potential risks such as single point of failure, while ExplorerChain is based on peer-to-peer architecture without such weaknesses. GloreChain14 and HierarchicalChain15 are based on blockchain, however, it is not resilient in situations in which a site leaves or joins the network. ModelChain16,17 was blockchain-based and proposed online learning, but it was not actually evaluated in practice. Finally, LearningChain18 is an online learning method on blockchain, but it focuses on improving privacy (ie, adopting differential privacy scheme) and security (ie, defending against the Byzantine attack) on stochastic gradient descent algorithm, and not on healthcare institutions’ needs for immutable ledgers, and transparency of use.
Table 1.
Comparison of the state-of-the-art distributed learning methods
| Method | Author | Reference | Architecture | Learning | Focus | Status |
|---|---|---|---|---|---|---|
| GLORE | Wu et al | 10 | Client-Server | Batch | Healthcare | Evaluated |
| EXPLORER | Wang et al | 11 | Client-Server | Online | Healthcare | Evaluated |
| WebGLORE | Jiang et al | 12 | Client-Server | Batch | Healthcare | Evaluated |
| SMAC-GLORE | Shi et al | 13 | Client-Server | Batch | Healthcare | Evaluated |
| GloreChain | Kuo et al | 14 | Peer-to-Peer | Batch | Healthcare | Evaluated |
| HierarchicalChain | Kuo et al | 15 | Peer-to-Peer | Batch | Healthcare | Evaluated |
| ModelChain | Kuo et al | 16 , 17 | Peer-to-Peer | Online | Healthcare | Proposed |
| LearningChain | Chen et al | 18 | Peer-to-Peer | Online | Privacy/Security | Evaluated |
| ExplorerChain | Kuo et al | – | Peer-to-Peer | Online | Healthcare | Evaluated |
OBJECTIVE
We developed and evaluated a decentralized blockchain-based predictive modeling method, EXpectation Propagation LOgistic REgRession on Permissioned BlockCHAIN (ExplorerChain), to support cross-institution healthcare/genomic research and quality improvements initiatives. This approach should, theoretically, remove certain risks introduced by the client-server architecture nature of distributed networks that rely on a central node. We aimed at empirically showing results of such a design on healthcare and genomic datasets.
MATERIALS AND METHODS
ExplorerChain
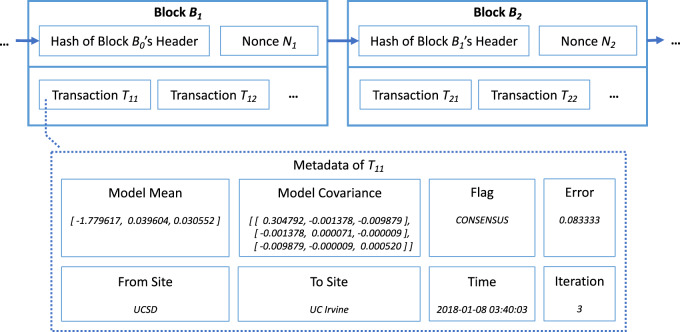
In developing the consensus protocols for predictive modeling blockchain networks, we adopt online distributed learning11,50,51 to train predictive models on the decentralized network architecture. Our working hypothesis was that online algorithms on a distributed blockchain-based network that does not rely on a coordinating node can provide better security/robustness than models that use a central node, without sacrificing predictive power.10,11,50–52 We chose the online logistic regression algorithm of EXPLORER11 as our predictive modeling method. In ExplorerChain, we utilize the metadata space of the transactions to disseminate the online machine learning models and integrate the blockchain network with distributed online learning (Figure 2).16,17 Intuitively, permissioned/private blockchain platforms22,46 are feasible for ExplorerChain, because the sites need to be permitted to participate in the predictive model learning process. We selected MultiChain28,39 as the underlying permissioned blockchain platform based on our prior review.46
Figure 2.
A simplified example of ExplorerChain. Only the aggregated data (ie, the machine learning model) and the meta information are stored on-chain, while the protected health information (PHI, the observation-level patient data) are stored off-chain. This design ensures that the institutions can share information to improve the predictive model without transmitting PHI. It should be noted that the amount of transactions is set to be zero, indicating that the blockchain serves purely as a nonfinancial distributed ledger.
Datasets
In our experiments, 3 clinical/genomic datasets were utilized to test ExplorerChain. The first dataset (“Edin”) is the Edinburg Myocardial Infarction (MI) data,53 which contains predictor features and one binary MI outcome (ie, presence of disease). The clinical question pursued by this data set is how accurate a prediction model for MI can be when only signs, symptoms and certain electrocardiographic data are available at presentation.53 The second dataset (“CA”) has cancer biomarkers (ie, CA-19 and CA-125) and a binary cancer outcome (ie, presence of cancer).54 The clinical question at hand is whether combining the level values of the 2 biomarkers helps identify pancreatic cancer. The third dataset, “THA,” was used to predict hospital length of stay following unilateral primary total hip arthroplasty (THA) surgery. Following the practice used in other anesthesia publications, the outcome to be predicted was a dichotomized prolonged hospital length-of-stay (LOS) outcome (ie, whether the actual LOS is greater than the expected LOS (3 days) for THA at our institution).55,56 The Institutional Review Board at University of California San Diego (UCSD) approved this study with Project Number 171344X on February 9, 2018, and the informed consent requirement was exempted by our institution’s Human Research Protections Program because the dataset was defined as Health Insurance Portability and Accountability Act (HIPAA) “deidentified.”57–64 The statistics/features of the test datasets are summarized in Table 2. Given the age and limited size of the Edin and CA datasets, they are presented for illustration purposes only. The THA dataset was recently used in a quality improvement project at an academic medical center.
Table 2.
Statistics and features of the datasets tested in our experiments. The class distribution (ie, the percentage of the positive/negative classes) is also included. The numerical covariates are labeled with an asterisk symbol (“*”), while the categorical ones are converted into binary through dummy coding. The values for the myocardial infarction and cancer biomarker datasets are adapted from11
| Dataset | MyocardialInfarction(Edin) | CancerBiomarker(CA) | Length of Hospitalization(THA) | |
|---|---|---|---|---|
| # of Covariates | 9 | 2 | 34 | |
| # of Samples | 1,253 | 141 | 960 | |
| Class Distribution | 0.219 / 0.781 | 0.638 / 0.362 | 0.278 / 0.722 | |
| Outcome | Presence of Disease | Presence of Cancer | Hospital Length of Stay is greater than 3 days | |
| Covariates | Pain in Right Arm | CA-19* | Male Sex | SA - Posterior |
| Nausea | CA-125* | Age ≥ 65 years old | SA - Anterolateral | |
| Hypo Perfusion | – | Preoperative METs < 4 | SA - Anterior | |
| ST Elevation | – | General Anesthesia (versus Neuraxial Anesthesia) | CM - Chronic Kidney Disease | |
| New Q Waves | – | Non-English Speaker | CM - Chronic Obstructive Pulmonary Disease | |
| ST Depression | – | OG - Mild | CM - Congestive Heart Failure | |
| T Wave Inversion | – | OG - Moderate | CM - Coronary Artery Disease | |
| Sweating | – | OG - Severe | CM - Hypertension | |
| Pain in Left Arm | – | OG - Avascular Necrosis | CM - Diabetes Mellitus | |
| – | – | CHD - No osteoarthritis | CM - Obstructive Sleep Apnea | |
| – | – | CHD - Mild osteoarthritis | CM - Dialysis | |
| – | – | CHD - Moderate osteoarthritis | CM - Psychiatric history (depression, anxiety, or bipolar disease) | |
| – | – | CHD - Severe osteoarthritis | CM - Active Smoker | |
| – | – | CHD - Previous Surgery (ie, hip replacement) | CM - Asthma | |
| – | – | CHD - Avascular Necrosis | CM - Thrombocytopenia (platelets < 150 000/uL) | |
| – | – | Obesity (BMI > 30kg/m2) | CM - Anemia | |
| – | – | Preoperative Opioid Use | CM - Dementia | |
Abbreviations: BMI, body mass index; CHD, Contralateral Hip Description; CM, comorbidities; METS, metabolic equivalents; OG, osteoarthritis grade (operative side); SA, surgical approach; THA, total hip arthroplasty.
Experiment settings
We aimed at determining whether ExplorerChain could reach equivalent predictive performance as state-of-the-art central server-based learning approaches, while taking advantage of some key benefits of blockchain technology. Specifically, we compared this approach with a central server-based method using an online machine learning method that we adapted for ExplorerChain. We selected the centralized version of EXPLORER11 as our baseline comparison method, and tested ExplorerChain on the iDASH private HIPAA-compliant computing environment network.65,66 Each dataset was evenly and randomly split into 2-, 4-, and 8-site combinations. Then, for each site, a training dataset was generated by randomly selecting 80% of records, while the test dataset was created using the remaining 20%. We adopted the area under the receiver operating characteristic curve (AUC)67,68 to evaluate whether the discrimination performance of ExplorerChain was comparable to that of EXPLORER. The averaged AUCtest among all N sites was used as the result for one trial. We repeated the above dataset splitting process (including both multiple-site and training/test splitting) over 30 trials. Also, we evaluated the Pearson Correlation Coefficient (PCC) between the AUC results of the 2 methods, to verify if their results were linearly correlated (the closer the PCC is to 1, the higher positive linear correlation between 2 methods). Furthermore, we used 2-sample t-test of AUCs between 2 methods with alpha = 0.05, and obtained a mean difference (delta) between the AUC results of the 2 methods that was not statistically significant (ie, P value > .05). We also computed iterations of ExplorerChain and compared the execution times with those of EXPLORER.
RESULTS
Correctness
The predictive performance results are shown in Table 3. The differences of the means (≤ 0.016) and the standard deviations (≤ 0.011) of the AUC values were relatively small. The PCC values (> 0.7) indicated high linear correlation between 2 methods. The P values for the 2-sample t-tests were above .05 for all datasets, demonstrating that the discrimination performance of ExplorerChain was very similar to that of EXPLORER (mean AUC difference ≤ 0.002). ExplorerChain approximates EXPLORER, and thus the latter may outperform the former; however, the difference was small and not statistically significant. Also, for both methods, the mean AUC values of the Edin data set are the highest and the ones of the THA are the lowest among all 3 datasets. The standard deviations of AUC of the CA data set are the highest and the ones of the Edin data set are the lowest. This pattern appears repeatedly for simulations involving N = 2, 4, and 8 sites, suggesting that the performance of ExplorerChain is consistent with that of EXPLORER.
Table 3.
The experiment results for the myocardial infarction (Edin), the cancer biomarker (CA), and the length of hospitalization (THA) datasets. The evaluation metric is the averaged full area under the receiver operating characteristic curve (AUC) among N sites, for 30 trials. The Pearson Correlation Coefficient (PCC) was computed to evaluate the linear correlation between 2 methods. Finally, the alpha in the 2-sample t-test was 0.05, and the p-values larger than 0.05 (shown in bold italic) indicate no statistically significant difference between the AUC results of EXPLORER and ExplorerChain
| EXPLORER |
ExplorerChain |
Correlation |
Two-Sample t-Test |
||||||
|---|---|---|---|---|---|---|---|---|---|
| Dataset | N | Mean AUC | Standard Deviation | Mean AUC | Standard Deviation | PCC | Delta | Test Statistics | P-value |
| Edin | 2 | 0.965 | 0.013 | 0.965 | 0.013 | 0.999 | 0.000 | −1.559 | 0.130 |
| 4 | 0.962 | 0.010 | 0.960 | 0.011 | 0.867 | 0.000 | 1.868 | 0.072 | |
| 8 | 0.957 | 0.014 | 0.954 | 0.015 | 0.906 | 0.002 | 1.371 | 0.181 | |
| CA | 2 | 0.893 | 0.054 | 0.891 | 0.055 | 0.977 | 0.000 | 1.106 | 0.278 |
| 4 | 0.862 | 0.075 | 0.853 | 0.078 | 0.932 | 0.000 | 1.694 | 0.101 | |
| 8 | 0.892 | 0.060 | 0.876 | 0.071 | 0.746 | 0.000 | 1.811 | 0.080 | |
| THA | 2 | 0.734 | 0.035 | 0.733 | 0.036 | 0.995 | 0.000 | 1.622 | 0.116 |
| 4 | 0.738 | 0.047 | 0.735 | 0.047 | 0.975 | 0.000 | 1.529 | 0.137 | |
| 8 | 0.718 | 0.040 | 0.712 | 0.040 | 0.909 | 0.000 | 1.878 | 0.070 | |
Abbreviations: AUC, area under the receiver operating characteristic curve; CA, cancer biomarker; PCC, Pearson correlation coefficient; THA, total hip arthroplasty.
Iteration
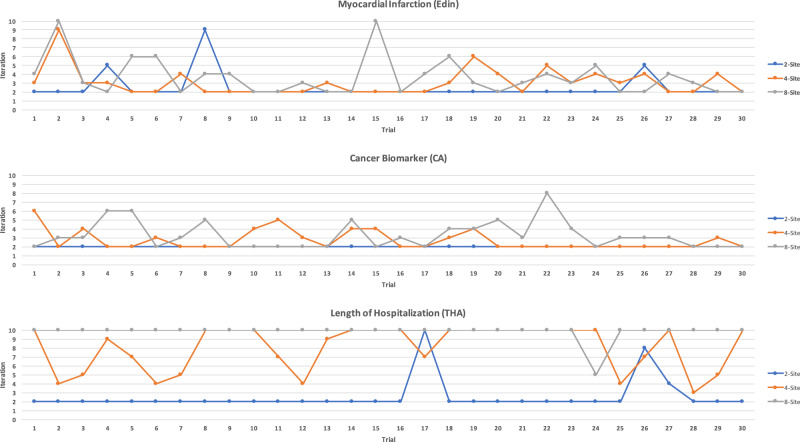
The iteration results (ie, rate of convergence) are shown in Table 4 and Figure 3. In general, ExplorerChain required a low number of iterations (2 or 3), indicating a fast rate of convergence. This held for N = 2, 4, 8 sites on the Edin and the CA data sets, as well as for N = 2 for the THA. For N = 4 and 8 sites on the THA, higher mean iterations were required (8 or 9). For the THA and N = 8, although the maximum number of iterations (10) was reached in almost every one of the 30 trials (Figure 3), the AUC was still comparable to that of EXPLORER (PCC = 0.909 and P value of the 2-sample t-test = .070 in Table 3), indicating that an increase in the limit of iterations may not be necessary. These speedy convergence results are consistent with EXPLORER (ie, < 10 iterations).11
Table 4.
Number of iterations and time results of ExplorerChain among N sites for 30 trials of the 3 datasets. All time measurements are averaged over N sites, and the total time includes both running time and synchronization time. For ExplorerChain, the per-iteration time is computed by dividing the time by the mean number of iterations, and the additional pausing time (240 seconds per trial) between trials for result collection was deducted
| # of Iterations |
Time (Seconds) |
||||||||
|---|---|---|---|---|---|---|---|---|---|
| ExplorerChain |
EXPLORER |
ExplorerChain |
|||||||
| Dataset | N | Mean | Standard Deviation | Total | Running | Total | Running | Total/ Iteration | Running/ Iteration |
| Edin | 2 | 2.433 | 1.455 | 2.477 | 2.426 | 144.519 | 7.609 | 59.391 | 3.127 |
| 4 | 3.033 | 1.542 | 2.451 | 2.399 | 165.890 | 9.939 | 54.689 | 3.277 | |
| 8 | 3.633 | 2.157 | 2.432 | 2.383 | 184.086 | 12.145 | 50.666 | 3.343 | |
| CA | 2 | 2.000 | 0.000 | 2.000 | 1.945 | 129.618 | 6.125 | 64.809 | 3.062 |
| 4 | 2.700 | 1.088 | 2.011 | 1.949 | 154.175 | 8.829 | 57.102 | 3.270 | |
| 8 | 3.233 | 1.547 | 1.996 | 1.947 | 172.654 | 10.938 | 53.398 | 3.383 | |
| THA | 2 | 2.533 | 1.814 | 2.399 | 2.348 | 147.259 | 8.045 | 58.128 | 3.176 |
| 4 | 8.000 | 2.533 | 2.366 | 2.315 | 317.266 | 23.865 | 39.658 | 2.983 | |
| 8 | 9.833 | 0.913 | 2.364 | 2.314 | 365.355 | 32.713 | 37.155 | 3.327 | |
Figure 3.
Number of iterations of ExplorerChain on 3 datasets and 2-, 4- and 8-site settings for each of the 30 trials. The number of iterations increases with the number of sites, with an upper limit to the maximum number of required iterations (10 in our experiment) to reach consensus. However, the relatively large standard deviation, especially when the number of sites is small (eg, N = 2), suggests that the number of required iterations is highly dependent on the dataset.
Time
The time results are also shown in Table 4. Compared to EXPLORER, our proposed ExplorerChain required significantly longer total time because of the synchronization time required to collect errors from all other sites, and also longer total times as the number of sites increased. Such a synchronization time is related to hyper-parameters of ExplorerChain and blockchain/network speed. Regarding running time, ExplorerChain was slower than EXPLORER because of the additional waiting time to find the consensus model. On the other hand, the per-iteration time results showed that, in each iteration, the running time of ExplorerChain was similar to that of EXPLORER. ExplorerChain required about 60 seconds of synchronization time on average. Specifically, the 30 seconds of waiting time period contributed to about half of the 60-second average synchronization time per iteration. The results of the per-iteration total time show variations (30–70 seconds) among the 3 datasets, while the results of the per-iteration running time remained stable (around 3 seconds).
DISCUSSION
Findings
Our study suggests that the proposed blockchain-based ExplorerChain method can learn cross-institution predictive models without transferring observation-level patient data and without relying on a central coordination node. Because of the distributed architecture that is not based on a central node, this approach may avoid concerns encountered in architectures that rely on a central node. The discrimination of the model learned using ExplorerChain was comparable to that of EXPLORER, a central server-based online learning method. ExplorerChain added a layer of protection, provided by the blockchain technology, that helped improve the security/robustness of the learning process. The cost for such improvement was paid mainly in terms of synchronization time, since, without a central server, it took a larger number of iterations for all sites to find a final consensus model (a similar trade-off noted for databases19,30,69,70). As the number of sites increased, the number of iterations and time grew linearly. We used relatively small but real-world data for our experiments. The THA data set is contemporary and motivated by medical center needs that address quality of care from the clinical and administrative perspectives. Although surgical outcomes registries such as American Joint Replacement Registry (AJRR)71 are helpful to develop predictive models for orthopedic surgery outcomes, they suffer from the following problems: registry fees, required data entry and maintenance, and institutional data privacy restrictions which may limit the inter-institution data comparisons. With machine learning and block chain technology, there exists the possibility to process sensitive data, such as that related to complications and cost, without human intervention in a decentralized, more private fashion.
Potential clinical applications
Gabriel et al56 showed that it is possible to build accurate models for patient LOS after total hip arthroplasty. Their model was based on a single institution and performed well, but if more institutions could participate in a way that preserves individual and institutional privacy, the model would be more robust. This is particularly valuable in the era of value-based care where the focus is on incentivizing higher quality care at lower costs. Given the sensitive nature of administrative processes and the fact that many institutions are competing with each other, institutions may want to keep their data hidden from other institutions and insurance companies, while still contributing to the development of a better predictive model. The method presented here enables this participation, while preserving institutional privacy, using a novel strategy based on blockchain technology. Future applications of this technology may be seen in development of a risk-stratification model for reimbursement. At present, in the bundled care model, there is a single reimbursement fee for all lower extremity arthroplasties, for the entire episode of care, regardless of patient factors that can affect complication rates. This has led to the practice of “cherry-picking” younger, healthy patients who are less likely to have complications. Such a decentralized model with protection of institutional privacy through the blockchain technology presented could encourage participation from a diverse group of practices to build an accurate model. The results of the THA data set show that the differences in AUC from the blockchain-based model and a benchmark centralized model do not have clinical significance: the models’ performances are essentially the same (AUC difference of 1%–2%).
Limitations
Two major concerns for the heavy use of blockchain includes scalability of the approach and acceptability by users. The scalability issue consists of several aspects, such as (S1) the number of users, (S2) the number of records in the datasets, and (S3) the number of the covariates in the datasets. For (S1), we expect a small number of users in most of the research networks, and therefore the impact to the blockchain network should be negligible. For (S2), the increment of the number of the records will not affect the size of transactions. This is because the size of the transaction is only related to the number of covariates. Additionally, the blockchain transaction speed will still be fast enough. Thus, this issue may not be a serious barrier for a well-designed blockchain network. For (S3), the transaction metadata size for the largest dataset (THA with 34 covariates) was small when compared to the default upper limit transaction metadata size for MultiChain.
Regarding the acceptability by users, one feasible solution is to start from a small number of pilot institutions within a system, and build the blockchain network in parallel with current operating platforms. Users should not be concerned about the technology underlying the network as long as it is reliable. Misconceptions about blockchain technology need to be overcome at the design, development, and dissemination levels. Given the hype surrounding blockchain and some unwarranted claims that it is an ideal solution for various problems, it is not surprising that the healthcare sector is exercising caution before adopting the technology. At the same time, it would be unwise not to consider the technology for situations in which its built-in advantages overcome its limitations.
We presented here a specific situation in which blockchain technology features such as disintermediation, immutable logging capability, resilience to node failure, and transparency of transactions match the needs of machine learning models being built across healthcare and research institutions. We also presented the practical drawbacks in terms of additional time for synchronization and commented on potential vulnerabilities. We anticipate that this information, when coupled with direct comparisons of platforms and use cases that are now emerging in the healthcare industry46 will help decision-makers choose among various technologies available for distributed machine learning.
CONCLUSION
ExplorerChain integrates 2 important technologies, artificial intelligence (online machine learning) and a distributed ledger (blockchain), without a central authority, to learn a predictive model across institutions in a distributed architecture (ie, without the need for sharing the patient-level data nor the need for a central coordinating node). We discussed the advantages/disadvantages of adopting blockchain, and showed empirically on 3 datasets that ExplorerChain produces results that are equivalent to the level of the state-of-the-art central server-based method. Additionally, through a traceable and immutable “path,” and as a peer-to-peer blockchain network without a central intermediary, ExplorerChain can improve security and robustness. These are useful for healthcare and genomics applications, as bottlenecks associated with local servers and network downtimes are eliminated. While ExplorerChain requires more iterations and time to reach a consensus without a central intermediary, the extra time may prove irrelevant for the development of machine learning models. ExplorerChain’s promising results warrant further evaluation and refinement, while suggesting that it is feasible to adopt blockchain-based artificial intelligence methods to build decentralized learning models across multiple sites. There are still mixed opinions and profound misunderstandings with corresponding questions about blockchain technology and its potential applications in healthcare/genomics. Examples, to name a few, include whether blockchain technology will prove to be practical and useful for healthcare; whether blockchain will only become practical as networks get faster and computer speeds increase; and whether the turnkey solution of blockchain will become available to all biomedical informatics researchers. ExplorerChain, as an early stage feasibility study, may serve as a step towards answering these open yet critical questions about the future of blockchain technology for applications in healthcare and genomics.
FUNDING
The authors were funded by the US National Institutes of Health (NIH) (OT3OD025462, R00HG009680, R01HL136835, R01GM118609, and U01EB023685) and a UCSD Academic Senate Research Grant (RG084150). The content is solely the responsibility of the authors and does not necessarily represent the official views of the NIH. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
AUTHOR CONTRIBUTIONS
T-TK contributed in conceptualization, data curation, formal analysis, funding acquisition, investigation, methodology, project administration, resources, software, validation, visualization, and writing (original draft). RAG contributed in data curation and writing (review and editing). KRC contributed in critical discussion and writing (review and editing). LO-M contributed in conceptualization, funding acquisition, project administration, resources, supervision, and writing (review and editing).
ACKNOWLEDGMENTS
The authors would like to thank the Office of the National Coordinator for Health Information Technology (ONC) and the National Institute of Standards and Technology (NIST) for awarding ModelChain, from which ExplorerChain originated, at the 2016 Use of Blockchain in Health IT and Health-related Research Challenge and Workshop. The authors would also like to thank Xiaoqian Jiang, PhD and Shuang Wang, PhD for very helpful discussions, as well as UCSD Health Department of Biomedical Informatics and Altman Clinical & Translational Research Institute teams for the technical support of the iDASH cloud infrastructure.
CONFLICT OF INTEREST STATEMENT
None declared.
REFERENCES
- 1. Navathe AS, Conway PH.. Optimizing health information technology’s role in enabling comparative effectiveness research. Am J Manag Care 2010; 16 (12 Suppl HIT): SP44–7. [PubMed] [Google Scholar]
- 2. Wicks P, Vaughan TE, Massagli MP, Heywood J.. Accelerated clinical discovery using self-reported patient data collected online and a patient-matching algorithm. Nat Biotechnol 2011; 29 (5): 411–4. [DOI] [PubMed] [Google Scholar]
- 3. Grossman JM, Kushner KL, November EA.. Creating Sustainable Local Health Information Exchanges: Can Barriers to Stakeholder Participation be Overcome? Washington, DC: Center for Studying Health System Change, 2008. [PubMed]
- 4. Rice S. Most hospitals face 30-day readmissions penalty in fiscal 2016. 2015. http://www.modernhealthcare.com/article/20150803/NEWS/150809981 Accessed August 23, 2019
- 5. Vecchione A. Predictive analytics lowers readmissions: how one health system dropped its rate from 21 percent to 14 percent. 2014. https://www.healthcareitnews.com/news/predictive-analytics-lowers-readmissions Accessed August 23, 2019
- 6. El Emam K, Hu J, Mercer J, et al. A secure protocol for protecting the identity of providers when disclosing data for disease surveillance. J Am Med Inform Assoc 2011; 18 (3): 212–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Loukides G, Denny JC, Malin B.. The disclosure of diagnosis codes can breach research participants’ privacy. J Am Med Inform Assoc 2010; 17 (3): 322–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Vaszar LT, Cho MK, Raffin TA.. Privacy issues in personalized medicine. Pharmacogenomics 2003; 4 (2): 107–12. [DOI] [PubMed] [Google Scholar]
- 9. Calloway SD, Venegas LM.. The new HIPAA law on privacy and confidentiality. Nurs Adm Q 2002; 26 (4): 40–54. [DOI] [PubMed] [Google Scholar]
- 10. Wu Y, Jiang X, Kim J, Ohno-Machado L.. Grid binary LOgistic REgression (GLORE): building shared models without sharing data . J Am Med Inform Assoc 2012; 19 (5): 758–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Wang S, Jiang X, Wu Y, Cui L, Cheng S, Ohno-Machado L.. Expectation propagation logistic regression (explorer): distributed privacy-preserving online model learning. J Biomed Inform 2013; 46 (3): 480–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Jiang W, Li P, Wang S, et al. WebGLORE: a web service for Grid LOgistic REgression. Bioinformatics 2013; 29 (24): 3238–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Shi H, Jiang C, Dai W, et al. Secure Multi-pArty computation grid LOgistic REgression (SMAC-GLORE). BMC Med Inform Decis Mak 2016; 16 (S3): 89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Kuo T-T, Gabriel RA, Ohno-Machado L.. Fair compute loads enabled by blockchain: sharing models by alternating client and server roles. J Am Med Inform Assoc 2019; 26 (5): 392–403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Kuo T-T, Kim J, Gabriel RA.. Privacy-preserving model learning on blockchain network-of-networks. J Am Med Inform Assoc 2020; 27 (3): 343–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Kuo T-T, Ohno-Machado L. ModelChain: Decentralized privacy-preserving healthcare predictive modeling framework on private blockchain networks. arXiv preprint arXiv: 1802.01746 2018.
- 17. Kuo T-T, Hsu C-N, Ohno-Machado L. ModelChain: Decentralized privacy-preserving healthcare predictive modeling framework on private blockchain networks. ONC/NIST Use of Blockchain for Healthcare and Research Workshop; September 26–September 27, 2016; Gaithersburg, MD.
- 18. Chen X, Ji J, Luo C, Liao W, Li P. When machine learning meets blockchain: A decentralized, privacy-preserving and secure design. In proceedings of the 2018 IEEE International Conference on Big Data (Big Data); December 10–December 13, 2018; Seattle, WA: IEEE; 2018:1178–87. [Google Scholar]
- 19. McConaghy T, Marques R, Müller A, et al. BigchainDB: A scalable blockchain database. 2016. https://www.bigchaindb.com/whitepaper/ Accessed July 1, 2017
- 20. Pilkington M. Blockchain technology: Principles and applications In: Xavier Olleros F., Majlinda Zhegu, eds. Research Handbook on Digital Transformations. Rochester, NY: Edward Elgar; 2016: 1–39. [Google Scholar]
- 21. Xu X, Pautasso C, Zhu L, et al. The blockchain as a software connector. In: proceedings of the 13th Working IEEE/IFIP Conference on Software Architecture (WICSA); April 5–8, 2016. Venice, Italy.
- 22. Kuo T-T, Kim H-E, Ohno-Machado L.. Blockchain distributed ledger technologies for biomedical and health care applications. J Am Med Inform Assoc 2017; 24 (6): 1211–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Luu L, Narayanan V, Baweja K, Zheng C, Gilbert S, Saxena P. SCP: a computationally-scalable Byzantine consensus protocol for blockchains. 2015. https://www.weusecoins.com/assets/pdf/library/SCP%20-%20%20A%20Computationally-Scalable%20Byzantine.pdf Accessed December 18, 2019.
- 24. Fromknecht C, Velicanu D, Yakoubov S.. A decentralized public key infrastructure with identity retention. IACR Cryptol ePrint Arch 2014; 2014: 803. [Google Scholar]
- 25. Bissias G, Ozisik AP, Levine BN, Liberatore M. Sybil-resistant mixing for bitcoin. In: proceedings of the 13th Workshop on Privacy in the Electronic Society; November 03, 2014; Scottsdale, AZ.
- 26. Lamport L, Shostak R, Pease M.. The Byzantine general’s problem. ACM Trans Program Lang Syst 1982; 4(3): 382–401. [Google Scholar]
- 27. Douceur JR. The Sybil attack In: International Workshop on Peer-to-Peer Systems. March 7–March 8, 2002; Cambridge, MA: Springer-Verlag Berlin Heidelberg; 2002: 251–60. [Google Scholar]
- 28. Greenspan G. MultiChain Private Blockchain - White Paper. 2015. http://www.multichain.com/download/MultiChain-White-Paper.pdf Accessed July 5, 2019
- 29. Nakamoto S. Bitcoin: A peer-to-peer electronic cash system. 2008. https://bitcoin.org/bitcoin.pdf Accessed July 3, 2019
- 30. Martin L. Blockchain vs. relational database: Which is right for your application? https://techbeacon.com/Blockchain-relational-database-which-right-for-your-application Accessed August 23, 2019
- 31. Miller A, LaViola JJ, Jr Anonymous byzantine consensus from moderately-hard puzzles: A model for bitcoin. 2014. https://nakamotoinstitute.org/static/docs/anonymous-byzantine-consensus.pdf Accessed August 23, 2019
- 32. Garay J, Kiayias A, Leonardos N.. The bitcoin backbone protocol: Analysis and applications In: proceedings of the Annual International Conference on the Theory and Applications of Cryptographic Techniques; April 26–April 30, 2015; Sofia, Bulgaria. [Google Scholar]
- 33. Goodfellow I, Bengio Y, Courville A.. Deep Learning. Cambridge, MA: MIT Press, 2016. [Google Scholar]
- 34. Abbas Q, Shafiq H, Ahmad I, Tharanidharan S. Concurrency control in distributed database system. In: proceedings of the 2016 International Conference on Computer Communication and Informatics (ICCCI);January 7–January 9, 2016; Coilmbatore, India: IEEE, New York City, NY; 2016: 1–4. [Google Scholar]
- 35.McConaghy T. Blockchain, Throughput, and Big Data Berlin: Bitcoin Startups; 2014. http://trent.st/content/2014-10-28%20mcconaghy%20-%20blockchain%20big%20data.pdf Accessed August 23, 2019
- 36. Meiklejohn S, Pomarole M, Jordan G, et al. A fistful of bitcoins: characterizing payments among men with no name. In: proceedings of the 2013 Internet Measurement Conference; October 23–25, 2013; Barcelona, Spain.
- 37. Vukolić M. The quest for scalable blockchain fabric: Proof-of-work vs. BFT replication In: proceedings of the International Workshop on Open Problems in Network Security; October 29, 2015; Zurich, Switzerland: Springer Nature Switzerland AG; 2015: 112–25. [Google Scholar]
- 38. Mainelli M, Smith M.. Sharing ledgers for sharing economies: an exploration of mutual distributed ledgers (aka blockchain technology). J Financ Perspect 2015; 3 (3): 38–69. [Google Scholar]
- 39.CoinSciencesLtd. MultiChain open platform for blockchain applications. http://www.multichain.com Accessed July 5, 2019
- 40.BigchainDB. BigchainDB The scalable blockchain database. https://www.bigchaindb.com Accessed July 1, 2017
- 41. Buterin V. A next-generation smart contract and decentralized application platform. 2014. https://github.com/ethereum/wiki/wiki/White-Paper Accessed July 3, 2019
- 42. Wood G. Ethereum: A secure decentralised generalised transaction ledger. 2014. https://gavwood.com/paper.pdf Accessed July 3, 2019
- 43.TheEthereumFoundation. The Ethereum Project. https://www.ethereum.org Accessed July 3, 2019
- 44.TheLinuxFoundation. Hyperledger. https://www.hyperledger.org/ Accessed July 5, 2019
- 45.TheLinuxFoundation. Hyperledger Architecture, Volume I: Introduction to Hyperledger Business Blockchain Design Philosophy and Consensus. https://www.hyperledger.org/wp-content/uploads/2017/08/HyperLedger_Arch_WG_Paper_1_Consensus.pdf Accessed July 5, 2019
- 46. Kuo T-T, Zavaleta Rojas H, Ohno-Machado L.. Comparison of blockchain platforms: a systematic review and healthcare examples. J Am Med Inform Assoc 2019; 26 (5): 462–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Boyd S, Ghosh A, Prabhakar B, Shah D.. Randomized gossip algorithms. IEEE Trans Inform Theory 2006; 14 (SI): 2508–30. [Google Scholar]
- 48. Boyd S, Ghosh A, Prabhakar B, Shah D. Gossip algorithms: Design, analysis and applications. In: proceedings of the IEEE 24th Annual Joint Conference of the IEEE Computer and Communications Societies. March 13–17, 2005; Miami, FL.
- 49. Shah D. Gossip algorithms. FNT Netw 2007; 3 (1): 1–125. [Google Scholar]
- 50. Fontenla-Romero Ó, Guijarro-Berdiñas B, Martinez-Rego D, Pérez-Sánchez B, Peteiro-Barral D. Online machine learning. In: Efficiency and Scalability Methods for Computational Intellect Hershey, PA: IGI Global; 2013: 27–54.
- 51. Shalev-Shwartz S. Online learning and online convex optimization. FNT Mach Learn 2011; 4 (2): 107–94. [Google Scholar]
- 52. Yan F, Sundaram S, Vishwanathan S, Qi Y.. Distributed autonomous online learning: Regrets and intrinsic privacy-preserving properties. IEEE Trans Knowl Data Eng 2013; 25 (11): 2483–93. [Google Scholar]
- 53. Kennedy R, Fraser H, McStay L, Harrison R.. Early diagnosis of acute myocardial infarction using clinical and electrocardiographic data at presentation: Derivation and evaluation of logistic regression models. Eur Heart J 1996; 17 (8): 1181–91. [DOI] [PubMed] [Google Scholar]
- 54. Zou KH, Liu A, Bandos AI, Ohno-Machado L, Rockette HE.. Statistical Evaluation of Diagnostic Performance: Topics in ROC Analysis. Boca Raton, FL: CRC Press; 2011. [Google Scholar]
- 55. Sharma BS, Swisher MW, Doan CN, Khatibi B, Gabriel RA.. Predicting patients requiring discharge to post-acute care facilities following primary total hip replacement: Does anesthesia type play a role? J Clin Anesth 2018; 51: 32–6. [DOI] [PubMed] [Google Scholar]
- 56. Gabriel RA, Waterman RS, Kim J, Ohno-Machado L.. A predictive model for extended postanesthesia care unit length of stay in outpatient surgeries. Anesth Analg 2017; 124 (5): 1529–36. [DOI] [PubMed] [Google Scholar]
- 57. Benitez K, Malin B.. Evaluating re-identification risks with respect to the HIPAA privacy rule. J Am Med Inform Assoc 2010; 17 (2): 169–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. El Emam K, Jonker E, Arbuckle L, Malin B.. A systematic review of re-identification attacks on health data. PLoS One 2011; 6 (12): e28071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. El Emam K, Brown A, AbdelMalik P.. Evaluating predictors of geographic area population size cut-offs to manage re-identification risk. J Am Med Inform Assoc 2009; 16 (2): 256–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Jiang X, Sarwate AD, Ohno-Machado L.. Privacy technology to support data sharing for comparative effectiveness research: A systematic review. Med Care 2013; 51 (8 0 3): S58–S65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Gardner J, Xiong L, Xiao Y, et al. SHARE: System design and case studies for statistical health information release. J Am Med Inform Assoc 2013; 20 (1): 109–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Baumer D, Earp JB, Payton FC.. Privacy of medical records: IT implications of HIPAA. Sigcas Comput Soc 2000; 30 (4): 40–7. [Google Scholar]
- 63. McGraw D. Building public trust in uses of Health Insurance Portability and Accountability Act de-identified data. J Am Med Inform Assoc 2013; 20 (1): 29–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Kim KK, McGraw D, Mamo L, Ohno-Machado L.. Development of a privacy and security policy framework for a multistate comparative effectiveness research network. Med Care 2013; 51: S66–S72. [DOI] [PubMed] [Google Scholar]
- 65. Ohno-Machado L, Bafna V, Boxwala AA,. et al. iDASH. Integrating data for analysis, anonymization, and sharing. J Am Med Inform Assoc 2012; 19 (2): 196–201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Ohno-Machado L. To share or not to share: That is not the question. Sci Transl Med 2012; 4 (165): 165cm15. [DOI] [PubMed] [Google Scholar]
- 67. Lasko TA, Bhagwat JG, Zou KH, Ohno-Machado L.. The use of receiver operating characteristic curves in biomedical informatics. J Biomed Inform 2005; 38 (5): 404–15. [DOI] [PubMed] [Google Scholar]
- 68. Hanley JA, McNeil BJ.. The meaning and use of the area under a receiver operating characteristic (ROC) curve. Radiology 1982; 143 (1): 29–36. [DOI] [PubMed] [Google Scholar]
- 69. Bauerle N. What is the difference between a blockchain and a database? https://www.coindesk.com/information/what-is-the-difference-blockchain-and-database Accessed August 23, 2019
- 70. Meunier S. Blockchain technology — a very special kind of Distributed Database. 2016. https://medium.com/@sbmeunier/blockchain-technology-a-very-special-kind-of-distributed-database-e63d00781118 Accessed August 23, 2019
- 71. Smith MA, Smith WT.. The American joint replacement registry. Orthop Nurs 2012; 31 (5): 296–99. [DOI] [PubMed] [Google Scholar]