Abstract
Respiratory disease caused by a novel coronavirus, COVID-19, has been labeled a pandemic by the World Health Organization. Very little is known about the infection mechanism for this virus. More importantly, there are no drugs or vaccines that can cure or prevent a person from getting COVID-19. In this study, the binding affinity of 2692 protease inhibitor compounds that are known in the protein data bank, are calculated against the main protease of the novel coronavirus with docking and molecular dynamics (MD). Both the docking and MD methods predict the macrocyclic tissue factor–factor VIIa (PubChem ID: 118098670) inhibitor to bind strongly with the main protease with a binding affinity of −10.6 and −10.0 kcal/mol, respectively. The TF-FVIIa inhibitors are known to prevent the coagulation of blood and have antiviral activity as shown in the case of SARS coronavirus. Two more inhibitors, phenyltriazolinones (PubChem ID: 104161460) and allosteric HCV NS5B polymerase thumb pocket 2 (PubChem ID: 163632044) have shown antiviral activity and also have high affinity towards the main protease of COVID-19. Furthermore, these inhibitors interact with the catalytic dyad in the active site of the COVID-19 main protease that is especially important in viral replication. The calculated theoretical dissociation constants of the proposed COVID-19 inhibitors are found to be very similar to the experimental dissociation constant values of similar protease-inhibitor systems.
Communicated by Ramaswamy H. Sarma
Keywords: COVID-19, molecular docking, SARS-CoV-2, molecular dynamics (MD) simulations, protease inhibitors, virtual screening, Mpro protease
Introduction
An outbreak of respiratory disease caused by a novel coronavirus has now been labeled a pandemic by the World Health Organization (WHO). The disease is now formally known as coronavirus disease 2019 (COVID-19). COVID-19 is responsible for the death of about 322,000 people worldwide as of May 19, 2020 (Dong et al., 2020). COVID-19 virus is a positive-stranded RNA virus with a crown-like appearance under an electron microscope (Fehr & Perlman, 2015). The spike glycoproteins on the envelope of the virus cause this crown-like appearance. Other coronaviruses, such as the severe acute respiratory syndrome coronavirus (SARS-CoV) and Middle East respiratory syndrome coronavirus (MERS-CoV), have similar structures with crown-like appearance. When the COVID-19 virus infects a cell, two replicase polyproteins, pp1a and pp1ab are synthesized by the RNA genome (Ziebuhr & Siddell, 1999). These polyproteins include a replication/transcription complex, several structural proteins and two proteases. One of these proteases is the main protease of coronavirus (Mpro) which cuts the polyproteins into individual functional pieces that is responsible for the replication of new viruses (Thiel et al., 2003; Ziebuhr et al., 1997). The Mpro has been identified as an attractive target for anti-coronavirus drugs (Al-Khafaji et al., 2020; R. J. Khan et al., 2020; S. A. Khan et al., 2020) since it mediates viral replication and transcription (Dougherty & Semler, 1993; Thiel et al., 2003; Xue et al., 2007; Yang et al., 2003). Finding suitable inhibitors of the main protease can prevent the COVID-19 virus from multiplying in the host cell.
In addition to the main protease, coronaviruses contain at least three viral proteins, a spike glycoprotein (S), membrane protein (M), and envelope protein (E) (Belouzard et al., 2012; Boopathi et al., 2020). E and M proteins have been found to be important in viral assembly, trafficking, and release of viral-like particles, (Ruch & Machamer, 2012; Siu et al., 2008) while coronaviruses rely on the S protein to mediate virus entry into the host cell (Belouzard et al., 2012; Bosch et al., 2003). The novel coronavirus uses the S protein to bind to the human receptor containing angiotensin-converting enzyme 2 (ACE2) cells (Hoffmann et al., 2020; Wrapp et al., 2020) and uses the serine protease, TMPRSS2 (transmembrane protease serine 2), for S protein priming. Multiple studies have shown that COVID-19 has a higher affinity to ACE2, (Tian et al., 2020; Wan et al., 2020; Wrapp et al., 2020), which may help to explain why COVID-19 has been more infectious.
Some antiviral drugs have been successfully developed to treat patients who are infected with HIV/AIDS and hepatitis C viruses. Most of these drugs are developed after successfully finding the protease inhibitor compounds that work by stopping the activity of protease enzymes; therefore, preventing the viruses from multiplying. Currently, there are over 16 protease inhibitors approved by the FDA to treat both HIV/AIDS and hepatitis C (Scholar, 2007). A number of initial treatments for COVID-19 have included giving those infected the HIV protease inhibitors lopinavir and ritonavir, (Chu et al., 2004) as well as Ebola antivirals (Hendaus, 2020; Routh, 2020).
Due to the importance of the coronavirus main protease (Mpro) in viral replication, our study utilized the crystal structure of 6LU7 (Liu et al., 2020) from the PDB, (Berman et al., 2000) which is the 312 amino acid main protease of the novel COVID-19 that is responsible for viral replication. The 6LU7 X-ray crystal structure is one of the first deposited main protease structures of the novel COVID-19 and this protein is a potential target for the inhibition of COVID-19. Protease inhibitors are compounds that block the action of proteases and can be used as antiviral drugs. A novel approach has been devised to identify the best five inhibitors from 2692 possible protease inhibitor compounds, that are known to bind to different proteases and their PDB (Berman et al., 2000) structures are known, against the COVID-19 main protease.
Firstly, molecular docking studies were carried out on all 2692 protease inhibitor compounds to obtain the binding affinity of these compounds against the COVID-19 main protease. Then, 10 best inhibitor-protease complex systems obtained from docking were selected for further study with molecular dynamics (MD) simulations. Each MD simulations was carried out for 100 ns in solution with proper physiological conditions. Some inhibitors were found to leave the protease; therefore, these inhibitors are removed from further study. For the remaining inhibitor-protease systems, molecular docking calculations were again performed to understand the binding affinity of these inhibitor compounds against the protease derived from MD simulation. In our approach, the best inhibitors are chosen when inhibitor compounds are found to have high binding affinity in both stand-alone docking studies using the PDB structure and the structure obtained from MD simulation in solution. MD simulations of the inhibitor-protease systems were also used to further study the stability, structure and dynamics of these systems. We propose possible protease inhibitor compounds that can be investigated further with experiments for the development of an antiviral drug against COVID-19.
Methods
The SDF files for 2692 protease inhibitor compounds were downloaded from the Protein Data Bank (PDB) (Berman et al., 2000) to use in molecular docking. All 2692 compounds were uploaded into the virtual screening program PyRx (Dallakyan & Olson, 2015). Hydrogen atoms were added and Gasteiger charges were computed (Gasteiger & Marsili, 1980). All compounds were minimized using the UFF force field with the conjugate gradient algorithm for 200 steps (Rappé et al., 1992). Molecular docking was performed with AutoDoc Vina (Trott & Olson, 2010) to predict the binding affinity and the best binding mode of each of the 2692 protease inhibitors. AutoDoc Vina uses a hybrid scoring function that is inspired by X-score (Wang et al., 2002), which accounts for van der waals forces, hydrogen bonding, deformation penalty, and hydrophobic effect. In addition, AutoDoc Vina combines both the conformational preferences of the receptor–ligand complex and experimental affinity measurements to compute its binding energy (Trott & Olson, 2010). To perform molecular docking, the grid box was centered on the 6LU7 crystal structure and grid dimensions were set to 51.3737 × 66.9738 × 59.60 Å3 to cover the entirety of the protein. All other AutoDoc Vina parameters were left as default. MD simulations were performed using the AMBER 18 package (Case et al., 2018). 11 systems made of COVID-19 main protease and potential inhibitors (Pubchem IDs: 118098670, 104161460, 137349331, 44228999, 163632044, 656932, 5289412, 90176081, 25141820 and PDB chemical ID: 10Q and N3) were prepared using the CHARMM36 additive force field (Brooks et al., 2009; Lee et al., 2016) for proteins and CHARMM General Force Filed (CGenFF)(Vanommeslaeghe et al., 2010) for ligands. The systems were solvated with TIP3P water molecules and the waterbox size was fit to the size of the protein complex within a radius of 10 Å from the surface of the complex. To mimic physiological conditions, 0.15 M KCl ions were added to the systems using the Monte-Carlo ion placing method. A steepest decent energy minimization was carried out for 2500 cycles and then the conjugate gradient algorithm was used for 5000 cycles. Next, the systems were equilibrated for 2 ns under (canonical ensemble) NVT conditions where amount of substance (N), volume (V), and temperature (T) are conserved. The temperature of 303.15 K was controlled using Langevin dynamics (Adelman & Doll, 1976). A restraint constant of 1 kcal/mol was used to restrain the complexes in equilibration. After minimization and equilibration of the systems were conducted, long MD simulations of 100 ns were performed. Long MD simulations were performed under NPT conditions where the temperature was kept at 303.15 K and pressure at 1 atm to mimic experimental conditions. A friction coefficient, γ, of 1.0 ps−1 was used for the Langevin thermostat and the pressure was held constant with the Monte Carlo barostat. Integration was done using a leap-frog algorithm with a 2-fs time step. All bonds involving hydrogen atoms were constrained to their equilibrium values using SHAKE (Axelsen & Li, 1998).
ProTox-II (Banerjee et al., 2018) (http://tox.charite.de/protox_II/index.php?site=home) was used to predict the hepatotoxicity of five main inhibitors found in this study. ProTox-II predicts toxicity based on the compound’s structure using molecular similarity, fragment propensity, machine learning, pharmacophores and is trained on real data to compute the toxic potential of virtual and existing compounds (Banerjee et al., 2018).
Results and discussion
From stand-alone docking calculations of 2692 inhibitors, the ten highest binding affinity inhibitors are presented in Table 1. The highest binding affinity inhibitor is found to be a macrocyclic tissue factor–factor VIIa inhibitor (PubChem ID: 118098670), which has a binding affinity of −10.6 kcal/mol. The binding affinity of the inhibitor 118098670 to the protease structure obtained from the MD simulation of inhibitor-protease complex was also found to have the highest binding affinity, −10.0 kcal/mol. A complex between factor VIIa (FVIIa) proteases and its tissue factor (TF) trigger blood coagulation. Within the TF-FVIIa pathway exits a trypsin-like serine protease factor Xa. The identified macrocyclic tissue factor–factor VIIa inhibitor works by binding to the active site of factor Xa. Factor Xa and its pathway inhibitor complex cause a feedback inhibition of TF-FVIIa, preventing the coagulation of blood. Therefore, TF-FVIIa inhibitors are thought to be promising compounds for treating many thrombotic diseases due to their anticoagulation properties (Al-Horani & Desai, 2016; Ladziata et al., 2016; Lazarus et al., 2004).
Table 1.
Binding affinity of inhibitors with COVID-19 main protease using molecular docking approach and the structures obtained from X-ray crystallography and 100 ns MD simulation.
| PubChem ID | IUPAC name | Binding affinity using PDB structure (kcal/mol) | Binding affinity using MD structure (kcal/mol) |
|---|---|---|---|
| 118098670 | (2R,15R)-2-[(1-Aminoisoquinolin-6-yl)amino]-4,15,17-trimethyl- 7-[1-(2H-tetrazol-5-yl)cyclopropyl]-13-oxa-4,11-diazatricyclo [14.2.2.16,10]henicosa-1(18),6,8,10(21),16,19-hexaene-3,12-dione | −10.6 | −10.0 |
| 104161460 | (R)-6-(2″-((3-hydroxypyrrolidin-1-yl)methyl)biphenyl-4-yl)-1-(3- (5-oxo-4,5-dihydro-1h-1,2,4-triazol-3-yl)phenyl)-3-(trifluoromethyl)- 5,6-dihydro-1H-pyrazolo[3,4-c]pyridin-7(4H)-one | −10.2 | −9.4 |
| 5289412 | N-[(5S,9S,10S,13S)-9-Hydroxy-5,10-bis(2-methylpropyl)-4,7,12,16- tetraoxo-3,6,11,17-tetrazabicyclo[17.3.1]tricosa-1(22),19(23), 20-trien-13-yl]-3-naphthalen-1-yl-2-(naphthalen-1-ylmethyl)propanamide | −9.4 | −9.7 |
| 137349331 | [(2′S,5R,8S,10R,14S)-2′-ethenyl-26-fluoro-14-[[(3S)-1-methyl- 2-oxopiperidine-3-carbonyl]amino]-2,2,4,7,13-pentaoxospiro[2lambda6 -thia-3,6,12,22-tetrazatricyclo[21.4.0.08,12]heptacosa-1(23),24,26-triene-5, 1′-cyclopropane]-10-yl] 4-fluoro-1,3-dihydroisoindole-2-carboxylate | −10.0 | −9.3 |
| 44228999 | 2-[6-[3-[3-(aminomethyl)phenyl]phenoxy]-4-[(3R)-3-(dimethylamino) pyrrolidin-1-yl]-3,5-difluoropyridin-2-yl]oxy-4-(dimethylamino)benzoic acid | −9.8 | – |
| 163632044 | 6-[3-([1,3]oxazolo[4,5-B]pyridin-2-Yl)-2-(Trifluoromethyl)phenoxy]-1- (2,4,6-Trifluorobenzyl)quinazolin-4(1H)-One | −9.6 | −8.8 |
| 656932 | [(1R)-2-[3-[methyl-[1-(naphthalene-2-carbonyl)piperidin-4-yl]carbamoyl] naphthalen-2-yl]-1-naphthalen-1-yl-2-oxoethyl]phosphonic acid | −9.6 | – |
| N3* | N-[(5-methylisoxazol-3-yl)carbonyl]alanyl-l-valyl-n∼1∼-(1R,2Z)-4-(benzyloxy)- 4-oxo-1-{[(3R)-2-oxopyrrolidin-3-yl]methyl}but-2-enyl)-l-leucinamide | −7.3 | – |
| 90176081 | (11S)-4,9-Dioxo-N-[(2S)-1-oxo-3-phenylpropan-2-yl]-17,22-dioxa-10, 30-diazatetracyclo[21.2.2.213,16.15,8]triaconta-1(26),5,7,13,15,23(27), 24,28-octaene-11-carboxamide | −9.3 | −9.2 |
| 10Q* | 2-{(2E,4aR,7aR)-7a-[4-(3-cyanophenyl)thiophen-2-yl]-2-imino-3-methyl- 4-oxooctahydro-6H-pyrrolo[3,4-d]pyrimidin-6-yl}pyridine-3-carbonitrile | −9.5 | – |
| 25141820 | (1R,18R,22R,26S,29S)-26-cyclopentyl-N-[(1R,2S)-1-(cyclopropylsulfonylcarbamoyl)-2- ethenylcyclopropyl]-24,27-dioxo-2,23-dioxa-11,25,28-triazapentacyclo[26.2.1.03, 12.05,10.018,22]hentriaconta-3,5,7,9,11-pentaene-29-carboxamide | −9.6 | – |
*Represents chemical ID from PDB.
Du and company (2007) found that the protease Xa, which is involved in the coagulation pathway described above, is also associated with viral infectivity of the SARS coronavirus (Du et al., 2007). The protease factor Xa cleaved the SARS-CoV S protein into two functional units S1 and S2 subunits, which facilitated viral infection. In addition, factor Xa was expressed in ACE2 cells, the host enzyme that the SARS S protein binds to infect human cells (Du et al., 2007). The macrocyclic tissue factor–factor VIIa inhibitor is a plausible inhibitor for COVID-19 because it inhibits factor Xa, which may prevent the novel coronavirus from entering its host. The COVID-19 main protease has 97% sequence identity with the SARS-CoV main protease (Zhang et al., 2020). Therefore, factor-factor VIIa inhibitors may also block the viral replication in the novel coronavirus. Additionally, the proposed inhibitor has high selectivity towards the novel coronavirus main protease which may help to prevent viral replication.
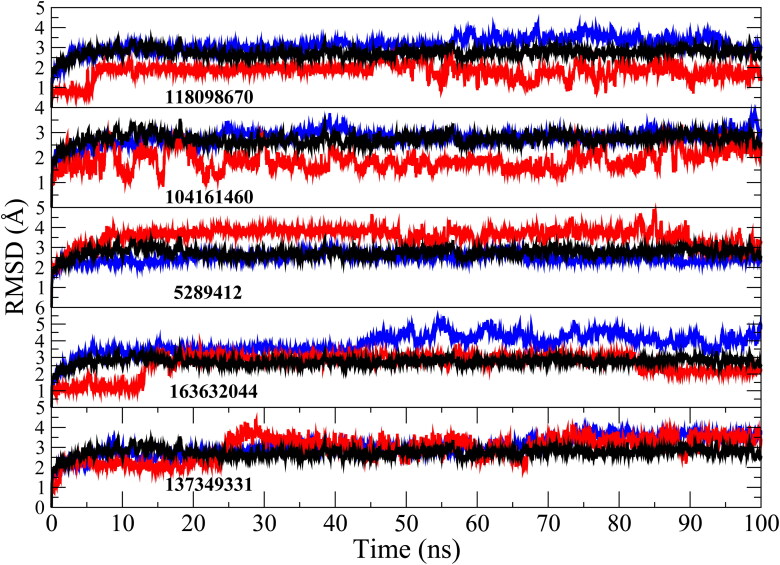
A MD simulation of the inhibitor (118098670)-protease system shows that the system deviates very little from the x-ray crystal structure (Figure 1), with root mean square deviation (RMSD) of 3 Å for the protease from crystal structure and 1.5 Å for the inhibitor (118098670) from its starting structure.
Figure 1.
RMSD of inhibitors (red), main protease (black) and protease-inhibitor complex (blue) obtained from 100 ns MD simulation.
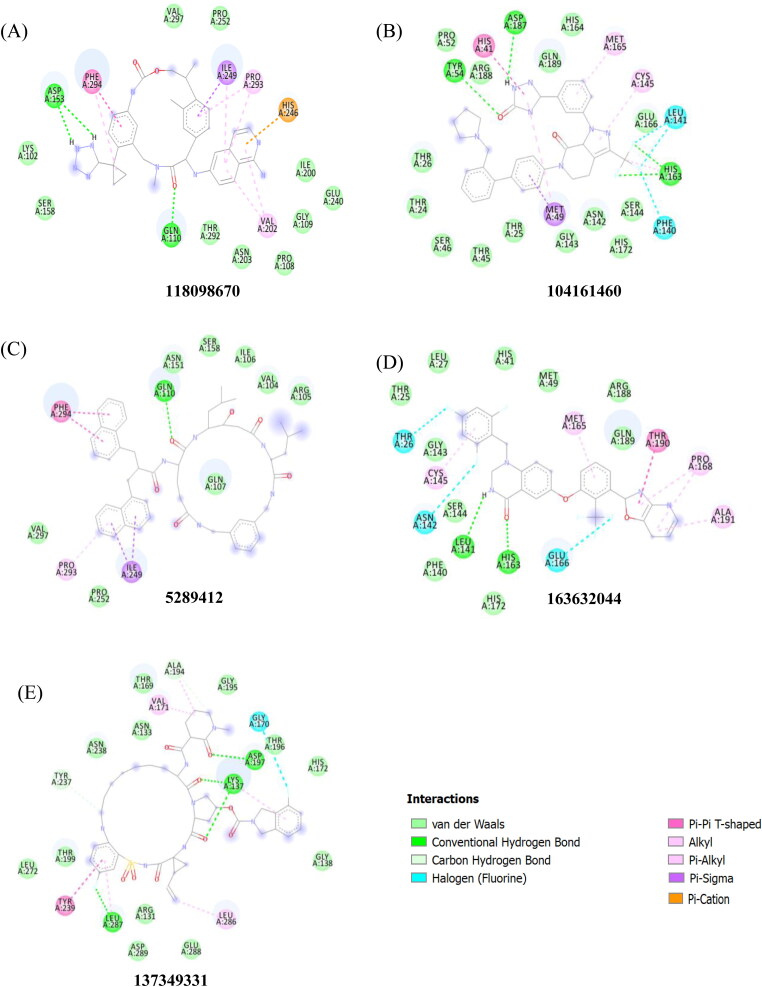
Both MD (Table 2) and docking studies (Figure 2) show significant hydrogen bonding, mainly between the inhibitor and the ASP-153 and GLN-110 amino acids in the COVID-19 main protease. In addition, a number of pi interactions were found between benzene rings and HIS-246 and VAL-202 before MD simulation and ILE-249, PRO-293 and PHE-294 after MD simulation (Figure 2 and Table 3). In general, pi interactions play a key role in the stability of ligands in the binding site (Arthur & Uzairu, 2019). The key residues in the active site of the COVID-19 main protease are the PHE-294 and ILE-249 residues, with occupancies of 99% and 99%, respectively and it appears the 118098670 inhibitor prefers to bind in the hydrophobic active site of the main protease (Figure 2A).
Table 2.
Average lengths (r), angles (θ), and occupancy (f) of unique residues in COVID-19 main protease that form intermolecular H-bonds with the five proposed inhibitor complexes from 100 ns MD simulation.a,b
| Donor/acceptor pair | f (%) | r (Å) | θ (°) | ||
|---|---|---|---|---|---|
| 118098670 Inhibitor | |||||
| GLN-110NE2/LIGO3 | 31 | 3.06 | 156.7 | ||
| ASP-153OD2/LIGN7 | 30 | 2.75 | 162.6 | ||
| ILE-249HA/LIGC19 | 27 | 3.25 | 158.2 | ||
| ASP-248O/LIGN3 | 25 | 3.14 | 152.9 | ||
| ASN-151CB/LIGH33 | 18 | 3.27 | 154.4 | ||
| ASP-245OD2/LIGN4 | 16 | 2.87 | 156.4 | ||
| PHE-294CB/LIGH25 | 16 | 3.31 | 154.6 | ||
| 104161460 Inhibitor | |||||
| GLU-166N/LIGO2 | 42 | 3.03 | 157.2 | ||
| HIS-41NE2/LIGN6 | 36 | 3.09 | 160.3 | ||
| TYR-54OH/LIGN7 | 24 | 3.07 | 155.4 | ||
| ASP-187O/LIGN7 | 21 | 2.98 | 161.4 | ||
| MET-49O/LIGN7 | 20 | 2.95 | 161.8 | ||
| MET-165CA/LIGO2 | 15 | 3.36 | 145.6 | ||
| ARG-188NH1/LIGO2 | 13 | 2.81 | 162.2 | ||
| THR-45CA/LIGO2 | 12 | 3.25 | 158.6 | ||
| SER-46CA/LIGH14 | 11 | 3.26 | 147.2 | ||
| 5289412 Inhibitor | |||||
| VAL-297HB/LIGC26 | 32 | 3.23 | 152.7 | ||
| ASP-153CG /LIGO6 | 27 | 2.80 | 161.4 | ||
| PHE-294CA/LIGH30 | 26 | 3.28 | 157.8 | ||
| ASP-248O/LIGN3 | 25 | 3.14 | 152.9 | ||
| ILE-249HA/LIGC31 | 12 | 3.31 | 152.3 | ||
| 137349331 Inhibitor | |||||
| THR-169OG1/LIGO3 | 26 | 2.80 | 163.2 | ||
| ASP-197N/LIGO9 | 20 | 3.04 | 162.5 | ||
| 163632044 Inhibitor | |||||
| ASP-187O/LIGN2 | 49 | 2.94 | 149.0 | ||
| GLU-47CD LIGN4 | 43 | 3.23 | 147.9 | ||
| HIS-41HB2 /LIGC27 | 17 | 3.22 | 148.1 | ||
| GLN-189NE2/LIGO1 | 14 | 3.12 | 155.1 | ||
| MET-165HB2/ LIGC22 | 12 | 3.25 | 151.3 | ||
| GLU-166N/LIGO1 | 11 | 2.95 | 161.7 | ||
aOnly occupancies (f) up to 10% are reported.
b3.5 Å hydrogen bond distance is given with respect to the heavy atoms.
Figure 2.
Binding interactions sites of main protease with inhibitors, 118098670 (A), 104161460 (B), 5289412 (C), 1636044(D) and 137349331 (E), obtained from molecular docking.
Table 3.
Native contacts of unique residues in COVID-19 main protease with five proposed inhibitors from 100 ns MD simulation.a
| 1180986670 |
104161460 |
5289412 |
137349331 |
163632044 |
|||||
|---|---|---|---|---|---|---|---|---|---|
| Residues | f (%) | Residues | f (%) | Residues | f (%) | Residues | f (%) | Residues | f (%) |
| PHE-294 | 99 | HIS-41 | 99 | PRO-252 | 99 | THR-169 | 71 | HIS-41 | 99 |
| ILE-249 | 99 | ASP-187 | 99 | ILE-249 | 81 | ALA-194 | 59 | THR-25 | 97 |
| PRO-293 | 90 | SER-46 | 96 | GLY-251 | 81 | VAL-171 | 53 | MET-49 | 97 |
| VAL-297 | 84 | THR-54 | 96 | LEU-253 | 78 | HIS-164 | 96 | ||
| PRO-252 | 81 | ARG-188 | 95 | LEU-250 | 78 | MET-165 | 96 | ||
| ASN-151 | 58 | MET-49 | 95 | ASP-258 | 77 | GLN-189 | 93 | ||
| ASP-153 | 57 | THR-45 | 91 | PRO-293 | 75 | SER-46 | 93 | ||
| SER-158 | 56 | THR-25 | 90 | ASP-248 | 74 | GLU-166 | 87 | ||
| GLN-110 | 54 | CYS-44 | 87 | VAL-297 | 68 | CYS-145 | 78 | ||
| ILE-152 | 53 | GLN-189 | 86 | PHE-294 | 66 | VAL-42 | 76 | ||
| THR-292 | 53 | LEU-50 | 85 | CYS-44 | 66 | ||||
| THR-111 | 53 | THR-24 | 85 | LEU-27 | 57 | ||||
| VAL-104 | 53 | ARG-40 | 82 | ||||||
| GLU-166 | 70 | ||||||||
| PRO-52 | 64 | ||||||||
| HIS-164 | 62 | ||||||||
| ASN-142 | 61 | ||||||||
| THR-190 | 56 | ||||||||
| MET-165 | 55 | ||||||||
aOnly occupancies (f) up to 50% are reported.
Our in silico approach also finds the factor Xa protease inhibitors, phenyltriazolinones (PubChem ID 104161460) (Quan et al., 2010), to have the second highest binding affinity of −10.2 kcal/mol from stand-alone docking calculations and −9.4 kcal/mol from the MD structure of the COVID-19 main protease (Table 1). Experimental data shows that cleavage of the SARS-CoV S protein into functional units increased with the amount of concentration of factor Xa and cleavage of the S protein can be prevented with factor Xa inhibitors (Du et al., 2007). Additionally, the high binding affinity of phenyltriazolinone for the COVID-19 main protease may be able to inhibit viral replication of the novel coronavirus. MD simulation shows a number of hydrogen bonds, mainly between the inhibitor and the GLU-166, HIS-41, TYR-54 and ASP-187 residues in the COVID-19 binding pocket (Table 2). Furthermore, a number of pi-interactions are formed with HIS-41, MET-165 and MET-49 before and after MD simulation (Figure 2 and Table 3). This inhibitor is especially important as the inhibitor binds to the active site consisting of a cysteine amino acid and a nearby histidine that cuts polyproteins into functional proteins to facilitate viral replication (Figure 5B) (Anand et al., 2002; Xue et al., 2008). In addition, the MD simulation between the inhibitor (104161460)-protease system shows that COVID-19 main protease deviates very little from the original X-ray crystal structure with a RMSD of 3 Å for the protease (Figure 1).
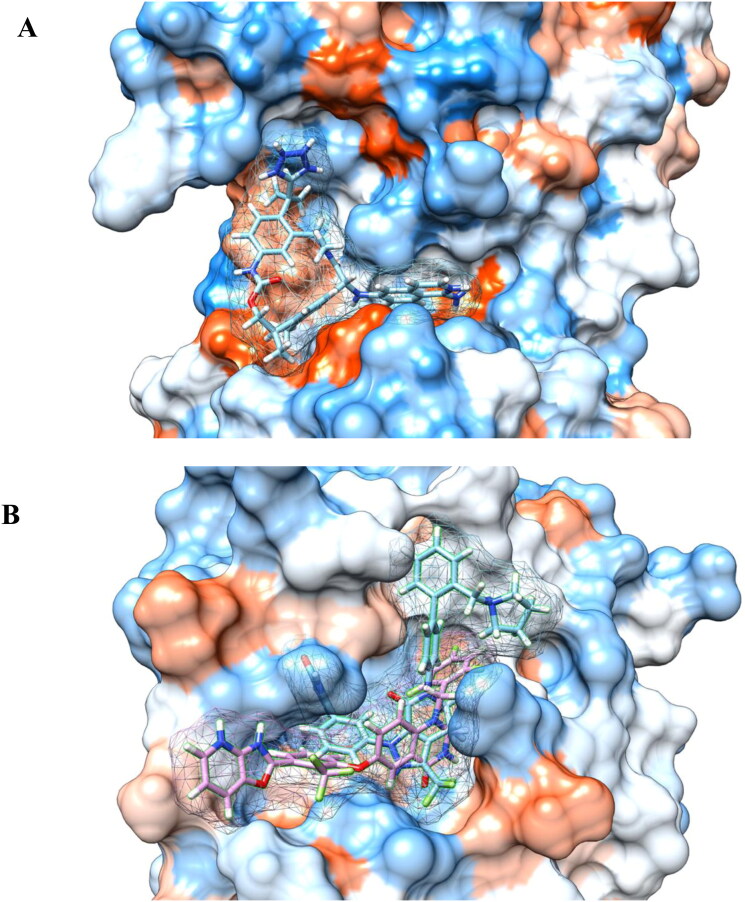
Figure 5.
Binding modes showing (A) 118098670 inhibitor interacts with the active site consisting of Phe-294 and Ile-249 residues of main protease and (B) the 104161460 and 163632044 inhibitors interacts with the catalytic dyad His-41 and Cys-145 residues of main protease. Blue color represents the hydrophilic residues, while orange-red color represents hydrophobic residues.
The third potential protease inhibitor compound, with a binding affinity of −9.4 kcal/mol obtained from standalone docking calculations and −9.7 kcal/mol from the MD structure of the COVID-19 main protease (Table 1), is an endothiapepsin inhibitor (PubChem ID 5289412) (Coates et al., 2002). Endothiapepsin is a member of the aspartic proteinase enzymes which are found in HIV retrovirus and they also play major roles in amyloid disease, fungal infections and malaria (Coates et al., 2002). The inhibition of these aspartic proteinases with inhibitors have been effective in the treatment of AIDS (dos Santos, 2010; Nguyen et al., 2008) and these class of inhibitors are targets for many therapeutic drugs (Hartman et al., 2015). MD simulation of the inhibitor-protease system indicate that the protease structure remains very similar to that of the x-ray structure (RMSD of 2.3 Å), but the inhibitor reorganizes into the binding pocket of the protease (Figure 1). MD simulations showed H-bonding between the inhibitor and the VAL-297, ASP-153, PHE-294 and ASP-248 residues (Table 2). In addition, a number of intermolecular interactions were also observed, mainly between the protease and PRO-252, ILE-249, GLY-251, LEU-253 and LEU-250 (Table 3).
A fourth potential protease inhibitor compound that was identified, with a binding affinity of −10 kcal/mol from standalone docking calculations and −9.3 kcal/mol using the MD-derived structure of the COVID-19 main protease (Table 1), is a macrocyclic HCV NS3/4A protease inhibitor (PubChem ID 137349331). The HCV NS3/4A protease in hepatitis C has been a key target for antiviral drugs (McGivern et al., 2015). HCV NS3/4A proteases cleaves the hepatitis C polyprotein at four junctions, releasing NS proteins 4A, 4B, 5A and 5B, which is important in viral replication (Bartenschlager et al., 1995; Brass et al., 2008). Both hepatitis C and the coronavirus contain a positive-stranded genome (Schiering et al., 2011) and rely on a similar mechanism to replicate their RNA (Schiering et al., 2011; Thiel et al., 2003). Furthermore, both COVID-19 and HCV NS3/4A proteases have a double β-barrel fold with similar orientation, regions of structural similarity, and a very similar substrate binding pocket with active site catalytic residues His41 and Cys145 and His57 and Ser139 (Khushboo et al., 2020). These similarities make the HCV NS3/4A protease inhibitor a plausible drug candidate to inhibit the COVID-19 main protease to block its viral and transcription function. The interactions between the ligand and ALA-194 and VAL-171 residues are maintained after MD simulation (Figure 2 and Table 3). Furthermore, hydrogen bonding is maintained after MD simulation with the ligand and ASP-197 (Figure 2 and Table 2). A RMSD analysis shows that the RMSD deviates very little from the original X-ray crystal structure (RMSD 3 Å). In addition, the RMSD shows that the ligand seems to reorganize into the binding pocket and displays a similar pattern to the protease (Figure 1).
Additionally, we find the binding affinity of an allosteric HCV NS5B polymerase thumb pocket 2 protease inhibitor (PubChem ID 163632044 to have −9.6 kcal/mol to the COVID-19 main protease in stand-alone docking calculations and −8.8 kcal/mol using the MD simulation derived structure (Table 1). The class of HCV NS3/4A protease inhibitors have shown to have antiviral activity with hepatitis C patients and a few drugs have already been approved by the FDA (Hinrichsen et al., 2004). The designed HCV NS5B polymerase thumb pocket 2 inhibitor created by Hucke et al. (2014) has a greater potency to the most prevalent hepatitis C genotypes (Hucke et al., 2014). Furthermore, the success of HCV protease inhibitors to prevent viral replication in hepatitis C patients could very well translate in the treatment of COVID-19. COVID-19 contains a non-structural protein 12 (nsp12) that catalyzes and synthesizes viral RNA that plays a critical role in viral replication and transcription (Gao et al., 2020). The HCV ns5b polymerase, in which the allosteric HCV NS5B polymerase thumb pocket 2 protease inhibitor targets, has been found to have high similarity to nsp12 (Gao et al., 2020). Therefore, ns5b polymerase inhibitors are promising drug targets to prevent viral replication of the novel coronavirus. A RMSD calculation of MD simulation finds that the 163632044 inhibitor does not deviate very much from its initial structure after 15 ns, and the protease also does not deviate much from the initial x-ray crystal structure (Figure 1). The 163632044 inhibitor seems to also be especially important, as it forms native contacts with the His-41 and Cys-145 residues in the COVID-19 main protease active site, with occupancies of 99% and 78%, respectively (Table 3 and Figure 5B). In addition, hydrogen bonding is observed between the inhibitor and HIS-41, ASP-187 and GLU-47 residues (Table 2). The His-41 and Cys-145 residues are especially important as they make up the catalytic dyad in the COVID-19 main protease and cut polyproteins into functional proteins to facilitate viral replication. Thus, their inhibition may be especially important in the development of antiviral drugs that target COVID-19.
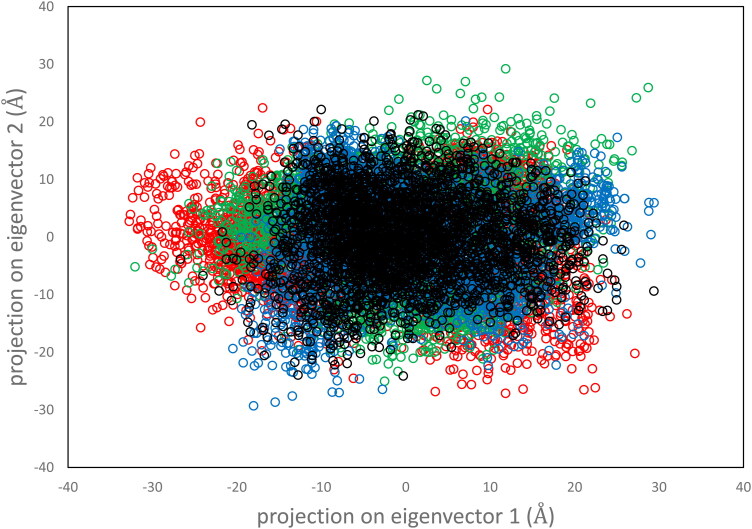
To further assess the stability and dynamics of the top three proposed protease-inhibitor complexes, the radius of gyration (Rg), solvent accessible surface area (SASA), and a principal component analysis (PCA) were calculated and compared to the main protease crystal structure obtained from PDB 6LU7. Eigenvectors is a PCA method to determine the motion of the protein-inhibitor complexes along two principal components. Figure 3 shows the eigenvector for a 100 ns simulation with a 2ps time step.
Figure 3.
Projection of motion along first two principal eigenvectors for Cα atoms of main protease (black), protease-118098670 complex (red), protease-104161460 complex (green), protease-5289412 complex (blue).
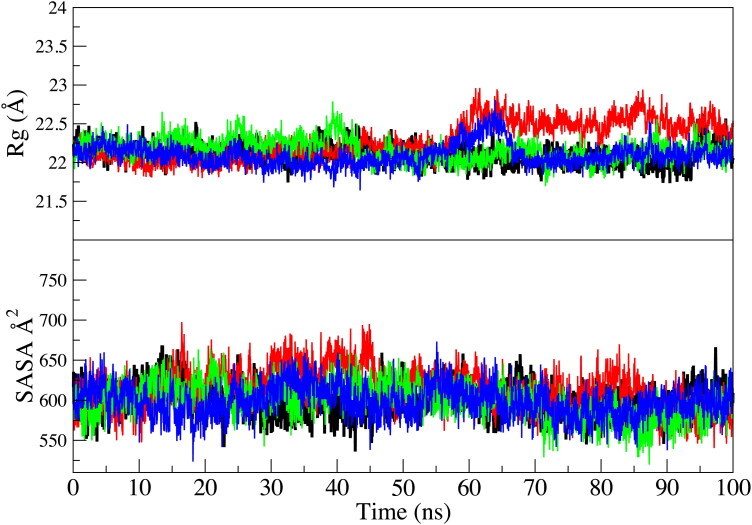
Overall, the distribution of motion is very similar for the three proposed inhibitor-protease systems compared to the Mpro. The internal motion of the protease-inhibitor systems seems to be defined along the first eigenvector (Figure 3). The radius of gyration and solvent accessible surface area analysis also agree with the PCA calculation (Figure 4).
Figure 4.
Radius of gyration (Rg) and solvent accessible surface area (SASA) of Cα atoms of main protease (black), protease-118098670 complex (red), protease-104161460 complex (green), protease-5289412 complex (blue).
The radius of gyration (Rg) of all protease-inhibitor complexes are found very similar to the Mpro crystal structure with Rg values between 21.75 and 22.75 Å. In addition, the solvent accessible surface area analysis shows that all three proposed inhibitor-protease systems are relatively similar and the values for the Mpro overlap well with the proposed protease-inhibitor systems.
Furthermore, the binding affinity of lopinavir, one of the HIV-protease inhibitors, and hydroxychloroquine, a drug to treat malaria, with the COVID-19 main protease are found to be −8.2 and −6.3 kcal/mol, respectively. The binding site for lopinavir is found to be similar to the 118098670, 137349331 and 5289412 inhibitors. The 118098670 and 5289412 inhibitors share the same native contacts with ILE-249, VAL-297 and PHE-294 in the main protease. Furthermore, both the 118098670 and 5289412 inhibitors form hydrogen bonds with the ASP-153, PHE-294, ASP-248 and the ILE-249 residues in the main protease. Interestingly, hydroxychloroquine’s binding site is found to be similar to the 104161460 and 163632044 inhibitors (see supporting info). Early data suggests that hydroxychloroquine may be effective in the treatment of COVID-19 by preventing symptoms from worsening and improving recovery times (Z. Chen et al., 2020). The early data from hydroxychloroquine may further exemplify the importance of targeting the catalytic dyad of His-41 and Cys-145 in the COVID-19 active site. Since hydroxychloroquine and the 104161460 and 163632044 inhibitors both interact with His-41 and Cys-145 residues in the active site of the COVID-19 main protease, they may very well be potential drug candidates to inhibit the viral replication abilities of the COVID-19 main protease. The catalytic dyad active site of the 104161460 and 163632044 inhibitors in the COVID-19 main protease are shown in Figure 5B. In addition, the 104161460 and 163632044 inhibitors share many of the same contacts with SER-46, MET-49, GLN-189, GLU-166, HIS-164 and MET-65. Hydrogen bonding is shared between residues ASP-187, HIS-41 and MET-165 and the main protease. The His and Cys residues appear in a mostly hydrophilic binding pocket and it seems that both hydroxychloroquine and the 104161460 and 163632044 inhibitors prefer this hydrophilic active site (Figure 5B).
Next, the theoretical dissociation calculations, Kd, obtained from the MD derived crystal structures were compared to the experimental dissociation constant, Kd, values of similar protease-inhibitor systems (Table 4). The theoretical Kd values are calculated from MD-derived binding free energy (ΔG) values as shown in Table 1 using the equation ΔG=RTln Kd. The five proposed inhibitors, 118098670, 5289412, 104161460, 137349331 and 163632044, have theoretical dissociation constants (Kd) of 5.18 × 10−8, 8.48 × 10−8, 1.40 × 10−7, 1.66 × 10−7, and 3.84 × 10−7 M, respectively (Table 4). Experimentally Kd values were determined for similar protease-inhibitor systems. Theoretical Kd values of five proposed inhibitors to the COVID-19 main protease are found to agree very well with the experimental Kd values for similar protease-inhibitor systems. The 118098670 inhibitor is found to have an experimental Kd value of 1.60 × 10−9 M against some serine proteases FIXa, FXa, thrombin, and trypsin (Ladziata et al., 2016), which agree with the theoretical Kd value, 5.18 × 10−8 M. In addition, the 5289412 inhibitor is found to have an experimental Kd of 4.00 × 10−9 M against saccharopepsin, an aspartic protease (Cronin et al., 2000). This binding affinity is also in agreement with the theoretical dissociation constant, 8.48 × 10−8 M. Furthermore, the 104161460 inhibitor has an experimental Kd of 1.90 × 10−10 M against factor Xa proteases (Quan et al., 2010), which also agrees with our theoretical calculations (Table 4).
Table 4.
Theoretical and experimental dissociation constants for the five proposed inhibitors.
| PubChem ID | Theoretical Kd (M) | Experimental Kd (M) |
|---|---|---|
| 118098670 | 5.18 × 10−8 | 1.60 × 10−9 |
| 5289412 | 8.48 × 10−8 | 4.00 × 10−9 |
| 104161460 | 1.40 × 10−7 | 1.90 × 10−10 |
| 137349331 | 1.66 × 10−7 | 1.00 × 10−10, 4.60 × 10−10 |
| 163632044 | 3.84 × 10-7 | 6.00 × 10−8 |
The use of FDA approved drugs or already existing experimental drugs against COVID-19 can dramatically improve the time required to provide therapeutic benefit against the novel coronavirus. The 137349331 inhibitor identified from molecular docking is similar to the hepatitis C macrocyclic NS3/4A inhibitors Vaniprevir and Danoprevir, which have already been approved for human use. Danoprevir, in particular, is already being used in clinical trials in China to treat COVID-19 patients (H. Chen et al., 2020). The experimental binding affinity (Kd) of Vaniprevir and Danoprevir against the HCV NS3/4A serine protease is found to be 4.60 × 10−10 and 1.00 × 10−10 M, respectively (Ali et al., 2013). These values are in agreement with the Kd values found from our theoretical calculations (Table 4). Furthermore, the proposed 163632044 inhibitor is very similar to Gilead’s hepatitis C drug Sofosbuvir that is currently being investigated as a potential drug for COVID-19. Sofosbuvir has an experimental Kd value of 6.00 × 10−8 M against the NS5B polymerase of the hepatitis C virus (‘Sovaldi (sofosbuvir),’ 2015). This experimental Kd value is very similar to that obtained from calculation of the 163632044 HCV NS5B polymerase inhibitor against the COVID-19 main protease, with a Kd value of 3.84 × 10−7 M.
Moreover, the calculated dissociation constants of the five proposed COVID-19 protease inhibitors were plotted against the experimental dissociation constant values (see supporting info). The correlation coefficient R2 is 0.81, indicating that there is a substantial correlation between our theoretical dissociation constant values and experimental dissociation constant values (see supporting info.)
Toxicity is a major issue in drug development. Most drug candidates fail in clinical trials due to toxicity that the body cannot tolerate. ProTox-II (Banerjee et al., 2018) was used to check the toxicity of the five proposed inhibitors, namely 118098670, 5289412, 104161460, 137349331 and 163632044, found in this study, to assess the safety profile of these chemical compounds. ProTox-II (Banerjee et al., 2018) shows that all these inhibitors may be tolerable by the body, since the hepatotoxicity profile of these compounds were found inactive with probabilities of 54%, 80%, 55%, 50% and 55%, respectively.
Conclusions
To summarize, this study identified the best inhibitor that binds with the main protease of COVID-19 to be the macrocyclic tissue factor-factor VIIa inhibitor (PubChem ID: 118098670). Both molecular docking and MD derived binding affinities were found to be −10.6 and −10.0 kcal/mol, respectively. In some people, the increased inflammation caused by the COVID-19 virus had led to increased clotting (Willyard, 2020). The TF-FVIIa inhibitors are known to prevent the coagulation of blood and have antiviral activity as shown in the case of SARS coronavirus (Du et al., 2007). Also, TF-VIIa coagulation cascade inhibitors are viewed as promising treatments for malaria, (Kendrick et al., 2006; Ruf, 2004), as studies indicate increased coagulation activity in malaria (Gérardin et al., 2002; Ladhani et al., 2002). Furthermore, this study finds the phenyltriazolinones (PubChem ID: 104161460) and allosteric HCV NS5B polymerase thumb pocket 2 (PubChem ID: 163632044) inhibitors to be plausible inhibitors for the inhibition of the COVID-19 main protease, as these inhibitors interact with the His-41 and Cys-145 catalytic dyad in the COVID-19 main protease that has been found to be especially important in viral replication (Zhu et al., 2011). This study also found the endothiapepsin (PubChem ID: 5289412) and macrocyclic HCV NS3/4A (PubChem ID: 137349331) protease inhibitors to be plausible inhibitors of the COVID-19 main protease, and clinically they have also shown great antiviral activity. The results obtained from our theoretical dissociation constant values of five proposed inhibitors to the COVID-19 main protease agree very well with the experimental values for similar protease-inhibitor systems. Future studies may employ more computationally intensive approaches such as molecular mechanics/generalized born surface area (MM/GBSA) to predict binding free energy of coronavirus inhibitors. In addition, docking studies and MD may also be used to target the coronavirus spike protein to prevent it from entering ACE2 host cells.
Supplementary Material
Funding Statement
The authors acknowledge the Advanced Cyberinfrastructure for Education and Research (ACER) at The University of Illinois at Chicago that have contributed to the research results reported within this paper. We also thank Dr. Justin Lorieau and Dr. Akhteruzzaman Molla for their insightful discussion.
Disclosure statement
No potential conflict of interest was reported by the authors.
References
- Adelman S. A., & Doll J. D. (1976). Generalized Langevin equation approach for atom/solid-surface scattering: General formulation for classical scattering off harmonic solids. The Journal of Chemical Physics, 64(6), 2375 10.1063/1.432526 [DOI] [Google Scholar]
- Al-Horani R. A., & Desai U. R. (2016). Factor XIa inhibitors: A review of the patent literature. Expert Opinion on Therapeutic Patents, 26(3), 323–345. 10.1517/13543776.2016.1154045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ali A., Aydin C., Gildemeister R., Romano K. P., Cao H., Özen A., Soumana D., Newton A., Petropoulos C. J., Huang W., & Schiffer C. A. (2013). Evaluating the role of macrocycles in the susceptibility of hepatitis C virus NS3/4A protease inhibitors to drug resistance. ACS Chemical Biology, 8(7), 1469–1478. 10.1021/cb400100g [DOI] [PMC free article] [PubMed] [Google Scholar]
- Al-Khafaji K., Al-Duhaidahawi D., & Tugba T. T. (2020). Using integrated computational approaches to identify safe and rapid treatment for SARS-CoV-2. Journal of Biomolecular Structure and Dynamics, 1–9. 10.1080/07391102.2020.1764392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anand K., Palm G. J., Mesters J. R., Siddell S. G., Ziebuhr J., & Hilgenfeld R. (2002). Structure of coronavirus main proteinase reveals combination of a chymotrypsin fold with an extra alpha-helical domain. The EMBO Journal, 21(13), 3213–3224. 10.1093/emboj/cdf327 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arthur D. E., & Uzairu A. (2019). Molecular docking studies on the interaction of NCI anticancer analogues with human Phosphatidylinositol 4,5-bisphosphate 3-kinase catalytic subunit. Journal of King Saud University - Science, 31(4), 1151–1166. 10.1016/j.jksus.2019.01.011 [DOI] [Google Scholar]
- Axelsen P. H., & Li D. (1998). Improved convergence in dual-topology free energy calculations through use of harmonic restraints. Journal of Computational Chemistry, 19(11), 1278–1283. [DOI] [Google Scholar]
- Banerjee P., Eckert A. O., Schrey A. K., & Preissner R. (2018). ProTox-II: A webserver for the prediction of toxicity of chemicals. Nucleic Acids Research, 46(W1), W257–263. 10.1093/nar/gky318 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartenschlager R., Lohmann V., Wilkinson T., & Koch J. O. (1995). Complex formation between the NS3 serine-type proteinase of the hepatitis C virus and NS4A and its importance for polyprotein maturation. Journal of Virology, 69(12), 7519–7528. 10.1128/JVI.69.12.7519-7528.1995 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belouzard S., Millet J. K., Licitra B. N., & Whittaker G. R. (2012). Mechanisms of coronavirus cell entry mediated by the viral spike protein. Viruses, 4(6), 1011–1033. 10.3390/v4061011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berman H. M., Westbrook J., Feng Z., Gilliland G., Bhat T. N., Weissig H., Shindyalov I. N., & Bourne P. E. (2000). The Protein Data Bank (www.rcsb.org). Nucleic Acids Research, 28(1), 235–242. 10.1093/nar/28.1.235 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boopathi S., Poma A. B., & Kolandaivel P. (2020). Novel 2019 coronavirus structure, mechanism of action, antiviral drug promises and rule out against its treatment. Journal of Biomolecular Structure and Dynamics, 30, 1–10. 10.1080/07391102.2020.1758788 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bosch B. J., van der Zee R., de Haan C. A. M., & Rottier P. J. M. (2003). The coronavirus spike protein is a class i virus fusion protein: structural and functional characterization of the fusion core complex. Journal of Virology, 77(16), 8801–8811. 10.1128/jvi.77.16.8801-8811.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brass V., Berke J. M., Montserret R., Blum H. E., Penin F., & Moradpour D. (2008). Structural determinants for membrane association and dynamic organization of the hepatitis C virus NS3-4A complex. Proceedings of the National Academy of Sciences of the United States of America, 105(38), 14545–14550. 10.1073/pnas.0807298105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brooks B. R., Brooks C. L., Mackerell A. D., Nilsson L., Petrella R. J., Roux B., Won Y., Archontis G., Bartels C., Boresch S., Caflisch A., Caves L., Cui Q., Dinner A. R., Feig M., Fischer S., Gao J., Hodoscek M., Im W., … Karplus M. (2009). CHARMM: The biomolecular simulation program. Journal of Computational Chemistry, 30(10), 1545–1614. 10.1002/jcc.21287 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Case D. A., Ben-Shalom I. Y., Brozell S. R., Cerutti D. S., Cheatham I. I. I., Cruzeiro T. E., Darden V. W. D., Duke T. A., Ghoreishi R. E., D., & Gilson M. K. (2018). Amber. University of California. [Google Scholar]
- Chen H., Zhang Z., Wang L., Huang Z., Gong F., Li X., Chen Y., & Wu J. J. (2020). First clinical study using HCV protease inhibitor danoprevir to treat naive and experienced COVID-19 patients. MedRxiv, 10.1101/2020.03.22.20034041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Z., Hu J., Zhang Z., Jiang S., Han S., Yan D., Zhuang R., Hu B., & Zhang Z. (2020). Efficacy of hydroxychloroquine in patients with COVID-19: Results of a randomized clinical trial. MedRxiv, 10.1101/2020.03.22.20040758 [DOI] [Google Scholar]
- Chu C. M., Cheng V. C. C., & Hung I. F. N. (2004). Role of lopinavir/ritonavir in the treatment of SARS: Initial virological and clinical findings. Thorax, 59(3), 252–256. 10.1136/thorax.2003.012658 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coates L., Erskine P. T., Crump M. P., Wood S. P., & Cooper J. B. (2002). Five atomic resolution structures of endothiapepsin inhibitor complexes: Implications for the aspartic proteinase mechanism. Journal of Molecular Biology, 318(5), 1405–1415. 10.1016/S0022-2836(02)00197-3 [DOI] [PubMed] [Google Scholar]
- Cronin N. B., Badasso M. O., Tickle I. J., Dreyer T., Hoover D. J., Rosati R. L., Humblet C. C., Lunney E. A., & Cooper J. B. (2000). X-ray structures of five renin inhibitors bound to saccharopepsin: Exploration of active-site specificity. Journal of Molecular Biology, 303(5), 745–760. 10.1006/jmbi.2000.4181 [DOI] [PubMed] [Google Scholar]
- Dallakyan S., & Olson A. J. (2015). Small-molecule library screening by docking with PyRx. Methods in Molecular Biology (Clifton, N.J.), 1263, 243–250. 10.1007/978-1-4939-2269-7_19 [DOI] [PubMed] [Google Scholar]
- Dong E., Du H., & Gardner L. (2020). An interactive web-based dashboard to track COVID-19 in real time. The Lancet. Infectious Diseases, 20(5), 533–534. 10.1016/S1473-3099(20)30120-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- dos Santos A. L. S. (2010). HIV aspartyl protease inhibitors as promising compounds against Candida albicans André Luis Souza dos Santos. World Journal of Biological Chemistry, 1(2), 21–30. 10.4331/wjbc.v1.i2.21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dougherty W. G., & Semler B. L. (1993). Expression of virus-encoded proteinases: Functional and structural similarities with cellular enzymes. Microbiological Reviews, 57(4), 781–822. 10.1128/MMBR.57.4.781-822.1993 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du L., Kao R. Y., Zhou Y., He Y., Zhao G., Wong C., Jiang S., Yuen K. Y., Jin D. Y., & Zheng B. J. (2007). Cleavage of spike protein of SARS coronavirus by protease factor Xa is associated with viral infectivity. Biochemical and Biophysical Research Communications, 359(1), 174–179. 10.1016/j.bbrc.2007.05.092 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fehr A. R., & Perlman S. (2015). Coronaviruses: An overview of their replication and pathogenesis. Methods in Molecular Biology (Clifton, N.J.), 1282, 1–23. 10.1007/978-1-4939-2438-7_1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao Y., Yan L., Huang Y., Liu F., Zhao Y., Cao L., Wang T., Sun Q., Ming Z., Zhang L., Ge J., Zheng L., Zhang Y., Wang H., Zhu Y., Zhu C., Hu T., Hua T., Zhang B., … Rao Z. (2020). Structure of the RNA-dependent RNA polymerase from COVID-19 virus. Science (New York, N.Y.), 368(6492), 779–782. 10.1126/science.abb7498 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gasteiger J., & Marsili M. (1980). Iterative partial equalization of orbital electronegativity-a rapid access to atomic charges. Tetrahedron, 36(22), 3219–3228. 10.1016/0040-4020(80)80168-2 [DOI] [Google Scholar]
- Gérardin P., Rogier C., Ka A. S., Jouvencel P., Brousse V., & Imbert P. (2002). Prognostic value of thrombocytopenia in African children with falciparum malaria. The American Journal of Tropical Medicine and Hygiene, 66(6), 686–691. 10.4269/ajtmh.2002.66.686 [DOI] [PubMed] [Google Scholar]
- Hartman A. M., Mondal M., Radeva N., Klebe G., & Hirsch A. K. H. (2015). Structure-based optimization of inhibitors of the aspartic protease endothiapepsin. International Journal of Molecular Sciences, 16(8), 19184–19194. 10.3390/ijms160819184 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hendaus M. A. (2020). Remdesivir in the treatment of coronavirus disease 2019 (COVID-19): A simplified summary. Journal of Biomolecular Structure and Dynamics, 10.1080/07391102.2020.1767691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hinrichsen H., Benhamou Y., Wedemeyer H., Reiser M., Sentjens R. E., Calleja J. L., Forns X., Erhardt A., Crönlein J., Chaves R. L., Yong C. L., Nehmiz G., & Steinmann G. G. (2004). Short-term antiviral efficacy of BILN 2061, a hepatitis C virus serine protease inhibitor, in hepatitis C genotype 1 patients. Gastroenterology, 127(5), 1347–1355. 10.1053/j.gastro.2004.08.002 [DOI] [PubMed] [Google Scholar]
- Hoffmann M., Kleine-Weber H., Krüger N., Müller M., Drosten C., & Pöhlmann S. (2020). The novel coronavirus 2019 (2019-nCoV) uses the SARS-coronavirus receptor ACE2 and the cellular protease TMPRSS2 for entry into target cells. BioRxiv, 10.1101/2020.01.31.929042 [DOI] [Google Scholar]
- Hucke O., Coulombe R., Bonneau P., Bertrand-Laperle M., Brochu C., Gillard J., Joly M. A., Landry S., Lepage O., Llinàs-Brunet M., Pesant M., Poirier M., Poirier M., McKercher G., Marquis M., Kukolj G., Beaulieu P. L., & Stammers T. A. (2014). Molecular dynamics simulations and structure-based rational design lead to allosteric HCV NS5B polymerase thumb pocket 2 inhibitor with picomolar cellular replicon potency. Journal of Medicinal Chemistry, 57(5), 1932–1943. 10.1021/jm4004522 [DOI] [PubMed] [Google Scholar]
- Kendrick B. J. L., Gray A. G., Pickworth A., & Watters M. P. R. (2006). Drotrecogin alfa (activated) in severe falciparum malaria. Anaesthesia, 61(9), 899–902. 10.1111/j.1365-2044.2006.04752.x [DOI] [PubMed] [Google Scholar]
- Khan R. J., Jha R. K., Amera G. M., Jain M., Singh E., Pathak A., Singh R. P., Muthukumaran J., & Singh A. K. (2020). Targeting SARS-CoV-2: A systematic drug repurposing approach to identify promising inhibitors against 3C-like proteinase and 2’-O-RIBOSEMETHYLTRANSFERASE. Journal of Biomolecular Structure & Dynamics, 1–14. 10.1080/07391102.2020.1753577 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khan S. A., Zia K., Ashraf S., Uddin R., & Ul-Haq Z. (2020). Identification of chymotrypsin-like protease inhibitors of SARS-CoV-2 via integrated computational approach. Journal of Biomolecular Structure and Dynamics, 1–10. 10.1080/07391102.2020.1751298 [DOI] [PubMed] [Google Scholar]
- Khushboo B., Robert M., K., & Gaetano M. (2020). Structural similarity of SARS-CoV2 Mpro and HCV NS3/4A proteases suggests new approaches for identifying existing drugs useful as COVID-19 therapeutics. ChemRxiv, 10.26434/chemrxiv.12153615.v1 [DOI] [Google Scholar]
- Ladhani S., Lowe B., Cole A. O., Kowuondo K., & Newton C. R. J. C. (2002). Changes in white blood cells and platelets in children with falciparum malaria: Relationship to disease outcome. British Journal of Haematology, 119(3), 839–847. 10.1046/j.1365-2141.2002.03904.x [DOI] [PubMed] [Google Scholar]
- Ladziata V., (Uladzimir, ), Glunz P. W., Zou Y., Zhang X., Jiang W., Jacutin-Porte S., Cheney D. L., Wei A., Luettgen J. M., Harper T. M., Wong P. C., Seiffert D., Wexler R. R., & Priestley E. S. (2016). Synthesis and P1' SAR exploration of potent macrocyclic tissue factor-factor VIIa inhibitors . Bioorganic & Medicinal Chemistry Letters, 26(20), 5051–5057. 10.1016/j.bmcl.2016.08.088 [DOI] [PubMed] [Google Scholar]
- Lazarus R., Olivero A., Eigenbrot C., & Kirchhofer D. (2004). Inhibitors of tissue factor • factor VIIa for anticoagulant therapy. Current Medicinal Chemistry, 11(17), 2275–2290. 10.2174/0929867043364568 [DOI] [PubMed] [Google Scholar]
- Lee J., Cheng X., Swails J. M., Yeom M. S., Eastman P. K., Lemkul J. A., Wei S., Buckner J., Jeong J. C., Qi Y., Jo S., Pande V. S., Case D. A., Brooks C. L., MacKerell A. D., Klauda J. B., & Im W. (2016). CHARMM-GUI input generator for NAMD, GROMACS, AMBER, OpenMM, and CHARMM/OpenMM simulations using the CHARMM36 additive force field. Journal of Chemical Theory and Computation, 12(1), 405–413. 10.1021/acs.jctc.5b00935 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X., Zhang B., Jin Z., Yang H., & Rao Z. (2020). The crystal structure of COVID-19 main protease in complex with an inhibitor N3. 6LU7, 10.1038/s41586.020-2223-y. [DOI] [Google Scholar]
- McGivern D. R., Masaki T., Lovell W., Hamlett C., Saalau-Bethell S., & Graham B. (2015). Protease inhibitors block multiple functions of the NS3/4A protease-helicase during the hepatitis C virus life cycle. Journal of Virology, 89(10), 5362–5370. 10.1128/JVI.03188-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen J. T., Hamada Y., Kimura T., & Kiso Y. (2008). Design of potent aspartic protease inhibitors to treat various diseases. Archiv Der Pharmazie, 341(9), 523–535. 10.1002/ardp.200700267 [DOI] [PubMed] [Google Scholar]
- Quan M. L., Pinto D. J. P., Rossi K. A., Sheriff S., Alexander R. S., Amparo E., Kish K., Knabb R. M., Luettgen J. M., Morin P., Smallwood A., Woerner F. J., & Wexler R. R. (2010). Phenyltriazolinones as potent factor Xa inhibitors. Bioorganic & Medicinal Chemistry Letters, 20(4), 1373–1377. 10.1016/j.bmcl.2010.01.011 [DOI] [PubMed] [Google Scholar]
- Rappé A. K., Casewit C. J., Colwell K. S., Goddard W. A., & Skiff W. M. (1992). UFF, a full periodic table force field for molecular mechanics and molecular dynamics simulations. Journal of the American Chemical Society, 114(25), 10024–10035. 10.1021/ja00051a040 [DOI] [Google Scholar]
- Routh J. (2020). NIH Clinical Trial of Remdesivir to Treat COVID-19 Begins NIH U.S National Library of Medicine. https://www.nih.gov/news-events/news-releases/nih-clinical-trial-remdesivir-treat-covid-19-begins
- Ruch T. R., & Machamer C. E. (2012). The coronavirus E protein: Assembly and beyond. Viruses, 4(3), 363–382. 10.3390/v4030363 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruf W. (2004). Emerging roles of tissue factor in viral hemorrhagic fever. Trends in Immunology, 25(9), 461–464. 10.1016/j.it.2004.07.002 [DOI] [PubMed] [Google Scholar]
- Schiering N., D'Arcy A., Villard F., Simic O., Kamke M., Monnet G., Hassiepen U., Svergun D. I., Pulfer R., Eder J., Raman P., & Bodendorf U. (2011). A macrocyclic Hcv Ns3/4A protease inhibitor interacts with protease and helicase residues in the complex with its full-length target. Proceedings of the National Academy of Sciences of the United States of America, 108(52), 21052–21056. 10.1073/pnas.1110534108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scholar E. (2007). xPharm: The Comprehensive Pharmacology Reference.
- Siu Y. L., Teoh K. T., Lo J., Chan C. M., Kien F., Escriou N., Tsao S. W., Nicholls J. M., Altmeyer R., Peiris J. S. M., Bruzzone R., & Nal B. (2008). The M, E, and N structural proteins of the severe acute respiratory syndrome coronavirus are required for efficient assembly, trafficking, and release of virus-like particles. Journal of Virology, 82(22), 11318–11330. 10.1128/JVI.01052-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sovaldi (sofosbuvir) (2015). FDA https://www.accessdata.fda.gov/drugsatfda_docs/label/2015/204671s004lbl.pdf
- Thiel V., Ivanov K. A., Putics Á., Hertzig T., Schelle B., Bayer S., Weißbrich B., Snijder E. J., Rabenau H., Doerr H. W., Gorbalenya A. E., & Ziebuhr J. (2003). Mechanisms and enzymes involved in SARS coronavirus genome expression. The Journal of General Virology, 84(Pt 9), 2305–2315. 10.1099/vir.0.19424-0 [DOI] [PubMed] [Google Scholar]
- Tian X., Li C., Huang A., Xia S., Lu S., Shi Z., Lu L., Jiang S., Yang Z., Wu Y., & Ying T. (2020). Potent binding of 2019 novel coronavirus spike protein by a SARS coronavirus-specific human monoclonal antibody. In Emerging Microbes and Infections, 9, 382–385. 10.1080/22221751.2020.1729069 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trott O., & Olson A. J. (2010). AutoDock Vina: Improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. Journal of Computational Chemistry, 31(2), 455–461. 10.1002/jcc.21334 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vanommeslaeghe K., Hatcher E., Acharya C., Kundu S., Zhong S., Shim J., Darian E., Guvench O., Lopes P., Vorobyov I., & Mackerell A. D. (2010). CHARMM general force field: A force field for drug-like molecules compatible with the CHARMM all-atom additive biological force fields. Journal of Computational Chemistry, 31(4), 671–690. 10.1002/jcc.21367 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wan Y., Shang J., Graham R., Baric R. S., & Li F. (2020). Receptor recognition by novel coronavirus from Wuhan: An analysis based on decade-long structural studies of SARS. Journal of Virology, 94(7), e00127–20. 10.1128/JVI.00127-20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang R., Lai L., & Wang S. (2002). Further development and validation of empirical scoring functions for structure-based binding affinity prediction. Journal of Computer-Aided Molecular Design, 16(1), 11–26. [DOI] [PubMed] [Google Scholar]
- Willyard C. (2020). Coronavirus blood-clot mystery intensifies. Nature, 581(7808), 250–250. 10.1038/d41586-020-01403-8 [DOI] [PubMed] [Google Scholar]
- Wrapp D., Wang N., Corbett K. S., Goldsmith J. A., Hsieh C.-L., Abiona O., Graham B. S., & McLellan J. S. (2020). Cryo-EM structure of the 2019-nCoV spike in the perfusion conformation. Science (Science.), 367(6483), 1260–1263. 10.1126/science.abb2507 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xue X., Yang H., Shen W., Zhao Q., Li J., Yang K., Chen C., Jin Y., Bartlam M., & Rao Z. (2007). Production of authentic SARS-CoV Mpro with enhanced activity: Application as a novel tag-cleavage endopeptidase for protein overproduction. Journal of Molecular Biology, 366(3), 965–975. 10.1016/j.jmb.2006.11.073 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xue X., Yu H., Yang H., Xue F., Wu Z., Shen W., Li J., Zhou Z., Ding Y., Zhao Q., Zhang X. C., Liao M., Bartlam M., & Rao Z. (2008). Structures of two coronavirus main proteases: implications for substrate binding and antiviral drug design. Journal of Virology, 82(5), 2515–2527. 10.1128/JVI.02114-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang H., Yang M., Ding Y., Liu Y., Lou Z., Zhou Z., Sun L., Mo L., Ye S., Pang H., Gao G. F., Anand K., Bartlam M., Hilgenfeld R., & Rao Z. (2003). The crystal structures of severe acute respiratory syndrome virus main protease and its complex with an inhibitor. Proceedings of the National Academy of Sciences of the United States of America, 100(23), 13190–13195. 10.1073/pnas.1835675100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L., Lin D., Sun X., Curth U., Drosten C., Sauerhering L., Becker S., Rox K., & Hilgenfeld R. (2020). Crystal structure of SARS-CoV-2 main protease provides a basis for design of improved α-ketoamide inhibitors. Science (New York, N.Y.), 368(6489), 409–412. 10.1126/science.abb3405 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu L., George S., Schmidt M. F., Al-Gharabli S. I., Rademann J., & Hilgenfeld R. (2011). Peptide aldehyde inhibitors challenge the substrate specificity of the SARS-coronavirus main protease. Antiviral Research, 92(2), 204–212. 10.1016/j.antiviral.2011.08.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ziebuhr J., Heusipp G., & Siddell S. G. (1997). Biosynthesis, purification, and characterization of the human coronavirus 229E 3C-like proteinase. Journal of Virology, 71(5), 3992–3997. 10.1128/JVI.71.5.3992-3997.1997 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ziebuhr J., & Siddell S. G. (1999). Processing of the human coronavirus 229E replicase polyproteins by the virus-encoded 3C-like proteinase: identification of proteolytic products and cleavage sites common to pp1a and pp1ab. Journal of Virology, 73(1), 177–185. 10.1128/JVI.73.1.177-185.1999 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.