Abstract
Objectives
To adapt and evaluate a deep learning language model for answering why-questions based on patient-specific clinical text.
Materials and Methods
Bidirectional encoder representations from transformers (BERT) models were trained with varying data sources to perform SQuAD 2.0 style why-question answering (why-QA) on clinical notes. The evaluation focused on: (1) comparing the merits from different training data and (2) error analysis.
Results
The best model achieved an accuracy of 0.707 (or 0.760 by partial match). Training toward customization for the clinical language helped increase 6% in accuracy.
Discussion
The error analysis suggested that the model did not really perform deep reasoning and that clinical why-QA might warrant more sophisticated solutions.
Conclusion
The BERT model achieved moderate accuracy in clinical why-QA and should benefit from the rapidly evolving technology. Despite the identified limitations, it could serve as a competent proxy for question-driven clinical information extraction.
Keywords: question answering, artificial intelligence, natural language processing, evaluation studies, clinical decision-making
INTRODUCTION
The reasoning and decision-making in clinical practice can be naturally framed as a series of questions and answers. Automated question answering (QA) has long been considered a feat in artificial intelligence and is vitally researched for clinical applications. Among the diverse information needs, why-QA is a distinct category that deals with cause, motivation, circumstance, and justification. In terms of prevalence, 20% of the top 10 question types asked by family physicians1 can actually be paraphrased into a why-question. Clinical why-QA is important because: (1) the ultimate task resembles expert-level explanatory synthesis of knowledge and evidence and (2) it would enable identifying reasons for the decisions documented in clinical text.
The current study concentrates on the second scenario above, a modest yet very useful task of reason identification. Essentially, the system has to identify the literal reason regarding certain decision specific to a patient, for example, why was his dobutamine stress test rescheduled? → “hypotension” (from note text). In non-medical domains, the counterpart to such document-based QA is known as reading comprehension QA (RCQA), with competitive open challenges and richly-annotated corpora. SQuAD 2.02 is an iconic RCQA corpus and challenge, which features the requirement for a system to refrain from answering when there is no suitable answer present in the text. A language model that has caught wide attention was the bidirectional encoder representations from transformers (BERT)3 and its evolving derivatives,4 for their high performance not only in SQuAD 2.0 but in multiple natural language understanding challenges.
As an initial step toward developing a clinical reason identification system, this study adapted the BERT model for clinical why-QA. We found domain customization was critical to performance, with a best achieved accuracy of 0.707 (or 0.760 by partial match). More importantly, our error analysis helped understand the data issues, the system behavior, and areas to improve on.
BACKGROUND
Related work
There have been QA systems developed in the medical domain, for example, AskHERMES,5 MiPACQ,6 and MEANS.7 These systems work mostly by consulting external knowledge sources to answer questions that are not patient-specific. In contrast, the RCQA task we target here deals with questions specific to the patient at hand by extracting answers directly from her/his clinical notes. The closest work was the emrQA, a large clinical RCQA corpus developed by Pampari et al.,8 in which they also evaluated a couple of machine learning systems.
Relevant resources
In the following, we introduce several existing resources that are important to our methods.
SQuAD
The Stanford Question Answering Dataset (SQuAD)9 was created to promote RCQA research and application development. We followed the SQuAD 2.0 task setting, because it can be critical to have the system refrain from making false suggestions especially in some clinical applications. Figure 1 illustrates a typical training instance in the SQuAD 2.0 format, consistent with that used in our experiments.
Figure 1.
Illustration of a SQuAD-style training instance for clinical why-QA. The format conforms to JSON syntax, where the gray highlight here is for helping readers locate the answer. For readability, some of the field names have been modified and are different from that in the original SQuAD 2.0.
emrQA
The emrQA8 is a large training set annotated for RCQA in the clinical domain. It was generated by template-based semantic extraction from the i2b2 NLP challenge datasets.10 The current emrQA release includes more than 400 000 QA pairs, of which 7.5% involve a why-question.
BERT
BERT3 represents a state-of-the-art language model that leverages deep bidirectional self-attention learning. The pre-training phase of BERT learns a transferrable representation, which can be followed by a fine-tuning phase where the actual task-specific (eg, RCQA) training takes place. Due to the immense memory demand for training BERTlarge, we used BERTbase for our experiments without losing conceptual generality.
Clinical BERT
Alsentzer et al. used approximately 2 million clinical notes from the MIMIC-III v1.4 database11 and pre-trained a Clinical BERT model.12 They made it publicly available; otherwise, it originally took about 17 days of computational runtime by a single GeForce GTX TITAN X 12 GB GPU.
MATERIALS AND METHODS
The experiments started with preparing the different data of progressive domain customization into BERT-compatible training format. Accordingly, models of incremental customization levels were trained. Lastly, accuracy metrics were computed against an independent test set, and qualitative error analysis was performed.
Preparation of the training data
emrQAwhy
This was our core training data, by selecting emrQA entries with a why-question. Additional processes included: (1) removing the “heart-disease-risk” subset due to problematic answer line number in the “evidence_start” field, (2) retaining QAs where the question had one and only one answer, in conformity with SQuAD 2.0 setting, (3) merging in a small set of our previously annotated clinical why-QAs,13 and (4) programmatically constructing a set of unanswerable QAs, for example, from an answerable instance “why was the patient on nitroglycerin?” → “substernal chest pain”, we searched over a random pool of other notes until hitting one that contained neither “nitroglycerin” nor “substernal chest pain” in text to make an unanswerable case. We obtained 27 762 answerable QAs and 2839 unanswerable QAs, all formatted like Figure 1. Lastly, the data were split into train/dev[elopement]/test partitions with 250/90/250 disjoint clinical notes, corresponding to 12 741/4 315/13 545 QAs. The dev partition was set aside to learn the optimal cutoff threshold for the system to refrain from answering.
i2b2notespre
A set of 1474 i2b2 notes with 106 952 pre-chunked sentences was available to us and amenable to BERT pre-training. Given the manageable size, we undertook this pre-training and evaluated its usefulness for domain customization in comparison to the more heavily trained Clinical BERT.
SQuADwhy
A phenomenon commonly observed in biomedical NLP is the smoothing effect introduced by inclusion of off-domain training data, that is, supplementing background language examples that compensate any under-sampled patterns in the domain-specific examples. To experiment with this aspect, we extracted 1833 why-QAs from SQuAD 2.0 (hence SQuADwhy). SQuADwhy was run as a “pre”-fine-tuning step to prime BERT into the why-QA genre.
Training and tuning of the QA models
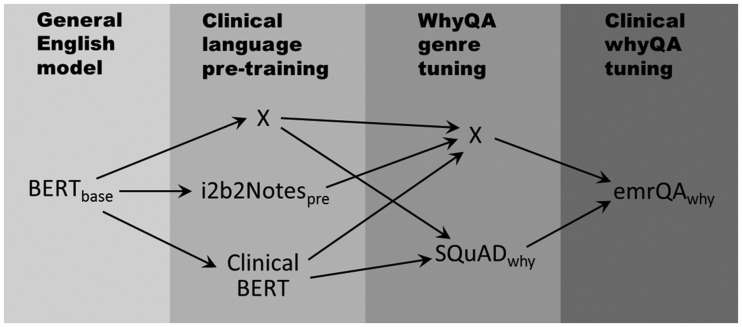
To assess the benefits of different data sources, we experimented with five models trained by incremental levels of domain and task customization. Figure 2 illustrates the five paths for configuring these models, where each column shows the alternative data sources used (or an “X” indicating nothing was used). The purpose of each data source is elaborated as follows: BERTbase was the original general-purpose model. The emrQAwhy served as the core task-specific training set and (when used alone) as the baseline for benchmarking other enhancements. Clinical BERT and i2b2Notespre represented domain adaptation; the former used a much larger corpus and more diverse note types. The SQuADwhy (1833 general English why-QAs) was an optional fine-tuning step to assess how an off-domain training set might benefit the model. Each fine-tuning experiment was run with 5 epochs (number of training iterations), batch_train_size = 32 (number of training instances fed into each optimization step), learning_rate = 3e-5 (the magnitude of each piecemeal weight-updating in the optimization), and max_seq_length = 128 (number of tokens in the sliding window by which the training instances were processed). The jobs were run on a Tesla V100 with compute capability 7.0 and 18 GB of memory.
Figure 2.
Different learning configurations to explore the effect of domain-, genre-, or task-specific customization. Each “X” means no enhancement performed. The grayscale roughly corresponds to the level of customization by the training data, from general (lighter) to specific (darker).
Evaluation and error analysis
The evaluation was based on the standard SQuAD 2.0 metrics, comparing the system answer to the gold by token-wise exact and partial matches. Each partial match was weighted by using f1-measure between the predicted and the gold bags of tokens.14 We computed the accuracies and included a break-down summary of the answerable versus unanswerable QAs. After the optimal configuration emerged, we doubled the epochs to 10 and trained a separate model for the final precision-recall and error analysis. The error analysis focused on false negatives (FNs), by randomly sampling 100 QAs where the system answer had completely no overlap tokens (including those refrained) with the gold answer. A cardiologist (M.Y.E.) manually reviewed the set and recorded his assessment.
RESULTS
Accuracy of the why-QA models
The performance metrics of the differently trained models are summarized in Table 1. In general, we can see the refraining mechanism worked well (the NoAns column). The “pre”-fine-tuning by SQuADwhy appeared beneficial (almost 3% accuracy increase from the baseline), suggesting that BERT learned certain genre characteristics even from a non-medical corpus. Pre-training using the 1474 notes of i2b2notespre lifted the accuracy up about 3%, but could not beat the 6% boost by the lavishly trained ClinBERTpre using 2 million notes. In the end, we combined the best configurations into training a single model, which achieved an accuracy of 0.700 (or 0.757 with partial match). The extended training with 10 epochs resulted in a marginal increase in accuracy. Figure 3 shows the precision-recall tradeoff of the final 10-epoch model. Overall the system appeared to conservatively favor higher precision versus recall, while maintaining the precision above or around 0.8 until the upper-bound recall due to the refraining. As for the cost of time, the best configuration (fine-tuned by SQuADwhy then emrQAwhy, on top of ClinBERTpre) took 53 minutes to train with 5 epochs and 64 minutes with 10 epochs.
Table 1.
Accuracy of differently trained models on the test set
| Model | Full test set: 13 545 QAs |
Test HasAns: 12 376 QAs |
Test NoAns: 1169 QAs |
|||
|---|---|---|---|---|---|---|
| Exact | Partial | Exact | Partial | Exact | Partial | |
| BERTbase + emrQAwhy | 0.633 | 0.688 | 0.599 | 0.659 | 0.995 | 0.995 |
| BERTbase + SQuADwhy + emrQAwhy | 0.660 | 0.728 | 0.628 | 0.703 | 0.994 | 0.994 |
| BERTbase + i2b2Notespre + emrQAwhy | 0.663 | 0.718 | 0.631 | 0.692 | 0.997 | 0.997 |
| BERTbase + ClinBERTpre + emrQAwhy | 0.695 | 0.744 | 0.666 | 0.720 | 0.994 | 0.994 |
| BERTbase + ClinBERTpre + SQuADwhy + emrQAwhy | 0.700 | 0.757 | 0.672 | 0.735 | 0.995 | 0.995 |
| BERTbase + ClinBERTpre + SQuADwhy + emrQAwhy (10 epochs) | 0.707 | 0.760 | 0.679 | 0.737 | 0.999 | 0.999 |
Abbreviations: HasAns: answerable according to the gold; NoAns: unanswerable according to the gold.
Figure 3.
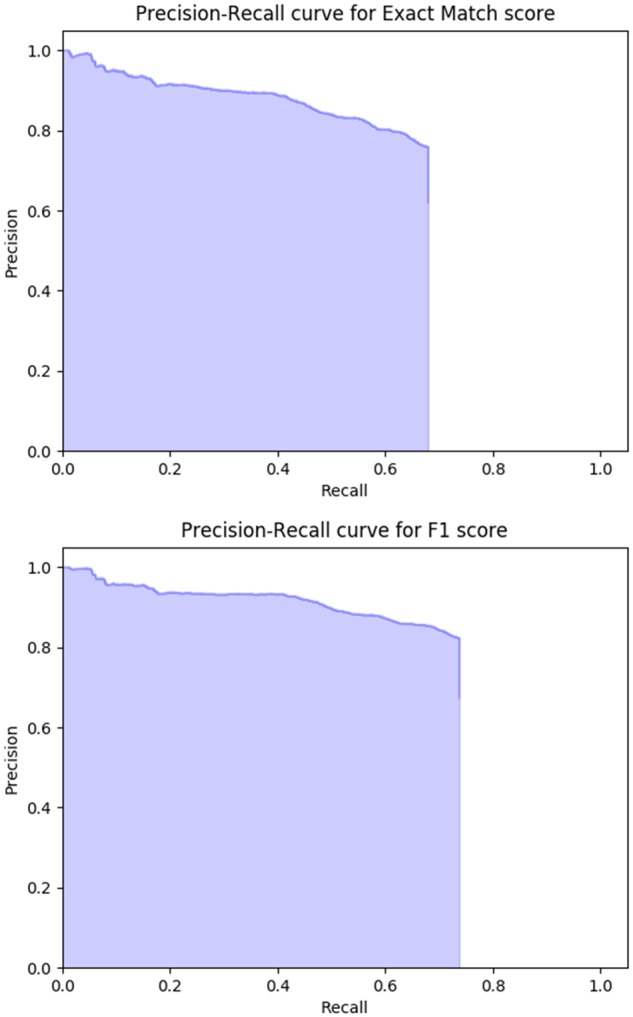
Precision-recall curves of the best model (upper: exact match, lower: partial match).
Error analysis with a focus on the false negatives
Given that the system performed decently on the NoAns QAs, we paid attention to those where the gold indeed had an answer. Based on review by the physician (M.Y.E.), we categorized the 100 FNs missed by our final best model as in Table 2 and elaborated below.
Table 2.
Error analysis of 100 FNs by the best model
| Main category | Subcategory | Count | Subtotala |
|---|---|---|---|
| Unanswerable | (a) Vague question | 6 | 14 |
| (b) Expert deemed unanswerable using only text | 8 | ||
| System answered | (c) Expert judged the system acceptable as the gold | 6 | 18 |
| (d) Expert sided with the system against the gold | 12 | ||
| (e) Real FN | 18 | 68 | |
| (f) Expert disagreed with both the system and gold | 7 | ||
| System refrained | (g) Real FN | 24 | |
| (h) Correct answer ranked second place | 19 |
aThis column stands for a “redemption” perspective: 14% that the system was not supposed to make it, 18% where the system answer was actually right, and 68% that the system was truly attributed for the FN.
Unanswerable
(a) Vague question (6%)
The question did not make sense, likely due to the fact that emrQA synthetically derived the questions from i2b2 NLP annotations. For example, “why did the patient have removal?”
(b) Expert deemed unanswerable using only text (8%)
There was no clear trace of reasoning mentioned in the text to support the answer without preexisting medical knowledge, or the correct answer was not even present.
System answered
(c) Expert judged the system as acceptable as the gold (6%)
There were two scenarios here. The first was that the system picked a conceptually synonymous answer, for example, “bacteremia” versus “sepsis”. The second revealed incompleteness in some gold annotations. For example, the reasons why one patient was on nitroglycerin actually included both “shortness of breath” and “chest pain”, but the gold had only the former.
(d) Expert sided with the system against the gold (12%)
The domain expert considered the system’s answer to be more suitable than the gold. For example, when asked to identify the reason behind a Flagyl prescription, the system picked “aspiration pneumonia” instead of the “elevated white count” by the gold annotation.
(e) Real FN (18%)
The system did not appear to really understand the nuances such as the indication versus the target effect. For example, in one case “diuresis” was picked as the reason for Lasix drip, while the gold had “decreased urine output”. In the other example, the mentioned side effect of Celebrex was mistaken as the reason for prescription, seemingly because they co-occurred within the same sentence.
(f) Expert disagreed with both the system and gold (7%)
The physician judged that neither the system nor the gold picked the correct answer. In one case “why was the patient on lantus insulin?”, the physician picked “DKA” (diabetic ketoacidosis) instead of the gold “right thigh cellulitis”. The system marked “gastroparesis”, apparently mistook a context that mentioned about insulin tolerance.
System refrained
(g) Real FN (24%)
The system not only refrained from answering but even its top candidate answer did not appear to be viable. In one case “why was the patient prescribed lactulose?” (with correct answer being “hepatic encephalopathy”), the system had no chance given its top answer “cardiac side effects”.
(h) Correct answer ranked second place (19%)
As a rescue investigation, we found that in 19% of the FNs the system actually had the correct answer but ranked it secondary to the refraining decision.
DISCUSSION
Our incremental training source comparisons proved to be informative. The 3% accuracy increase by the SQuADwhy pre-tuning suggested that a close-genre corpus, even off-domain, could benefit the end task. In alignment with intuition, Clinical BERT as an extensively pre-trained in-domain model was a vital booster to accuracy (6% improvement). Noteworthy, on the other hand, the 6% was earned by a hefty 2 million training notes. Given the auxiliary finding that even the 1474 notes of i2b2notespre made a 3% increase, it raised the question whether the benefit could have been saturated much earlier before the training data was increased into the millions.
The results revealed much room for improvement both in terms of the data preparation and model optimization. Even though erring conservatively is desirable for many precision-oriented applications, the over-refraining tendency implied that our heuristically constructed unanswerable instances were noisy. Besides, the error analysis discovered various issues (eg, nonsense questions and inaccurate answers) in emrQA, which was efficient for producing massive training data but regrettably could not match the quality of manually-authored QAs like the SQuAD corpus.
Despite the rapidly evolving field, our study exposed issues and challenges that are general to deep learning language models as applied in clinical why-QA. Some off-the-mark answers by the system showed that BERT might have just leveraged adjacent cues or recurring associations, instead of true understanding. In that same vein, those deemed by the physician as unanswerable by the text indicated that why-QA probably should not be framed simply as an RCQA task, that is, much richer contexts (including external knowledge sources) are needed both at the training and the reasoning phases. Furthermore, the long documents and existence of multiple viable answers distinguish clinical why-QA as a unique challenge that warrants redesign in the annotation and evaluation approaches.
CONCLUSION
The BERT language model was evaluated for the task of clinical why-QA. The best accuracy was 0.707 (or 0.760 with partial match), specifically benefiting from domain- and genre-customization. The error analysis indicated improvements to be needed in the training data preparation and even redesign of the fundamental task. Although at its current state the model is premature for truly intelligent why-reasoning, we propose to use it practically as a question-driven clinical information extraction tool for detecting reasons with explicit cues in text.
Author Contributors
J.F. conceived the study, performed the experiments, and drafted the manuscript. A.W. implemented the tooling that facilitated the data analysis. M.Y.E. reviewed the system’s and gold answers in the error analysis. All authors participated in the data interpretation, critically revised the manuscript, approved the final version, and agreed to be accountable for resolving any questions related to the integrity of their work.
ACKNOWLEDGMENTS
De-identified clinical records used in this research were provided by the i2b2 National Center for Biomedical Computing funded by U54LM008748 and were originally prepared for the Shared Tasks for Challenges in NLP for Clinical Data organized by Dr Ozlem Uzuner, i2b2 and SUNY.
CONFLICT OF INTEREST STATEMENT
None declared.
REFERENCES
- 1. Ely JW, Osheroff JA, Ebell MH, et al. Analysis of questions asked by family doctors regarding patient care. BMJ 1999; 319 (7206): 358–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Rajpurkar P, Jia R, Liang P. Know what you don't know: Unanswerable questions for SQuAD. In: Proceedings of the 56th Annual Meeting of the Association for Computational Linguistics 2018. Melbourne, Australia.
- 3. Devlin J, Chang M-W, Lee K, Toutanova K. BERT: Pre-training of deep bidirectional transformers for language understanding. arXiv e-prints 2018: 1810.04805.
- 4. Liu Y, Ott M, Goyal N, et al. RoBERTa: A robustly optimized BERT pretraining approach. arXiv e-prints 2019: 1907.11692.
- 5. Cao Y, Liu F, Simpson P, et al. AskHERMES: An online question answering system for complex clinical questions. J Biomed Inform 2011; 44 (2): 277–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Cairns BL, Nielsen RD, Masanz JJ, et al. The MiPACQ clinical question answering system. AMIA Annu Symp Proc 2011; 2011: 171–80. [PMC free article] [PubMed] [Google Scholar]
- 7. Abacha AB, Zweigenbaum P.. MEANS: A medical question-answering system combining NLP techniques and semantic web technologies. Inform. Process. Manag. 2015; 51 (5): 570–94. [Google Scholar]
- 8. Pampari A, Raghavan P, Liang J, Peng J. emrQA: A large corpus for question answering on electronic medical records. Proceedings of the Conference on Empirical Methods in Natural Language Processing 2018. Brussels, Belgium.
- 9. Rajpurkar P, Zhang J, Lopyrev K, Liang P. SQuAD: 100, 000+ questions for machine comprehension of text. Proceedings of the Conference on Empirical Methods in Natural Language Processing 2016. Austin, TX, USA.
- 10.The i2b2 NLP Research Data Sets. Secondary The i2b2 NLP Research Data Sets. https://www.i2b2.org/NLP/DataSets/Main.php (Accessed April, 2019).
- 11. Johnson AE, Pollard TJ, Shen L, et al. MIMIC-III, a freely accessible critical care database. Sci Data 2016; 3 (1): 160035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Alsentzer E, Murphy JR, Boag W, et al. Publicly available clinical BERT embeddings. Proceedings of the 2nd Clinical Natural Language Processing Workshop 2019. Minneapolis, MN, USA.
- 13. Fan J. Annotating and characterizing clinical sentences with explicit why-QA cues. Proceedings of the 2nd Clinical Natural Language Processing Workshop 2019. Minneapolis, MN, USA.
- 14. Hripcsak G, Rothschild AS.. Agreement, the f-measure, and reliability in information retrieval. J Am Med Inform Assoc 2005; 12 (3): 296–8. [DOI] [PMC free article] [PubMed] [Google Scholar]