Abstract
At present, the world is facing a pandemic named as COVID-19, caused by SARS-CoV-2. Traditional Chinese medicine has recommended the use of liquorice (Glycyrrhiza species) in the treatment of infections caused by SARS-CoV-2. Therefore, the present investigation was carried out to identify the active molecule from the liquorice against different protein targets of COVID-19 using an in-silico approach. The molecular docking simulation study of 20 compounds along with two standard antiviral drugs (Lopinavir and Rivabirin) was carried out with the help of Autodock vina software using two protein targets from COVID-19 i.e. spike glycoprotein (PDB ID: 6VSB) and Non-structural Protein-15 (Nsp15) endoribonuclease (PDB ID: 6W01). From the observed binding energy and the binding interactions, glyasperin A showed high affinity towards Nsp15 endoribonuclease with uridine specificity, while glycyrrhizic acid was found to be best suited for the binding pocket of spike glycoprotein and also prohibited the entry of the virus into the host cell. Further, the dynamic behavior of the best-docked molecules inside the spike glycoprotein and Nsp15 endoribonuclease were explored through all-atoms molecular dynamics (MD) simulation study. Several parameters from the MD simulation have substantiated the stability of protein-ligand stability. The binding free energy of both glyasperin A and glycyrrhizic acid was calculated from the entire MD simulation trajectory through the MM-PBSA approach and found to high binding affinity towards the respective protein receptor cavity. Thus, glyasperin A and glycyrrhizic acid could be considered as the best molecule from liquorice, which could find useful against COVID-19.
Communicated by Ramaswamy H. Sarma
Keywords: COVID-19, glycyrrhizic acid, glyasperin, liquiritin, liquorice
Graphical Abstract
Introduction
The recent pandemic 2019 novel coronavirus, also known as severe acute respiratory syndrome coronavirus-2 (SARS-CoV-2) (Zhou, Yang, et al., 2020) has resulted in 295,101 deaths with a fatality rate of 6.85% and has infected a total of 4,307,287 individuals around the globe, as of 9:28 am CEST, 15 May 2020 (World Health Organization, 2020). The spectrum of SARS-CoV-2 infection is found to have a very wide range of severity, from asymptomatic carriage to mild respiratory tract infection and severe to fatal pneumonia (Singhal, 2020). Besides this, SARS-CoV-2 is characterized by very high morbidity and high mortality, which emphasizes the urgent medical as well as a public health need for validation, development and approval of effective prophylactic and therapeutic interventions against the COVID-19 (Zhu et al., 2020).
The coronaviruses (CoV) are single-stranded enveloped, positive-sense RNA with the largest genome size that ranges approximately from 26 to 32 kilobases as observed till date. The CoV uses its spike (S) glycoprotein (180 kDa) to bind its receptor, and mediate membrane fusion and virus entry. Indeed, S proteins are typical homo trimeric class I fusion proteins, and protease cleavage is required for activation of the fusion potential of S protein. A two-step sequential protease cleavage starts with activation of S proteins CoVs, which involves prime cleavage between two subunits (i.e. S1 and S2) and activates cleavage on S2’ site (Belouzard et al., 2009; Cotten et al., 2013; Ou et al., 2020; Wan et al., 2020). Depending on virus strains and cell types, CoV S proteins may be cleaved by one or several host proteases, including furin, trypsin, cathepsins, transmembrane protease serine protease-2 (TMPRSS-2), TMPRSS-4, or human airway trypsin-like protease (HAT). Availability of these proteases largely mediating attachment and membrane fusion on target cells (Kang et al., 2020). Upon the entry, the viral particle is encoded and ready for translation ORF 1a and 1b into polyproteins pp1a (4382 amino acids) and pp1ab (7073 amino acids) that are processed by proteases 3-C-like protease (3CLpro) and papain-like protease (PLpro). Subsequently, these polyproteins are cleaved into at least 16 non-structural proteins (Nsps), which assembles and form the replication-transcription complex (Pillaiyar, Meenakshisundaram, & Manickam, 2020). Importantly, the homology modelling revealed that SARS-CoV-2 S and SARS-CoV S share the same functional host cell receptor i.e. angiotensin-converting enzyme 2 (ACE-2) (Zhang et al., 2020). Besides, the SARS-CoV-2 RBD-SD1 106 fragment (S residues from 319-591) have reported having ACE-2 binding site (Wrapp et al., 2020). The affinity with which ACE-2 bounds to SARS-CoV-2 S ectodomain is 10- to 20-fold higher than ACE-2 binding to SARS-CoV S. Due to the high binding affinity of SARS-CoV-2 S for human ACE-2, the SARS-CoV-2 exhibits greater ability to transmit from person-to-person (Wan et al., 2020). Furthermore, the genomic RNA, acting as a messenger RNA (mRNA) has been found to play a critical role in the initial RNA synthesis of the infectious cycle, template for replication, transcription and also act as a substrate for packaging (Ricagno et al., 2006; Snijder et al., 2016). The Nsp15 is a nidoviral RNA uridylate-specific endoribonuclease (NendoU) and possess C-terminal catalytic domain, which is specific for uridine acting on both, single- and double-stranded RNA (Elfiky, 2020a). Reports reveal that the NendoU activity of Nsp15 is the main contributor for the protein interference with innate immune response and thus, it is said that Nsp15 plays a pivotal role in the biological progression of coronavirus (Bhardwaj et al., 2008). Therefore, the drugs under investigation, if targets the conserved sites can be likely to block the entry, replication and proliferation of the virus and thus may exhibit a wide spectrum of activity (Adeoye et al., 2020; Elfiky, 2020b; Joshi et al., 2020; Pan et al., 2020; Prajapat et al., 2020; Tian et al., 2020; Wu et al., 2020).
Although, scientific fraternity and interdisciplinary research groups are putting their sincere efforts to combat pathogenic SARS-CoV-2 by applying variable holistic approaches to understand the possible features of pathogenesis, for the effective prognosis and early identification/detection of cases along with the development of effective therapeutic interventions including prevention, prophylactics, vaccines and treatment measures against COVID-19, a public health emergency of international concern. In principle, the cost-effective and time-efficient computational technique provides a powerful network-based tool (Cheng et al., 2018) to test a novel hypotheses conceptualizing systematic drug repositioning strategy for rapid identification of therapeutic lead and/or potential leads combinations effective against 2019-nCoV/SARS-CoV-2 from existing approved and/or preclinical drugs (Kandeel & Al-Nazawi, 2020; (Pillaiyar, Meenakshisundaram, Manickam, et al., 2020; Serafin et al., 2020; Sinha et al., 2020; Zhou, Hou, et al., 2020).
A promising published report on glycyrrhizin, a bioactive constituent of liquorice (Glycyrrhiza glabra L. Family: Fabaceae), which is a key medicine of Traditional Chinese Medicine (TCM) system, indicates its role in the treatment of patients suffering from an infection caused by SARS-CoV (Cinatl et al., 2003). Moreover, according to TCM theory, liquorice is primarily effective for fatigue and debilitation, asthma with coughing, excessive phlegm, respiratory infections and for relieving drug toxicity (Grienke et al., 2014; Wang et al., 2013). Further, bioactive compounds of liquorice have been reported with antimicrobial, antiviral and immunoregulatory features. In brief, glycyrrhizic acid has been reported for its efficacy against HIV-1 chronic hepatitis C virus, coxsackievirus A16 and enterovirus 71 and Kaposi sarcoma-associated herpesvirus (Curreli et al., 2005; De Clercq, 2000; Sabouri Ghannad et al., 2014; Wang et al., 2013). The licochalcone A and isoliquiritigenin has been effective in preventing the acute lung injury induced of pathogenic origin (Liu et al., 2019). Liquiritin has also been reported for their anti-viral potential against HIV (Grienke et al., 2010).
Application of computational resource and power in drug discovery research to screen molecular databases has become faster and less expensive approach towards the scientific community worldwide (Okimoto et al., 2009). The structure-based screening of chemical or phytochemical databases already reached at a new height due to the availability of large numbers of crystal target. With the help of excellent algorithms and pharmacoinformatics tools molecular docking can predict the almost true conformational state of a protein-ligand complex and predicts the binding affinity of the small molecules about correctly (Meng et al., 2011). In the current investigation, we have conceptualized the structure-based antiviral screening of natural products from liquorice with the aim to obtained structurally potential inhibitors by targeting the crystal structure of prefusion SARS-CoV-2 spike glycoprotein and Nsp15 endoribonuclease from SARS-CoV-2, which expedites the discovery of leads in treatment against COVID-19. Initially, the phytochemicals are screened through molecular docking approach. Further, the molecular dynamics (MD) simulation study is carried out to explore the dynamic behavior of the molecules. Finally, the affinity of the small molecules is checked through the MM-PBSA (Molecular Mechanics Poisson–Boltzmann Surface Area) based binding energy calculation.
Material and methods
Molecular docking simulation
Virtual screening of phytochemicals through pharmacoinformatics approach has become a pivotal tool in the drug discovery research. Availability of crystal structure of protein molecules has given a new dimension towards the structure-based screening of small to large chemical databases. To explore promising anti-COVID molecules, a set of 20 reported bioactive compounds of liquorice (Glycyrrhiza species) were retrieved from the PubChem database (Kim et al., 2016). In brief, the names search for bioactive compounds of Glycyrrhiza glabra which has been documented for their antitumor, antimicrobial, antiviral and immunoregulatory efficacies viz. glycyrrhizinic acid (CID_14982), licochalcone A (CID_5318998), licochalcone B (CID_5318999), licochalcone C (CID_9840805), licochalcone D (CID_10473311), licochalcone E (CID_46209991), licochalcone F (CID_44130137), licochalcone G (CID_49856081), glyasperin A (CID_5481963), glyasperin B (CID_480784), glyasperin C (CID_480859), glyasperin D (CID_480860), isoliquiritinapioside (CID_6442433), 1-methoxyphaseollidin (CID_480873), dehydroglyasperin C (CID_480775), kanzonol Q (CID_11253965), liquiritin (CID_503737), hedysarimcoumestan B (CID_11558452), 5,6,7,8-tetrahydro-2,4-dimethylquinoline (CID_5321849), 5,6,7,8-tetrahydro-4-methylquinoline (CID_185667) (Bode & Dong, 2015; Jiang et al., 2020; Kim & Ma, 2018; Liu et al., 2019; Prajapati & Patel, 2015; Yang et al., 2017); was carried out in PubChem database. The resulting 3D structures in sdf file format were directly used in the generation of input pdbqt files for Autodock vina. The 2D format of all molecules were downloaded and transformed to 3D coordinates and subsequently optimized with Openbabel2.3.2 GUI. The molecular docking simulation study was performed on the Autodock vina program (Trott & Olson, 2010), which is the most widely used and trusted steadfast open-source docking simulation tool. Recently submitted two crystal structure of SARS-CoV-2 i.e. spike glycoprotein [PDB ID: 6VSB (Wrapp et al., 2020)] and non-structural protein 15 [PDB ID: 6W01 (Kim et al., 2020)] were downloaded from the Research Collaboratory for Structural Bioinformatics-Protein Databank (RCSB-PDB). Both crystal structures were prepared using AutoDock Tools (ADT) (Morris et al., 2009) by repairing the missing atoms, the addition of polar hydrogens and the Gastigier charges. Finally, both structures were saved in the pdbqt format after assigning the AD4 (AutoDock 4) (Morris et al., 2009) atom type. To select the binding site of both proteins the already available information was considered. The region around the bound citric acid in the Nsp15 was considered as the active site. In the case of Spike glycoprotein, it is mentioned that amino acid residues in a range of 319 to 591 recombinantly expressed and measures the ACE2 binding using biolayer interferometry (Wrapp et al., 2020). Hence the coordinate of Ser325 amino residue was considered as the position of the grid box. To confine the active site of the protein molecule, the grid box information was obtained from the ADT with the help of above information. In case of PDB ID: 6W01, grid box was generated around the bound citric acid having co-ordinate of −63.624, 72.524 and 28.280 for -x, -y and -z-axis respectively. On the other hand, for PDB ID: 6VSB, the grid coordinate was considered as 204.457, 199.799 and 246.898 along the x-, y- and z-axis respectively. The grid box size for both crystal structures was determined by manual inspection and set as 50x50x50. The configuration file was created by adding the grid, receptor and ligand information before the execution of molecular docking. To adjudge the molecular docking study, two standard drug molecules, Lopinavir and Rivabirin (Cao et al., 2020) were considered as control molecules throughout the study. In recent past, a number of findings have been reported the efficacy of Lopinavir, a protease inhibitor and Ribavirin, a nucleoside analogue in suppressing the shedding of SARS-CoV-2 (Hung et al., 2020; Lim et al., 2020; Sheahan et al., 2020). All 20 phytochemicals along with above two drug molecules were also prepared by adding polar hydrogen and Gasteiger charges in ADT. Each molecule was saved as pdbqt format for Autodock vina input after detection and set of torsion angles. The molecular docking outcomes were analyzed based on binding energy and binding interactions profile. Highest binding energy among the drugs was used as a threshold to select the best phytochemicals for both the receptors. The binding interactions between ligands and the receptor were assessed through the protein-ligand interaction profile (PLIP) (Salentin et al., 2015).
Molecular dynamics simulation
Molecular dynamics (MD) simulation study is one the crucial and effective tools to explore the dynamic nature of the protein-ligand complex and the relative stability. The highest binding affinity scorer ligand in respect to each protein i.e. Glycyrrhizic acid for spike glycoprotein and glyasperin A for Nsp15 endoribonuclease complex with respective protein was selected for further MD simulation study. The MD simulation was done on Gromacs-2018.2 software tool (www.gromacs.org) installed at the Lengau CHPC server, Cape Town, South Africa. This study was performed for 100 ns with a 2fs of time step at a 1 atm constant pressure and 300 K constant temperature. To generate the protein topology the all-atoms CHARMM36 force field was considered. During the study, each protein-ligand complex system was created within the cubic box of 1 Å from the center of the system. The system was solvated by the TIP3P water model. Sufficient numbers of Na+ and Cl− ions were added to neutralize the simulation system and further minimized the system by employing the steepest descent algorithm of 10,000 steps. To address the long-range distances the van der Waals and electrostatic interaction cutoff range were set to 0.9 and 1.4 nm respectively. The SwissParam tool (Daina et al., 2017) was used to generate ligand topology. After each of 1ps interval, the trajectory information was saved for further analysis. The entire system was equilibrated through the NVT (constant number of particles, volume, and temperature) followed by NPT (constant number of particles, pressure, and temperature) ensemble approaches to distribute the solvent and ions equally around the protein-ligand complex. Further root-mean-square deviation (RMSD), root-mean-square fluctuation (RMSF) and radius of gyration (Rg) were calculated to determine the molecular complex stability in terms of conformation and performance. The binding free energy was then determined through the Molecular Mechanics Poisson-Boltzmann Surface Area (MM-PBSA) method with the help of g_mmpbsa tool (Islam & Pillay, 2019, 2020). Theory and details of this method have been given in our previous publication (Bhowmick et al., 2019; Parida et al., 2020).
Result and discussion
Virtual screening
The structure-based screening is one of the crucial approaches to explore small to large molecular databases to find promising molecules for a specific target. In the current study, an effort was taken to find out anti-spike glycoprotein and Nsp15 endoribonuclease molecules from a set of 20 phytochemicals through molecular docking and MD simulation studies. The flow diagram of the work is given in Figure 1. The binding site of the SARS-CoV-2 spike glycoprotein was considered around the Ser325 which is already reported as active site residue (Yu et al., 2020). In the case of Nsp15 endoribonuclease the binding site was selected around the co-crystal ligand. All 20 phytochemicals along with Lopinavir and Ribavirin were docked in the spike glycoprotein and Nsp15 endoribonuclease.
Figure 1.
Work flow of screening of phytochemicals against spike glycoprotein and Nsp15 endoribonuclease.
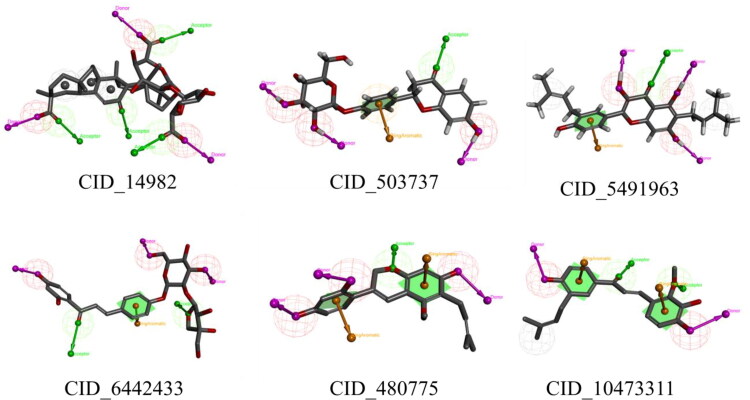
Details of each of phytochemicals and binding energies against the spike glycoprotein and Nsp15 endoribonuclease are given in Table S1 (Supplementary file). The binding energy of Lopinavir and Ribavirin was found to be −7.4 and −5.6 Kcal/mol respectively when docked in the spike glycoprotein. Moreover, the Lopinavir and Ribavirin were docked in the Nsp15 endoribonuclease and binding energy was found to be −8.3 and −6.6 Kcal/mol respectively. To select best phytochemical, the threshold binding energy for spike glycoprotein and Nsp15 endoribonuclease was considered as −7.4 and −8.3 Kcal/mol, respectively. From Table S1 (Supplementary file) it can be seen that CID_14982, CID_503737, CID_5481963 and CID_6442433 were satisfied above criteria for both spike glycoprotein and Nsp15 endoribonuclease. Moreover, CID_480775 and CID_10473311 were found to have binding energy less than −8.30 on docking in the Nsp15 endoribonuclease. Hence, CID_14982, CID_503737, CID_5481963 and CID_6442433 were found to be promising molecules for both enzymes. In addition to the above CID_480775 and CID_10473311 were found promising against Nsp15 endoribonuclease. Two-dimensional representation of all six promising molecules is given in Figure 2.
Figure 2.
Two-dimensional representation of final molecules. #Spike glycoprotein inhibitors; *Nsp15 endoribonuclease inhibitors.
All the above proposed final molecules were found structurally diverse. All molecules consist of several pharmacophoric features including hydrogen bond (HB) acceptor (HBA), HB donor (HBD), hydrophobic (Hy), etc., those might be crucial for potential binding interactions with the catalytic amino acids at the active site. In brief, the HBD and HBA features in CID_14982 (Figure 3) were found crucial in hydrogen bond interactions with Gln298, His625, Gln321 and Arg319, while the Hy region was found to be important in the interaction with Ile624 and Val620 residues in the spike glycoprotein. In the case of Nsp15 endoribonuclease of the SARS-COV-2, the HBD and HBA features were revealed as important in interactions with residues Tyr343, His235, His250, while the hydrophobic interactions formed with residues Tyr343. Likewise, the HBD, HBA, aromatic ring features in the other five compounds (Figure 3) were found to be crucial for the interactions with similar residues at the binding site of both the proteins. To further assess the binding interactions of each of the molecule were explored with respective enzymes. The stability of the molecules inside the protein was explored through all-atoms MD simulation. Finally, the binding free energy was calculated through the MM-PBSA approach.
Figure 3.
Two-dimensional representation of the pharamacophoric features of final molecules.
Binding interaction analysis
The binding interaction profile of all proposed molecules was explored to analyze the catalytic amino acid residues responsible for holding the molecules inside the receptor cavity of Spike glycoprotein and Nsp15 endoribonuclease. The Protein-Ligand Interaction Profiler (PLIP) webserver (Salentin et al., 2015) was used to explore two-dimensional binding interactions. The binding mode in surface view of proposed phytochemicals and considered drug molecules was explored using the PyMol. The binding energies of all six proposed molecules and considered drug molecules are given in Table 1.
Table 1.
Binding energy of best docked compounds with the active site of spike glycoprotein and Nsp15 endoribonuclease of SARS-CoV-2.
| S.No. | PubChem CID | Name | Binding energy (Kcal/mol) |
|
|---|---|---|---|---|
| Spike glycoprotein | Nsp15 endoribonuclease | |||
| 1 | 14982 | Glycyrrhizic acid | −9.2 | −8.3 |
| 2 | 480775 | Dehydroglyasperin C | −6.9 | −8.4 |
| 3 | 503737 | Liquiritin | −7.7 | −8.8 |
| 4 | 5481963 | Glyasperin A | −7.9 | −9.2 |
| 5 | 6442433 | Isoliquiritinapioside | −7.4 | −9.0 |
| 6 | 10473311 | Licochalcone D | −7.1 | −8.3 |
| 7 | 92727 | Lopinavir | −7.4 | −8.3 |
| 8 | 37542 | Ribavirin | −5.6 | −6.6 |
Binding interactions analysis of the natural bioactive ligand with SARS-CoV-2 spike glycoprotein
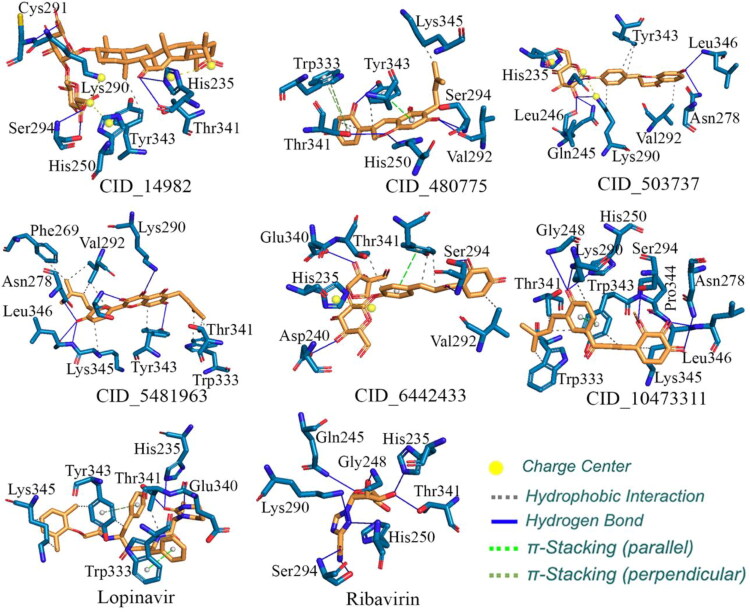
All four proposed spike glycoprotein inhibitors along with Lopinavir and Ribavirin were docked in the spike glycoprotein and binding interactions are given in Figure 4. On successful docking, the complex between spike glycoprotein and Lopinavir was explored and found that the Lopinavir properly positioned into the catalytic site assembled by Arg273, Asp627, His625, Ala292, Val320, Val595, Val620, Gln619, Tyr612, Ile624, Phe318 and Pro295 amino acids with binding energy −7.4 Kcal/mol. Amine group of tetrahydro pyrimidine of lopinavir showed H-bonding with Asp627, carboxyl with Arg273 and amide with His625, Ala292. The phenyl ring of Lopinavir formed π-π parallel stacking with His625. The triazole nitrogen of Ribavirin showed H-bonding with His625, hydroxymethyl group of tetrahydrofuran with Glu298, Ser316 and the hydroxyl group of tetrahydrofuran with Ala623 amino acid with binding energy −5.6 Kcal/mol. The triazole ring of Ribavirin formed π-π perpendicular stacking with His625. Docking study on the SARS-CoV-2 spike glycoprotein revealed that the glycyrrhizic acid (CID_14982) exhibited the best binding mode among all the ligand and standard drugs. It was properly positioned into the binding pocket of spike glycoprotein constructed by His625, Glu298, Arg319, Gln321, Gln321, Val320, Tyr343, Thr323, Ile624 and Asn317 amino acids with a binding energy of −9.2 Kcal/mol. The glycyrrhizic acid has two oxane (tetrahydropyran) ring substituted with five hydroxyl and two carboxyl groups buried properly into the binding pocket of S1 subunit and showed H-bonding with His625, Glu298, Arg319, Gln321, Val320 and three π-cation interaction with His625. It also has 7 methyl groups which provide a proper grip between hydrophobic pockets. The equal distribution of polar (i.e. 5 -OH and 2 -COOH group) and non-polar (i.e. 7 methyl group) provide a good balance between hydrophilicity and hydrophobicity (CID_14982 in Figure 4).
Figure 4.
The binding interaction of glycyrrhizic acid (CID_14982), liquiritin (CID_503737), glyasperin A (CID_5481963), isoliquiritinapioside (CID_6442433), Lopinavir and Ribavirin in the spike glycoprotein.
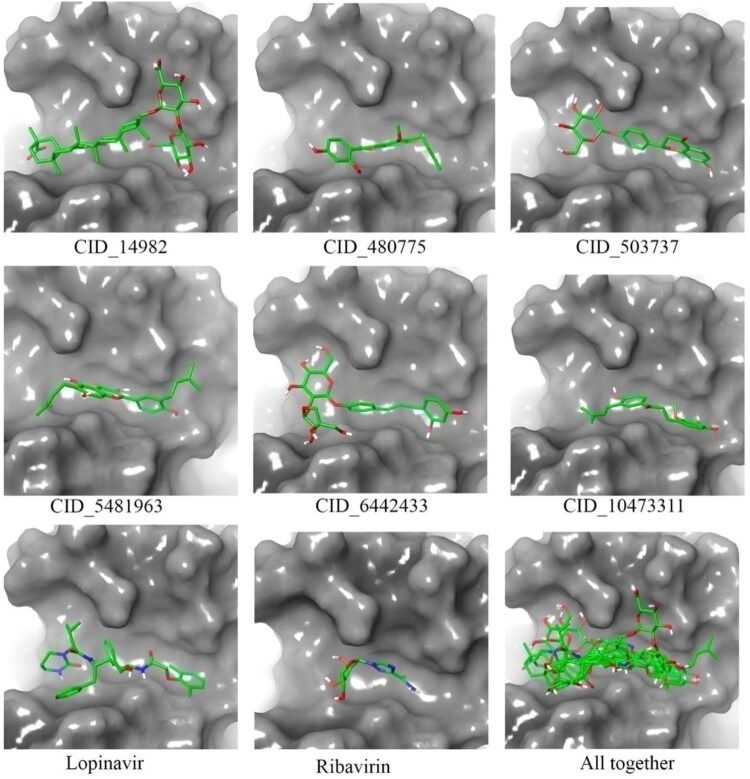
The hydroxyl group of chromenone ring of glyasperin A (CID_5481963) showed H-bonding with Val620, Val320, carboxy of chromenone ring with Gln321 and other hydroxyl groups of the phenyl ring with Glu298 amino acid residue with binding energy −7.9 Kcal/mol (CID_5481963 in Figure 4). The liquiritin (CID_503737) has chromenone ring substituted by hydroxyl group showed H-bonding with Gln321, the carboxyl group of chromenone ring with Val620 and the hydroxyl group of oxane moiety with Cys291, His625 amino acid residues with a binding energy of −7.7 Kcal/mol (CID_503737 in Figure 4). The phenyl ring substituted by hydroxyl group showed H-bonding with Gln321, Val320, the hydroxyl group of oxane ring with Glu298, His625, the hydroxyl group of a furan ring with Glu298 and carboxyl group of isoliquiritinapioside (CID_6442433) showed H-bonding with Val620 amino acid residue with a binding energy of −7.4 Kcal/mol (CID_6442433 in Figure 4). From the above observations it can be explained that all proposed molecules for the spike glycoprotein hold similar or better binding interactions in comparison to the both Lopinavir and Ribavirin. The binding mode in three-dimensional space of all molecules were extracted which are given in Figure 5. It can be seen that all molecules perfectly fitted inside the receptor cavity of spike glycoprotein. The important amino residues and their involvement in binding interactions with spike glycoprotein inhibitors are given in Table 2.
Figure 5.
Demonstrates the binding mode in 3D-surface-image of glycyrrhizic acid (CID_14982), liquiritin (CID_503737), glyasperin A (CID_5481963), isoliquiritinapioside (CID_6442433), Lopinavir and Ribavirin in the active site of spike glycoprotein of the SARS-CoV-2.
Table 2.
List of bioactive compounds with binding interaction parameters with the active site of spike glycoprotein of SARS-CoV-2.
| S. No. | Ligand | Binding interaction with the active site of spike glycoprotein |
||||||
|---|---|---|---|---|---|---|---|---|
| Binding energy (Kcal/mol) | H-Bonding | Hydrophobic | Charge center | π-Stacking (Parallel) | π- Stacking (Perpendicular) | π- cation (Interaction) | ||
| 1 | CID_14982 | −9.2 | His625, Glu298, Arg319, Gln321, Val320 | Gln321, Val320, Tyr343, The323, Ile624 | His625 | – | – | His625 |
| 2 | CID_503737 | −7.7 | His625, Val620 Gln321, Cys291 | Gln321, Ile624, Val320 | – | – | – | – |
| 3 | CID_5481963 | −7.9 | Val620, Val320 Glu298, Gln321 | Val320, Val620, Val595, Gln321, Thr323, Phe318, Ile624 | – | – | – | – |
| 4 | CID_6442433 | −7.4 | His625, Val320, Gln321, Glu298, | Val620, Gln321, Phe318, Ile624, Ty 612 | – | – | – | – |
| 5 | Lopinavir | −7.4 | Arg273, Asp627, His625, Ala292 | Val320, Val595, Val620, Gln619, Tyr612, Ile624, Ala292 Phe 318, Pro 295 | His625 | His625(π cation Interaction) | – | – |
| 6 | Ribavirin | −5.6 | His625, Glu298, Ser316, Ala623 | – | – | – | His625 | – |
Binding interactions analysis of natural bioactive ligand with SARS-CoV-2 Nsp15 endoribonuclease
Best six phytochemicals found through virtual screening were docked in the Nsp15 endoribonuclease and binding energy is given in Table 1. Important amino acids found in binding interaction formation with proposed molecules are given in Table 3. Both standard Lopinavir and Ribavirin was docked and binding energy found to be −8.3 and −6.6 Kcal/mol respectively. Lopinavir was properly positioned into the catalytic site constructed by His235, Thr341, Glu340, Tyr343, Trp333 and Lys345 amino acids with a binding energy of −8.3 Kcal/mol. Pyrimidine of Lopinavir showed H-bonding with Glu340 and carboxyl with His235, Thr341. The phenyl ring of Lopinavir formed π-π parallel stacking with Trp333 and π-π perpendicular stacking with Tyr343 (Figure 6). Ribavirin was found to interact into the binding pocket with a binding energy of −6.6 Kcal/mol (Figure 6). The simulation study on the crystal structure of Nsp15 endoribonuclease revealed that glyasperin A (CID_5481963) have chromenone ring substituted with three hydroxyl and 3-methylbut-2-enyl group and a phenyl ring which is substituted with a hydroxyl and a 3-methylbut-2-enyl group showed H-bonding with Tyr343, Lys290 Leu346, Asn278 and Lys345 with a binding energy of −9.2 Kcal/mol (Figure 6). Glyasperin A showed the highest stable binding mode i.e. lowest binding energy among all the docked test ligand and standard drug Lopinavir and Ribavirin. Glyasperin B, C and D have 2, 4-dihydroxy phenyl ring attached to chromen ring but in Glyasperin A one hydroxy of phenyl is substituted with a 3-methylbut-2-enyl group at 3rd position which showed additional hydrophobic interaction with Phe269 and Val292, due to which glyasperin A exhibited better binding affinity. The isoliquiritinapioside (CID_6442433) have furan ring substituted with hydroxyl group showed H-bonding with Glu340, Asp240 and Ser294 showed π stacking and hydrophobic interaction with isoliquiritinapioside (Figure 6) (−9.0 Kcal/mol). The hydroxyl group of chromenone ring of liquiritin (CID_503737) showed H-bonding with Leu346, Asn278, Leu246, Lys290, Gln245, two π-cation interactions with Lys290, His235 amino acid residues (−8.8 Kcal/mol) (Figure 6). The hydroxyl group of chromene ring of dehydroglyasperin C (CID_480775) showed H-bonding with Ser294, Val292 and Thr341, His250 with a binding energy of −8.4 Kcal/mol. The phenyl ring of dehydroglyasperin C formed π-π parallel stacking with Tyr343 and π-π perpendicular stacking with Trp333 (Figure 6). Structure of dehydroglyasperin C has resembled with glyasperin A, but due to lack of one 3-methylbut-2-enyl group on phenyl ring decreases its hydrophobic interaction and also a binding affinity with protein.
Table 3.
List of bioactive compounds with binding interaction parameters in the active site of Nsp15 endoribonuclease of the SARS-CoV-2.
| S.No. | Ligand | Binding interactionwith the active site of Nsp15 endoribonuclease |
||||||
|---|---|---|---|---|---|---|---|---|
| Binding energy (Kcal/mol) | H-Bonding | Hydrophobic | Charge center | π-Stacking (Parallel) | π- Stacking (Perpendicular) | π-cation (Interaction) | ||
| 1 | CID_14982 | −8.3 | His235, Ser294, Cys291, Thr341 | Tyr343 | His235, His250, Lys290 | – | – | His235, His250, Lys290 |
| 2 | CID_480775 | −8.4 | His250, Ser294, Tyr343, Thr341, Val292 | Lys345, Tyr343, Thr341 | – | Tyr343 | Trp333 | |
| 3 | CID_503737 | −8.8 | Lys290, Gln245, Leu246, Leu346, Asn278 | Lys290, Tyr343, Val292 | Lys290, His235 | – | – | Lys290, His235 |
| 4 | CID_5481963 | −9.2 | Lys290, Leu346, Asn278, Lys345, Tyr343 | Tyr343, Val292, Lys345, Phe269 | – | – | – | – |
| 5 | CID_6442433 | −9.0 | Asp240, Glu340, Ser294 | Thr341, Ser294, Val292 | His235 | Thr341 | – | His235 |
| 6 | CID_10473311 | −8.3 | His250, Lys290, Asn278, Gly248, Leu346, Ser294, Pro 344 | Trp343, Trp333, Thr341, Lys290, Lys345 | – | Trp343 | – | – |
| 7 | Lopinavir | −8.3 | His235, Thr341, Glu340 | Thr341, Tyr343, Lys345 | – | Trp333 | Tyr343 | – |
| 8 | Ribavirin | −6.6 | His235, His250, Thr341, Gln245, Gly248, Lys290, Ser294 | – | – | – | – | – |
Figure 6.
The binding interaction of glycyrrhizic acid (CID_14982), dehydroglyasperin C (CID_480775), liquiritin (CID_503737), glyasperin A (CID_5481963), isoliquiritinapioside (CID_6442433), licochalcone D (CID_10473311), Lopinavir and Ribavirin in the active site of Nsp15 endoribonuclease of the SARS-CoV-2.
The glycyrrhizic acid (CID_14982) have oxane ring-substituted with hydroxyl and carboxyl groups exhibited H-bonding with Ser294, Cys291, His250 and Thr341 amino acids with binding energy −8.3 Kcal/mol and three π-cation interactions with Lys290, His235, His250 (Figure 6). Licochalcone D showed H-bonding with Lys290, Asn278, Gly248, Leu346, His250, Pro344 (−8.3 Kcal/mol) and phenyl ring formed π-π parallel stacking with Trp343 (Figure 6). The isoliquiritinapioside and glycyrrhizic acid have oxan ring, and liquiritin and dehydroglyasperin C have chromenone ring showed good binding with active site located between the two β-sheets, carries residues Lys290, Tyr343, His235. The Lys290 in which His235 has been proposed to constitute the catalytic triad and Tyr343 is believed to govern Uridyl specificity. Moreover, a US patent (US005843990A) signifies the use of pyran-chromenone compounds in inhibiting the growth or replication of a viruses, which is not limited to herpes Simplex virus (types 1 and 2), HIV-1, HIV-2, cytomegalovirus, Varicella Zoster virus, papillomavirus, feline leukaemia virus, avian sarcoma viruses like hepatitis types A-E, Rous sarcoma virus, influenza virus, measles, rubella and mumps viruses (Baker, 1998). The position of the binding pose in three-dimensional space (Figure 7) was clearly explained all six molecules perfectly fitted inside the receptor cavity of Nsp15 endoribonuclease.
Figure 7.
The binding mode of glycyrrhizic acid (CID_14982), dehydroglyasperin C (CID_480775), liquiritin (CID_503737), glyasperin A (CID_5481963), isoliquiritinapioside (CID_6442433), licochalcone D (CID_10473311), Lopinavir and Ribavirin in the active site of Nsp15 endoribonuclease of the SARS-CoV-2.
Molecular dynamics simulation
In order to check the stability of glycyrrhizic acid-glycoprotein and glyasperin A-Nsp15 Endoribonuclease complexes, an all-atoms MD simulation of time span of 100 ns was performed. On successful completion of the MD simulation the entire trajectory of each complex was considered to explore the RMSD, RMSF and Rg. The average, maximum and minimum value of RMSD, RMSF and Rg were obtained from the entire frames and given in Table 4.
Table 4.
Maximum, minimum and average values of RMSD, RMSF, Rg and MM-PBSA based binding free energy of final proposed molecules complex with SARS-CoV-2 protein(s).
| Values | Glycyrrhizic acid (CID_14982) with spike glycoprotein |
Glyasperin A (CID_5481963) with Nsp15 endoribonuclease |
||||||
|---|---|---|---|---|---|---|---|---|
| Binding energy (kJ/mol) | RMSD (nm) | RMSF (nm) | Rg (nm) | Binding energy (kcal/mol) | RMSD (nm) | RMSF (nm) | Rg (nm) | |
| Maximum | 665.065 | 1.531 | 1.730 | 4.877 | −22.508 | 0.350 | 0.419 | 2.320 |
| Minimum | −1164.460 | 0.396 | 0.271 | 4.104 | −260.376 | 0.136 | 0.057 | 2.193 |
| Average | −331.723 | 1.149 | 0.634 | 4.403 | −124.036 | 0.231 | 0.148 | 2.250 |
Root mean-squares deviation (RMSD)
The protein backbone RMSD is one of the critical parameters obtained from the protein-ligand complex which gives the overall information about the stability and insight into the structural conformation in the dynamic states during the MD simulation. The system equilibration in terms of stability can be explained through the RMSD analysis. The lower range of RMSD along with consistent variation throughout the simulation can be inferred the stability of the protein backbone. On the other hand, the higher RMSD and (or) high fluctuation to the native structure indicates comparatively low stability of the protein-ligand complex. It is always preferable to accept the protein biomolecule with the lower range of RMSD but higher deviated RMSD can also be acceptable which might indicate that the protein is probably undergoing large or some sort of conformational change during the simulation. The RMSD of each frame for both complexes was calculated and it is given in Figure 8. The average, maximum and minimum RMSD values of both complexes are given in Table 4.
Figure 8.
Protein backbone vs time of MD simulation. A: Spike (S) glycoprotein bound with glycyrrhizic acid. B: Nsp15 endoribonuclease bound with of glyasperin A.
The average RMSD value was found to be 1.149 and 0.231 nm for the backbone of spike glycoprotein and Nsp15 endoribonuclease respectively. It is also important to note that not a single frame of glycoprotein and Nsp15 endoribonuclease backbone was deviated higher than 1.531 and 0.350 nm in comparison to the respective native structure when bound with glycyrrhizic acid and glyasperin A, respectively. On close inspection, it can be seen that spike glycoprotein backbone bound with glycyrrhizic acid fluctuated from beginning to about 20 ns and afterwards it was equilibrated until the end of the simulation. A similar pattern was also observed in case of Nsp15 endoribonuclease backbone bound with glyasperin A. Above observation clearly explained the stability of the protein-ligand complexes during the all-atoms MD simulation.
Root mean-squares deviation (RMSF)
The individual amino residue in the protein-ligand complex plays a critical role in complex stability. The fluctuation of the amino residues can be inferred by the RMSF parameter which explains the average deviation of each amino residue over time from the reference position. More precisely it can be said that it analyzes the specific part of the protein structure that are fluctuating from its mean structure. The amino acid or group of amino acids with high RMSF value indicate the greater flexibility attained by the complex, whereas lower RMSF indicates lesser flexibility for the complex. The RMSF of individual amino residues of both glycoprotein and Nsp15 endoribonuclease bound glycyrrhizic acid and glyasperin A is given in Figure 9. The average, maximum and minimum RMSF values are given in Table 4.
Figure 9.
The RMSF of individual amino acids. A: Spike (S) glycoprotein bound with glycyrrhizic acid. B: Nsp15 endoribonuclease bound withglyasperin A.
The average RMSF was found to be 0.634 and 0.148 nm for spike glycoprotein and Nsp15 endoribonuclease, respectively. In case of spike glycoprotein bound with glycyrrhizic, the amino acids were found consistent except in the range 200 to 500. The fluctuation of the above amino acids might be due to lack of inter- and intra-molecular binding interactions. On the other hand, the RMSF plot of amino acids belongs to the Nsp 15 endoribonuclease bound with glyasperin A was found consistent. Due to the free end of both proteins, the end amino acid was found to fluctuate highest in comparison to others.
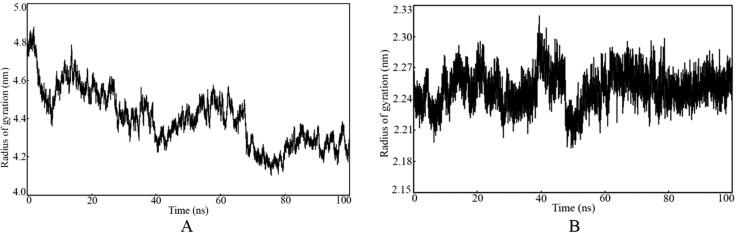
Radius of gyration (Rg)
The compactness and rigidity of the protein-ligand complexes can be assessed through the Rg parameter obtained from MD simulation trajectory. It can be defined as the mass-weighted root-mean-square distance of a collection of atoms from their common center of mass (Baig et al., 2014). Therefore, the overall dimensions and the alteration in the macromolecular structure during the MD simulation can be explored by the Rg parameter. The Rg values for each frame of both complexes were calculated and plotted against the time simulation, and it is given in Figure 10. Moreover, to explore in more details the average, maximum and minimum Rg values were calculated and showed in Table 4.
Figure 10.
The Rg values of each frame plotted against the time of simulation. A: Spike (S) glycoprotein bound with glycyrrhizic acid. B: Nsp15 endoribonuclease bound with glyasperin A.
The Rg value was varied from 4.104 and 4.877 nm, and 2.193 and 2.320 nm for the spike glycoprotein and Nsp15 endoribonuclease, respectively. The Rg parameter of the spike glycoprotein system bound with glycyrrhizic acid was gradually decreasing from about 4.8 nm and finally equilibrated around 2 nm. In the case of Nsp15 endoribonuclease bound with glyasperin A, the Rg was consistent throughout the simulation except for few fluctuations around 40 ns. Hence, the trend of Rg plot against the time simulation and low fluctuation of the same undoubtedly explain that residual backbone and folding of both spike glycoprotein and Nsp15 endoribonuclease have remained consistent after binding with glycyrrhizic acid and glyasperin A, respectively.
Binding free energy through MM-PBSA approach
The binding energy calculated through the MM-PBSA approach is considered to be more accurate than binding energy calculated by any other means including molecular docking. The binding energy of any small molecule can give an idea about the affection towards the macromolecule. To explore the affinity of both glycyrrhizic acid and glyasperin A towards the glycoprotein and Nsp15 endoribonuclease respectively, the binding free energy was calculated from the entire trajectory of MD simulation through the MM-GPBSA approach. Higher negative binding energy explains more affinity towards the receptor. The binding free energy of each molecule was plotted against the time of simulation and it is given in Figure 11. The average, minimum and maximum binding energy of all frames were also calculated and it is given in Table 4.
Figure 11.
A: Binding free energy of glycyrrhizicacidbound with Spike (S) glycoprotein. B: Binding free energy of glyasperin Abound with Nsp15 endoribonuclease.
From Table 4, it can be seen that average binding free energy was found to be −331.723 kJ/mol and −124.036 kJ/mol for glycyrrhizic and glyasperin A, respectively. In the case of glycyrrhizic, few frames at the beginning of the MD simulation were found with positive binding energy but after about 7 ns all frames showed high negative value. New orientation and conformation gained by the complex after 7 ns might be the reason for change binding energy of the molecule. The binding free energy of glyasperin A was found to be almost constant throughout the simulation. Not a single frame was found to have positive binding free energy. Hence, the binding energy of both molecules suggests that glycyrrhizic and glyasperin A possess a strong affinity to inhibit the spike glycoprotein and Nsp15 endoribonuclease, respectively.
Conclusion
The pharmacoinformatics approaches such as molecular docking and MD simulation studies were carried out to explore a set of molecules belong to the natural products. All the selected molecules including two anti-viral drugs, Lopinavir and Rivabirin were docked in the COVID-19 targets such as spike glycoprotein and Nsp15 endoribonuclease. The binding energies from the molecular docking study and binding interaction were explored in details. Several crucial amino residues were found to interact with all the molecules. A total of six phytochemicals were found promising compounds based on comparative analysis of binding interactions and binding energy with Lopinavir and Ribavirin against spike glycoprotein and Nsp15 endoribonuclease. Further, high binding energy scored one molecule from each of spike glycoprotein (glycyrrhizic acid) and Nsp15 endonuclease (glyasperin A) were used for the MD simulation in complex with the respective target molecule. Many parameters were calculated from the MD simulation and found that both molecules were retained inside the protein in the dynamic states. Finally, the binding free energy of both molecules was calculated from the MD simulation trajectories. High negative binding free energy value of both molecules substantiated their strong affinity towards the target molecule. It can be concluded that that glyasperin A might block the Nsp15 endoribonuclease activity with uridine specificity and glycyrrhizic acid connect well with the widespread binding pocket of spike glycoprotein due to its bulky nature. Moreover, it can be said that the glycyrrhizic acid disturbed the connection of the virus with ACE-2 receptor at entry-level and after entry into host cell glyasperin A inhibits the replication process of the virus. Hence, both proposed molecules might be important molecules to control the COVID-19 subjected to experimental validation.
Supplementary Material
Glossary
Abbreviations
- 2019-nCoV
2019 Novel Coronavirus
- 3CLpro
Main protease
- ACE-2
Human Angiotensin Conversion Enzyme-2
- AD4
AutoDock 4
- ADT
AutoDock Tools
- CoV
Corona Virus
- COVID-19
Coronavirus Disease 2019
- E Protein
Enveloped Protein
- HB
Hydrogen Bond
- M Protein
Membrane Protein
- MD
Molecular Dynamics
- MERS
Middle East Respiratory Syndrome
- MM-PBSA
Molecular Mechanics Poisson–Boltzmann Surface Area
- mRNA
messenger RNA
- N Protein
Nucleocapsid Protein
- NPT
Constant Number of Particles, Pressure, and Temperature
- Nsp
Nonstructural Protein
- NVT
Constant Number of Particles, Volume, and Temperature
- ORF
Open Reading Frame
- PLIP
Protein-Ligand Interaction Profile
- PLpro
Papain-like Protease
- RBD
Receptor-Binding Domain
- RCSB-PDB
Research Collaboratory for Structural Bioinformatics-Protein Databank
- RdRp
RNA-dependent RNA polymerase
- Rg
Radius of Gyration
- RMSD
Root Mean Square Deviation
- RMSF
Root-Mean-Square Fluctuation
- S Glycoprotein
Spike Glycoprotein
- SARS
Severe Acute Respiratory syndrome
- SARS-CoV-2
Severe Acute Respiratory Syndrome Coronavirus-2
- TCM
Traditional Chinese Medicine
- TMPRSS-2
Type-II Transmembrane Serine Protease-2
- WHO
World Health Organization
Acknowledgements
The CHPC (www.chpc.ac.za) is thankfully acknowledged for computational resources and tools.
Disclosure statement
The author declares that there are no known conflicts of interest with regards to this work.
References
- Adeoye A. O., Oso B. J., Olaoye I. F., Tijjani H., & Adebayo A. I. (2020). Repurposing of chloroquine and some clinically approved antiviral drugs as effective therapeutics to prevent cellular entry and replication of coronavirus. Journal of Biomolecular Structure and Dynamics, 1–14. 10.1080/07391102.2020.1765876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baig M. H., Sudhakar D. R., Kalaiarasan P., Subbarao N., Wadhawa G., Lohani M., Khan M. K., & Khan A. U. (2014). Insight into the effect of inhibitor resistant S130G mutant on physico-chemical properties of SHV type beta-lactamase: A molecular dynamics study. PLoS One, 9(12), e112456 10.1371/journal.pone.0112456 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker D. (1998). Pyran-chromenone compounds, their synthesis and anti-HIV activity (United States Patent 5,843,990). The University of Tennessee Research Corporation, Knoxville.
- Belouzard S., Chu V. C., & Whittaker G. R. (2009). Activation of the SARS coronavirus spike protein via sequential proteolytic cleavage at two distinct sites. Proceedings of the National Academy of Sciences of the United States of America, 106(14), 5871–5876. 10.1073/pnas.0809524106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhardwaj K., Palaninathan S., Alcantara J. M., Yi L. L., Guarino L., Sacchettini J. C., & Kao C. C. (2008). Structural and functional analyses of the severe acute respiratory syndrome coronavirus endoribonuclease Nsp15. Journal of Biological Chemistry, 283(6), 3655–3664. 10.1074/jbc.M708375200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhowmick S., Chorge R. D., Jangam C. S., Bharatrao L. D., Patil P. C., Chikhale R. V., & Islam M. A. (2019). Identification of potential cruzain inhibitors using de novo design, molecular docking and dynamics simulations studies. Journal of Biomolecular Structure and Dynamics, 1–11. 10.1080/07391102.2019.1664334 [DOI] [PubMed] [Google Scholar]
- Bode A. M., & Dong Z. (2015). Chemopreventive effects of licorice and its components. Current Pharmacology Reports, 1(1), 60–71. 10.1007/s40495-014-0015-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao B., Wang Y., Wen D., Liu W., Wang J., Fan G., Ruan L., Song B., Cai Y., Wei M., Li X., Xia J., Chen N., Xiang J., Yu T., Bai T., Xie X., Zhang L., Li C., … Wang C. (2020). A trial of lopinavir-ritonavir in adults hospitalized with severe Covid-19. The New England Journal of Medicine, 382(19), 1787–1799. 10.1056/NEJMoa2001282 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng F., Desai R. J., Handy D. E., Wang R., Schneeweiss S., Barabási A. L., & Loscalzo J. (2018). Network-based approach to prediction and population-based validation of in silico drug repurposing. Nature Communications, 9(1), 2691 10.1038/s41467-018-05116-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cinatl J., Morgenstern B., Bauer G., Chandra P., Rabenau H., & Doerr H. W. (2003). Glycyrrhizin, an active component of liquorice roots, and replication of SARS-associated coronavirus. The Lancet, 361(9374), 2045–2046. 10.1016/S0140-6736(03)13615-X [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cotten M., Watson S. J., Kellam P., Al-Rabeeah A. A., Makhdoom H. Q., Assiri A., Al-Tawfiq J. A., Alhakeem R. F., Madani H., AlRabiah F. A., Al Hajjar S., Al-Nassir W. N., Albarrak A., Flemban H., Balkhy H. H., Alsubaie S., Palser A. L., Gall A., Bashford-Rogers R., … Memish Z. A. (2013). Transmission and evolution of the Middle East respiratory syndrome coronavirus in Saudi Arabia: A descriptive genomic study. Lancet (London, England), 382(9909), 1993–2002. 10.1016/S0140-6736(13)61887-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Curreli F., Friedman-Kien A. E., & Flore O. (2005). Glycyrrhizic acid alters Kaposi sarcoma-associated herpesvirus latency, triggering p53-mediated apoptosis in transformed B lymphocytes. The Journal of Clinical Investigation, 115(3), 642–652. 10.1172/JCI23334 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daina A., Michielin O., & Zoete V. (2017). SwissADME: A free web tool to evaluate pharmacokinetics, drug-likeness and medicinal chemistry friendliness of small molecules. Scientific Reports, 7, 42717 10.1038/srep42717 [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Clercq E. (2000). Current lead natural products for the chemotherapy of human immunodeficiency virus (HIV) infection. Medicinal Research Reviews, 20(5), 323–349. [DOI] [PubMed] [Google Scholar]
- Elfiky A. A. (2020. a). SARS-CoV-2 RNA dependent RNA polymerase (RdRp) targeting: An in silico perspective. Journal of Biomolecular Structure and Dynamics, 1–9. 10.1080/07391102.2020.1761882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elfiky A. A. (2020. b). Natural products may interfere with SARS-CoV-2 attachment to the host cell. Journal of Biomolecular Structure and Dynamics, 1–10. 10.1080/07391102.2020.1761881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grienke U., Braun H., Seidel N., Kirchmair J., Richter M., Krumbholz A., von Grafenstein S., Liedl K. R., Schmidtke M., & Rollinger J. M. (2014). Computer-guided approach to access the anti-influenza activity of licorice constituents. Journal of Natural Products, 77(3), 563–570. 10.1021/np400817j [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grienke U., Schmidtke M., Kirchmair J., Pfarr K., Wutzler P., Dürrwald R., Wolber G., Liedl K. R., Stuppner H., & Rollinger J. M. (2010). Antiviral potential and molecular insight into neuraminidase inhibiting diarylheptanoids from Alpinia katsumadai. Journal of Medicinal Chemistry, 53(2), 778–786. 10.1021/jm901440f [DOI] [PubMed] [Google Scholar]
- Hung I. F., Lung K. C., Tso E. Y., Liu R., Chung T. W., Chu M. Y., Ng Y. Y., Lo J., Chan J., Tam A. R., Shum H. P., Chan V., Wu A. K., Sin K. M., Leung W. S., Law W. L., Lung D. C., Sin S., Yeung P., Yip C. C., … Yuen K. Y. (2020). Triple combination of interferon beta-1b, lopinavir-ritonavir, and ribavirin in the treatment of patients admitted to hospital with COVID-19: An open-label, randomised, phase 2 trial. Lancet, 395(10238), 1695–1704. 10.1016/S0140-6736(20)31042-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Islam M. A., & Pillay T. S. (2019). Pharmacoinformatics-based identification of chemically active molecules against Ebola virus. Journal of Biomolecular Structure & Dynamics, 37(15), 4104–4119. 10.1080/07391102.2018.1544509 [DOI] [PubMed] [Google Scholar]
- Islam M. A., & Pillay T. S. (2020). Identification of promising anti-DNA gyrase antibacterial compounds using de novo design, molecular docking and molecular dynamics studies. Journal of Biomolecular Structure & Dynamics, 38(6), 1798–1809. 10.1080/07391102.2019.1617785 [DOI] [PubMed] [Google Scholar]
- Jiang M., Zhao S., Yan S., Li X., He X., Wei X., Song Q., Li R., Fu C., Zhang J., & Zhang Z. (2020). An “essential herbal medicine”-licorice: A review of phytochemicals and its effects in combination preparations. Journal of Ethnopharmacology, 249, 112439 10.1016/j.jep.2019.112439 [DOI] [PubMed] [Google Scholar]
- Joshi R. S., Jagdale S. S., Bansode S. B., Shankar S. S., Tellis M. B., Pandya V. K., Chugh A., Giri A. P., & Kulkarni M. J. (2020). Discovery of potential multi-target-directed ligands by targeting host-specific SARS-CoV-2 structurally conserved main protease. Journal of Biomolecular Structure and Dynamics, 1–16. 10.1080/07391102.2020.1760137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kandeel M., & Al-Nazawi M. (2020). Virtual screening and repurposing of FDA approved drugs against COVID-19 main protease. Life Sciences, 251, 117627 10.1016/j.lfs.2020.117627 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang S., Peng W., Zhu Y., Lu S., Zhou M., Lin W., Wu W., Huang S., Jiang L., Luo X., & Deng M. (2020). Recent progress in understanding 2019 novel Coronavirus associated with human respiratory disease: Detection, mechanism and treatment. International Journal of Antimicrobial Agents, 55(5), 105950 10.1016/j.ijantimicag.2020.105950 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim A., & Ma J. Y. (2018). Isoliquiritin apioside suppresses in vitro invasiveness and angiogenesis of cancer cells and endothelial cells. Frontiers in Pharmacology, 9, 1455 10.3389/fphar.2018.01455 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim S., Thiessen P. A., Bolton E. E., Chen J., Fu G., Gindulyte A., Han L., He J., He S., Shoemaker B. A., Wang J., Yu B., Zhang J., & Bryant S. H. (2016). PubChem substance and compound databases. Nucleic Acids Research, 44(D1), D1202–D1213. 10.1093/nar/gkv951 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim Y., Jedrzejczak R., Maltseva N., Endres M., Godzik A., Michalska K., & Joachimiak A. (2020). The 1.9 A crystal structure of NSP15 endoribonuclease from SARS CoV-2 in the complex with a citrate. Protein Science, 1–11. 10.1101/2020.03.02.968388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim J., Jeon S., Shin H. Y., Kim M. J., Seong Y. M., Lee W. J., Choe K. W., Kang Y. M., Lee B., & Park S. J. (2020). Case of the index patient who caused tertiary transmission of COVID-19 infection in Korea: The application of lopinavir/ritonavir for the treatment of COVID-19 infected pneumonia monitored by quantitative RT-PCR. Journal of Korean Medical Science, 35(6), e79 10.3346/jkms.2020.35.e79 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y., Hong Z., Qian J., Wang Y., & Wang S. (2019). Protective effect of Jie-Geng-Tang against Staphylococcus aureus induced acute lung injury in mice and discovery of its effective constituents. Journal of Ethnopharmacology, 243, 112076 10.1016/j.jep.2019.112076 [DOI] [PubMed] [Google Scholar]
- Meng X. Y., Zhang H. X., Mezei M., & Cui M. (2011). Molecular docking: A powerful approach for structure-based drug discovery. Current Computer-Aided Drug Design, 7(2), 146–157. 10.2174/157340911795677602 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morris G. M., Huey R., Lindstrom W., Sanner M. F., Belew R. K., Goodsell D. S., & Olson A. J. (2009). AutoDock4 and AutoDockTools4: Automated docking with selective receptor flexibility. Journal of Computational Chemistry, 30(16), 2785–2791. 10.1002/jcc.21256 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okimoto N., Futatsugi N., Fuji H., Suenaga A., Morimoto G., Yanai R., Ohno Y., Narumi T., & Taiji M. (2009). High-performance drug discovery: Computational screening by combining docking and molecular dynamics simulations. PLoS Computational Biology, 5(10), e1000528 10.1371/journal.pcbi.1000528 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ou X., Liu Y., Lei X., Li P., Mi D., Ren L., Guo L., Guo R., Chen T., Hu J., Xiang Z., Mu Z., Chen X., Chen J., Hu K., Jin Q., Wang J., & Qian Z. (2020). Characterization of spike glycoprotein of SARS-CoV-2 on virus entry and its immune cross-reactivity with SARS-CoV. Nature Communications, 11(1), 1620 10.1038/s41467-020-15562-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan Y., Guan H., Zhou S., Wang Y., Li Q., Zhu T., Hu Q., & Xia L. (2020). Initial CT findings and temporal changes in patients with the novel Coronavirus pneumonia (2019-nCoV): A study of 63 patients in Wuhan, China. European Radiology, 30, 3306–3309. 10.1007/s00330-020-06731-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parida P., Bhowmick S., Saha A., & Islam M. A. (2020). Insight into the screening of potential beta-lactamase inhibitors as anti-bacterial chemical agents through pharmacoinformatics study. Journal of Biomolecular Structure and Dynamics, 1–20. 10.1080/07391102.2020.1720819. [DOI] [PubMed] [Google Scholar]
- Pillaiyar T., Meenakshisundaram S., & Manickam M. (2020). Recent discovery and development of inhibitors targeting coronaviruses. Drug Discovery Today, 25(4), 30041–30046. 10.1016/j.drudis.2020.01.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pillaiyar T., Meenakshisundaram S., Manickam M., & Sankaranarayanan M. (2020). A medicinal chemistry perspective of drug repositioning: Recent advances and challenges in drug discovery. European Journal of Medicinal Chemistry, 195, 112275 10.1016/j.ejmech.2020.112275 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prajapat M., Sarma P., Shekhar N., Avti P., Sinha S., Kaur H., Kumar S., Bhattacharyya A., Kumar H., Bansal S., & Medhi B. (2020). Drug targets for corona virus: A systematic review. Indian Journal of Pharmacology, 52(1), 56–65. 10.4103/ijp.IJP_115_20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prajapati S. M., & Patel B. R. (2015). A comparative clinical study of Jethimala (Taverniera nummularia Baker.) and Yashtimadhu (Glycyrrhiza glabra Linn.) in the management of Amlapitta. Ayu, 36(2), 157–162. 10.4103/0974-8520.175551 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ricagno S., Egloff M. P., Ulferts R., Coutard B., Nurizzo D., Campanacci V., Cambillau C., Ziebuhr J., & Canard B. (2006). Crystal structure and mechanistic determinants of SARS coronavirus nonstructural protein 15 define an endoribonuclease family. Proceedings of the National Academy of Sciences of the United States of America, 103(32), 11892–11897. 10.1073/pnas.0601708103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sabouri Ghannad M., Mohammadi A., Safiallahy S., Faradmal J., Azizi M., & Ahmadvand Z. (2014). The effect of aqueous extract of Glycyrrhiza glabra on herpes simplex virus 1. Jundishapur Journal of Microbiology, 7(7), e11616 10.5812/jjm.11616 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salentin S., Schreiber S., Haupt V. J., Adasme M. F., & Schroeder M. (2015). PLIP: Fully automated protein-ligand interaction profiler. Nucleic Acids Research, 43(W1), W443–W447. 10.1093/nar/gkv315 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Serafin M. B., Bottega A., Foletto V. S., da Rosa T. F., Hörner A., & Hörner R. (2020). Drug repositioning an alternative for the treatment of coronavirus COVID-19. International Journal of Antimicrobial Agents, 9, 105969 10.1016/j.ijantimicag.2020.105969 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheahan T. P., Sims A. C., Leist S. R., Schäfer A., Won J., Brown A. J., Montgomery S. A., Hogg A., Babusis D., Clarke M. O., Spahn J. E., Bauer L., Sellers S., Porter D., Feng J. Y., Cihlar T., Jordan R., Denison M. R., & Baric R. S. (2020). Comparative therapeutic efficacy of remdesivir and combination lopinavir, ritonavir, and interferon beta against MERS-CoV. Nature Communications, 11(1), 222 10.1038/s41467-019-13940-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singhal T. (2020). A review of Coronavirus disease-2019 (COVID-19). The Indian Journal of Pediatrics, 87(4), 281–286. 10.1007/s12098-020-03263-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sinha S. K., Shakya A., Prasad S. K., Singh S., Gurav N. S., Prasad R. S., & Gurav S. S. (2020). An in-silico evaluation of different Saikosaponins for their potency against SARS-CoV-2 using NSP15 and fusion spike glycoprotein as targets. Journal of Biomolecular Structure and Dynamics, 1–13. 10.1080/07391102.2020.1762741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snijder E. J., Decroly E., & Ziebuhr J. (2016). The nonstructural proteins directing Coronavirus RNA synthesis and processing. Advances in Virus Research, 96, 59–126. 10.1016/bs.aivir.2016.08.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian X., Li C., Huang A., Xia S., Lu S., Shi Z., Lu L., Jiang S., Yang Z., Wu Y., & Ying T. (2020). Potent binding of 2019 novel coronavirus spike protein by a SARS coronavirus-specific human monoclonal antibody. Emerging Microbes & Infections, 9(1), 382–385. 10.1080/22221751.2020.1729069 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trott O., & Olson A. J. (2010). AutoDock Vina: improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. Journal of Computational Chemistry, 31(2), 455–461. 10.1002/jcc.21334 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wan Y., Shang J., Graham R., Baric R. S., & Li F. (2020). Receptor recognition by the novel Coronavirus from Wuhan: An analysis based on decade-long structural studies of SARS Coronavirus. Journal of Virology, 94(7), 20 10.1128/JVI.00127-20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J., Chen X., Wang W., Zhang Y., Yang Z., Jin Y., Ge H. M., Li E., & Yang G. (2013). Glycyrrhizic acid as the antiviral component of Glycyrrhiza uralensis Fisch. Against coxsackievirus A16 and enterovirus 71 of hand foot and mouth disease. Journal of Ethnopharmacology, 147(1), 114–121. 10.1016/j.jep.2013.02.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- World Health Organization (2020). Coronavirus Disease (COVID-2019) Retrieved May 15, 2020, from https://covid19.who.int/.
- Wrapp D., Wang N., Corbett K. S., Goldsmith J. A., Hsieh C. L., Abiona O., Graham B. S., & McLellan J. S. (2020). Cryo-EM structure of the 2019-nCoV spike in the prefusion conformation. Science (New York, N.Y.), 367(6483), 1260–1263. 10.1126/science.abb2507 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu C., Liu Y., Yang Y., Zhang P., Zhong W., Wang Y., Wang Q., Xu Y., Li M., Li X., Zheng M., Chen L., & Li H. (2020). Analysis of therapeutic targets for SARS-CoV-2 and discovery of potential drugs by computational methods. Acta Pharmaceutica Sinica B, 1–23. 10.1016/j.apsb.2020.02.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang R., Yuan B. C., Ma Y. S., Zhou S., & Liu Y. (2017). The anti-inflammatory activity of licorice, a widely used Chinese herb. Pharmaceutical Biology, 55(1), 5–18. 10.1080/13880209.2016.1225775 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu R., Chen L., Lan R., Shen R., & Li P. (2020). Computational screening of antagonist against the SARS-CoV-2 (COVID-19) coronavirus by molecular docking. International Journal of Antimicrobial Agents, 7, 106012 10.1016/j.ijantimicag.2020.106012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H., Penninger J. M., Li Y., Zhong N., & Slutsky A. S. (2020). Angiotensin-converting enzyme 2 (ACE2) as a SARS-CoV-2 receptor: Molecular mechanisms and potential therapeutic target. Intensive Care Medicine, 46(4), 586–590. 10.1007/s00134-020-05985-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou P., Yang X. L., Wang X. G., Hu B., Zhang L., Zhang W., Si H. R., Zhu Y., Li B., Huang C. L., Chen H. D., Chen J., Luo Y., Guo H., Jiang R. D., Liu M. Q., Chen Y., Shen X. R., Wang X., … Shi Z. L. (2020). A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature, 579(7798), 270–273. 10.1038/s41586-020-2012-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Y., Hou Y., Shen J., Huang Y., Martin W., & Cheng F. (2020). Network-based drug repurposing for novel coronavirus 2019-nCoV/SARS-CoV-2. Cell Discovery, 6, 14 10.1038/s41421-020-0153-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu N., Zhang D., Wang W., Li X., Yang B., Song J., Zhao X., Huang B., Shi W., Lu R., Niu P., Zhan F., Ma X., Wang D., Xu W., Wu G., Gao G. F., & Tan W. (2020). A novel Coronavirus from patients with pneumonia in China, 2019. New England Journal of Medicine, 382(8), 727–733. 10.1056/NEJMoa2001017 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.