Abstract
Functional co-delivery of plasmid DNA and RNA oligonucleotides in the same nanoparticle system is challenging due to differences in their physical properties as well as their intracellular locations of function. In this study, we synthesized a series of reducible branched ester-amine quadpolymers (rBEAQs) and investigated their ability to co-encapsulate and deliver DNA plasmids and RNA oligos. The rBEAQs are designed to leverage polymer branching, reducibility, and hydrophobicity to successfully co-complex DNA and RNA in nanoparticles at low polymer to nucleic acid w/w ratios and enable high delivery efficiency. We validate the synthesis of this new class of biodegradable polymers, characterize the self-assembled nanoparticles that these polymers form with diverse nucleic acids, and demonstrate that the nanoparticles enable safe, effective, and efficient DNA-siRNA co-delivery as well as non-viral CRISPR-mediated gene editing utilizing Cas9 DNA and sgRNA co-delivery.
Keywords: DNA, RNA oligonucleotides, reducible branched ester-amine quadpolymers, rBEAQs, nanoparticle, non-viral, co-delivery, CRISPR
Graphical Abstract
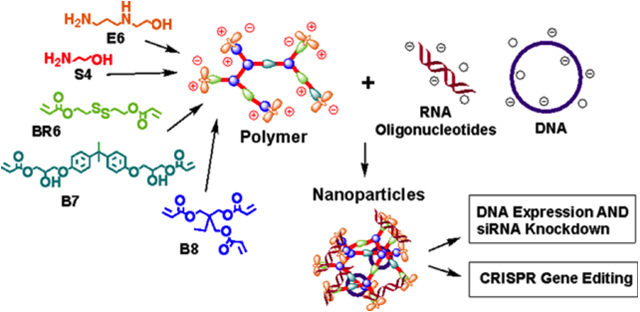
1. Introduction
The introduction of exogenous genetic material into mammalian cells has been widely used in the laboratory to modulate gene expression and induce cellular reprogramming,1 differentiation,2–3 and programmed cell death.4–6 Recently, these technologies have begun moving into the clinic and mark the beginning of a new paradigm for genetic medicine.7–8 Traditional gene therapies involve the delivery of DNA, often in the form of plasmids or mini-circle DNA9, into target cells. RNA oligonucleotides such as short interfering RNA (siRNA) can enable target-specific gene silencing,10–11 and single guide RNAs (sgRNAs) complex with Cas9 endonucleases to achieve site-specific gene editing via the CRISPR/Cas9 system.12–13 The biological functionality of these nucleic acids depend heavily on their successful intracellular delivery.14
Although non-viral vectors delivering either plasmid DNA or siRNA have been widely reported, very few studies have been able to functionally co-deliver both in the same nanoparticle system. This can be challenging as DNA and RNA oligonucleotides are vastly different in size (5,000 bp vs. 20 bp) and stiffness.15–16 In this study, we synthesized a series of reducible branched ester-amine quadpolymers (rBEAQs) and investigated their ability to form nanoparticles that could functionally co-deliver plasmid DNA and RNA oligonucleotides. The rBEAQs were designed based on recent studies that have demonstrated that hyperbranched cationic polymers are superior than their linear counterparts at DNA17–20 and oligonucleotide21–22 delivery in multiple polymeric vector systems. The branched polymer architecture could increase the charge density of each polymer molecule, allowing for stronger nucleic acid binding affinity.23 Disulfide bonds are another useful functionality as they can enable environmentally-triggered cargo release in the reducing cytosolic environment. They can be incorporated into delivery vectors as polymer side chains,24 crosslinking moieties between polymer chains,25 and part of the polymer backbone26, and have been used successfully in several siRNA delivery systems. Finally, increasing polymer hydrophobicity has been shown to improve nanoparticle stability and increase DNA27 as well as siRNA delivery efficacy.28
Using a facile one-pot Michael addition reaction, we were able to tune the reducibility and hydrophobicity of the polymers by simply adjusting the monomer ratios. We found that the nucleic acid binding affinity, release kinetics, nanoparticle uptake, and functional nucleic acid delivery could be modulated in a highly controlled manner. Our nanoparticle system enabled up to 77% DNA transfection and 66% siRNA-mediated knockdown. More importantly, delivery of Cas9 DNA and sgRNA enabled 40% gene knockout, further highlighting the robustness of this co-delivery system.
2. Materials and Methods
2.1. Materials
2-Hydroxyethyl disulfide (CAS 1892291), triethylamine (CAS 121448), acryloyl chloride (CAS 814686), bisphenol A glycerolate (1 glycerol/phenol) diacrylate (B7; CAS 4687949), trimethylolpropane triacrylate (B8; CAS 15625895), 2-(3-aminopropylamino)ethanol (E6; CAS 4461396), L-buthionine-sulfoximine (CAS 83730534), and solvents were purchased from Sigma Aldrich (St. Louis, MO). 4-Amino-1-butanol (S4; CAS 133251005) was purchased from Alfa Aesar (Tewksbury, MA). Plasmids pCAG-GFPd2 (14760) and piRFP670-N1 (45457) were purchased from Addgene (Cambridge, MA). PB-CMV-MCS-EF1a-RFP PiggyBac plasmid (PB512B-1) and PiggyBac transposase expression plasmid (PB200A-1) were purchased from System Biosciences (Palo Alto, CA). Negative control siRNA (1027281) was purchased from Qiagen (Germantown, MD). GFP siRNA targeting the sequence 5’-GCA AGC TGA CCC TGA AGT TC-3’ (P-002048-01) was purchased from Dharmacon (Lafayette, CO). Cy5-labeled siRNA (SIC005) was purchased from Sigma Aldrich.
2.2. Polymer Synthesis
Bioreducible monomer 2,2-disulfanediylbis(ethane-2,1-diyl) diacrylate (BR6) was synthesized using a method similar to Kozielski et al.26 Briefly, 2-hydroxyethyl disulfide was acrylated with acryloyl chloride (1:1.1 molar ratio in dichloromethane) in the presence of excess triethylamine. After filtering out the precipitate, the product was washed with water, dried with sodium sulfate, and the solvent was removed by rotary evaporation.
For polymer synthesis, monomers BR6, B7, B8, and S4 were dissolved in anhydrous dimethylsulfoxide (DMSO) according to the B monomer molar ratios listed in Table S1 for an overall vinyl: amine ratio of 2.2:1 at a concentration of 150 mg/mL. After overnight reaction at 90°C with stirring, the polymers were end-capped by reacting with monomer E6 (0.2 M final concentration in DMSO) at room temperature for 1 hr. The end-capped polymers were purified by 2 diethyl ether washes, after which remaining solvent was removed in a vacuum chamber. Polymers were dissolved in DMSO at 100 mg/mL and stored in aliquots at −20°C under desiccant.
2.3. Yo-Pro-1 Iodide Nucleic Acid Binding Assay
Yo-Pro-1 iodide fluorescent dye (Invitrogen) was mixed with siRNA at a final concentration of 0.5 μM Yo-Pro and 0.5 μM scRNA in 25 mM sodium acetate (NaAc, pH 5.0). Polymers were dissolved in NaAc, and 25 μL polymer solution was mixed with 75 μL RNA/Yo-Pro solution per well in 96 well black bottom plates. The solutions were incubated at 37°C for 20 minutes before fluorescence readings were taken on a fluorescence multiplate reader (Biotek Synergy 2). To measure siRNA binding in reducing conditions over time, polymer concentration was set at the lowest concentration at which each polymer achieved >80% quenching. The polymer/siRNA/Yo-Pro solution was mixed with 10 μL glutathione solution (final concentration 5 mM) and incubated at 37°C. Fluorescence readings were taken at the indicated time points.
2.4. Polymer Characterization: NMR and GPC
Polymer structure was characterized by nuclear magnetic resonance spectroscopy (NMR) via 1H NMR in CDCl3 (Bruker 500 MHz) and analyzed using TopSpin 3.5 software. To measure polymer molecular weight and polydispersity, polymers were dissolved in BHT-stabilized tetrahydrofuran with 5% DMSO and 1% piperidine, filtered through a 0.2 μm PTFE filter, and measured with gel permeation chromatography against linear polystyrene standards (Waters, Milford, MA).
2.5. Gel Retardation Assay
Nanoparticles were synthesized by dissolving polymer and siRNA separately in NaAc buffer at the desired concentrations. The solutions were mixed at a 1:1 volume ratio and nanoparticles were allowed to self-assemble at room temperature for 10 minutes, after which, nanoparticles were incubated in the presence of 5 mM glutathione or 150 mM PBS at 37°C. Samples were taken at various time points and frozen at −80°C to stop the reaction. For gel retardation assays of R6,7,8_64 nanoparticles co-encapsulating plasmid DNA and siRNA, nucleic acids were first pre-mixed at 1:1 volume ratio and then mixed with polymer to allow for nanoparticle self-assembly. Polymer dosage was varied from 10 w/w to 0 w/w (free nucleic acids). Samples were loaded onto a 1% agarose gel using 30% glycerol as loading buffer. Gel electrophoresis was performed in TAE buffer at 100 V for 15 min, after which the gel was imaged under UV.
2.6. Nanoparticle Characterization
Nanoparticles were prepared as described above and diluted in 150 mM PBS to determine particle size and surface charge in neutral isotonic buffer. Hydrodynamic diameter was measured via nanoparticle tracking analysis at 1:500 dilution in PBS using a NanoSight NS300, while zeta potential was measured at 1:6 dilution in PBS via electrophoretic light scattering on a Malvern Zetasizer NanoZS (Malvern Panalytical). To characterize nanoparticle stability over time in physiological conditions, nanoparticle size was also measured at 1:6 dilution in 10% serum-containing cell culture medium once per hour for 9 hours using a Malvern Zetasizer Pro (Malvern Panalytical). Transmission electron microscopy (TEM) images were acquired with a Philips CM120 (Philips Research). Nanoparticles were prepared at a polymer concentration of 1.8 mg/mL in 25 mM NaAc, and 30 μL were added to 400-square mesh carbon coated TEM grids and allowed to coat grids for 20 min. Grids were then rinsed with ultrapure water, counterstained with uranyl acetate (0.5% in distilled water), and allowed to fully dry before imaging.
2.7. Cell Culture and Cell Line Preparation
HEK-293T human embryonic kidney and Huh7 human hepatocellular carcinoma cells were cultured in Dulbecco’s Modified Eagle Medium (DMEM; ThermoFisher) supplemented with 10% FBS and 1% penicillin/streptomycin. A PiggyBac transponson/transposase system was used to generate cell lines constitutively expressing a destabilized form of GFP (GFPd229). The GFPd2 PiggyBac transposon plasmid was created by inserting the GFPd2 gene into the PB-CMV-MCS-EF1a-RFP PiggyBac plasmid through standard restriction enzyme cloning. The transposon plasmid was then co-transfected with the PiggyBac transposase expression plasmid into cells using the method described below. Cells underwent 2 transfections and were then grown out for 5 passages to allow fluorescence signal from transient transfections to fade. Positively-expressing cells were isolated via fluorescence-assisted cell sorting (FACS), and colonies grown from single cells were grown out to establish stably expressing cell lines.
2.8. Transfection
Cells were seeded onto 96 well tissue culture plates at a density of 15,000 cells per well in 100 μL complete medium and allowed to adhere overnight. Nanoparticles were formed immediately prior to transfection as described above. For experiments delivering siRNA only, each nanoparticle condition was formulated with a scrambled control RNA (scRNA) or an siRNA targeting GFP (siGFP) with a final RNA concentration of 100 nM per well. For experiments co-delivering siRNA and DNA, nanoparticles were formulated with a final dose of 200 ng DNA per well in addition to 100 nM scRNA or siGFP, respectively, for a final total nucleic acid dose of 400 ng per well. Nanoparticles co-encapsulating DNA and siRNA were formed by pre-mixing the nucleic acids at 1:1 volume ratio in NaAc buffer prior to mixing with polymer solution. Prior to the addition of nanoparticles, cell culture medium was replaced with 100 μL serum-free media. 20 μL of nanoparticles were added per well and incubated with cells for two hours, at which point the nanoparticle/media mixture was replaced with fresh complete media. Knockdown of GFPd2 fluorescence was assessed via flow cytometry one day post transfection using a BD Accuri C6 flow cytometer (BD Biosciences). Knockdown was quantified by normalizing the geometric mean of fluorescence of wells treated with siGFP to that of wells transfected using the same nanoparticle formulation delivering scRNA. For co-codelivery experiments, DNA transfection was quantified as the percentage of cells positively expressing iRFP when gated against untreated controls. (N = 4 +/− SEM.)
Transfections in which sodium bicarbonate (NaHCO3) was used to increase nanoparticle pH were done by forming nanoparticles in acidic NaAc buffer as previously described and then mixing with 50 mg/mL NaHCO3 buffer (pH 9) at 1:1 volume ratio before adding to cells. Transfections using commercially-available non-viral transfection reagents Lipofectamine 2,000™, Lipofectamine 3,000™ (ThermoFisher), and jetPrime® (Polyplus) were performed according to manufacturer instructions. 25 kD bPEI was used at 1 w/w in DNA-siRNA co-delivery experiments.
2.9. Cellular Uptake and Viability
Cy-5 labeled siRNA was diluted 1:5 in unlabeled siRNA and used to formulate nanoparticles as described above. Nanoparticles were added to cells in serum-free media and incubated for two hours, at which point cells were washed once with PBS and detached via trypsinization. Cells were further washed with heparin (50 μg/mL in PBS) to remove nanoparticles adhering to cells, resuspended in FACS buffer (2% FBS in PBS), and nanoparticle uptake was quantified by flow cytometry. Cell viability was assessed 24 hr post-transfection using MTS CellTiter 96 Aqueous One cell proliferation assay (Promega) following manufacturer’s instructions. Cell viability of treated cells were normalized to that of untreated cells; N = 4 +/− SEM.
2.10. Glutathione Inhibition with L-buthionine-sulfoximine (BSO)
L-buthionine-sulfoximine (BSO) was dissolved in cell culture media at 2,000 μM. Cells were allowed to settle for three hours after plating, at which time 50 μL of media was replaced by 50 μL BSO solution for 1,000 μM final BSO concentration, which has been shown to effectively inhibit intracellular glutathione levels.30 Cells were incubated for 24 hr with complete medium containing 1,000 μM BSO, which was replaced with serum-free BSO medium immediately before transfection. After two hours incubation with nanoparticles, cells were replenished with fresh BSO-containing complete medium and incubated for 24 hr, at which point cell viability and flow cytometry assays were performed.
2.11. Confocal microscopy.
HEK-293T cells were plated on Nunc Lab-Tek 8 chambered borosilicate coverglass well plates (155411; Thermo Fisher) at 30,000 cells/well one day prior to transfection in 300 μL phenol red free DMEM supplemented with 10% FBS and 1% penicillin/streptomycin. R6,7,8_64 nanoparticles were prepared as described above at 10 w/w ratio using pre-mixed Cy3 labeled siRNA and Cy5 labeled plasmid DNA at a 1 w/w ratio of nucleic acids. Cy5 labeled plasmid DNA was prepared as previously described31–32 and mixed at 4 w/w ratio with unlabeled eGFP-N1 plasmid DNA. Nanoparticles were then diluted into media and added to cells at a total nucleic acid dose of 1,000 ng/well and incubated for two hours. Prior to imaging, cells were then stained with Hoechst 33342 at a 1:5,000 dilution for nuclei visualization. Images were acquired over a 19,660 μm area at Nyquist limit resolution using a Zeiss LSM 780 microscope with Zen Blue software and 63x oil immersion lens. Specific laser channels used were 405 nm diode, 488 nm argon, 561 nm solid-state, and 639 nm diode lasers. Laser intensity and detector gain settings were maintained across all image acquisition.
2.12. CRISPR Gene Editing
The template used for in vitro transcription of sgRNA targeting GFP was synthesized as a gBlock from IDT (sequence listed in Table S2). In vitro transcription was performed using a MEGAshortscript T7 Transcription kit (Invitrogen) according to manufacturer’s instructions, and the sgRNA product was purified using MEGAclear Transcription Clean-up kit (Invitrogen). Cas9 plasmid DNA (41815)12 was purchased from Addgene and amplified by Aldeveron (Fargo, ND). For co-delivery transfections, DNA and sgRNA were delivered using R6,7,8_64 nanoparticles as described above. Gene knockout was assessed using flow cytometry 5 days post-transfection unless otherwise noted.
2.13. Statistics
Prism 6 (Graphpad, La Jolla, CA) was used for all statistical analyses and curve plotting. Statistical tests were performed with a global alpha value of 0.05. Unless otherwise stated, absence of statistical significance markings where a test was stated to have been performed signified no statistical significance. The statistical test used and the number of experimental replicates were listed in the captions for each figure. Statistical significance was denoted as follows: *p<0.05; **p<0.01, ***p<0.001, ****p<0.0001.
3. Results and Discussion
3.1. Polymer Synthesis and Characterization
Polymers were synthesized following a facile one-pot Michael addition reaction in which acrylate monomers BR6 and B8 were copolymerized with amine-containing monomer S4 (Scheme 1). After end-capping with monomer E6, this class of polymers is referred to as R6,8_N, where N denotes the branching B8 monomer content in the polymer backbone (i.e. R6,8_20 contains 20% B8). In the polymer series containing the additional diacrylate monomer B7, polymers are referred to as R6,7,8_M, where M denotes the B7 monomer content in the polymer backbone. For acrylate-terminated base polymer synthesis, B and S monomers were dissolved in anhydrous DMSO at 150 mg/mL (monomer concentrations >400 mg/mL resulted in complete gelation), and step-wise polymerization reaction was allowed to proceed overnight at 90°C with stirring. The chemical structures of base polymers were determined via NMR spectroscopy, which verified that the polymers were acrylate terminated by three distinct acrylate peaks at 5.5–6.5 ppm (Figure S1). Polymer end-capping with monomer E6 was performed at room temperature for 1 hr and confirmed by the disappearance of these peaks. Molecular weight data was obtained from GPC analysis, which showed that with increasing B8 content, both Mn and Mw values generally increased (Table 1). For R6,7,8-4-6 polymers, for which B8 content was fixed at 20%, molecular weight did not change significantly with varying B7 content, suggesting that molecular weight is largely controlled by polymer branching and crosslinking effects contributed by triacrylate monomer B8.
Scheme 1. Monomer structures and proposed mechanism for polymer function.
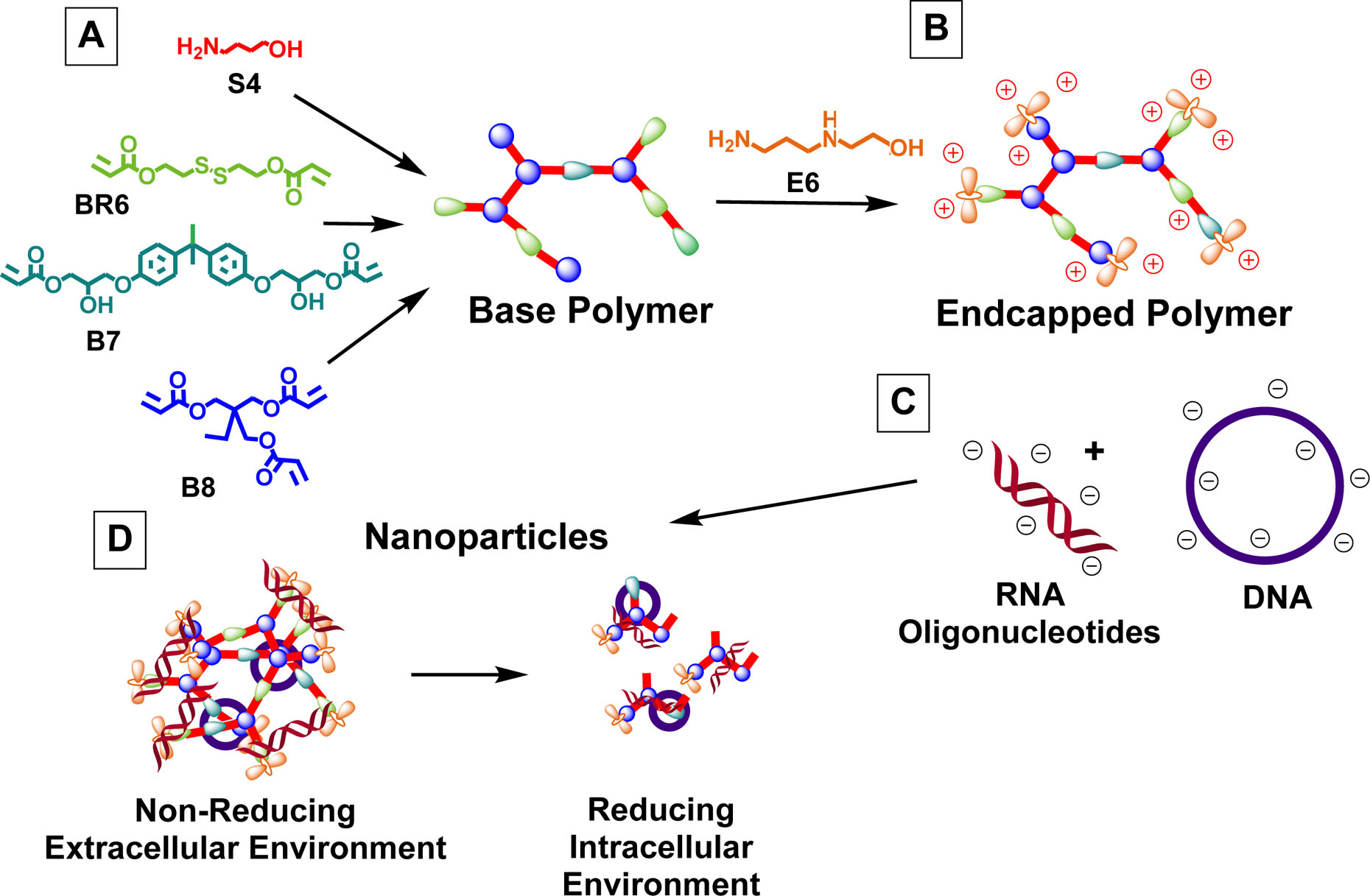
(A) B and S monomers were co-polymerized to form acrylate-terminated base polymers, which were then (B) end-capped with monomer E6. (C) These polymers self-assembled into nanoparticles with anionic nucleic acids and (D) partially degraded at reducible linkages in the reducing cytosolic environment, allowing for intracellular cargo release.
Table 1.
Molecular weight data from GPC characterization and monomer composition calculated from 1H NMR spectra.
| Polymer Name | Mn | Mw | PDI | Monomer Fraction in Polymer | |||
|---|---|---|---|---|---|---|---|
| BR6 | B7 | B8 | |||||
| R6,8-4-6 | R6,8_0 | 2,224 | 2,682 | 1.21 | 1.00 | -- | -- |
| R6,8_20 | 3,168 | 4,038 | 1.27 | 0.75 | -- | 0.25 | |
| R6,8_40 | 4,050 | 5,896 | 1.46 | 0.54 | -- | 0.46 | |
| R6,8_60 | 4,943 | 9,949 | 2.01 | 0.36 | -- | 0.64 | |
| R6,8_80 | 4,675 | 8,728 | 1.87 | 0.23 | -- | 0.77 | |
| R6,7,8-4-6 (20% B8) | R6,7,8_16 | 4,511 | 7,616 | 1.69 | 0.62 | 0.17 | 0.21 |
| R6,7,8_40 | 4,570 | 7,435 | 1.63 | 0.41 | 0.40 | 0.19 | |
| R6,7-8_64 | 4,438 | 7,066 | 1.59 | 0.17 | 0.63 | 0.20 | |
3.2. siRNA Delivery: Gene Knockdown, Cellular Uptake, and Cytotoxicity
R6,8-4-6 polymers were used to deliver siRNA targeting GFP (siGFP) in HEK-293T cells stably expressing a destabilized form of GFP with short half-life (GFPd2).29 At 100 nM siRNA dose and 180 polymer-siRNA w/w ratio, R6,8_20 achieved 75% knockdown with negligible cytotoxicity (Figure 1A). All branched polymers in the R6,8_N series with the exception of R6,8_80 achieved significantly higher knockdown than the linear polymer (R6,8_0). Knockdown levels peaked with R6,8_20 and R6,8_40 (Figure S2), and the same trend was observed for nanoparticle uptake (Figure 1B). Previous studies have demonstrated that nanoparticle uptake and transfection efficacy increased with increasing polymer molecular weight.33–34 This was not the case in our polymer system as R6,8_60 and R6,8_80 had the highest molecular weight but achieved relatively poor knockdown. This could in part be due to the fact that increasing polymer branching resulted in lower cell viability caused by decreasing reducible BR6 monomer content. Indeed, when cells were pre-treated with L-buthionine-sulfoximine (BSO) to inhibit cellular production of glutathione, the main intracellular reducing agent,35 nanoparticle-mediated cytotoxicity significantly increased (Figure 1C). This increased toxicity was beyond the additive effects of either nanoparticle or BSO treatment alone, indicating that the cell’s inability to reduce disulfide bonds after glutathione blockade induced higher levels of cell death and confirming our hypothesis that polymer reducibility attenuated cytotoxicity by enabling them to rapidly degrade to relatively non-toxic oligomers. Thus, the bio-reducibility of the rBEAQ nanoparticles is designed to both enable environmentally-triggered release upon entering the cytosol and as a mechanism to limit potential cytotoxicity of the branched polymers by quickly breaking them down into smaller components once they reach their target inside the cell.
Figure 1. R6,8-4-6 polymers enable efficient intracellular siRNA delivery.
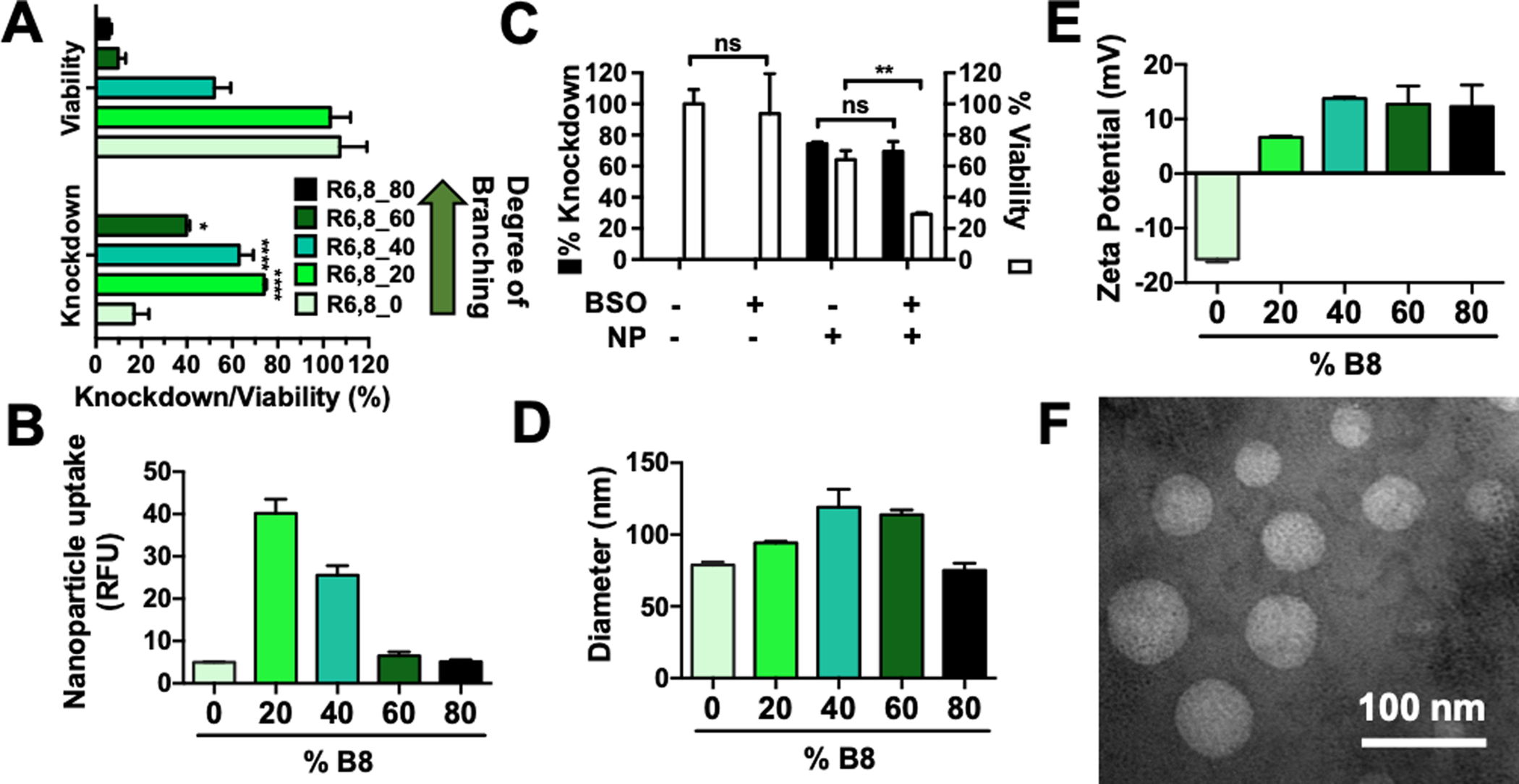
(A) Gene knockdown and cytotoxicity of nanoparticles delivering 100 nM siRNA at 180 w/w. Statistical analysis was assessed by one-way ANOVA with Tukey post-hoc tests. N = 4. (B) Nanoparticle uptake measured by flow cytometry after treatment of nanoparticles containing Cy5-labeled siRNA. N = 4. (C) Pre-treatment with 1,000 μM L-buthionine-sulfoximine (BSO) show that intracellular glutathione blockade did not change knockdown levels but significantly increased polymer-mediated cytotoxicity as assessed by Holm-Sidak corrected multiple t-tests; R6,8_20 nanoparticles (180 w/w) were used to deliver 100 nM siRNA. N = 4. (D) Nanoparticle hydrodynamic diameter and (E) zeta potential measured using dynamic light scattering. Bars show average + SEM; N = 3. (F) Representative TEM image of R6,8_40 nanoparticles.
To elucidate the mechanism by which moderately branched polymers achieved the highest levels of knockdown, we assessed the physical characteristics of the nanoparticles. All polymers in the series formed nanoparticles with hydrodynamic diameters around 100 nm (Figure 1D). Nanoparticles formed with the linear polymer had negative surface charge, and zeta potential generally became increasingly positive with increased polymer branching (Figure 1E). This is likely due to the fact that increased branching resulted in increasing numbers of secondary amine-containing end-groups per polymer molecule, which are positively charged in pH 5 NaAc buffer. The increased cationic charge of moderately-branched polymer nanoparticles likely contributes to nanoparticle uptake and siRNA-mediated knockdown in vitro, which is consistent with many published reports.36–38 This trend does not apply to very highly branched polymers, however, in part due to the high levels of cytotoxicity incurred by these nanoparticle formulations.
3.3. siRNA Binding and Environmentally-triggered Release
A competitive binding assay using Yo-Pro-1 iodide (Yo-Pro) was used to assess siRNA binding strength in R6,8-4-6 polymers. Yo-Pro dye fluoresces upon nucleic acid binding, and quenching of fluorescence after polymer outcompetes the dye for siRNA binding was used as a measure of binding strength. Increasing polymer branching increased siRNA binding strength, which was seen in both end-capped (Figure 2A) as well as acrylate-terminated polymers (Figure S3A). Plotting knockdown as a function of the polymer EC50 w/w of siRNA binding (where lower EC50 w/w corresponds to tighter siRNA binding and higher degree of polymer branching) revealed a biphasic response (Figure 2B). Binding affinity and degree of knockdown both increased approximately 4-fold from R6,8_0 to R6,8_20 and decreased steadily when B8 content exceeded 40%. This suggests that an optimal range for siRNA binding affinity exists, and polymers that bind too tightly cannot release siRNA to achieve efficient knockdown while those that do not bind tightly enough cannot form nanoparticles that effectively promote nanoparticle internalization.39–40 siRNA binding affinity, along with other nanoparticle biophysical and chemical properties such as the size, surface charge, and bio-reducibility discussed earlier all contribute to the differential gene silencing effects seen here. For polymers with the same B8 content, end-capped polymers exhibited stronger binding than their acrylate-terminated counterparts (Figure S3B). These results suggest that polymer branching increases siRNA binding via two mechanisms. The first is mediated by increased branching structure in the polymer backbone, which increases the molecular weight of the polymer and drives stronger binding through greater hydrophobic effects. The second is mediated by increased branching endpoints, which increases the number of end-capping molecules. As the secondary amines in the polymer end-groups are positively-charged in pH 5 NaAc buffer, they further increase siRNA binding through electrostatic interactions.
Figure 2. Polymer branching and reducibility can be modulated to control siRNA binding affinity and release kinetics.
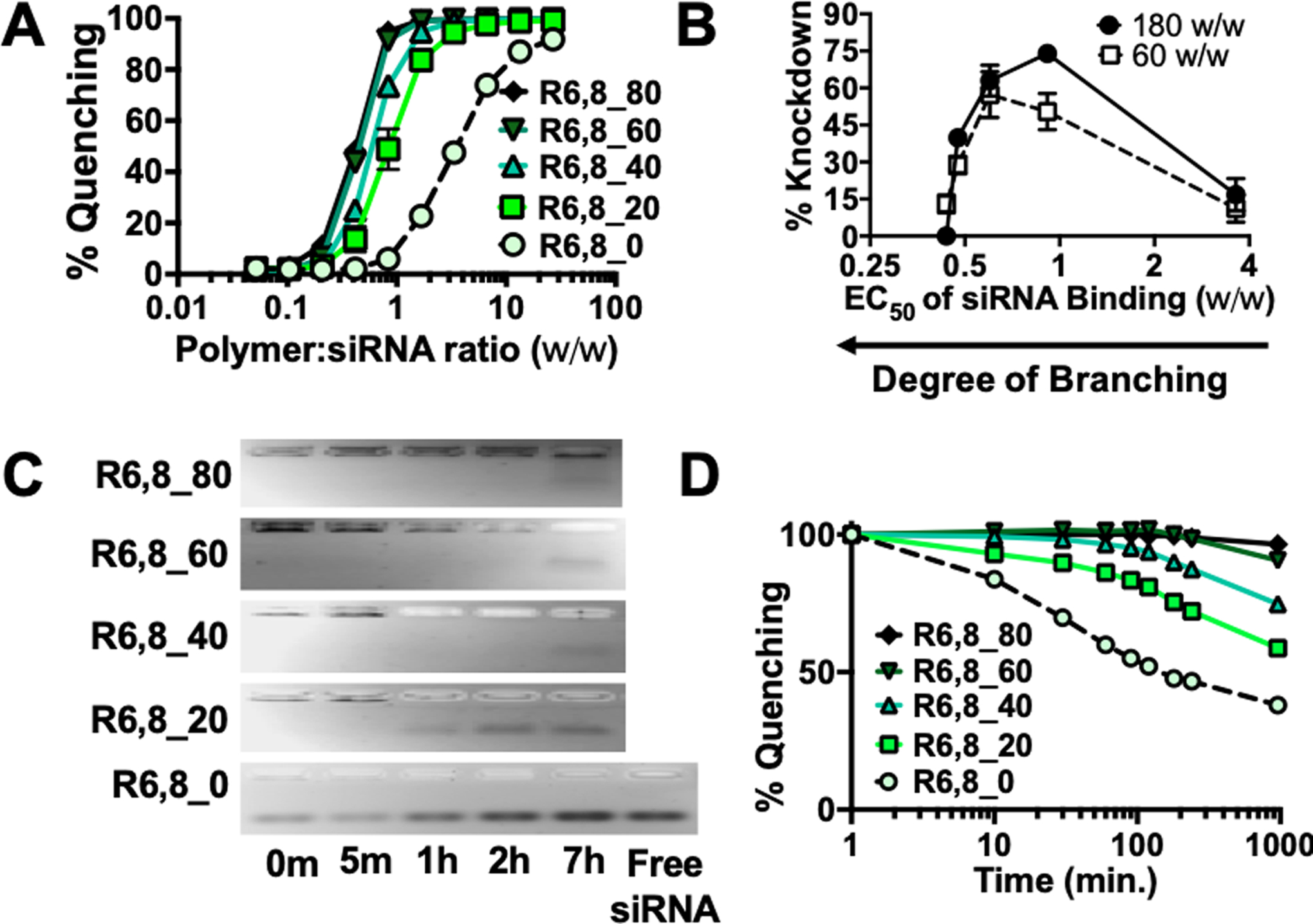
(A) Yo-Pro-1 iodide binding competition assay of R6,8-4-6 polymers to assess siRNA binding affinity. N = 4. (B) % knockdown plotted as a function of polymer EC50 w/w for siRNA binding. N = 4. (C) Gel retardation assays (N = 1) and (D) Yo-Pro binding assay (N = 4) were performed on nanoparticles incubated in 5 mM glutathione to mimic intracellular reducing environments and nucleic acid release slowed as the polymers became more branched with less frequent bio-reducible linkages.
We next investigated siRNA release kinetics of R6,8-4-6 nanoparticles in 5 mM glutathione to mimic the reducing intracellular environment.35 Nanoparticles were sampled at specific time points and standard gel electrophoresis was performed to assess siRNA release (Figure 2C). The linear polymer released siRNA almost instantaneously, and release was complete by 1 hr. Increased polymer branching slowed siRNA release considerably, with R6,8_20 beginning release at 1 hr and higher branching polymers at 7 hr. The same trend was observed when a Yo-Pro binding assay was performed with nanoparticles incubated over time in reducing buffer conditions (Figure 2D). These results indicate that siRNA binding and release can be modulated in a highly controlled manner by changing the ratio between branching and reducible monomers and that siRNA release can be designed to occur in an environmentally-triggered manner via reduction of disulfide bonds. However, we have also shown that blocking intracellular glutathione levels did not significantly decrease the observed level of siRNA-mediated knockdown (Figure 1C), which suggests that other polymer degradation mechanisms such as the hydrolysis of ester bonds over a period of 4–6 hr41 could also contribute to siRNA release from nanoparticles. Incorporation of disulfide linkages in the rBEAQ polymers helps ensure fragmentation of the polymers into small oligomers, reducing cytotoxicity, and enables higher doses, branching, or w/w formulation ratios of the polymers to be safely utilized.
3.4. Co-delivery of siRNA and DNA
As moderately branched polymers have been shown to maintain strong nucleic acid binding affinity while effectively releasing siRNA cargo in the reducing cytosolic environment, we hypothesized that they may be suitable for the co-delivery of plasmid DNA and siRNA. R6,8_20 (the top polymer for siRNA delivery) was used to encapsulate 200 ng each of siGFP siRNA and a plasmid DNA encoding iRFP670. R6,8_20 nanoparticles enabled efficient co-delivery to HEK-293T cells (Figure 3A), resulting in 66% siRNA-mediated knockdown and 77% DNA transfection with negligible cytotoxicity (Figure S4A). The same formulation achieved much lower delivery efficiency in harder-to-transfect Huh7 cells (23% knockdown and 5% transfection; Figure 3B), prompting the need to develop more effective polymers for co-delivery. To this end, we investigated the effect of polymer hydrophobicity by incorporating monomer B7 at ratios indicated in Table S1 to synthesize the R6,7,8-4-6 polymer series. B7 was chosen as it contains a bisphenol A group, which has been shown to bind DNA via hydrophobic effects42 and enable high efficiency DNA transfection.27, 43–44 B7-containing polymers effectively complexed nucleic acids at very low w/w, forming nanoparticles around 150 nm in diameter and +6 to +16 mV in zeta potential (Figure S5). R6,7,8_64 nanoparticles at a 10 w/w ratio were quite stable in complete cell culture medium mimicking physiological conditions for several hours with a hydrodynamic diameter doubling time >4 hours as assessed by DLS (Figure S5D). In contrast, R6,8-4-6 polymers with 0% B7 content formed much larger nanoparticles (270 nm) with −11 mV zeta potential at 10 w/w. B7-containing polymers were used at significantly lower w/w formulations compared to R6,8-4-6 polymers used for siRNA complexation earlier because R6,7,8-4-6 polymers incurred significantly higher cytotoxicity than the R6,8-4-6 polymers, limiting their use to very low w/w formulations (Figure S4B). Nevertheless, B7-containing polymers enabled higher levels of knockdown and transfection at 10 w/w in HEK-293T cells (Figure 3A and 3C), though the difference was less notable when R6,8-4-6 polymers were used at higher w/w. More strikingly, R6,7,8-4-6 polymers enabled significantly higher co-delivery in Huh7 cells compared to R6,8-4-6 at all w/w formulations, with the best formulation achieving 53% knockdown and 37% transfection (Figure 3B). A gel retardation assay demonstrated that R6,7,8_64 completely condensed both plasmid DNA and siRNA at 10 w/w, and decreasing polymer dose resulted in siRNA release at 5 w/w and DNA release at 1 w/w (Figure 3D).
Figure 3. Hydrophobic R6,7,8-4-6 polymer series enables efficient co-delivery of DNA and siRNA.
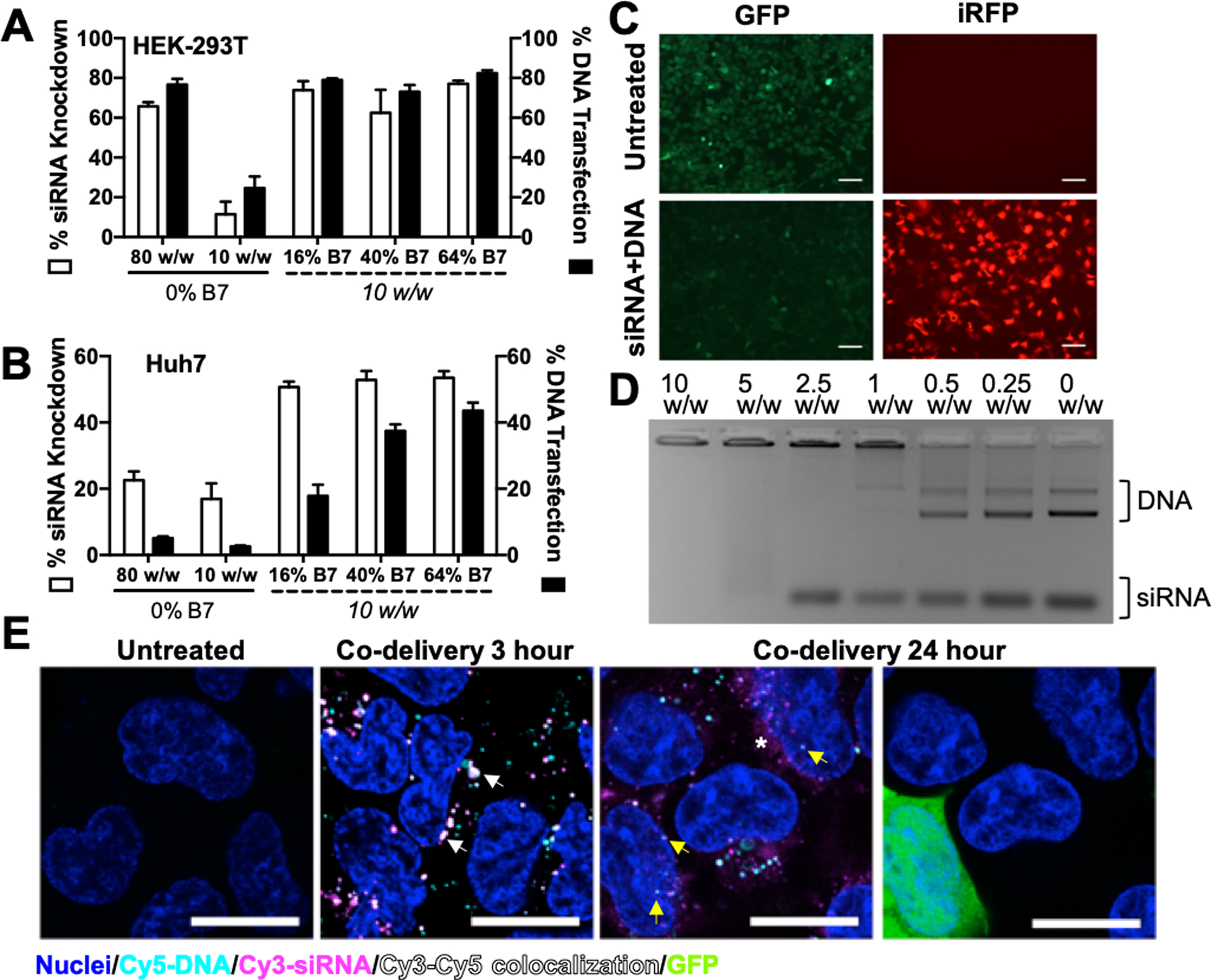
Co-delivery efficacy of R6,8-4-6 (0% B7) and R6,7,8-4-6 nanoparticles encapsulating 400 ng total nucleic acid in 293T (A) and Huh7 (B). N = 4. (C) Fluorescence microscopy images of HEK-293T cells treated with R6,7,8_16 nanoparticles co-delivering 200 ng siRNA and 200 ng DNA (10 w/w formulation). Scale bar 100 μm. (D) R6,7,8_64 completely encapsulated plasmid DNA and siRNA at 10 w/w as seen by a gel retardation assay. (E) Confocal microscopy images of 293T cells treated with R6,7,8_64 nanoparticles co-delivering Cy3-siRNA, Cy5-DNA, and unlabeled GFP plasmid DNA (0.5:0.4:0.1 composition by weight) at 3 hr and 24 hr post-uptake. Cy3 and Cy5 signal colocalization could be seen at 3 hours post-uptake (white arrows). At 24 hours post-uptake, diffuse Cy3-siRNA signal could be seen in the cytosol (white asterisk) while some Cy5-DNA signal was detected in the nucleus (yellow arrows) and some cells were visibly expressing GFP. Scale bar 20 μm.
We further explored the intracellular delivery location of siRNA and DNA using confocal laser scanning microscopy, which demonstrated different fates for internalized siRNA and DNA. At an early 3 hour timepoint following nanoparticle treatment, most endosomes possessed both siRNA and DNA, while at 24 hours post-treatment, diffuse cytosolic siRNA was detectable in most cells and the occasional z-slice revealed some Cy5 labeled plasmid DNA in the nucleus (Figure 3E). Using a mix of fluorescently labeled plasmid DNA and unlabeled plasmid DNA, we were also able to detect a fraction of the cells expressing a fluorescent reporter protein GFP at 24 hours post-transfection, which was undetectable in cells at 3 hours post-treatment (Figure S6).
Studies have shown that polymers optimized for DNA delivery may not be optimal for siRNA and vice versa.45 This may be due to the differences in size and charge density between DNA and siRNA as well as their intracellular sites of action. Bishop et al. approached this problem with a polymer-coated gold nanoparticle system where siRNA and DNA were adsorbed onto the nanoparticle using different polymers in a layer-by-layer synthesis scheme; the optimal formulation in this study resulted in 34% knockdown and 14% transfection in human brain cancer cells.46 Another study using poly(L-lysine) polyplexes for co-delivery of siRNA and DNA to HEK-293T cells showed >80% knockdown but achieved <10% DNA transfection.47 The delivery system reported herein achieved significantly higher co-delivery in both HEK-293T cells as well as harder-to-transfect Huh7 human liver cancer cells. These polymers are easy to formulate into nanoparticles via self-assembly in a single step, and enabled more efficient co-delivery of both DNA and siRNA compared to several leading commercially available non-viral transfection reagents (Figure S7).
We further compared the DNA-siRNA co-delivery efficacy of the system presented herein with that of using nanoparticle formulations previously optimized for the delivery of each nucleic acid separately (Figure S8). In the latter strategy, plasmid DNA was encapsulated using polymer 446 at 60 w/w (previously optimized for DNA delivery48) and siRNA was encapsulated using polymer R646 at 120 w/w (previously optimized for siRNA delivery49). The two nanoparticles were formulated separately and added to cells after nanoparticle formation. In the single nanoparticle strategy, the same amount of nucleic acids was pre-mixed and co-encapsulated in R6,7,8_64 nanoparticles (10 w/w). Our results show that using the dual nanoparticle delivery strategy, siRNA knockdown levels were significantly lower than that achieved by the single nanoparticle co-delivery strategy while DNA transfection levels were similar. Furthermore, when polymers 446 and R646 were used to formulate nanoparticles at 10 w/w for direct comparison with polymer R6,7,8_64, both siRNA and DNA delivery levels were significantly lower. These results indicate that co-encapsulation of multiple nucleic acid cargo types in the same nanoparticle system has the advantages of higher transfection efficiency as well as greater simplicity in formulation; this is especially important for potential clinical translation as it could greatly simplify the synthesis and regulatory approval processes.
3.5. Co-delivery of Cas9 DNA and sgRNA for CRISPR-mediated Gene Editing
Next, we co-encapsulated Cas9 plasmid DNA and sgRNA targeting GFP in our nanoparticles for intracellular delivery of the CRISPR/Cas9 gene editing system within one biodegradable nanoparticle. Gene knockout, which can be assessed by a decrease in GFP fluorescence, is contingent upon co-delivery of both components as the Cas9 endonuclease must assemble with sgRNA to form a functional ribonucleoprotein (RNP) complex. This is a rigorous test of co-delivery as the two components must be present in the same cell as well as remain bioactive at the same time in order for editing to occur. Our results showed that R6,7,8_64 nanoparticles enabled 40% gene knockout in HEK-293T cells (Figure 4A). Delivery of either component alone did not result in appreciable levels of knockout, confirming the need for co-delivery. The optimal sgRNA-Cas9 molar ratio was 33. Interestingly, we saw a distinct GFP-negative population (GFP null) in CRISPR-treated cells which was not observed in cells treated with GFP siRNA (Figure 4B). siRNA-mediated gene silencing downshifted the GFP fluorescence of the entire population of treated cells while CRISPR-mediated knockout completely turned off GFP in a fraction of cells. Kinetic studies showed that siRNA-mediated gene silencing faded rapidly, and fluorescence returned to pre-treatment levels after 11 days (Figure 4C). In contrast, CRISPR-mediated silencing peaked after 5 days and remained constant for the entirety of the period tested. Our results suggest that gene silencing mediated by siRNA knockdown or CRISPR knockout could be suitable for different therapeutic goals. The former has a faster onset and results in significant but transient downregulation in the entire population of treated cells. The latter takes longer to reach peak levels but can produce a sustained and binary downregulation in a smaller fraction of the population.
Figure 4. Co-delivery of anti-GFP sgRNA and Cas9 plasmid enables CRISPR-mediated gene knockout.
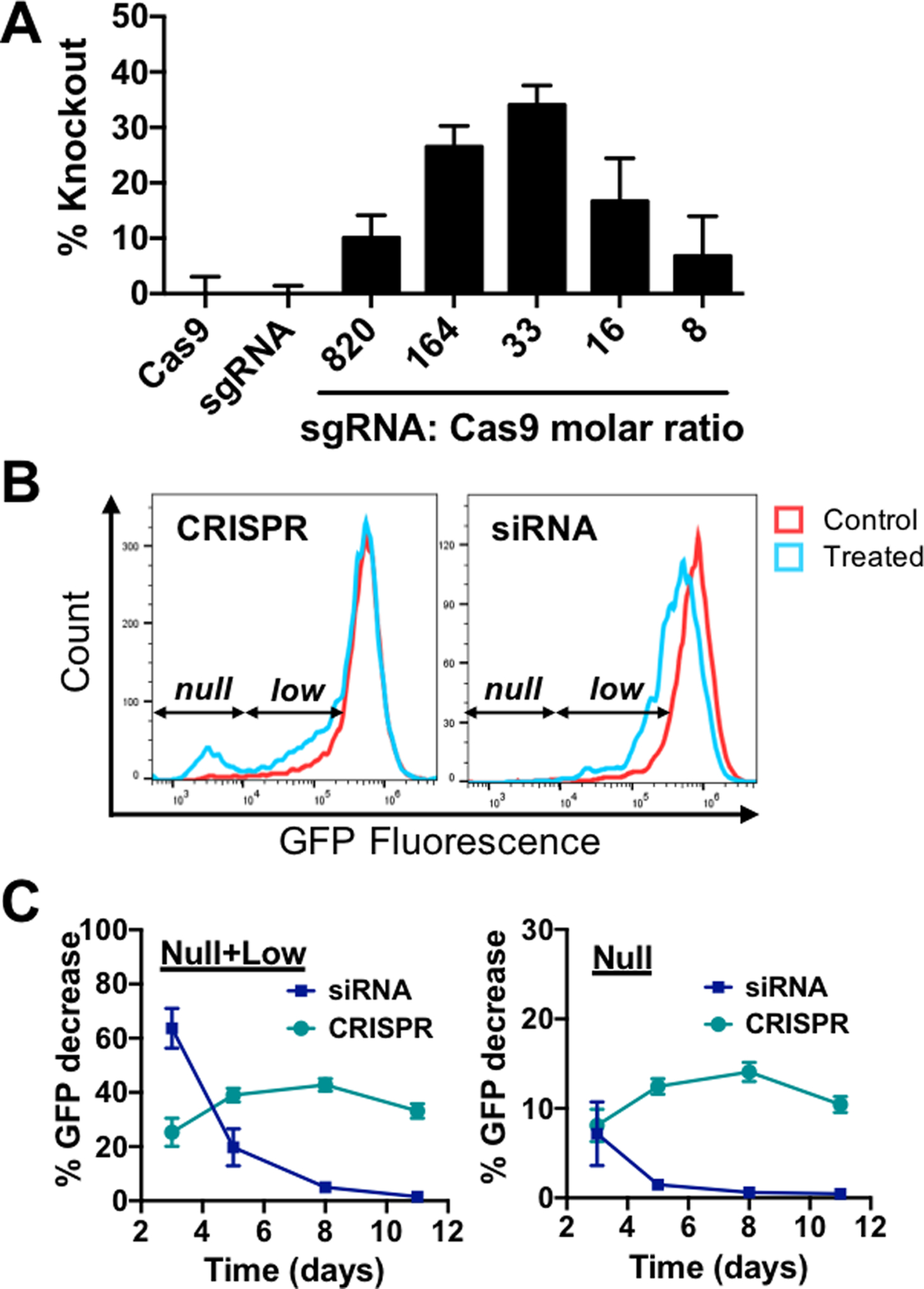
(A) HEK-293T cells were transfected with R6,7,8_64 10 w/w nanoparticles encapsulating Cas9 DNA and sgRNA at the indicated nucleic acid molar ratios. N = 4. (B) Flow cytometry histograms of CRISPR- or siRNA-treated cells. CRISPR treatment produced a completely GFP-negative population (null) while siRNA treatment mainly resulted in a general population shift to lower GFP fluorescence (low). (C) Gene suppression kinetics of CRISPR and siRNA treated cells. N = 4.
It is important to note that all transfection experiments so far have been performed in serum-free medium. It has been widely reported that the presence of serum may decrease transfection efficacy by inducing polyplex disruption and aggregation.50 On the contrary, some studies have also demonstrated that the presence of serum proteins may prevent disassembly of nanocomplexes.51 To investigate the performance of our nanoparticle system in serum conditions, R6,7,8_64 nanoparticles (10 w/w) were formulated with siRNA or Cas9 DNA and sgRNA and administered to cells in complete medium (10% serum). The presence of serum significantly reduced transfection efficacy in both cases (Figure S9). However, when NaHCO3 was added to the nanoparticle formulation to increase nanoparticle pH prior to addition to the cells, transfection in both cases increased back to similar levels as in serum-free conditions. The addition of anionic compounds to nanoparticles to increase transfection in serum conditions has been utilized in other delivery systems52 and is a viable strategy to stabilize polymeric polyplexes.
4. Conclusions
We synthesized a new series of reducible branched ester-amine quadpolymers (rBEAQs) that enabled co-delivery of plasmid DNA and RNA oligonucleotides in the same biodegradable self-assembled nanoparticle system. Our best formulation achieved 77% DNA transfection and 66% siRNA-mediated knockdown in HEK-293T cells, and 37% transfection and 53% knockdown in Huh7 cells. More importantly, co-delivery of Cas9 DNA and sgRNA in the same non-viral nanoparticles enabled 40% CRISPR/Cas9-mediated gene knockout. To our knowledge, this is the first time that CRISPR-mediated gene editing has been achieved through the co-delivery of Cas9 plasmid and sgRNA. The effective co-delivery of plasmid DNA and RNA oligonucleotides reported here, as well as the ability to leverage bio-reducibility, hydrophobicity, and polymer branching to enable effective co-delivery in different cell types, may prove useful for applications such as novel combinatorial gene therapies and genome editing.
Supplementary Material
Funding Sources
The authors would like to thank the following organizations for financial support: NSF Graduate Research Fellowship DGE-0707427 (DRW) and DGE-1232825 (YR), Microscopy Core Grant (S10 OD016374) and the United States NIH grants R01EB016721 (JJG), and R01EB022148 (JJG). JJG thanks the Bloomberg~Kimmel Institute for Cancer Immunotherapy and the Research to Prevent Blindness James and Carole Free Catalyst Award for support.
Footnotes
Supporting Information
The Supporting Information is available free of charge on the ACS Publications website.
Supplemental figures with additional results: polymer synthesis and characterization; additional knockdown and cytotoxicity data; DNA-siRNA co-delivery using standard non-viral transfection reagents.
References
- 1.Bhise NS; Wahlin KJ; Zack DJ; Green JJ, Evaluating the Potential of Poly(Beta-Amino Ester) Nanoparticles for Reprogramming Human Fibroblasts to Become Induced Pluripotent Stem Cells. International journal of nanomedicine 2013, 8, 4641–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Warren L; Manos PD; Ahfeldt T; Loh Y-H; Li H; Lau F; Ebina W; Mandal PK; Smith ZD; Meissner A, Highly Efficient Reprogramming to Pluripotency and Directed Differentiation of Human Cells with Synthetic Modified Mrna. Cell stem cell 2010, 7 (5), 618–630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Tzeng SY; Hung BP; Grayson WL; Green JJ, Cystamine-Terminated Poly(Beta-Amino Ester)S for Sirna Delivery to Human Mesenchymal Stem Cells and Enhancement of Osteogenic Differentiation. Biomaterials 2012, 33 (32), 8142–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Powers MV; Clarke PA; Workman P, Dual Targeting of Hsc70 and Hsp72 Inhibits Hsp90 Function and Induces Tumor-Specific Apoptosis. Cancer Cell 2008, 14 (3), 250–262. [DOI] [PubMed] [Google Scholar]
- 5.Caldas H; Jaynes FO; Boyer MW; Hammond S; Altura RA, Survivin and Granzyme B–Induced Apoptosis, a Novel Anticancer Therapy. Molecular cancer therapeutics 2006, 5 (3), 693–703. [DOI] [PubMed] [Google Scholar]
- 6.Mangraviti A; Tzeng SY; Kozielski KL; Wang Y; Jin Y; Gullotti D; Pedone M; Buaron N; Liu A; Wilson DR; Hansen SK; Rodriguez FJ; Gao G-D; DiMeco F; Brem H; Olivi A; Tyler B; Green JJ, Polymeric Nanoparticles for Nonviral Gene Therapy Extend Brain Tumor Survival in Vivo. ACS Nano 2015, 9 (2), 1236–1249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ledford H, Engineered Cell Therapy for Cancer Gets Thumbs up from Fda Advisers. Nature 2017, 547 (7663), 270. [DOI] [PubMed] [Google Scholar]
- 8.Fischer A Fda Approves Novel Gene Therapy to Treat Patients with a Rare Form of Inherited Vision Loss. FDA.gov.
- 9.Munye MM; Tagalakis AD; Barnes JL; Brown RE; McAnulty RJ; Howe SJ; Hart SL, Minicircle DNA Provides Enhanced and Prolonged Transgene Expression Following Airway Gene Transfer. Sci. Rep 2016, 6, 23125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fire A; Xu SQ; Montgomery MK; Kostas SA; Driver SE; Mello CC, Potent and Specific Genetic Interference by Double-Stranded Rna in Caenorhabditis Elegans. Nature 1998, 391 (6669), 806–811. [DOI] [PubMed] [Google Scholar]
- 11.Hannon GJ, Rna Interference. nature 2002, 418 (6894), 244–251. [DOI] [PubMed] [Google Scholar]
- 12.Mali P; Yang L; Esvelt KM; Aach J; Guell M; DiCarlo JE; Norville JE; Church GM, Rna-Guided Human Genome Engineering Via Cas9. Science 2013, 339 (6121), 823–826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cong L; Ran FA; Cox D; Lin S; Barretto R; Habib N; Hsu PD; Wu X; Jiang W; Marraffini LA; Zhang F, Multiplex Genome Engineering Using Crispr/Cas Systems. Science 2013, 339 (6121), 819–823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kozielski KL; Rui Y; Green JJ, Non-Viral Nucleic Acid Containing Nanoparticles as Cancer Therapeutics. Expert Opin Drug Deliv 2016, 1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kebbekus P; Draper DE; Hagerman P, Persistence Length of Rna. Biochemistry-Us 1995, 34 (13), 4354–4357. [DOI] [PubMed] [Google Scholar]
- 16.Hagerman PJ, Flexibility of Rna. Annu Rev Bioph Biom 1997, 26, 139–156. [DOI] [PubMed] [Google Scholar]
- 17.Tian H; Xiong W; Wei J; Wang Y; Chen X; Jing X; Zhu Q, Gene Transfection of Hyperbranched Pei Grafted by Hydrophobic Amino Acid Segment Pblg. Biomaterials 2007, 28 (18), 2899–2907. [DOI] [PubMed] [Google Scholar]
- 18.Kadlecova Z; Rajendra Y; Matasci M; Baldi L; Hacker DL; Wurm FM; Klok H-A, DNA Delivery with Hyperbranched Polylysine: A Comparative Study with Linear and Dendritic Polylysine. Journal of controlled release 2013, 169 (3), 276–288. [DOI] [PubMed] [Google Scholar]
- 19.Zhao T; Zhang H; Newland B; Aied A; Zhou D; Wang W, Significance of Branching for Transfection: Synthesis of Highly Branched Degradable Functional Poly (Dimethylaminoethyl Methacrylate) by Vinyl Oligomer Combination. Angewandte Chemie International Edition 2014, 53 (24), 6095–6100. [DOI] [PubMed] [Google Scholar]
- 20.Wilson DR; Rui Y; Siddiq K; Routkevitch D; Green JJ, Differentially Branched Ester Amine Quadpolymers with Amphiphilic and Ph Sensitive Properties for Efficient Plasmid DNA Delivery. Molecular pharmaceutics 2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rahbek UL; Nielsen AF; Dong M; You Y; Chauchereau A; Oupicky D; Besenbacher F; Kjems J; Howard KA, Bioresponsive Hyperbranched Polymers for Sirna and Mirna Delivery. Journal of drug targeting 2010, 18 (10), 812–820. [DOI] [PubMed] [Google Scholar]
- 22.Jia H-Z; Zhang W; Zhu J-Y; Yang B; Chen S; Chen G; Zhao Y-F; Feng J; Zhang X-Z, Hyperbranched–Hyperbranched Polymeric Nanoassembly to Mediate Controllable Co-Delivery of Sirna and Drug for Synergistic Tumor Therapy. Journal of Controlled Release 2015, 216, 9–17. [DOI] [PubMed] [Google Scholar]
- 23.Wilson DR; Rui Y; Siddiq K; Routkevitch D; Green JJ, Differentially Branched Ester Amine Quadpolymers with Amphiphilic and Ph-Sensitive Properties for Efficient Plasmid DNA Delivery. Molecular Pharmaceutics 2019, 16 (2), 655–668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chen G; Wang Y; Xie R; Gong S, Tumor-Targeted Ph/Redox Dual-Sensitive Unimolecular Nanoparticles for Efficient Sirna Delivery. Journal of Controlled Release 2017, 259, 105–114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tai Z; Wang X; Tian J; Gao Y; Zhang L; Yao C; Wu X; Zhang W; Zhu Q; Gao S, Biodegradable Stearylated Peptide with Internal Disulfide Bonds for Efficient Delivery of Sirna in Vitro and in Vivo. Biomacromolecules 2015, 16 (4), 1119–1130. [DOI] [PubMed] [Google Scholar]
- 26.Kozielski KL; Tzeng SY; Green JJ, A Bioreducible Linear Poly(Beta-Amino Ester) for Sirna Delivery. Chemical Communications 2013, 49 (46), 5319 – 5321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Eltoukhy AA; Chen D; Alabi CA; Langer R; Anderson DG, Degradable Terpolymers with Alkyl Side Chains Demonstrate Enhanced Gene Delivery Potency and Nanoparticle Stability. Advanced Materials 2013, 25 (10), 1487–1493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Nelson CE; Kintzing JR; Hanna A; Shannon JM; Gupta MK; Duvall CL, Balancing Cationic and Hydrophobic Content of Pegylated Sirna Polyplexes Enhances Endosome Escape, Stability, Blood Circulation Time, and Bioactivity in Vivo. ACS Nano 2013, 7 (10), 8870–8880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Matsuda T; Cepko CL, Controlled Expression of Transgenes Introduced by in Vivo Electroporation. Proceedings of the National Academy of Sciences 2007, 104 (3), 1027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yao C; Tai Z; Wang X; Liu J; Zhu Q; Wu X; Zhang L; Zhang W; Tian J; Gao Y, Reduction-Responsive Cross-Linked Stearyl Peptide for Effective Delivery of Plasmid DNA. International journal of nanomedicine 2015, 10, 3403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wilson DR; Mosenia A; Suprenant MP; Upadhya R; Routkevitch D; Meyer RA; Quinones-Hinojosa A; Green JJ, Continuous Microfluidic Assembly of Biodegradable Poly(Beta-Amino Ester)/DNA Nanoparticles for Enhanced Gene Delivery. J Biomed Mater Res A 2017, 105 (6), 1813–1825. [DOI] [PubMed] [Google Scholar]
- 32.Wilson DR; Routkevitch D; Rui Y; Mosenia A; Wahlin KJ; Quinones-Hinojosa A; Zack DJ; Green JJ, A Triple-Fluorophore-Labeled Nucleic Acid Ph Nanosensor to Investigate Non-Viral Gene Delivery. Molecular Therapy 2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Huang M; Khor E; Lim L-Y, Uptake and Cytotoxicity of Chitosan Molecules and Nanoparticles: Effects of Molecular Weight and Degree of Deacetylation. Pharmaceutical Research 2004, 21 (2), 344–353. [DOI] [PubMed] [Google Scholar]
- 34.Eltoukhy AA; Siegwart DJ; Alabi CA; Rajan JS; Langer R; Anderson DG, Effect of Molecular Weight of Amine End-Modified Poly(B-Amino Ester)S on Gene Delivery Efficiency and Toxicity. Biomaterials 2012, 33 (13), 3594–3603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Griffith OW, Biologic and Pharmacologic Regulation of Mammalian Glutathione Synthesis. Free Radical Biology and Medicine 1999, 27 (9–10), 922–935. [DOI] [PubMed] [Google Scholar]
- 36.Yu B; Zhang Y; Zheng W; Fan C; Chen T, Positive Surface Charge Enhances Selective Cellular Uptake and Anticancer Efficacy of Selenium Nanoparticles. Inorg. Chem 2012, 51 (16), 8956–8963. [DOI] [PubMed] [Google Scholar]
- 37.Harush-Frenkel O; Debotton N; Benita S; Altschuler Y, Targeting of Nanoparticles to the Clathrin-Mediated Endocytic Pathway. Biochemical and biophysical research communications 2007, 353 (1), 26–32. [DOI] [PubMed] [Google Scholar]
- 38.Liu X; Howard KA; Dong M; Andersen MØ; Rahbek UL; Johnsen MG; Hansen OC; Besenbacher F; Kjems J, The Influence of Polymeric Properties on Chitosan/Sirna Nanoparticle Formulation and Gene Silencing. Biomaterials 2007, 28 (6), 1280–1288. [DOI] [PubMed] [Google Scholar]
- 39.Schaffer DV; Fidelman NA; Dan N; Lauffenburger DA, Vector Unpacking as a Potential Barrier for Receptor-Mediated Polyplex Gene Delivery. Biotechnol. Bioeng 2000, 67 (5), 598–606. [DOI] [PubMed] [Google Scholar]
- 40.Bishop CJ; Ketola T.-m.; Tzeng SY; Sunshine JC; Urtti A; Lemmetyinen H; Vuorimaa-Laukkanen E; Yliperttula M; Green JJ, The Effect and Role of Carbon Atoms in Poly(B-Amino Ester)S for DNA Binding and Gene Delivery. Journal of the American Chemical Society 2013, 135 (18), 6951–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sunshine JC; Peng DY; Green JJ, Uptake and Transfection with Polymeric Nanoparticles Are Dependent on Polymer End-Group Structure, but Largely Independent of Nanoparticle Physical and Chemical Properties. Molecular pharmaceutics 2012, 9 (11), 3375–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zhang Y-L; Zhang X; Fei X-C; Wang S-L; Gao H-W, Binding of Bisphenol a and Acrylamide to Bsa and DNA: Insights into the Comparative Interactions of Harmful Chemicals with Functional Biomacromolecules. J. Hazard. Mater 2010, 182 (1–3), 877–885. [DOI] [PubMed] [Google Scholar]
- 43.Cutlar L; Zhou D; Gao Y; Zhao T; Greiser U; Wang W; Wang W, Highly Branched Poly (B-Amino Esters): Synthesis and Application in Gene Delivery. Biomacromolecules 2015, 16 (9), 2609–2617. [DOI] [PubMed] [Google Scholar]
- 44.Gao Y; Huang J-Y; O’Keeffe Ahern J; Cutlar L; Zhou D; Lin F-H; Wang W, Highly Branched Poly (B-Amino Esters) for Non-Viral Gene Delivery: High Transfection Efficiency and Low Toxicity Achieved by Increasing Molecular Weight. Biomacromolecules 2016, 17 (11), 3640–3647. [DOI] [PubMed] [Google Scholar]
- 45.Tzeng SY; Green JJ, Subtle Changes to Polymer Structure and Degradation Mechanism Enable Highly Effective Nanoparticles for Sirna and DNA Delivery to Human Brain Cancer. Advanced Healthcare Materials 2013, 2 (3), 468–480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Bishop CJ; Tzeng SY; Green JJ, Degradable Polymer-Coated Gold Nanoparticles for Co-Delivery of DNA and Sirna. Acta Biomaterialia 2015, 11, 393–403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Chang Kang H; Bae YH, Co-Delivery of Small Interfering Rna and Plasmid DNA Using a Polymeric Vector Incorporating Endosomolytic Oligomeric Sulfonamide. Biomaterials 2011, 32 (21), 4914–4924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wilson DR; Mosenia A; Suprenant MP; Upadhya R; Routkevitch D; Meyer RA; Quinones-Hinojosa A; Green JJ, Continuous Microfluidic Assembly of Biodegradable Poly(Beta-Amino Ester)/DNA Nanoparticles for Enhanced Gene Delivery. Journal of Biomedical Materials Research Part A 2017, n/a–n/a. [DOI] [PubMed] [Google Scholar]
- 49.Kozielski KL; Tzeng SY; Mendoza B. A. H. d.; Green JJ, Bioreducible Cationic Polymer-Based Nanoparticles for Efficient and Environmentally Triggered Cytoplasmic Sirna Delivery to Primary Human Brain Cancer Cells. ACS Nano 2014, 8 (4), 3232–3241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Moore TL; Rodriguez-Lorenzo L; Hirsch V; Balog S; Urban D; Jud C; Rothen-Rutishauser B; Lattuada M; Petri-Fink A, Nanoparticle Colloidal Stability in Cell Culture Media and Impact on Cellular Interactions. Chemical Society Reviews 2015, 44 (17), 6287–6305. [DOI] [PubMed] [Google Scholar]
- 51.Pezzoli D; Zanda M; Chiesa R; Candiani G, The Yin of Exofacial Protein Sulfhydryls and the Yang of Intracellular Glutathione in in Vitro Transfection with Ss14 Bioreducible Lipoplexes. Journal of controlled release 2013, 165 (1), 44–53. [DOI] [PubMed] [Google Scholar]
- 52.Guo W; Lee RJ, Efficient Gene Delivery Via Non-Covalent Complexes of Folic Acid and Polyethylenimine. Journal of controlled release : official journal of the Controlled Release Society 2001, 77 (1–2), 131–138. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


