Abstract
Leucoefdin an important constituent of various fruits such as banana, raspberry, etc. was explored to target MPro protease of SARS Co-V 2. Ligand was found to bind at active site of MPro with large negative binding energies in molecular docking and simulation study. The docking results showed that Leucoefdin interacted with the MPro by forming hydrogen bonds, at Leu 141, His163, His 164, and Glu 166. Other non-bonded interactions were seen at Met49, Pro52, Tyr54, Phe140, Leu141, Cys145 and Met165. Results of Leucoefdin was in coherence with the recently reported MPro protease-inhibitor complex. It even displayed better binding energies (kcal/mol) in HTVS (-6.28), SP (-7.28), XP (-9.29) and MMGBSA (-44.71) as compared to the reference ligand [HTVS (-4.87), SP (-6.79), XP (-5.75) and MMGBSA (-47.76)]. Leucoefdin-MPro complex on molecular dynamic simulation showed initial fluctuations in RMSD plot for a certain period and attained equilibrium which remained stable during entire simulation for 150 ns. RMSF of protein showed less secondary structure fluctuations and a greater number of H-bond formation with Leucoefdin during 150 ns simulation. Post simulation MMGBSA analysis showed binding energy of -45.98 Kcal/mol. These findings indicated the potential of Leucoefdin as lead compound in R&D for drug discovery and development against SARS CoV-2.
Communicated by Ramaswamy H. Sarma
Keywords: Leucoefdin, SARS CoV-2, MPro, docking, simulation
1. Introduction
Antiviral drugs target various stages of viral replication, like cell entry (fusion protein inhibitors), uncoating (ion channel blocker, capsid stabilizers), transcription of viral genome, translation of viral proteins, post-translational modification and release inhibitors. Till date, Food and Drug Administration (FDA) has not approved any specific drug against newly emerged SARS-CoV-2 virus (COVID-19). There are no clinical trial data supporting any prophylactic therapy. Many antiviral drugs are under clinical trial for the efficacy and safety against COVID-19. Several research and active clinical trials are underway especially on Protease inhibitors like Lopinavir, Ritonavir etc., RNA polymerase inhibitors such as Remdesivir, Favipiravir and Ribavirin etc, and antihelminths etc. Some other drugs like Baricitinib, Imatinib, Dasatinib, Cyclosporine are also undertaken for in vitro activity against SARS-CoV-2. Also, nonspecific anti-inflammatory and immunomodulators are being evaluated for its efficacy against COVID-19 (Agostini et al., 2018; Elfiky, 2020; Sarma et al., 2020; Wang et al., 2020; Wu et al., 2020). The replicase gene of COVID-19 virus encodes two overlapping polyproteins, ppa and ppab which are vital for viral replication and transcription. A main protease (MPro) leads proteolytic processing of polyproteins and generate functional polypeptides. Therefore, MPro can be an attractive target for antiviral drug design (Fearon et al., 2020; Jin et al., 2020). The present study intended to find inhibitor of MPro through molecular docking and simulation. The phytochemicals are rich source of many bioactive constituents which are essential for normal physiological functions, (USDA, 2017). Leucoefdin, a member of catechol, and a polyphenol is colourless and linked to leucoanthocyanidins found in the Arachis hypogaea (Earth nut in seeds), Musa sp. (Banana, in the fruit), Phyllanthus emblica (Indian gooseberry), Vicia faba (bell-bean, in the seed), Nelumbo nucifera (lotusleaf) etc. (Drewes & Roux,1966; Ganguly et al., 1958) have been selected for the docking and simulation analysis.
2. Methods
MPro (PDB ID:5R84) (Fearon et al., 2020) and ligand Leucoefdin (Pubchem ID: 3081374) were obtained from RCSB protein data bank and Pubchem respectively. Structure based virtual screening and molecular docking was conducted using the Schrödinger package to find the best docked pose (LigPrep, 2015). The docking was done with default parameters for grid and pose generation using HTVS, SP and XP mode. After XP study, the best interacting ligand was selected based on GlideScore (Friesner et al., 2004, 2006; Halgren et al., 2004; Jacobson et al., 2004; Sastry et al., 2013).
2.1. Active site prediction
SiteMap module of Schrödinger suite was used to forecast active sites of target protein structure. Potential sites of the target protein and site-score values were generated.
2.2. Molecular docking
Protein preparation wizard of Schrödinger suite was used to fix the drawbacks of protein. Further, protonation done by Epik module of Schrödinger suite. Water molecules more than 3 Å away from the ligand were removed before subjecting to protein minimization and hydrogen bond optimization. OPLSe force field employed in minimization studies. The structures of ligand were prepared with the LigPrep module, then energy minimisation was done in gas phase using Macromodel with the OPLSe force field. Glide-receptor grid generation module was used for docking studies (Halgren, 2007). Co-crystallized ligand was first selected and around binding site MPro ligand complex grid box was generated for molecular docking (Fearon et al., 2020). Leucoefdin and known inhibitors from published sources Z31792168 was then docked with protein. The docking was done for high throughput virtual screening (HTVS), standard precision (SP) and extra precision (XP) to evaluate the binding energies with default parameters (Glide Schrodinger suite, 2015). First the co-crystallized ligand was extracted from the MPro protein and re docked. Docking results of Leucoefdin and reference ligand was compared.
2.3. Molecular dynamics simulation
The ligand with best XP score was further used for MD simulation studies. Molecular dynamics (MD) of Leucoefdin-MPro complex was performed with Desmond module of Schrodinger Suite. The prepared complexes were solvated using orthorhombic solvent box and a solvent buffer (extended 10 Å outside the protein in all directions) and OPLS3 force field was applied. System was further neutralized by adding the desired number of counter ions with 0.15 M salt concentration. The six-stage system relaxation protocol of Schrodinger suite, was used before starting production runs (Harder et al., 2016; Macromodel, Schrodinger suite, 2015). 2000 steps of steepest descent minimization in initial two stages were conducted with and without a restraint of 50 kcal/mol/Å2 on the solute atoms, then other four short MD simulations done with 12ps in NVT ensemble at 10 K with solute heavy atoms restrained with a force constant of 50 kcal/mol/Å2. Further 12ps MD simulation in NPT ensemble at10 K with the same restraint followed by 12ps MD simulation in NPT ensemble at 300 K with the same restraint and finally 24ps MD simulation in NPT ensemble at 300 K without any restraints. Unrestrained production simulations in the NPT ensemble was conducted for 150 ns at 300 K temperature and 1.01325 bar pressure. Relaxation time of 1ps was used with Nosé-Hoover chain thermostat and the isotropic Martyna-Tobias-Klein barostat was used with a relaxation time of 2ps. To evaluate short range interactions 9 Å cut off was used, while long range coulombic interactions were estimated with smooth particle mesh Ewald method (PME). r-RESPA integrator was used to study non-bonded interaction where every step was updated for short range forces and after every three steps the long range forces were updated. MD simulations for 150 ns was performed for MPro-Leucoefdin complex using OPLS2005 (Optimized Kanhesia for Liquid Simulations) force fields. Trajectories were documented after every 4.8 ps, where 1.2 ps interval used for energy recording for 150 ns simulation. The trajectories were saved at 50.0 ps intervals for analysis (Macromodel, Schrodinger suite, 2015). Prime-MM/GBSA (Molecular Mechanics/Generalized Born Surface Area) was used for the binding free energy calculation (Mobley & Dill, 2009; Prime, version 4.5, 2016; Kevin et al., 2006; Shivakumar et al., 2010; Wang, et al., 2015).
3. Results and discussion
Almost all continents are having cases of SARS CoV-2, therefore, fast discovery of compounds against this problem is needed. Viral replication and transcription require a main protease MPro for proteolytic processing of polyproteins to generate functional polypeptides of SARS CoV-2 replicase gene. MPro being an attractive target for antiviral drug design as, it is not closely related to homologues in humans (Jin et al., 2020; Wu et al., 2020). We have used database of 250 compounds (compiled from published literature on phytochemicals with antimicrobial properties) against MPro protease and Leucoefdin was chosen for the present study based on its high binding energy and interaction property.
3.1. Active site prediction
Site Map analysis was made based on different parameters such as size, volume, enclosure, contact, amino acid exposure, hydrophobicity, hydrophilicity and donor/acceptor ratio for identification of potential sites. Results showed residues 41, 49,52, 53, 54, 140, 141, 142, 144, 145, 163, 164, 165, 166, 187, 188, 189, 190 with a site score of 0. 89.
3.2. Molecular docking
The 306 amino acids length with a resolution of 1.83 Å of MPro was taken for the study to find inhibitor from some selected phytochemicals. The MPro (PDB ID:5R84) structure of COVID-19 virus was used to produce receptor grid for docking (Fearon et al., 2020). Docking results showed ligand’s interaction to the target with large negative binding energies (Table 1). Table 1. displaying binding energies of High throughput virtual screening, standard precision and extra precision of Leucoefdin and reference compound with MPro protease. Leucoefding-MPro complex was displaying better binding energies than reference molecule. The post docking MMGBSA binding free energy was -41.71 and -44.76 Kcal/mol for Leucoefdin and reference compound (Z31792168) respectively (Table 1). The Leucoefdin produced significant MM-GBSA binding energy as a result of more favourable coulombic interactions, lipophilic interactions and van der Waals’ interactions than to reference ligand (Z31792168).
Table 1.
Molecular docking binding energy study of Leucoefdin and reference compound with MPro protease.
| Target (MPro Protease) | HTVS Kcal/mol | SP Kcal/mol | XP Kcal/mol | MMGBSA Kcal/mol | ||
|---|---|---|---|---|---|---|
| 1 | Leucoefdin (Pub chem i.d. 3081374) | −6.28 | −7.85 | −9.29 | −44.71 | |
| 2 | Co-crystalized ligand ( Z31792168) | −4.87 | −6.79 | −5.75 | −47.76 | |
Figure 1a, represented co-crystallized ligand Z31792168 interaction with MPro (5R84) with hydrogen bonds at His163 and His 164 residues in addition to other non-bonded interactions at Met 49, Pro 52, Tyr 54, Phe 140, Leu 141, Cys 145 and Met165 (Fearon et al., 2020; Jin et al., 2020). Similar binding pattern was seen in case of Leucoefdin-MPro protein complex as evident from Figure 1b, hydrogen bonds were formed with residues His163, His 164, Glu 166, Leucine 141 and non-bonded interactions at Met 49, Pro 52, Tyr 54, Phe 140, Leu 141, Cys 145 and Met 165. 6LU7 complex with inhibitor N3, showed hydrogen bond with residue number Phe140, Gly 143, His 163, His 164, Glu166 and Gln 189 and hydrophobic, ionic interactions at Met49, Pro52, Tyr54, Phe140, Leu141, Cys145 and Met165 (Liu et al., 2020). MProProtein 6W63-complex with inhibitor X77 showed hydrogen bond interaction with residues Gly143, and Glu166 and similar non-bonded interactions at Tyr54, Phe140, Met165, Met49 and Leu141. Cys145 was making pi interactions with X77 (Mesecar, 2020). Leucoefdin-MPro complex results was found to be comparable with other reported inhibitor.
Figure 1.
Protein ligand interaction study using Glide for (a) Reference (Z31792168) Ligand MPro complex; (b) MPro with Leucoefdin.
3.3. Molecular dynamics simulation
Molecular dynamics (MD) simulations represents dynamic fluctuations within the complex and interactions of ligand, lipid and water molecules. To study the dynamic interaction of viral protein with the docked Leucoefdin, molecular dynamics (MD) simulations of the MPro complexed with Leucoefdin was performed using Desmond. It was used to evaluate the structural constancy and binding site adaptations to the docked ligand. MD simulations was run for the protein and ligand for 150 ns. The stability and fluctuations of the protein and ligand alone and in complex during the simulation were investigated and the resulting trajectory for complex was made with the backbone root mean square deviations (RMSDs) which is the average displacement of atoms from particular frame to a reference frame.
Protein ligand binding overall was stable during entire simulation. Figure 2 displayed some fluctuation in Protein RMSD during initial 10 ns and remained stable for next 140 ns i.e. fluctuation remains within 1 A0. In case ligand initial fluctuation was observed till 70 ns thereafter it remained stable for next 80 ns that might be due to H-bond formation at Glu 166 (84% of the time) and other Hydrogen bonds formation as described in Figure 6a. It was evident from the Lig-fit-Prot of Figure 2 that the value was lower than the RMSD of the protein during entire simulation of 150 ns, thus it can be said that the ligand has not diffused away from the initial binding site and the interaction was stable (Figure 2).
Figure 2.
RMSD profile of M Pro and Leucoefdin during 150 ns MD simulation.
Figure 6.
(a) Protein ligand interaction study during 150 ns simulation(green-Hbond; pink-ionic; blue-water bridges; violet-Hydrophobic); (b) Schematic ligand interaction with protein red charged, green hydrophobic, blue polar,·−· salt bridge during simulation.
Root mean square fluctuations (RMSF) was studied to recognize the critical residues involved in the major interactions with a ligand. The essential residues forming hydrogen bond with ligands being displayed by green bar during 150 ns simulation (Figure 3). The two major fluctuations in local domain of protein was seen in loop regions, first fluctuation was in between 1 and 10 residues and second after 300 residues. This might be due to the reason that N and C terminal fluctuate more than any other part of the protein (Figure 3).
Figure 3.
RMSF study to show local changes around protein chain in 150 ns simulation (green bar shows ligand contact sites).
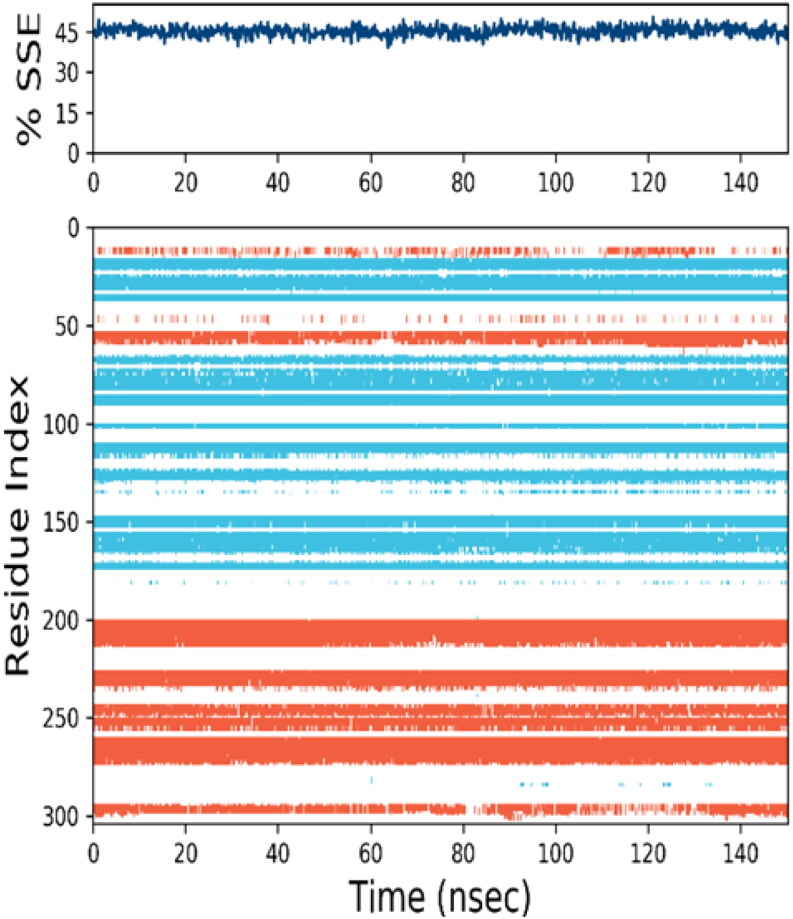
Figure 4, top panel indicated the stable pattern of secondary structure element (SSE) over 150 ns simulation. Bottom panel showed behaviour of each residue and its SSE over time during entire simulation (red-ɑ helices, blue-βstrands, white-loop). The secondary structures of protein were conserved during entire simulation. Some minor fluctuations in RMSF plot (Figure 3) or white region i.e. loop (Figure 4) were observed in 45-55 and 170-200 residues.
Figure 4.
Secondary structure composition during entire 150 ns simulation (red-ɑ helices, blue-βstrands, white-loop).
Figure 5 showed different types of contacts such as H bond, hydrophobic, ionic and water bridges between protein and ligand during simulation. Top panel indicated the sum of specific interaction protein makes with the ligand over the course of the trajectory. Specific residues interaction with the ligand in each trajectory frame was shown in bottom panel. Sometimes more than one specific contacts with ligands were formed with some residues which was represented by dark orange shade (Figure 5). Furthermore, after 40 ns of simulation dark orange horizontal bar has been seen at Glu 166 which remained in hydrogen bond interaction with leucoefdin for entire 150 ns simulation.
Figure 5.
Timeline representation of the interaction and contacts, top panel show specific contact of ligand and protein during 150 ns simulation.
Figure 6a demonstrated the interaction fraction analysis of Leucoefdin with MPro during 150 ns simulation period. Hydrogen bonding were seen at His 41, Gly143, Ser 144, His 163, His 164, Glu 166, Val 186, Asp187, Arg188 and Gln189 and other major non-bonded interactions at His 41, Ser 46, Asn 142, Cys 145, His 164, Met 165, Glu 166, Ala 173, Val186, Arg 188, Gln 189, Thr 190, Ala 191, and Gln 192. The Figure 6a clearly indicated that Leucoefdin was interacting with protein throughout 150 ns simulation. This was further substantiated by the Figure 6b where the actual percentage of interaction was established between protein and ligand during simulation. Glu 166 was seen to interact 84% times with three hydrogen bonds formation. Ala 191 form 2 H bonds, other H bonds were also seen with Thr 190, Gln 192. Pro 168, Val 186 and Arg 188 were coordinated with water bridges. These interactions suggested that Leucoefdin-MPro complex was stable throughout simulation. Complex was further subjected to Post simulation MM-GBSA analysis and binding energy found to be -45.98Kcal/mol.
4. Conclusion
COVID-19 was first time reported in China, December 2019 subsequently more than 170 countries are affected with this disease. WHO declared COVID-19 as a pandemic. Lots of efforts being made by various researchers to develop drug against SARS CoV-2 virus. There are various targets like viral MPro protease, RNA dependent RNA polymerase, host protease like TMPRSS2 and ACE2. We have performed computational study to find out inhibitor of MPro protease. Study revealed that Leucoefdin has potential to inhibit MPro protein, which is essential protease responsible for formation of functional viral polyprotein. Ligand protein complex showed good negative binding energies in HTVS, SP, XP and MMGBSA studies. MD simulations, study showed stable protein-ligand interaction. Thus, the present study gives an insight on a mechanism through which it interacts with viral protease. The finding also strengthens our understanding on these phytochemicals and their mechanism of action at molecular level.
Disclosure statement
No potential conflict of interest was reported by the author(s).
References
- Agostini M. L., Andres E. L., Sims A. C., Graham R. L., Sheahan T. P., Lu X., Smith E. C., Case J. B., Feng J. Y., Jordan R., Ray A. S., Cihlar T., Siegel D., Mackman R. L., Clarke M. O., Baric R. S., & Denison M. R. (2018). Coronavirus susceptibility to the antiviral remdesivir (GS-5734) is mediated by the viral polymerase and the proofreading Exoribonuclease. mBio, 9(2), 6 10.1128/mBio.00221-18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drewes S. E., & Roux D. G. (1966). A new flavan-3,4-diol from Acacia auriculiformis by paper ionophoresis. The Biochemical Journal, 98(2), 493–500. 10.1042/bj0980493 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elfiky A. A. (2020). SARS-CoV-2 RNA dependent RNA polymerase (RdRp) targeting: An in silico perspective. J of Biomolecular Structure and Dynamics, 1–9. 10.1080/07391102.2020.1761882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fearon D., Powell A. J., Douangamath A., Owen C. D., Wild C., Krojer T., Lukacik P., Strain-Damerell C. M., Walsh M. A., & von Delft F. (2020). PanDDA analysis group deposition—Crystal structure of COVID-19 main protease in complex with Z31792168. DOI: 10.2210/pdb5R84/pdb. [DOI]
- Friesner R. A., Banks J. L., Murphy R. B., Halgren T. A., Klicic J. J., Mainz D. T., Repasky M. P., Knoll E. H., Shaw D. E., Shelley M., Perry J. K., Francis P., & Shenkin P. S. (2004). Glide: A new approach for rapid, accurate docking and scoring. 1. Method and assessment of docking accuracy. Journal of Medicinal Chemistry, 47(7), 1739–1749. 10.1021/jm0306430 [DOI] [PubMed] [Google Scholar]
- Friesner R. A., Murphy R. B., Repasky M. P., Frye L. L., Greenwood J. R., Halgren T. A., Sanschagrin P. C., & Mainz D. T. (2006). Extra precision glide: docking and scoring incorporating a model of hydrophobic enclosure for protein-ligand complexes. Journal of Medicinal Chemistry, 49(21), 6177–6196. 10.1021/jm051256o [DOI] [PubMed] [Google Scholar]
- Ganguly A. K., Seshadri T. R., & Subramanian P. (1958). A study of leucoanthocyanidins of plants—I : Isomers of leucodelphinidin from karada bark and eucalyptus gum. Tetrahedron, 3(3), 225–229. 10.1016/0040-4020(58)80017-4 [DOI] [Google Scholar]
- Glide, version 6.7 (2015).Schrödinger, LLC. [Google Scholar]
- Halgren T. (2007). New method for fast and accurate binding-site identification and analysis. Chemical Biology & Drug Design, 69(2), 146–148. 10.1111/j.1747-0285.2007.00483.x [DOI] [PubMed] [Google Scholar]
- Halgren T. A., Murphy R. B., Friesner R. A., Beard H. S., Frye L. L., Pollard W. T., & Banks J. L. (2004). Glide: A new approach for rapid, accurate docking and scoring. 2. Enrichment factors in database screening. Journal of Medicinal Chemistry, 47(7), 1750–1759. 10.1021/jm030644s [DOI] [PubMed] [Google Scholar]
- Harder E., Damm W., Maple J., Wu C., Reboul M., Xiang J. Y., Wang L., Lupyan D., Dahlgren M. K., Knight J. L., Kaus J. W., Cerutti D. S., Krilov G., Jorgensen W. L., Abel R., & Friesner R. A. (2016). OPLS3: A force field providing broad coverage of drug-like small molecules and proteins. Journal of Chemical Theory and Computation, 12(1), 281–296. 10.1021/acs.jctc.5b00864 [DOI] [PubMed] [Google Scholar]
- Jacobson M. P., Pincus D. L., Rapp C. S., Day T. J. F., Honig B., Shaw D. E., & Friesner R. A. (2004). A hierarchical approach to all-atom protein loop prediction. Proteins, 55(2), 351–367. 10.1002/prot.10613 [DOI] [PubMed] [Google Scholar]
- Jin Z., Du X., & Xu Y. (2020). Structure of MPro from SARS-CoV-2 and discovery of its inhibitors. Nature, 582, 289–293. 10.1038/s41586-020-2223-y [DOI] [PubMed] [Google Scholar]
- LigPrep, version 3.4 (2015). Schrödinger, LLC. [Google Scholar]
- Liu X., Zhang B., Jin Z., Yang H., & Rao Z. (2020). The crystal structure of COVID-19 main protease in complex with an inhibitor N3. Protein DataBank; DOI: 10.2210/pdb6LU7/pdb [DOI] [Google Scholar]
- MacroModel, version 10.8 (2015). Schrödinger, LLC. [Google Scholar]
- Mesecar A. D. (2020). Center for Structural Genomics of Infectious Diseases (CSGID) Structure of COVID-19 main protease bound to potent broad-spectrum non-covalent inhibitor X77. DOI: 10.2210/pdb6W63/pdb. [DOI]
- Mobley D. L., & Dill K. A. (2009). Binding of small-molecule ligands to proteins: “What you see” is not always “what you get”. Structure (London, England : 1993)), 17(4), 489–498. 10.1016/j.str.2009.02.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prime, version 4.0. (2015). Schrödinger, LLC. [Google Scholar]
- Sarma P., Sekhar N., Prajapat M., Avti P., Kaur H., Kumar S., Singh S., Kumar H., Prakash A., Dhibar D. P., & Medhi B. (2020). In-silico homology assisted identification of inhibitor of RNA binding against 2019-nCoV N-protein (N terminal domain. Journal of Biomolecular Structure & Dynamics. 10.1080/07391102.2020.1753580 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sastry G. M., Adzhigirey M., Day T., Annabhimoju R., & Sherman W. (2013). Protein and ligand preparation: Parameters, protocols, and influence on virtual screening enrichments. Journal of Computer-Aided Molecular Design, 27(3), 221–234. 10.1007/s10822-013-9644-8 [DOI] [PubMed] [Google Scholar]
- Shivakumar D., Williams J., Wu Y., Damm W., Shelley J., & Sherman W. (2010). Prediction of absolute solvation free energies using molecular dynamics free energy perturbation and the OPLS force field. Journal of Chemical Theory and Computation, 6(5), 1509–1519. 10.1021/ct900587b [DOI] [PubMed] [Google Scholar]
- US Department of Agriculture. (2017). Why is it important to eat vegetables? Nutrients. ChooseMyPlate.gov, USDA Center for Nutrition Policy & Promotion. [Google Scholar]
- Wang L., Wu Y., Deng Y., Kim B., Pierce L., Krilov G., Lupyan D., Robinson S., Dahlgren M. K., Greenwood J., Romero D. L., Masse C., Knight J. L., Steinbrecher T., Beuming T., Damm W., Harder E., Sherman W., Brewer M., … Abel R. (2015). Accurate and reliable prediction of relative ligand binding potency in prospective drug discovery by way of a modern free-energy calculation protocol and force field. Journal of the American Chemical Society, 137(7), 2695–2703. 10.1021/ja512751q [DOI] [PubMed] [Google Scholar]
- Wang M., Cao R., Zhang L., Yang X., Liu J., Xu M., Shi Z., Hu Z., Zhong W., & Xiao G. (2020). Remdesivir and chloroquine effectively inhibit the recently emerged novel coronavirus (2019-nCoV) in vitro. Cell Research, 30(3), 269–271. doi: 10.1038/s41422020-0282-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu F., Zhao S., Yu B., Chen Y.-M., Wang W., Song Z.-G., Hu Y., Tao Z.-W., Tian J.-H., Pei Y.-Y., Yuan M.-L., Zhang Y.-L., Dai F.-H., Liu Y., Wang Q.-M., Zheng J.-J., Xu L., Holmes E. C., & Zhang Y.-Z. (2020). A new coronavirus associated with human respiratory disease in China. Nature, 579(7798), 265–269. 10.1038/s41586-020-2008-3 [DOI] [PMC free article] [PubMed] [Google Scholar]