Abstract
Iron is an essential nutrient that is actively acquired by bacterial pathogens during infections. Clinically important Staphylococcus aureus obtains iron by extracting heme from hemoglobin (Hb) using the closely related IsdB and IsdH surface receptors. In IsdH, extraction is mediated by a conserved tridomain unit that contains its second (N2) and third (N3) NEAT domains joined by a helical linker, called IsdHN2N3. Leveraging the crystal structure of the IsdHN2N3:Hb complex, we have probed the mechanism of heme capture using NMR, stopped-flow transfer kinetics measurements, and molecular dynamics (MD) simulations. NMR studies of the 220 kDa IsdHN2N3:Hb complex reveal that it is dynamic, with persistent interdomain motions enabling the linker and N3 domains in the receptor to transiently engage Hb to remove its heme. An alanine mutagenesis analysis reveals that two receptor subsites positioned ~20 Å apart trigger heme release by contacting Hb’s F-helix. These subsites are located within the N3 and linker domains and appear to play distinct roles in stabilizing the heme transfer transition state. Linker domain contacts primarily function to destabilize Hb–eheme interactions, thereby lowering ΔH‡, while contacts from the N3 subsite play a similar destabilizing role, but also form a bridge through which heme moves from Hb to the receptor. Interestingly, MD simulations suggest that within the transiently forming interface, both the F-helix and receptor bridge are in motion, dynamically sampling conformations that are suitable for heme transfer. Thus, IsdH triggers heme release from Hb via a flexible, low-affinity interface that forms fleetingly in solution.
Keywords: Iron-regulated surface determinant system, Staphylococcus aureus hemoglobin receptor, Stopped-flow spectrophotometry, Molecular dynamics, Nuclear magnetic resonance spectroscopy
Introduction
Staphylococcus aureus is a major cause of life-threatening hospital- and community-acquired infections in the United States [1,2]. Highly virulent methicillin-resistant S. aureus (MRSA) strains are now common, creating the need to discover new antibiotics to treat this pathogen [3,4]. During infections, S. aureus actively acquires iron, which functions as a versatile metal cofactor in microbial enzymes that mediate a wide range of essential cellular processes [5–8]. Heme (protoporphyrin IX + iron) present within human hemoglobin (Hb) accounts for ~75–80% of the total iron found in the human body and is preferentially used by S. aureus as an iron source [8]. S. aureus scavenges heme using the iron-regulated surface determinant (Isd) system, a network of bacterial proteins that collectively function to extract heme from Hb and remove its iron for use by the microbe [9–11]. An understanding of the molecular mechanism of heme scavenging could lead to new antiinfective agents that function by limiting microbial access to iron, as S. aureus mutant strains lacking key components of this system have reduced virulence [12–14].
The S. aureus Isd system contains nine proteins that capture the heme-bound iron from Hb. In this process, Hb is first bound to receptors on the microbial surface that extract its heme molecules. Liberated heme is then transferred across the cell wall to the membrane and pumped into the cytoplasm where it is degraded to release free iron [9–11]. Four Isd proteins (IsdA, IsdB, IsdC, and IsdH/HarA) are covalently linked to the bacterial cell wall via sortase transpeptidases [15–17]. Heme binding and Hb binding by these proteins are mediated by NEAT (NEAr iron Transporter) domains, highly conserved modules that are present in Gram-positive bacteria [18]. Heme acquisition is initiated by the closely related IsdB and IsdH proteins, which bind Hb and extract its heme [19–25]. Heme is then passed from these receptors to IsdA, which is partially buried in the cell wall [14]. Holo-IsdA subsequently transfers heme to the fully buried IsdC protein via an ultra-weak handclasp complex [26–29]. Lastly, holo-IsdC delivers the heme to the bacterial ABC transporter complex, IsdDEF, which pumps heme into the cytoplasm where it is degraded to release free iron by the heme oxygenases IsdG or IsdI [30,31]. Many other bacterial pathogens scavenge heme using related systems that are important for virulence [32–38].
Biochemical and structural studies have shown that the IsdH receptor removes heme from Hb using a tridomain unit (IsdHN2N3, residues Ala326-Asp660) [19]. IsdHN2N3 contains the second (N2) and third (N3) NEAT domains in IsdH, which are joined by a helical linker (L) domain (Fig. 1). The NEAT domains have distinct functions; N2 binds Hb, while the N3 domain binds to heme [39,40].Interestingly, the domains function synergistically and must be part of the same polypeptide in order to effectively induce heme release from Hb [19], which occurs ~1250-fold faster than the rate at which Hb spontaneously releases heme into the solvent [23]. Atomic structures of the isolated domains within IsdHN2N3 have been determined [19,24,25,40,41]. NMR studies indicate that prior to engaging Hb, IsdHN2N3 adopts an elongated and semiflexible state in which the linker and N3 domains form a rigid structure (hereafter called LN3) that can reorient relative to the N2 domain [22]. A low-resolution (4.24 Å) crystal structure of the IsdHN2N3:Hb complex revealed that the receptor engages the globin chains within Hb via two distinct binding interfaces (Fig. 1) [24]. The first interface is formed by the N2 domain, which interacts with the A and E helices of the globin that are positioned distal to the heme-binding pocket (called the Hb-N2 interface). In the second interface, the heme pocket of the same globin chain is engaged by the linker and N3 domains (called the Hb-LN3 interface) through which heme is transferred to the receptor. A subsequent higher resolution (2.55 Å) crystal structure of an IsdHN2N3 mutant bound to only the α-globin chains revealed that Hb-LN3 interfacial interactions distorted Hb’s F-helix [25], but their functional role in heme transfer remains to be determined. The related receptor IsdB shares significant primary sequence homology with IsdH and also contains the conserved tridomain unit, suggesting that IsdH and IsdB extract heme from Hb using a similar mechanism [20,21,42,43].
Fig. 1. Overview of the Hb: αIsdH interface.
Crystal structure (PDB: 4XS0) of the αHb: αIsdH complex with each interface highlighted. Cartoon representations are shown for the alpha subunit of Hb and the receptor (coloring: Hb, copper; N2 domain, dark brown; linker and N3 domains, light brown). Atoms for the heme molecule are shown as spheres.
In this study, we have used a combination of computational and experimental approaches, to investigate how IsdH extracts ferric heme from Hb. Using a newly developed stopped-flow assay that selectively measures heme removal from the α-globin chain and alanine mutagenesis, we show that two spatially distinct receptor subsites within the Hb-LN3 interface trigger heme release from Hb. Molecular dynamic (MD) simulations and Erying analyses of the transfer data indicate that the subsites have divergent, energetically distinct functions in the transfer reaction. A receptor subsite located on the linker domain lowers ΔH‡ by stabilizing a distorted form of Hb’s F-helix that houses the FeIII-His87 coordination bond, while contacts from a distally positioned N3 subsite destabilize the F-helix and form a platform for heme egress from Hb. Interestingly, the results of MD and NMR experiments indicate that the IsdHN2N3:Hb complex is highly dynamic, with persistent receptor interdomain motions in the complex causing the transient formation of the Hb-LN3 interface and unfurling of the heme egress platform. A working model of this dynamic extraction process is presented.
Results
NMR and MD studies reveal that the IsdH receptor transiently engages Hb’s heme pocket
Prior to engaging Hb, the IsdHN2N3 receptor does not adopt a single conformation because of interdomain motions that occur between its N2 and LN3 domains [22]. In contrast, in crystal structures of the IsdHN2N3:Hb complex, a single conformation of the receptor is observed in which its N2 and LN3 portions engage with different sites on Hb (Fig. 1) [24,25]. To determine if Hb binding quenches interdomain receptor motions, we used NMR to study the solution properties of the IsdHN2N3:Hb complex (M.W. 220 kDa). In this complex IsdHN2N3 is uniformly labeled with 2H, 15N, and 13C, except for the methyl groups in the side chains of Leu, Val, and Ile (δ1 position only), which are labeled with 1H and 13C (ILV-IsdHN2N3). This protein also contains a Y642A mutation in the N3 domain that prevents heme binding, but otherwise the receptor binds to Hb with similar affinity as the wild-type receptor [22–25]. Selective methyl labeling yields site-specific information, while simplifying the NMR spectra and facilitating signal detection in high-molecular-weight systems [44]. Previously, we have assigned the 1H and 13C chemical shifts in ILV-IsdHN2N3 in its Hb-free form [22]. To probe how the receptor interacts with Hb, we performed a Hb titration study that tracked receptor methyl spectral changes by recording 1He–13C methyl-TROSY spectra (Fig. 2a and b) [45]. A tetramer-stabilized recombinant form of Hb, called Hb0.1, was used in the titration in its ferrous carbonmonoxy form to limit heme release [46]. Adding Hb0.1 causes dramatic spectral changes in methyl groups located at the Hb-N2 interface (Fig. 1), with signals in the N2 domain of ILV-IsdHN2N3 broadening beyond detection when Hb is present at twofold molar excess (N2 signals that are broadened are labeled in Fig. 2a and are shown as red spheres in Fig. 2c). In contrast, signals originating from methyl groups located in the linker and N3 domains exhibit only minimal chemical shift perturbations or broadening when Hb is added (Fig. 2b and gray spheres in Fig. 2c). This is surprising, since in the crystal structure of the IsdHN2N3:Hb complex both the Hb-LN3 and Hb-N2 interfaces are fully formed and bury comparable solvent-exposed surface areas, 1235 Å2 and 926 Å2, respectively (Fig. 1). Moreover, in the crystal structure of the complex, many of the methyl groups in LN3 portion of the receptor are in close proximity to Hb, such that broadening and/or chemical shift changes in their signals would be expected if this interface were as significantly populated in solution as it is in the crystal.
Fig. 2. NMR studies of the 220 kDa Hb:IsdHN2N3 complex.
A. 2D 1H–13C methyl-TROSY spectra of apo-ILV-IsdHN2N3 (black) and after titration (purple) with 2 eq. of Hb0.1-CO B. Zoomed-in portions of 1H–13C methyl-TROSY spectra highlighting peak broadening/chemical shift perturbations of resonances associated with N2 and relatively intact signals from LN3. C. Crystal structure of Hb:αIsdH complex (PDB: 4XS0) with Ile, Leu, and Val methyl groups shown as spheres. Methyl signals that show peak broadening/chemical shift perturbations upon titration with Hb0.1-CO are shown in red, whereas intact signals are shown in gray.
The discrepancy between the crystal structure and NMR data suggests that in solution, the Hb-N2 interface forms with higher affinity than the Hb-LN3 interface and that packing interactions help stabilize the Hb-LN3 interface in crystals of the complex. To investigate this issue, we performed a series of steered MD (sMD) simulations [47] to determine the relative energies needed to “pull” the N2 or LN3 portions of the receptor from their respective sites on Hb’s α-subunit. Using the coordinates of the IsdHN2N3 :Hb crystal structure as a starting point, two sets of ten 50 ns simulations were performed in which interactions with either the N2 or LN3 portions were selectively disrupted by pulling them from Hb. For both sets of simulations, pulling a distance of 15 Å was sufficient to completely disrupt each interface, while Hb contacts at the nonpulled interface remained intact (Fig. 3a and b). The force required to separate each interface was integrated as a function of the distance pulled, which demonstrated that when the domains were pulled 15 Å, the work required to separate N2 from Hb was 1.5 times more than the work required to separate LN3 domains from Hb’s heme pocket (Fig. 3c). We note that the total work estimated computationally is greater than the true binding affinity due to the nonequilibrium component introduced by the fast pulling speeds that are required in the simulations. Regardless, the fact that LN3 separation consistently required significantly less work than N2 separation strongly suggests that the former has a weaker interface with Hb than the latter. Interestingly, Hb contacts formed by the N2 domain broke fairly evenly as a function of distance, while contacts formed by the LN3 domain broke unevenly. The cause of this unevenness originates from contacts made by the β7-β8 turn (residues Val636-Glu643), which consistently were the last to separate from Hb within the Hb-LN3 interface (Fig. 3b, dotted line). This suggests that contacts to Hb from this region are most important for forming the weaker Hb-LN3 interface. We conclude from the MD and NMR data that the receptor retains its dynamic nature when bound to Hb: the receptor is affixed to the globin chain via contacts originating from the N2 domain that cause its NMR resonances to broaden, while intradomain motions at the hinge between the N2 and linker domains enable the LN3 domains to transiently engage Hb’s heme pocket via weaker interactions causing the Hb-LN3 interface to be sparsely populated in the complex.
Fig. 3. Results from steered MD calculations.
Each set is distinguished by color. (a,b) Contacts between N2 or N3 and Hb as a function of separation distance. Hb:N3 contacts involving β7–β8 are shown as a dotted line in (b). (c) The total work required to separate the IsdH domain and Hb as a function of distance. The black dotted line in each panel represents a separation distance of 15 Å; at this distance, the pulled domain has lost nearly all contact with Hb. The work required to pull N2 15 Å is ~1.5x greater than to pull N3.
Two spatially distinct subsites within the Hb-LN3 interface trigger heme release from Hb
The Hb-LN3 interface in the crystal structure of the IsdHN2N3:Hb complex presumably resembles a pretransfer complex in which heme bound to Hb is poised for transfer to the receptor. We determined the functional importance of receptore–Hb contacts within this interface using a recently developed stopped-flow UV-Vis transfer assay that quantitatively measures the rate of ferric heme extraction from the α-subunit within tetrameric Hb [23]. Single alanine mutations at the Hb-LN3 interface were introduced into a variant of IsdHN2N3 that preferentially binds to the α-subunit of Hb because it contains alterations in the N2 domain that selectively weaken its affinity for the β-subunit (αIsdH, IsdHN2N3 containing F365Y and A369F substitutions within its N2 domain) [25]. The assay also employs the aforementioned Hb0.1 variant that has a significantly lower propensity to dissociate into αβ Hb dimers whose presence complicates kinetic analyses. For each αIsdH mutant, the kinetics of heme capture were measured using a 30-fold molar excess of receptor to ensure saturation (5 and 150 μM of Hb and receptor, respectively) [25]. Representative transfer data recorded upon adding heme-free receptor (hereafter called apo-αIsdH) to Hb0.1 shows an expected rapid shift in Hb’s UV/Vis spectrum indicating heme transfer from Hb0.1 to the receptor (Fig. 4a). The spectral changes exhibit biphasic kinetics, with approximately half of the total magnitude of the absorbance change at 405 nm characterized by a rapid event defined by the rate constant kfast and the remaining, much slower changes, described by kslow (Fig. 4b). As described previously, kfast is a direct measure of the rate of heme transfer from Hb0.1 to the receptor, while kslow measures the much slower indirect process of heme transfer in which Hb first releases heme into the solvent before it is subsequently bound by the receptor [25].
Fig. 4. Stopped-flow measurements of heme transfer.
A) Representative stopped-flow UV/Vis spectra collected with 2 mm pathlength over 1000 s from rapid mixing of 20 μM Hb0.1 and 150 μM apo-αIsdH in 20 mM NaH2PO4, 150 mM NaCl, 0.45 M sucrose, pH 7.5. Bold traces in blue and orange represent the reaction at 0.003 s (mainly holo-Hb0.1) and 1000 s (mainly holo-αIsdH), respectively. B) Single-wavelength kinetic time courses of αIsdHN2N3 and representative transfer-impaired alanine mutants. Segments of the time courses pertaining to kfast and kslow are highlighted. C) Histogram of kfast values (shown as a % relative to wild-type (WT) αIsdH) obtained for each alanine mutant on the Hb:LN3 interface, assayed at 150 μM receptor:5 μM Hb0.1. Error bars represent standard deviation of kfast from triplicate trials. D) Representative Eyring plot obtained for variant V637A from data collected in triplicate.
The process of heme removal monitored by the assay can be represented by Scheme 1, where k1, k−1, and ktrans are the micro-rate constants of the individual reaction steps that describe the process of heme removal from the α-subunit. During the reaction, the receptor first binds to Hb’s α-subunits via the N2 domain (k1 and k-1) and heme is then removed from Hb via formation of the Hb-LN3 interface (ktrans). Because IsdH binds ferric heme with much greater affinity than Hb, transfer is effectively irreversible. Our studies have shown that kfast increases hyperbolically with increasing receptor concentrations, such that ktrans describes the rate-limiting step in the transfer reaction when saturating amounts of receptor are used [23]. In particular, at the Hb:receptor ratio used in our experiments, the contribution of k−1/k1 to kfast becomes negligible, such that kfast ≈ ktrans.
Scheme 1.
Kinetic scheme for the process of direct αIsdH-mediated heme transfer from the α-subunit of Hb.
Heme transfer kinetics were measured for 14 single amino acid mutants of the αIsdH receptor that alter residues within Hb-LN3 interface: residues whose side chains are located within 5 Å of Hb’s heme pocket in the crystal structure of the IsdHN2N3 :Hb complex. The mutated residues are located either within the N3 domain and form a ring that surrounds Hb’s heme pocket or within helix H2 of the linker domain that contacts Hb’s F-helix. The results of these experiments are summarized in Fig. 4c and Table S1. In general, with the exception of the S561A and Y646A mutants, all of the alterations in the receptor decrease the rate of heme transfer. However, four receptor mutants exhibit pronounced reductions in kfast as compared to αIsdH: R492A, Y495A, V637A, and I640A mutants. Interestingly, these functionally important residues cluster to two subsites within the Hb-LN3 interface that are separated from one another by approximately 20 Å (highlighted in Fig. 1). The first subsite contains the R492 and Y495 receptor side chains located in the linker domain that contact the center of Hb’s F-helix (linker subsite), while the second is located in the N3 domain and contains residues V637 and I640 that contact the C-terminal end of the F-helix (N3 subsite). Mutating both residues within each subsite does not substantially reduce the rate of heme capture as compared to the corresponding single amino acid mutants. However, a quadruple αIsdH mutant (R492A/Y495A/V637A/I640A, dubbed QM) collectively alters all of the side chains within the two subsites and is significantly impaired in capturing heme from Hb0.1 (~20-fold more slowly than αIsdH at 25 °C) (Table S1). As expected, when extraction from native adult hemoglobin (HbA) is probed, the effects of the subsite mutations are even more pronounced, with the rate of heme capture by QM slowed ~50-fold relative to αIsdH (Fig. 4c). Importantly, all mutations in QM only alter surface residues that are located distal to the main Hb-N2 binding site and do not alter its capacity to bind heme. This is substantiated by the 1H–15N TROSY spectrum of the transfer impaired QM that reveals that it is folded and adopts a structure that is similar to the wild-type receptor (Fig. S4). Thus, our results show that contacts originating from two distally positioned subsites within the transient Hb-LN3 interface act to trigger heme release.
Fig.5. Summary of Hb: LN3 interface alanine scan and two subsites identified.
A)Surfacerepresentationof aIsdHY642A in complex with aHb (dotted line). Mutated residues on the LN3:Hb interface are colored according to their impact on heme transfer rate relative to wild-type aIsdHN2N3, from blue (minimal impact) to red (high impact). B) Close-up of subsites identified by alanine scanning mutagenesis. The linker (purple) and N3 (green) subsites flank either end of the aHb F-helix (L86-L91, shown in pink), which adopts a distorted conformation stabilized by interactions from each of these surfaces.
An Erying analysis indicates that the two subsites have energetically distinct roles in heme transfer
To gain insight into how the receptor subsites trigger heme release, the stopped-flow experiments were performed at temperatures ranging from 15 to 37 °C. The data were then analyzed using the linearized form of the Eyring equation,
| (1) |
where kB, h, and R are the Boltzmann, Planck, and ideal gas constants, respectively, while ΔH‡ and ΔS‡ are the activation enthalpy and entropy, respectively. Representative data is presented in Fig. 4d. Transfer to native αIsdH is primarily limited by an enthalpic barrier with ΔH‡ and ΔS‡ values of 19.7 ± 0.8 kcal/mol and 5.4 ± 0.3 cal/mol,K, respectively [23,25]. Activation energy measurements were made for receptor mutants that altered either one or both amino acids within each subsite: R492A, Y495A, and R492A/Y495A in the linker subsite, as well as V637A, I640A, and V637A/I640A in the N3 subsite (Table 1). Interestingly, these studies revealed that contacts originating from the linker and N3 contact subsites affect the transfer activation energy in fundamentally different ways. Altering contacts from the N3 subsite primarily increases the activation free energy by causing an unfavorable decrease in ΔS‡ that is incompletely counterbalanced by a slight reduction in ΔH‡. In marked contrast, single-site alterations to the linker subsite (R492A and Y495A mutations) substantially increase ΔH‡ relative to αIsdH, while ΔS‡ remains virtually unchanged. As expected, a V637/I640A double mutant of the N3 subsite primarily decreases ΔS‡, an effect that is similar to the single parent mutants. However, an R492/Y495A double mutant that alters both contacts originating from the linker subsite has a distinct effect as compared to the parent mutants; the R492/Y495A double mutant exhibits starkly unfavorable decreases in ΔS‡ and favorable decreases ΔH‡ relative to αIsdH. A discussion of the potential physical basis of these effects is presented in the Discussion section.
Table 1. Summary of heme transfer energetic parameters.
Temperature-dependent kinetics were measured in triplicate and linear fits obtained from the Eyring equation. Values obtained from fitting to a double-exponential expression from 0.003 – 1000 s. Plots were generated from mean triplicate values. Linear regression of 1/T vs. ln(kfast/T) yielded values for slope (ΔH‡) and y-intercept (ΔS‡) for each variant tested. The differences between the Eyring plots were evaluated in GraphPad PRISM by one-way ANOVA. The calculated lines are significantly different (P <0.0001).
| Mutant | ΔH‡ (kcal/mol) | ΔS‡ (cal/mol•K) | ΔG‡37°C (kcal/mol) |
|---|---|---|---|
| WT αIsdH | 19.7 ± 0.8 | 5.4 ± 0.3 | 18.0 |
| R492A | 20.9 ± 0.5 | 6.1 ± 0.2 | 19.0 |
| Y495A | 20.5 ± 0.8 | 6.0 ± 0.3 | 18.6 |
| V637A | 14.2 ± 0.5 | −14.8 ± 0.7 | 18.8 |
| I640A | 13.1 ± 0.9 | −19.5 ± 2.2 | 19.1 |
| [R492A + Y495A] | 12.0 ± 0.8 | −23.5 ± 2.5 | 19.3 |
| [V637A + I640A] | 15.5 ± 1.5 | −11.6 ± 1.6 | 19.1 |
| QMa | 11.9 ± 1.3 | −26.5 ± 5.7 | 19.2 |
Values for quadruple mutant were obtained by constraining the plateau absorbance value to that obtained at 37°C to account for incomplete reaction at 1000 s at lower temperatures.
MD simulations probe how the subsites promote heme release
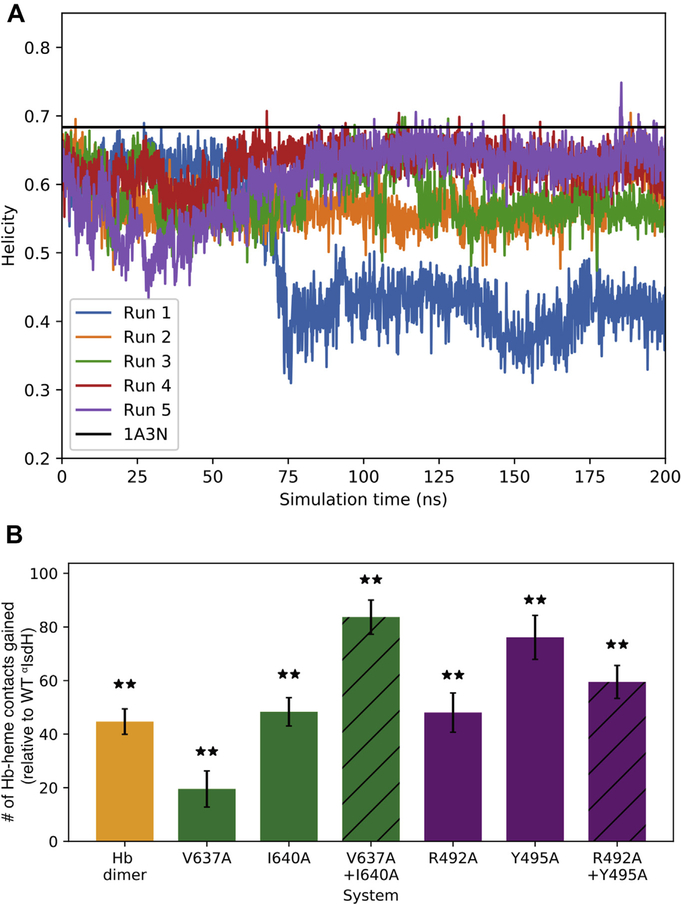
To learn how the receptor subsites participate in heme capture, we performed MD simulations of dimeric αβ Hb bound to either αIsdH or mutant αIsdH receptor proteins. As a starting point, we created a structure of αIsdH (taken from PDB code 4XS0) bound to a native, undistorted αβ dimer Hb (taken from PDB code 1A3N). Five 200 ns MD simulations were performed and the helicity of Hb’s F-helix during the MD simulation was calculated (described in Methods and Materials). As shown in Fig. 6a, in each of the five trajectories, the C-terminal end of the F-helix near the subsites became distorted, with significant loss of helicity occurring within 100 ns. However, the F-helix did not distort in the same manner each time–multiple disordered states for residues in the F-helix were observed when the receptor is bound. This may mean that full receptor-stabilized distortion occurs on microsecond or longer times cales and/or that multiple isoenergetic configurations of the distorted F-helix exist when the receptor is bound. Interestingly, the F-helix appears to be intrinsically unstable in dimeric αβ Hb, since simulations of this protein in the receptor-free state also revealed helix unwinding (Fig. S1). We therefore sought to more rigorously define the impact of receptor binding by determining how it affects the conformation of the hydrophobic cage in Hb that shields the FeIII–His bond from solvent and autooxidation [48]. The cage in the α-subunit is formed by six residues: Leu83, Leu86, Leu91, Val93, Phe98, and Leu136 (shown in Fig. S2). The simulations were analyzed to determine the number of contacts between the six-residue cage and heme in either the αIsdH:Hb complex or the receptor-free αβ Hb dimer (Fig. 6b). Consistent with experimental kinetic data that show that receptor binding accelerates heme release, fewer heme–Hb contacts were observed in the αIsdH:Hb complex as compared to native Hb; the native Hb dimer (yellow, Fig. 6b) has ~45 more contacts than when αIsdH is bound. Thus, even though the F-helix is unstable in the native Hb dimer, αIsdH binding further compromises the hydrophobic cage and exposes the FeIII–His bond to solvent. This exposure is presumably required for axial bond rupture during transfer and agrees with our previous study on αIsdH:Hb (PDB code: 4XS0), which found αIsdH allows water to favorably approach the FeIII–His bond [23]. We wondered if receptor mutants that exhibit slowed heme capture in our experimental studies are also impaired in their ability to disrupt cage–heme contacts. To address this issue, simulations were performed as described earlier for six distinct αIsdH receptor mutants (R492A, Y495A, R492A/Y495A, V637A, I640A, V637A/I640A). Indeed, for each mutant, fewer cage–heme interactions are disrupted by the receptor explaining why they exhibit slower capture kinetics (Fig. 6b).
Fig. 6. Structural effects of IsdH on Hb.
A) Measured helicity of the eight C-terminus F-helix residues for the WT system. Each of the three trajectories is drawn, as well as the helicity value for the used apo-Hb crystal structure (PDB: 1A3N). Reduction in the helicity occurs quickly (<25 ns) and equilibrates around 100 ns. B) Difference in the hydrophobic packing for the cage (shown in Fig. S3) surrounding Fe–His bond after F-helix distortion compared to the WT αIsdH system. Significantly different values (p < 0.01) are marked with a double asterisk.
To gain physical insight into why receptor mutations at different subsites have distinct effects on the entropic and enthalpic components of the activation energy of transfer, we analyzed the results of our simulations to answer two questions: i) how do the mutations affect receptor structure and motions? ii) how do the mutations alter receptor–Hb interactions? To determine how interfacial mutations affected the structure and dynamics of each subsite, we calculated the root-mean-square fluctuation (RMSF) of their α-carbon atoms. Interestingly, the RMSF of residues within the N3 subsite reveals that they exhibit the highest degree of flexibility within the N3 domain (excluding its C-terminus), and this mobility increases when residues in the N3 subsite are mutated (Fig. S3). To gain a detailed picture of this flexibility, PCA was performed yielding two eigenvectors that describe a bending and twisting motion of the β7–β8 turn (first and second components, respectively), while the rest of the N3 structure remains static (Fig. 7a, top; SI Movie 1). A histogram of these components (Fig. 7a, bottom) shows that the native β7–β8 turn in αIsdH (blue) primarily undergoes two types of orthogonal motions when bound to Hb. In contrast, mutations in the N3 subsite result in an adjustment in the β7–β8 turn configuration and flexibility, as demonstrated by the flatter and broader histograms. Interestingly, mutating both N3 subsite residues did not further enhance flexibility compared to mutating a single site–suggesting that flexibility is increased by a single disruption of hydrophobic packing interactions within the N3 subsite. As described in greater detail in the Discussion section, this finding suggests that mutations in this region increase the inherent flexibility and disorder of the ground state of the receptor leading to increases in ΔS‡. In contrast, mutants that alter residues within the linker subsite do not exhibit increased localized flexibility as evidenced by their small-magnitude RMSF values (data not shown). This is compatible with single-site mutations in this region not significantly affecting ΔS‡.
Fig. 7. Effects of mutations on Hb: αIsdH system.
A) Results from principal component analysis on the N3 structure before and after N3 mutations. Structures along the first two components are shown (top) as well as the histogram of each component for the mutated N3 systems (bottom) (see also SI Movie 1). In the figure, cartoon images of Hb and the receptor are colored pink and brown, respectively, while residues in the N3 subsite are colored green. Similar analysis for mutated linker systems is shown in Fig. S2. B) Difference in total (blue) and all hydrophobic (orange) contacts between Hb and LN3 compared to the WT system. Standard error is shown in the black error bars. Each mutation reduces the total number of interprotein contacts, while N3 mutants also reduce hydrophobic contacts. Significantly different values (p < 0.05) are marked with an asterisk while highly different values (p < 0.01) are marked with a double asterisk. Standard error for the WT measurements were 17.0 and 5.5 pairs for total and hydrophobic contacts, respectively. C) Average contact between residues in the linking domain (abscissa) and Hb (ordinate) for the WT (top, left) and mutated linker (remaining panels) systems. A line is drawn to indicate the location of the single/double mutation. The total number of contacts between these two regions as well as the number of pairings is provided in the top right of each panel.
Supplementary video related to this article can be found at https://doi.org/10.1016/j.jmb.2019.12.023
To better understand how receptor mutations affect Hb–receptor interactions, a contact analysis of the simulation data was performed. As shown in Fig. 7b, during the simulations, the average number of contacts between Hb and the LN3 portion of the receptor is reduced in each mutant relative to the wild-type. This suggests that mutations in either subsite have a destabilizing effect on the IsdH:Hb complex as a whole; indeed, this destabilization is also seen in estimated Hb-LN3 interaction energies from MM/GBSA (Table S2). While mutating either subsite reduces the number of hydrophobic contacts to Hb, the largest reductions occur when the N3 subsite is altered. This is presumably because N3 subsite mutations disrupt packing interactions in the β7–β8 turn, which contribute significantly to the Hb-LN3 interaction based on MD analyses (Figs. 7a,3b). A more detailed contact map between the linker subsite and Hb is shown in Fig. 7c, which enumerates specific residue–residue interactions that occur as the simulations progress for systems containing either the wild-type or N3 mutant receptors. Contact effects are most pronounced for the R492A mutant, since its mutation causes surrounding residues in the linker to form new, less frequently sampled contacts with Hb (compare wild-type with R492A data in Fig. 7c). For example, D85 and L86 form a strong contact with R492; when R492 is mutated, these two residues form new interactions with the remaining Y495, but also with E498 and K499. Note that in this case, the total number of contacts between the linker and F-helix remains consistent with wild-type αIsdH; this is not the case when Y495 is mutated. Moreover, when both residues are mutated to alanine, the region as a whole fails to strongly interact with Hb’s distorted F-helix.
Discussion
Many bacterial pathogens display or secrete Hb receptors that are important virulence factors because they enable microbes to gain access to vital iron located within Hb’s heme molecules [36,49]. While the structures of several microbial receptors have now been determined, it is not known how they extract Hb’s heme, which must involve surmounting a significant energetic barrier as Hb binds to ferric heme with very high affinity (0.8 pM and 4.2 pM for tetrameric α and β Hb, respectively) [50]. The S. aureus IsdH protein has become a model system in which to understand the mechanism of extraction, as atomic structures of this Hb receptor in its free state and in complex with Hb have been determined [19,24,25,40,41]. In this study, we employed a newly developed stopped-flow assay, molecular simulations, and NMR methods to identify specific receptor interactions that trigger heme release, to learn how they function to stabilize the heme-transfer transition state and whether these interactions occur within a static or dynamic complex.
Our results obtained using NMR spectroscopy reveal that IsdH forms a dynamic complex with Hb that retains interdomain motions that cause the Hb-LN3 heme transfer interface to form transiently in solution (Fig. 8b). This is evident from Hb NMR titration data using a 13C-methyl labeled sample of IsdHN2N3, which revealed selective chemical shift and line broadening changes for methyl groups located only in the N2 domain and minimal changes within the receptor’s LN3 portion (Fig. 2). As saturating amounts of Hb did not cause significant spectral changes, we conclude that the Hb-LN3 interface present in crystallo is only minimally populated in solution [25]. It seems likely that even when bound to Hb, intradomain motions between the N2 and LN3 portions of the receptor persist. This would enable the Hb-LN3 interface to transiently engage Hb’s heme pocket, while the N2 domain stayed fully engaged with Hb’s A-helix. This notion is actually compatible with the crystal structure of the IsdHN2N3:Hb complex as electron density for eight residues that connect the N2 to LN3 segments is missing (residues Asn465–Glu472) [25]. This suggests that they could function as hinge point for domain reorientation when the receptor is bound to Hb and that crystal packing interactions are likely responsible for holding the LN3 portion near Hb. The idea that a dynamic receptor engages Hb via high- and low-affinity interfaces is supported by our steered MD simulations that show that within the context of the Hb-IsdHN2N3 complex, it takes 1.5-times more work to separate the N2 portion of the receptor from Hb versus the LN3 portion (Fig. 3c). It is also compatible with experimental Hb-binding affinity measurements that employed polypeptide fragments of the receptor that contained either the N2 or LN3 portions of the receptor [19]; polypeptides containing the LN3 portion bind the receptor with weaker affinity as compared to N2, presumably because binding is accompanied by unfavorable endothermic distortions of Hb’s F-helix. Based on our NMR finding that minimal perturbations are observed in LN3 when excess Hb is added to IsdH, we estimate that in solution even when the N2 domain of the receptor engages Hb via its A-helix, less than 5% of the bound receptors engage and distort the F-helix via the weaker Hb-LN3 interface. Additional studies are needed to define the time scales associated with receptor engagement via the Hb-LN3 interface and the role of interdomain motions in this process.
Fig. 8. Mechanistic hypothesis of IsdH heme extraction.
A) Interactions from IsdH subsites that interact with the αHb F-helix (pink) to stabilize its distorted conformation. B) Multistep reaction of IsdH with Hb:αIsdH associates with Hb through the high-affinity interface of N2, while LN3 remains dynamic in solution. Association of LN3 with Hb is concurrent with F-helix distortion, which is promoted through interactions with each subsite (linker and N3 subsites shown in purple and green, respectively). LN3 association and F-helix distortion allows for heme transfer into the IsdH pocket. C) Conceptual schematic of the heme transfer step: The linker subsite provides interactions that disrupt Hb:heme affinity, while the N3 subsite provides a transiently formed bridging platform over which direct heme transfer occurs.
Stopped-flow experiments reveal that contacts originating from two distally positioned subsites within the receptor trigger heme release from Hb. In the crystal structure of the IsdHN2N3:Hb complex, the heme pockets within each protein are joined via an extensive Hb-LN3 interface that buries 1235 Å2 of solvent-exposed surface area. Because Hb’s F-helix is distorted and heme remains bound to Hb in the crystal structure, we reasoned that the structure may resemble the high-energy transition state that is stabilized by the receptor to promote heme release. Indeed, using alanine scanning mutagenesis and a newly developed transfer assay designed to monitor only heme extraction from only the α-chain of tetrameric Hb [23], we identified two functionally important contact points within the Hb-LN3 interface. They are located at distinct ends of Hb’s F-helix (Figs. 1 and 8). A subsite on the linker domain of the receptor contacts residues in the F-helix positioned immediately N-terminal to the FeIII coordinating His87 residue, while a N3 subsite contacts the C-terminal end of the F-helix. Mutation of these sites can have a dramatic effect, reducing the rate of heme capture from native Hb up to 50-fold. The rate reductions exhibited by these mutants can be attributed to defects in their ability to stabilize the heme transfer transition state, as the altered side chains are solvent-exposed and located distal to the Hb-N2 interface that mediates high-affinity receptor binding and because they do not significantly affect the structure of the receptor (Fig. S4). Collectively, our mutagenesis data are consistent with the idea that the crystal structure of the IsdHN2N3:Hb complex resembles the heme transfer transition state and suggest that F-helix distortion precedes, or is concurrent with, the rate-limiting step in this reaction [25].
Eyring and MD analyses suggest that contacts from the linker and N3 subsites have divergent roles in the heme transfer reaction. The primary function of the linker subsite is presumably to weaken Hb’s affinity for heme. This is evident from single alanine point mutations (R492A and Y495A), which reduce the rate of transfer by increasing ΔH‡ (20.9 ± 0.5 and 20.5 ± 0.8 kcal/mol, respectively) (Table 1). A comparison of the coordinates of Hb in its free and receptor-bound states reveals that residues within the linker subsite (R492 and Y495) likely stabilize a distorted state of the F-helix by interacting with the side chains of D85 and L86 (Fig. 8a). When the receptor is bound, the side chain of L86 in Hb is rotated away from its helical position, presumably weakening Hb’s affinity for heme because it disrupts van der Waals interactions between the L86 side chain and the porphyrin ring, as well as packing interactions that maintain Hb’s hydrophobic cage that retains heme (L86 is part of the cage) [25]. Rotation of the L86 side chain also disrupts interactions with the H87 side chain, likely perturbing the coordination geometry of the FeIII–H87 bond to reduce its strength. Therefore, because the R492A or Y495A mutants truncate interactions with D85 and L86, they exhibit an increase in ΔH‡ because they are less able to destabilize heme–Hb interactions. This is supported by our molecular simulations, which show that the R492A or Y495A mutations significantly reduce the contacts formed with D85 and L86 (Fig. 7c), while generally not affecting the level of structural disorder in this portion of the interface.
Interprotein contacts localized at the N3 subsite appear to have a dual function in heme transfer, destabilizing Hb–heme interactions and creating a heme transfer platform. Complex formation causes significant conformational changes in both the receptor and Hb near the N3 subsite. A comparison of the structure of Hb in its free and receptor-bound states reveals that the C-terminal end of the F-helix partially unravels, enabling its H89 and L91 side chains to form a bridge with the N3 subsite (Fig. 8c). The N3 subsite is located within the turn connecting strands β7 and β8 (β7–β8 turn) and is formed by the side chains of V637 and I640 that contact one another. During transfer, heme likely moves across the hydrophobic surface formed by these side chains as it slides into the receptor where it forms a FeIII–Y642 bond. The side chains of V637 and I640 may also stabilize heme after it has arrived at the receptor, as their side chains pack against the edge of the porphyrin ring in the crystal structure of isolated N3eheme complex [40]. Interestingly, when the structures of N3 domain are compared in its heme-free and Hb-bound states, it is apparent that the β7–β8 turn unfurls, undergoing a large 7 Å displacement toward Hb to form a heme transfer bridge. MD simulations suggest the β7–β8 turn is dynamic within the IsdHN2N3:Hb complex, sampling the unfurled state via rigid body motions that are described by two principal components. Our results show that single alanine substitutions in the N3 subsite cause large unfavorable effects on ΔS‡; V637A and I640A mutants have ΔS‡ = −14.8 ± 0.7 and −19.5 ± 2.2 cal/mol•K, respectively, which is significantly lower than the wild-type protein (ΔS‡ = 5.4 ± 0.3 cal/mol•K) (Table 1). We reason that these unfavorable changes in ΔS‡ are caused by an increase in the entropy of the ground state of the N3 subsite–in the mutants there is a larger conformational ensemble prior to forming the ordered bridge through which heme moves from Hb to the receptor. Thus, a larger entropic penalty needs to be paid in the V637A and I640A mutants as they transition from a disordered-to-ordered conformation during the transfer reaction. Simulation data support this assertion; both hydrophobic and overall interactions with Hb are decreased in simulations where V637 and/or I640 are mutated (Fig. 7b). Moreover, PCA of the V637A and I640A mutations reveals they have significantly increased flexibility in this region, as evidenced by larger RMSF values and flatter PCA histograms (Fig. S3, Fig. 7a). N3 subsite interactions that unravel the C-terminal end of F-helix also promote transfer by destabilizing heme–Hb interactions, as the L91 side chain that is contacted by this subsite is also part of Hb’s hydrophobic cage and is supported by an Hb–heme contact analysis of the simulation data (Fig. 6b). Alternative explanations for the unfavorable changes in ΔS‡ in the V637A and I640A mutants are also possible. In particular, as alanine substitution reduces the size of the hydrophobic platform formed by these residues, they could decrease ΔS‡ by reducing the number of transfer competent bridging geometries within the transition state, or potentially they could leave gaps in the bridge, resulting in unfavorable increased solvation of nonpolar portions of heme in the transition state.
The Hb-LN3 interface in the IsdHN2N3:Hb complex is dynamic. In addition to the aforementioned transient unfurling of the N3 subsite to form the heme transfer bridge, MD simulations suggest that Hb’s F-helix is dynamic, sampling a range of partially unwound conformations that are stabilized by contacts to the receptor subsites (Fig. 6). The notion that the receptor stabilizes a distorted state for the F-helix is supported by our simulations of the native and mutant forms of the receptor bound to native Hb. These results show that the F-helix interchanges between fully helical and partially unwound states on the nanosecond time scale when it is positioned within the Hb-LN3 interface (Fig. 6a). One of these distorted conformers has presumably been visualized in the crystal structure of the IsdHN2N3:Hb complex, since as compared to the structure of unbound Hb, in this complex the angle of the F-helix is altered by ~10° and its contacts with heme are disrupted. Two analyses of the simulation data indirectly indicate that receptor subsites stabilize distorted forms of Hb’s F-helix to promote heme release. First, mutant receptors that truncate side chains within each subsite form fewer contacts with Hb during the simulations, suggesting that they are less capable of stabilizing its distorted structure (Fig. 7b and c). Second, as compared to the wild-type receptor, the mutant receptors are impaired in their ability to disrupt Hb’s hydrophobic cage–a key structural element that promotes heme binding to Hb and shields the axial FeIII–H87 bond from solvent (Fig. 6 and S2) [23,25,48]. We conclude that Hb contacts from the functionally important subsites promote heme release by stabilizing distorted conformers of the F-helix that are dynamically sampled within the Hb-LN3 interface.
It seems likely that the S. aureus IsdB protein uses a similar mechanism to extract ferric heme from Hb as it shares significant primary sequence homology with IsdH and it also employs a similar tridomain unit [12]. Notably, residues within the linker and N3 subsites that we have identified in IsdH as being functionally important are conserved in IsdB, suggesting that they play similar roles in heme extraction. Interestingly, a recent structure of IsdB bound to Hb may have visualized a later step in the heme transfer reaction in which the FeIII–H87 bond has been broken and the heme molecule is partially transferred to the receptor [43]. Consistent with our transfer model for IsdH, in the structure of the IsdB-Hb complex, the analogous N3 subsite located in the β7–β8 turn of IsdB presents a hydrophobic platform for accepting heme, with N3 subsite residue I438 (analogous to I640 in IsdH) contacting the porphyrin plane. Moreover, the linker subsite in IsdB directly engages the side chain of H87 in Hb, which is rotated out of the heme pocket because the FeIII–H87 bond has been broken. Intriguingly, in the Hb-IsdB crystal structure, heme forms an atypical hemichrome, exhibiting bis-His metal coordination with the His58 and His89 imidazole rings that are typically positioned at the edge of the Hb’s heme binding pocket. Hemichromes exhibit distinct UV-Vis absorbance signals, which were not observed during active transfer in the IsdB study and could be not detected by us in our studies of the IsdH heme transfer reaction. Thus, it is possible that the hemichrome may be too short lived during the transfer reaction to be detectable or that it is an artifact that was generated during the crystallization of the Hb-IsdB complex [43].
To our knowledge, the only heme-transfer reaction other than IsdH/B that has been studied at atomic detail is the HasA-HasR system, which transfers heme from the HasA hemophore to the HasR receptor embedded in the outer membrane of S. marescens [51]. Similar to IsdH, the HasR receptor must overcome the high heme affinity of its donor HasA, which like Hb releases heme slowly into the solvent unless prompted by a receptor [52].However, the IsdH/B and HasR proteins accomplish this task through fundamentally distinct ways. In the HasA-HasR system, the transfer reaction is endergonic, as heme is transferred from a high-affinity binding site in HasA to a lower affinity site in HasR. To drive this thermodynamically unfavorable transfer reaction, HasR leverages a strong binding interface with HasA to sterically disrupt the axial coordination of heme by HasA, presumably weakening HasA’s heme affinity [52]. In contrast, heme transfer in the IsdH system is exergonic, as the receptor (IsdH) has higher affinity for ferric heme than the donor (Hb). As a result, IsdH does not need to use high-affinity protein–protein interactions to drive transfer of heme, but rather forms a transient Hb-LN3 interface whose function is to increase the rate at which heme is released from the donor and to form a conduit through which heme can move to the higher affinity binding site in the receptor. Thus, heme is transferred between distinct static and dynamic complexes that serve different biological purposes. The high-affinity static HasA-HasR complex delivers heme to the outer membrane where it remains in HasR until the TonB system powers heme transfer into the periplasm. In contrast, formation of the dynamic IsdH:Hb complex enables surface-attached IsdH receptor to extract heme and then release Hb into the milieu. IsdH then holds heme on the cell wall exterior, acting like a sponge that releases heme very slowly until it is contacted by downstream NEAT containing hemoproteins in Isd-system (IsdA and IsdC) that take the heme and transfer it across the expanse of cell wall to the membrane for import into the cytoplasm [14,26,28,29].
The findings in this study provide new insight into the mechanism of heme extraction. NMR, MD pulling experiments, and affinity measurements of isolated components of the receptor indicate that the Hb-N2 interface acts to tether the receptor to Hb’s globin via contacts that are distal to the site of heme transfer. This enables the weaker Hb-LN3 interface to form over the heme pocket in which Hb’s F-helix adopts a range of distorted conformations that interchange on the nanosecond time scale. NMR data are consistent with the receptor forming a dynamic complex with Hb, with interdomain motions between the N2 and LN3 portions of receptor persisting after Hb is bound via the N2 domain, such that the weaker Hb-LN3 interface forms transiently and is sparsely populated. Based on the structural, kinetic, and MD analyses, formation of a transfer-competent Hb-LN3 interface requires two key steps: 1) destabilization of the Hb–heme pocket to promote heme removal (mediated by both the linker and N3 subsites), and 2) creation of a temporary “transfer platform” to permit heme entry into the receptor’s heme-binding pocket (mediated through the N3 subsite). At the heart of this mechanism is the transient uncoiling of the F-helix, stabilized at one end by the linker subsite and at the other by the N3 subsite. These events set the stage for transfer, sliding of heme into the N3 pocket where the FeIII–Y642 bond forms. The sliding mechanism likely entails additional conformational changes in the β7–β8 turn, which based on modeling studies needs to be displaced by several angstroms to accommodate heme movement, as well as rotation of the heme molecule as it transits to enable its propionate groups to project into the solvent after it is bound by the receptor. Interestingly, this heme transfer bridge is also in motion in the complex, transiently sampling conformations that are presumably competent for heme transfer. Transfer is effectively unidirectional as IsdH binds ferric heme with significantly higher affinity than Hb based on competition studies with apo-Mb [23]. Our mechanism is consistent with other systems that have shown that protein–protein induced disruptions that reduce affinity are a primary method to mobilize heme [53]. Further, they expand upon this concept by demonstrating that destabilizing interactions that trigger heme release can occur within flexible, low-affinity interfaces. Such dynamic interfaces are capable of reconfiguring into productive geometries for heme extraction and may be pervasive in biology.
Experimental
Cloning and protein expression
IsdHN2N3 variants were expressed from pET28b-based plasmids containing an N-terminal hexahistidine small ubiquitin-like modifier (SUMO) fusion tag to facilitate purification. Previously reported protein-expressing plasmids used herein include the following: pRM216 (Ala326–Asp660 IsdHN2N3) that contains a Y642A substitution (IsdHN2N3–Y642A); pMMS322 (Ala326-Asp660) containing a365FYHYA 369 → 365YYHYF369 substitution to confer specificity for αHb subunits (αIsdH). QuikChange site-directed mutagenesis (Agilent Technologies) was used to introduce mutations into the αisdh gene. Crude plasmid mixtures from QuikChange protocols were purified by Zymo Clean & Concentration (Zymo Research) kit and transformed into chemically competent E. coli BL21 (DE3) cells. Protein expression and purification procedures have been described previously [23]. Briefly, 2 L LB/kanamycin expression cultures of E. coli containing αIsdH-expression plasmid were grown to OD600 = 0.6 and expression was induced at 37 °C by adding 1 mM isopropyl-β-D-thiogalactoside (IPTG) to cell cultures. Expression was conducted overnight at 37 °C. The bacterial cells were then harvested by centrifugation and lysed by sonication, and the protein was purified using a HisPure Co2+-chelating column (Thermo Fisher Scientific). The N-terminal His6-SUMO tag was then cleaved using the ULP1 protease, and the mixture was reapplied to the Co2+-chelating column to remove the protease and cleaved SUMO tag. The heme-free forms for the IsdH proteins were generated by heme extraction with methyl ethyl ketone followed by buffer exchange into 20 mM NaH2PO4, 150 mM NaCl, pH 7.5.
Recombinant genetically stabilized human hemoglobin was expressed from plasmid pSGE1.1-E4 in E.coli BL21 (DE3) cells as described previously [23]. Cells were grown in M9 minimal media containing 3 g/liter glucose and 1 g/liter NH4Cl as the sole source of carbon and nitrogen, respectively. Prior to induction, exogenous hemin was added to the cell cultures to a final concentration of 8 μM. Expression was induced at OD600 = 0.6 and performed at 16 °C overnight by adding IPTG to cell cultures to achieve a final concentration of 1 mM. Cells were then harvested by centrifugation and washed with lysis buffer (50 mM Tris-HCl, 17 mM NaCl, pH 8.5) twice to remove excess hemin. The cell suspension was then purged with a steady stream of CO for 15 min in an ice-water bath. All buffers used during the purification were purged with CO to prevent heme oxidation. The cells containing recombinant Hb0.1 were lysed by sonication and purified in a single step using a Co2+-chelating column (Thermo Fisher Scientific) via interactions with the naturally occurring His residues on the surface of Hb0.1. The column was washed using three buffers prior to elution with imidazole: 5 column volumes (CV) of lysis buffer (see above), 8 CVs of wash buffer 1 (20 mM Tris-HCl, 500 mM NaCl, pH 8.5), and 8 CVs of wash buffer 2 (20 mM Tris-HCl, pH 8.5). Column-bound Hb was then eluted with five CVs of elution buffer (20 mM Tris-HCl, 100 mM imidazole, pH 8.5). Ferric Hb0.1 (metHb0.1) was used in the heme transfer experiments. To oxidize the protein, CO-Hb0.1 was diluted to 50 mM concentration (heme units) in 20 mM Tris-HCl, 500 mM NaCl, pH 8.5 and treated with a fivefold molar excess of potassium ferricyanide. This mixture was incubated overnight at 4 °C followed by centrifugal concentration with an Amicon 10 K MWCO filter (Millipore Sigma) and application of the concentrated sample to a Sephadex G-25 column (GE Healthcare) to remove the excess ferricyanide. Ferric Hb0.1 was then buffer exchanged into 20 mM NaH2PO4, 150 mM NaCl, pH 7.5. The concentration of hemin within each form of the protein was determined using the extinction coefficient of 179 Mm−1cm−1 at a wavelength of 405 nm.
NMR titration experiments
A sample of IsdHN2N3(Y642A) with 13C–1H3 methyl labeling at Ile, Leu, and Val (ILV, U-[15N, 13C, 2H], Ile-[13CH3, δ1], Leu, Val-[13CH3, CD3] IsdHN2N3(Y642A)) was produced as described previously [54]. Briefly, labeled protein was produced in 99% D2O M9 minimal media using 1 g/L15NH4Cl as the nitrogen source and 3 g/L U-[13C, 2H] glucose as the carbon source. One hour prior to induction, 60 mg/L of [U–13C4,3,3–2H2] α-ketobutyrate and 100 mg/L if [1,2,3,4–13C4,3,4′,4′,4′−2H4] α-ketoisovalerate (Cambridge Isotope Laboratories, Inc.) were added. Purification of IsdH was performed as previously described [22]. The final sample was exchanged into NMR buffer composed of 20 mM sodium phosphate, 150 mM NaCl, 8% D2O, pH 7.5.
The resonances of methyl groups were assigned in a previous study by a combination of 15N-NOESY-HSQC, 13C-NOESY-HSQC, and CC(CO)NH spectra [54]. The chemical shifts and peak intensities of methyl groups were monitored over the course of a titration of Hb0.1 in its carbonmonoxy (CO) form using 13C–1H HMQC (methyl-TROSY) experiments [45]. The initial concentration of ILV-IsdHN2N3(Y642A) was 200 μM, and Hb0.1 was added such that molar ratios of Hb0.1 (heme basis):IsdH were 0:1, 0.25:1, 0.5:1, 2:1, 5:1, and 10:1. Additional isotopically labeled (U-[15N, 10% 13C]) samples of αIsdH and QM (αIsdH R492A/Y495A/V637A/I640A) variants were prepared as previously described [22]. Briefly, E. coli BL21 (DE3) cells containing αIsdH or QM expression plasmids were grown at 37 °C in M9 minimal media using 1 g/L 15NH4Cl, 2.7 g/L glucose, 0.3 g/L U–13C-glucose containing 50 μg/mL kanamycin. Expression was induced at OD600 = 0.6 with 1 mM IPTG and cultures were incubated at 37 °C for 20 h. Purification of αIsdH variants was carried out as previously described [22]. 2-D 15N–1H TROSY-HSQC spectra were collected on both samples to confirm retention of the appropriate global fold in the QM variant [55]. NMR data were collected at 310 K on Bruker DRX 500 MHz and AVANCE NEO 800 MHz spectrometers equipped with cryogenic probes. Spectra were processed using NMRPipe [56] and analyzed/visualized in NMRFAM-Sparky [57].
Heme transfer experiments
The kinetics of heme transfer from ferric Hb0.1 (donor) to apo-αIsdH proteins (acceptor) were measured using an Applied Photophysics SX20 stopped-flow spectrophotometer (Applied Photophysics). Acceptor and donor proteins were dissolved in 20 mM NaH2PO4, 150 mM NaCl, pH 7.5, supplemented with 0.45 M sucrose to prevent absorbance changes caused by apoprotein aggregation. Ferric Hb was mixed with apo-acceptor present at 30- or 40-fold excess (in heme units) to maintain pseudo-first-order conditions. After rapid mixing (dead time 3 ms), absorbance changes at 405 nm were monitored for 1000 s. Experiments were performed in triplicate, and the data were analyzed by fitting the observed time courses to a double-phase exponential expression using the GraphPad Prism program (GraphPad Software, version 5.01).
Molecular dynamics simulations
The initial, undistorted Hb:IsdH structure was built using the distorted Hb: αIsdHY642A complex (PDB: 4XS0) and replacing the Hb chains in the complex with coordinates from native Hb determined in the absence of the receptor that does not contain a distorted F-helix (PDB: 1A3N). The heme and bound histidine were parameterized with GAFF [58] atom types using MCPB.py [59,60]. The apo-Hb: αIsdHY642A was parameterized, neutralized with Na+, and solvated in LEAP with the Amber14 and TIP3P forcefields [61]. A 150 mM NaCl environment was created by replacing randomly selected water molecules with sodium and chloride ions. The mass of each solute hydrogen atom was repartitioned using ParmEd [62]. The system was then minimized for 5000 steps (2500 with solute restraints followed by 2500 without). The Y642A mutation was reversed on the minimized Hb: αIsdHY642A; this Hb: αIsdH structure was then reminimized, heated under constant volume to 300 K and solute restraints over 20 ps, and restraints were slowly removed for 4.6 ns under constant pressure at 1 atm. Temperature was kept constant using Langevin dynamics and a collision frequency of 2 ps−1; pressure was kept constant using the Monte Carlo barostat. All the preparation and following production runs were calculated using the CUDA implementation of PMEMD in Amber18 [63] with a timestep of 4 fs; electrostatic energies were calculated using particle-mesh Ewald [64] while van der Waals energies were calculated with a cutoff of 10 Å.
The apo-Hb: αIsdH structure was used in steered MD simulations. In these simulations a harmonic potential on the distance between Hb and a selection of IsdH was introduced. Two sets of simulations were produced: one set biasing the distance between Hb and N2, the other biasing between Hb and N3. For each set the center of mass of the alpha carbons in each group was calculated. The equilibrium position (i.e., the minimum of the harmonic bias) was set to the initial distance between the two groups, then increased by 25 Å over 50 ns with a force constant of 10 kcal mol−1 Å−2. Ten pulling trajectories for each bias were produced; the average and standard error of observed quantities are presented here.
To produce the mutated systems, each alanine substitute was built by introducing the substitution as well as reversing the Y642A mutation, then following the same minimization, heating, and relaxation scheme detailed earlier. These substitute and Hb: αIsdH systems were run for 200 ns. The helicity was measured for the C-terminus end of the F-helix using the alpha-helix content collective variable [65], which uses the angle formed by neighboring alpha-carbons and the hydrogen bonding between the backbones of different residues. RMSF calculation on alpha-carbons and contact analysis were done using CPPTRAJ [66]. Contacts throughout this work were defined as a heavy atom pairing (i.e., hydrogens are excluded) with 4 Å or less distance; each atom pairing was normalized by the number of frames, then pairings were summed by residue. Principal component analysis (PCA) was performed on all N3 heavy backbone atoms using GROMACS 2018.3 [67]. N3:Hb interaction energy was estimated using MMGBSA.py [60]. For each analysis, 100 ns was removed from analysis for equilibration; helicity was measured over the entire simulation in order to measure F-helix distortion. One-tail p-values were obtained using the standard independent two sample tests; number of uncorrelated points was determined by calculating the statistical inefficiency using pymbar [68].
Supplementary Material
Acknowledgments
We thank members of the Clubb and Wereszczynski laboratories for useful discussions. We acknowledge Dr. Megan Sjodt and Dr. Ramsay Macdonald for helpful correspondence and experimental advice, and Yirui Hong for technical assistance. Experimental work in the Clubb group was funded by the National Science Foundation MCB-1716948 and by the National Institutes of Health/National Institute of Allergy and Infectious Diseases Grants R01 AI121360 and AI052217. Work in the Wereszczynski group was funded by the National Science Foundation MCB-1716099 and the National Institutes of Health 1R35GM119647. This work used the Extreme Science and Engineering Discovery Environment (XSEDE), which is supported by National Science Foundation ACI-1548562 [69] and Comet at the San Diego Supercomputer Center at UC San Diego through allocation TG-MCB140081.
Footnotes
Appendix A. Supplementary data
Supplementary data to this article can be found online at https://doi.org/10.1016/j.jmb.2019.12.023.
References
- [1].Klevens RM, et al. , Invasive methicillin-resistant Staphylococcus aureus infections in the United States, J. Am. Med. Assoc 298 (15) (2007) 1763–1771. [DOI] [PubMed] [Google Scholar]
- [2].Lee AS, et al. , Methicillin-resistant Staphylococcus aureus, Nat Rev Dis Primers 4 (2018). [DOI] [PubMed] [Google Scholar]
- [3].Dukic VM, Lauderdale DS, Wilder J, Daum RS, David MZ, Epidemics of community-associated methicillin-resistant Staphylococcus aureus in the United States: a meta-analysis, PLoS One 8 (1) (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Klein EY, et al. , National costs associated with methicillin-susceptible and methicillin-resistant Staphylococcus aureus hospitalizations in the United States, 2010–2014, Clin. Infect. Dis 68 (1) (2019) 22–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Ong ST, Ho JZ, Ho B, Ding JL, Iron-withholding strategy in innate immunity, Immunobiology 211 (4) (2006) 295–314. [DOI] [PubMed] [Google Scholar]
- [6].Haley KP, Skaar EP, A battle for iron: host sequestration and Staphylococcus aureus acquisition, Microb. Infect 14 (3) (2012) 217–227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Farrand AJ, Reniere ML, Ingmer H, Frees D, Skaar EP, Regulation of host hemoglobin binding by the Staphylococcus aureus clp proteolytic system, J. Bacteriol 195 (22) (2013) 5041–5050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Skaar EP, Humayun M, Bae T, DeBord KL, Schneewind O, Iron-source preference of Staphylococcus aureus infections, Science 305 (5690) (2004) 1626–1628. [DOI] [PubMed] [Google Scholar]
- [9].Mazmanian SK, et al. , Passage of heme-iron across the envelope of Staphylococcus aureus, Science 299 (5608) (2003) 906–909. [DOI] [PubMed] [Google Scholar]
- [10].Maresso AW, Schneewind O, Iron acquisition and transport in Staphylococcus aureus, Biometals 19 (2) (2006) 193–203. [DOI] [PubMed] [Google Scholar]
- [11].Reniere ML, Torres VJ, Skaar EP, Intracellular metalloporphyrin metabolism in Staphylococcus aureus, Biometals 20 (3–4) (2007) 333–345. [DOI] [PubMed] [Google Scholar]
- [12].Torres VJ, Pishchany G, Humayun M, Schneewind O, Skaar EP, Staphylococcus aureus IsdB is a hemoglobin receptor required for heme iron utilization, J. Bacteriol 188 (24) (2006) 8421–8429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Pishchany G, et al. , IsdB-dependent hemoglobin binding is required for acquisition of heme by Staphylococcus aureus, JID (J. Infect. Dis.) 209 (11) (2014) 1764–1772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Pishchany G, Dickey SE, Skaar EP, Subcellular localization of the Staphylococcus aureus heme iron transport components IsdA and IsdB, Infect. Immun 77 (7) (2009) 2624–2634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Mazmanian SK, Ton-That H, Su K, Schneewind O, An iron-regulated sortase anchors a class of surface protein during Staphylococcus aureus pathogenesis, Proc. Natl. Acad. Sci. U. S. A 99 (4) (2002) 2293–2298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Ton-That H, Marraffini LA, Schneewind O, Protein sorting to the cell wall envelope of Gram-positive bacteria, Biochim. Biophys. Acta 1694 (1–3) (2004) 269–278. [DOI] [PubMed] [Google Scholar]
- [17].Jacobitz AW, Kattke MD, Wereszczynski J, Clubb RT, Sortase transpeptidases: structural biology and catalytic mechanism, Adv. Protein. Chem. Str 109 (2017) 223–264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Andrade MA, Ciccarelli FD, Perez-Iratxeta C, Bork P, NEAT: a domain duplicated in genes near the components of a putative Fe3+ siderophore transporter from Gram-positive pathogenic bacteria, Genome Biol. 3 (9) (2002), RESEARCH0047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Spirig T, et al. , Staphylococcus aureus uses a novel multidomain receptor to break apart human hemoglobin and steal its heme, J. Biol. Chem 288 (2) (2013) 1065–1078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Zhu H, Li DF, Liu MY, Copie V, Lei BF, Non-hemebinding domains and segments of the Staphylococcus aureus IsdB protein critically contribute to the kinetics and equilibrium of heme acquisition from methemoglobin, PLoS One 9 (6) (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Fonner BA, et al. , Solution structure and molecular determinants of hemoglobin binding of the first NEAT domain of IsdB in Staphylococcus aureus, Biochemistry 53 (24) (2014) 3922–3933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Sjodt M, Macdonald R, Spirig T, Chan AH, Dickson CF, Fabian M, et al. , The PRE-derived NMR model of the 38.8kDa tri-domain IsdH protein from Staphylococcus aureus suggests that it adaptively recognizes human hemoglobin, J. Mol. Biol 428 (2016) 1107–1129, 10.1016/j.jmb.2015.02.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Sjodt M, et al. , Energetics underlying hemin extraction from human hemoglobin by Staphylococcus aureus, J. Biol. Chem 293 (18) (2018) 6942–6957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Dickson CF, et al. , Structure of the hemoglobin-IsdH complex reveals the molecular basis of iron capture by Staphylococcus aureus, J. Biol. Chem 289 (10) (2014) 6728–6738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Dickson CF, Jacques DA, Clubb RT, Guss JM, Gell DA, The structure of haemoglobin bound to the haemoglobin receptor IsdH from Staphylococcus aureus shows disruption of the native alpha-globin haem pocket, Acta. Crystallogr. D. Biol. Crystallogr 71 (6) (2015) 1295–1306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Villareal VA, et al. , Transient weak protein-protein complexes transfer heme across the cell wall of Staphylococcus aureus, J. Am. Chem. Soc 133 (36) (2011) 14176–14179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Abe R, Caaveiro JMM, Kozuka-Hata H, Oyama M, Tsumoto K, Mapping ultra-weak protein-protein interactions between heme transporters of Staphylococcus aureus, J. Biol. Chem 287 (20) (2012) 16477–16487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Muryoi N, et al. , Demonstration of the iron-regulated surface determinant (Isd) heme transfer pathway in Staphylococcus aureus, J. Biol. Chem 283 (42) (2008) 28125–28136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Zhu H, et al. , Pathway for heme uptake from human methemoglobin by the iron-regulated surface determinants system of Staphylococcus aureus, J. Biol. Chem 283 (26) (2008) 18450–18460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Matsui T, et al. , Heme degradation by Staphylococcus aureus IsdG and IsdI liberates formaldehyde rather than carbon monoxide, Biochemistry 52 (18) (2013) 3025–3027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Reniere ML, et al. , The IsdG-family of haem oxygenases degrades haem to a novel chromophore, Mol. Microbiol 75 (6) (2010) 1529–1538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Macdonald R, Mahoney BJ, Ellis-Guardiola K, Maresso A, Clubb RT, NMR experiments redefine the hemoglobin binding properties of bacterial NEAr-iron Transporter domains, Protein Sci. 28 (8) (2019) 1513–1523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Zhu H, Liu M, Lei B, The surface protein Shr of Streptococcus pyogenes binds heme and transfers it to the streptococcal heme-binding protein Shp, BMC Microbiol. 8 (2008) 15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Nobles CL, Maresso AW, The theft of host heme by Gram-positive pathogenic bacteria, Metall 3 (8) (2011) 788–796. [DOI] [PubMed] [Google Scholar]
- [35].Sheldon JR, Heinrichs DE, Recent developments in understanding the iron acquisition strategies of gram positive pathogens, FEMS Microbiol. Rev 39 (4) (2015) 592–630. [DOI] [PubMed] [Google Scholar]
- [36].Skaar EP, The battle for iron between bacterial pathogens and their vertebrate hosts, PLoS Pathog. 6 (8) (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Huang WL, Wilks A, Extracellular heme uptake and the challenge of bacterial cell membranes, Annu. Rev. Biochem 86 (2017) 799–823. [DOI] [PubMed] [Google Scholar]
- [38].Palmer LD, Skaar EP, Transition metals and virulence in bacteria, Annu. Rev. Genet 50 (2016) 67–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Pilpa RM, et al. , Functionally distinct NEAT (NEAr Transporter) domains within the Staphylococcus aureus IsdH/HarA protein extract heme from methemoglobin, J. Biol. Chem 284 (2) (2009) 1166–1176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Watanabe M, et al. , Structural basis for multimeric heme complexation through a specific protein-heme interaction: the case of the third neat domain of IsdH from Staphylococcus aureus, J. Biol. Chem 283 (42) (2008) 28649–28659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Vu NT, et al. , Selective binding of antimicrobial porphyrins to the heme-receptor IsdH-NEAT3 of Staphylococcus aureus, Protein. Sci 22 (7) (2013) 942–953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Bowden CFM, Verstraete MM, Eltis LD, Murphy MEP, Hemoglobin binding and catalytic heme extraction by IsdB near iron transporter domains, Biochemistry 53 (14) (2014) 2286–2294. [DOI] [PubMed] [Google Scholar]
- [43].Bowden CFM, et al. , Structure-function analyses reveal key features in Staphylococcus aureus IsdB-associated unfolding of the heme-binding pocket of human hemoglobin, J. Biol. Chem 293 (1) (2018) 177–190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Tugarinov V, Kay LE, An isotope labeling strategy for methyl TROSY spectroscopy, J. Biomol. NMR 28 (2) (2004) 165e172. [DOI] [PubMed] [Google Scholar]
- [45].Ollerenshaw JE, Tugarinov V, Kay LE, Methyl trosy: explanation and experimental verification, Magn. Reson. Chem 41 (2003) 843–852. [Google Scholar]
- [46].Looker D, et al. , A human recombinant haemoglobin designed for use as a blood substitute, Nature 356 (6366) (1992) 258–260. [DOI] [PubMed] [Google Scholar]
- [47].Isralewitz B, Gao M, Schulten K, Steered molecular dynamics and mechanical functions of proteins, Curr. Opin. Struct. Biol 11 (2) (2001) 224–230. [DOI] [PubMed] [Google Scholar]
- [48].Gelin BR, Karplus M, Mechanism of tertiary structuralchange in hemoglobin, P Natl. Acad. Sci. USA 74 (3) (1977) 801–805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [49].Pishchany G, Skaar EP, Taste for blood: hemoglobin as a nutrient source for pathogens, PLoS Pathog. 8 (3) (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [50].Hargrove MS, Whitaker T, Olson JS, Vali RJ, Mathews AJ, Quaternary structure regulates hemin dissociation from human hemoglobin, J. Biol. Chem 272 (28) (1997) 17385–17389. [DOI] [PubMed] [Google Scholar]
- [51].Krieg S, et al. , Heme uptake across the outer membrane as revealed by crystal structures of the receptor-hemophore complex, Proc. Natl. Acad. Sci. U. S. A 106 (4) (2009) 1045–1050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [52].Izadi-Pruneyre N, et al. , The heme transfer from the soluble HasA hemophore to its membrane-bound receptor HasR is driven by protein-protein interaction from a high to a lower affinity binding site, J. Biol. Chem 281 (35) (2006) 25541–25550. [DOI] [PubMed] [Google Scholar]
- [53].Donegan RK, Moore CM, Hanna DA, Reddi AR, Handling heme: the mechanisms underlying the movement of heme within and between cells, Free Radic. Biol. Med 133 (2019) 88–100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [54].Spirig T, Clubb RT, Backbone 1H, 13C and 15N resonance assignments of the 39 kDa staphylococcal hemoglobin receptor IsdH, Biomol NMR Assign 6 (2) (2012) 169–172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [55].Pervushin K, Riek R, Wider G, Wuthrich K, Attenuated T-2 relaxation by mutual cancellation of dipole-dipole coupling and chemical shift anisotropy indicates an avenue to NMR structures of very large biological macromolecules in solution, P Natl Acad Sci USA 94 (23) (1997) 12366–12371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [56].Delaglio F, et al. , Nmrpipe - a multidimensional spectral processing system based on unix pipes, J. Biomol. NMR 6 (3) (1995) 277e293. [DOI] [PubMed] [Google Scholar]
- [57].Lee W, Tonelli M, Markley JL, NMRFAM-SPARKY: enhanced software for biomolecular NMR spectroscopy, Bioinformatics 31 (8) (2015) 1325–1327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [58].Wang J, Wolf RM, Caldwell JW, Kollman PA, Case DA, Development and testing of a general amber force field, J. Comput. Chem 25 (9) (2004) 1157–1174. [DOI] [PubMed] [Google Scholar]
- [59].Li P, Merz KM, MCPB.py: a Python based metal center parameter builder, J. Chem. Inf. Model 56 (4) (2016) 599–604. [DOI] [PubMed] [Google Scholar]
- [60].Miller BR, et al. , MMPBSA.py: an efficient program for endstate free energy calculations, J. Chem. Theory Comput 8 (9) (2012) 3314–3321. [DOI] [PubMed] [Google Scholar]
- [61].Jorgensen WL, Chandrasekhar J, Madura JD, Impey RW, Klein ML, Comparison of simple potential functions for simulating liquid water, J. Chem. Phys 79 (1983) 926–935. [Google Scholar]
- [62].Hopkins CW, Le Grand S, Walker RC, Roitberg AE, Longtime-step molecular dynamics through hydrogen mass repartitioning, J. Chem. Theory Comput 11 (4) (2015) 1864–1874. [DOI] [PubMed] [Google Scholar]
- [63].Case DA, et al. , AMBER 2018, University of California, San Francisco: ), 2018. [Google Scholar]
- [64].Darden T, York D, Pedersen L, Particle mesh Ewald: an N,log(N) method for Ewald sums in large systems, J. Chem. Phys. 98 (12) (1993) 10089–10092. [Google Scholar]
- [65].Fiorin G, Klein ML, Henin J, Using collective variables t o drive molecular dynamics simulations, Mol. Phys 111 (22e23) (2013) 3345–3362. [Google Scholar]
- [66].Roe DR, Cheatham TEI, PTRAJ and CPPTRAJ: software for processing and analysis of molecular dynamics trajectory data, J. Chem. Theory Comput 9 (2013) 3084–3095. [DOI] [PubMed] [Google Scholar]
- [67].Abraham MJ, et al. , GRO- MACS: high performance molecular simulations through multi-level parallelism from lap- tops to supercomputers, Software 1–2 (2015) 19–25. [Google Scholar]
- [68].Shirts MR, Chodera JD, Statistically optimal analysis of samples from multiple equilibrium states, J. Chem. Phys 129 (12) (2008) 124105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [69].Towns J, et al. , XSEDE: accelerating scientific Discovery, Comput. Sci. Eng 16 (5) (2014) 62–74. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.