SUMMARY
The rational design of dengue virus (DENV) vaccines requires a detailed understanding of the molecular basis for antibody-mediated immunity. The durably protective antibody response to DENV following primary infection is serotype-specific. However, there is an incomplete understanding of the antigenic determinants for DENV type-specific (TS) antibodies, especially for DENV serotype 3, which has only one well-studied strongly neutralizing human monoclonal antibody (mAb). Here, we investigated the human B cell response of children following natural DENV infection in the endemic area of Nicaragua and isolated 15 DENV3 TS mAbs recognizing the envelope (E) glycoprotein. Functional epitope mapping of these mAbs and small animal prophylaxis studies revealed a complex landscape with protective epitopes clustering in at least six to seven antigenic sites. Potently neutralizing TS mAbs recognized sites principally in E glycoprotein domains I and II, with patterns suggesting frequent recognition of quaternary structures on the surface of viral particles.
Keywords: Flavivirus, Dengue virus serotype 3, Envelope protein, Neutralizing monoclonal antibodies, Antigenic site, Functional epitopes
In Brief
The repertoire of type-specific neutralizing sites in dengue virus serotype 3 (DENV3) is poorly defined. Young et al. identify human monoclonal antibodies to multiple previously unidentified antigenic sites on DENV3 envelope protein and show that they neutralize DENV3 potently in vitro and reduce viral infection in mice.
Graphical abstract
INTRODUCTION
Dengue viruses (DENV) are positive-sense RNA viruses belonging to the Flavivirus genus and are transmitted to humans by Aedes aegypti or Aedes albopictus mosquitoes (Diamond and Pierson, 2015). The four serotypes of DENV (DENV1–4) are estimated to cause ~390 million infections and 100 million cases each year (Bhatt et al., 2013), ranging from mild fever to severe Dengue Hemorrhagic Fever and Dengue Shock Syndrome (Halstead et al., 1973). Infection with one DENV serotype does not confer lasting protective immunity to the other three serotypes, which has complicated vaccine design. After a primary infection, type-specific (TS) antibodies to the infecting serotype provide durable, essentially life-long, protection (de Alwis et al., 2012; Murphy and Whitehead, 2011). While cross-reactive (CR) antibodies to the other three serotypes develop during a primary infection, these responses although initially protective, often wane over time in the absence of secondary exposures, and low to intermediate levels of CR antibodies may contribute to enhanced viral replication and an increased risk of severe disease (de Alwis et al., 2014; Halstead, 2015; Katzelnick et al., 2017a; Salje et al., 2018; Sangkawibha et al., 1984). Following a secondary infection, individuals typically develop a durable serotype cross-protective immunity (Dejnirattisai et al., 2015a; Patel et al., 2017). The only licensed dengue vaccine, Denvaxia, caused increased risk in dengue-naive children for severe dengue after natural infection, and breakthrough infections with DENVs were common, including DENV3 (Ferguson et al., 2016). Another tetravalent vaccine, TAK-003 failed to protect against DENV3, at 18 months in dengue-naive populations (Biswal et al., 2019). The basis for DENV3 vaccine failure is uncertain; however, the full repertoire of antibodies and the epitopes targeted following primary or secondary DENV infections remains partially characterized, preventing a full understanding of the mechanisms of protective immunity and immune enhancement (Gallichotte et al., 2018a; Katzelnick et al., 2017b).
The DENV envelope (E) glycoprotein mediates viral entry into cells and is the main target of neutralizing antibodies after infection and vaccination (Kuhn et al., 2015; Pierson and Diamond,2008). The four DENV serotypes vary by 25–40% in the amino acid sequence of the E protein (Fleith et al., 2016). Within each serotype, the E protein of different genotypes varies by 6–9% (Chen and Vasilakis, 2011; Flipse and Smit, 2015), and genotypic variation plays an underappreciated role in antibody immune escape (Brien et al., 2010; Sukupolvi-Petty et al., 2013; Wahala et al., 2010). The DENV E protein consists of three domains (designated E protein domain I [EDI], EDII, and EDIII), and two protomers form head-to-tail dimers on the surface of viral particles. Three dimers lie parallel to each other and form thirty rafts in a herringbone pattern on the mature virion. Several human TS neutralizing monoclonal antibodies (mAbs) against DENV1, DENV2 and DENV4 have been mapped, many of which recognize quaternary epitopes that span different E protein molecules and are therefore only present on the assembled virion (de Alwis et al., 2012; Fibriansah et al., 2015a; Teoh et al., 2012). The human antibody response to DENV3 is less studied at the clonal level than the other DENV serotypes. A single potent TS neutralizing human mAb (hmAb, 5J7) was described in detail, which recognizes a complex quaternary epitope spanning three E protomers in viral particles. Using viral reverse genetics, we demonstrated that residues in the DENV3-specific hmAb 5J7 epitope can be transplanted onto the E protein from DENV1 or DENV4 to generate chimeric infectious virions displaying the 5J7 epitope (Andrade et al., 2017; Messer et al., 2016; Widman et al., 2017). Interrogation of these chimeric viruses with panels of hmAbs and primary sera revealed that a highly variable fraction of the polyclonal serum DENV3-reactive neutralizing antibody response targets the hmAb 5J7 epitope, suggesting that major neutralizing epitopes of DENV3 remained undiscovered.
Here, we report the isolation and characterization of fifteen TS DENV3 neutralizing hmAbs from the B cells of three individuals in a long-standing, prospective pediatric cohort in Nicaragua (Katzelnick et al., 2017a; Kuan et al., 2009). Using wild-type and recombinant DENV3 genotype E glycoprotein variants, coupled with detailed molecular mapping studies using epitope transplant gain-or loss-of-function recombinant viruses and other mapping techniques, we identified six to seven TS antigenic sites in the DENV3 E protein recognized by neutralizing mAbs. Finally, selected hmAbs were protective against DENV3 infection when given as prophylaxis in mice. These results indicate that multiple antigenic sites on the DENV3 E protein are recognized by neutralizing and protective human antibodies.
RESULTS
Isolation of TS hmAbs from children previously infected with DENV3.
To define the antigenic landscape of DENV3, we immortalized memory B cells from children with previous laboratory-confirmed DENV3 infection in a Nicaraguan cohort study. Peripheral blood mononuclear cells (PBMCs) were collected from one individual who had a primary DENV3 infection (followed by an inapparent DENV1 infection) and from two DENV2-immune individuals who experienced a secondary DENV3 infection (Figure 1A, Table S1). We transformed B cells and isolated DENV3 hmAbs from these PBMC samples as described (Nivarthi et al., 2017; Smith et al., 2012) (Figure S1, Tables S1 and S2). The percentages of DENV E protein-reactive B cells in circulation were estimated as 1.8, 1.1 or 1.6% of transformable B cells for subjects 3243, 985, or 1791, respectively.
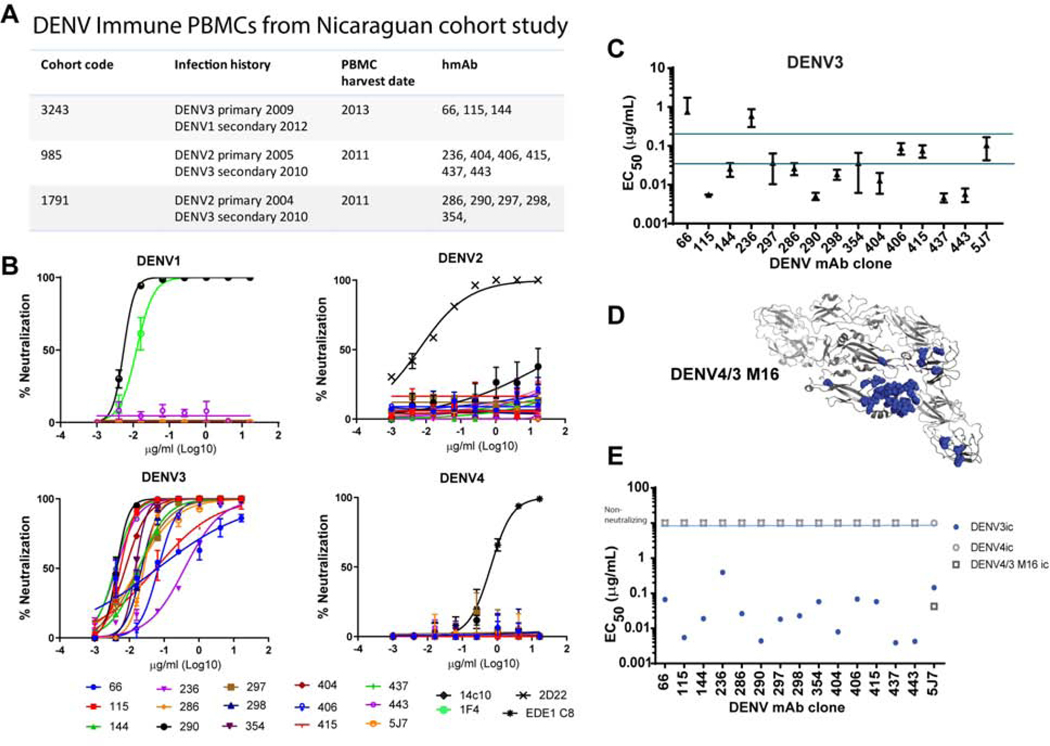
Figure 1. Fifteen hmAbs isolated from 3 children post DENV3 infection are DENV3-specific.
Memory B cells were immortalized from children experiencing primary or secondary DENV3 infections in Nicaragua. TS hmAbs were isolated that bound and neutralized DENV3.
A) DENV Immune PBMCs from children in a Nicaraguan Cohort study experienced DENV3 as either a primary infection or secondary infection after a DENV2 primary infection.
B) Fifteen hmAbs were tested against DENV1–4 by Vero-81 focus reduction neutralization test (FRNT). Positive controls were as follows, DENV1-specific hmAbs 1F4 and 14c10 for DENV1, DENV2-specific hmAb 2D22 for DENV2, DENV3-specific hmAb 5J7 for DENV3 and crossreactive hmAb EDEI C8 for DENV4. All assays were performed twice in duplicate.
C) DENV 3 neutralization by hmAbs was evaluated by FRNT using wildtype and recombinant viruses. Comparison of DENV3-specific hmAbs to 5J7. Using wild-type DENV3, EC50 values from repeat experiments performed on different days were averaged to show reproducibility. EC50 value denotes amount of hmAb needed to neutralize 50% of the virus in a Vero-81 FRNT. Error bars indicate standard deviation.
D) DENV4/3 M16 ic-PyMOL representation of DENV3 residues (blue) that capture the hmAb 5J7 epitope transplanted into a DENV4 backbone.
E) DENV3-specific hmAbs do not use 5J7 epitope. DENV4/3 M16 ic is neutralized by hmAb 5J7 but not by the panel of 15 DENV3-specific hmAbs. EC50 values in a FRNT of hmAbs of DENV4/3 M16 ic, and parental DENV3 ic and DENV4ic.
The hmAbs that bound only to DENV3 (and not to DENV1, 2 or 4) were evaluated for neutralizing activity against serotypes DENV1–4 (Figure 1B). Fifteen DENV3-specific neutralizing hmAbs were isolated from the 3 subjects. DENV3 neutralizing antibody potencies varied, but included hmAbs that were: (a) equally potent as the previously described hmAb 5J7 (e.g., hmAbs DENV-297, −354, −406, and −415) (EC50 of 0.2–0.35 μg/mL), (b) more potent than hmAb 5J7 (e.g., hmAbs DENV-115, −144, −286, −290, −298, −404, −419, −437, and −443) (EC50 < 0.035μg/mL), or (c) less potent than hmAb 5J7 (e.g., hmAbs DENV-66 and −236) (EC50 of > 0.2 μg/mL) (Figure 1C). A low level of cross-neutralization against some strains of DENV1 was detected with hmAb DENV-144 (Table S2), derived from subject 3243, who had been infected sequentially with DENV3 followed by DENV1 (Figure 1A, Table S1). To determine whether any of these hmAbs targeted the hmAb 5J7 quaternary epitope, each was tested for its ability to neutralize the DENV4/3 M16 recombinant virus, which presents the DENV3 hmAb 5J7 epitope in the structural context of a DENV4 E glycoprotein backbone (Widman et al., 2017). Although all newly generated hmAbs neutralized the parental DENV3, none neutralized the DENV4/3 M16 recombinant virus, indicating that the identified DENV3 hmAbs do not recognize the hmAb 5J7 epitope (Figure 1D, 1E).
Epitope mapping using DENV3 loss-of-function recombinant viruses.
We previously used recombinant chimeric DENVs to map epitopes in DENV1, 2, 3 or 4 viruses recognized by murine or human mAbs (Gallichotte et al., 2018a; Gallichotte et al., 2018b; Gallichotte et al., 2018c; Gallichotte et al., 2017; Swanstrom et al., 2019). To identify the epitopes recognized by the 15 DENV3 TS hmAbs identified in this study, we first generated a panel of DENV3 loss-of-function (LOF) mutant viruses. Starting with a previously described DENV3/1 EDI-A chimeric virus that incorporates 22 residues of the DENV1 EDI hmAb 1F4 footprint in the DENV3 E protein (Swanstrom et al., 2018), we introduced progressively larger portions of DENV1 TS hmAbs 1F4 and 14c10 antigenic region residues, designated DENV3/1 EDI-B, DENV3/1 EDI/III-C and DENV3/1 EDI/III-D (Figure 2A, Table 1a). These epitope domains, which mostly reside in EDI and/or a portion of EDIII of DENV1, were transplanted into the DENV3 backbone (Messer et al., 2016). All recombinant viruses replicated efficiently in Vero cells (Figure 2A–C, Figure S2).
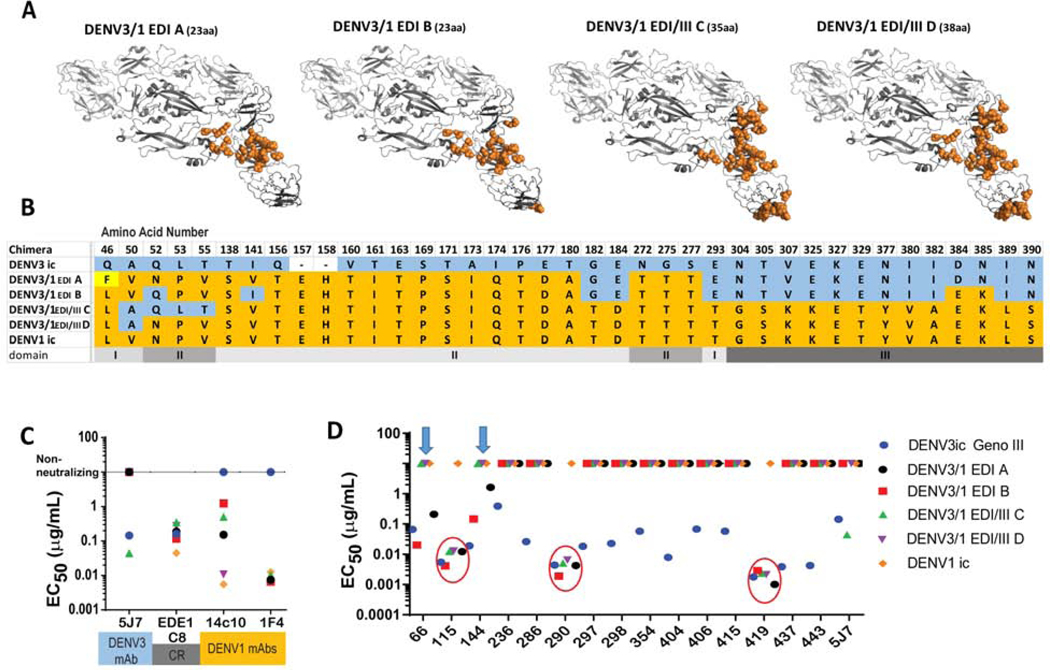
Figure 2. DENV3/1 recombinant EDI loss-of-function chimeras reveal previously unidentified DENV3 hmAb characteristics.
Chimeric DENV3 viruses encoding progressively larger blocks of DENV1 E glycoprotein sequence were used to interrogate the role of these transplanted regions in loss of DENV3 antibody function.
A) Four DENV3/1 chimeric viruses with increasing portions of DENV1 residues in a DENV3 backbone. DENV3/1 chimera PyMOL representations of DENV1 residues (orange) transplanted into DENV3 backbone (grey). Number of amino acids changed is in parenthesis.
B) Amino acid alignment of changed residues in DENV3/1 chimeras. DENV3 residues are shown in blue. DENV1 residues are shown in orange. A tissue culture adaptation in DENV3/1 A is shown in yellow. Blank spaces in DENV3 indicate residues not present in DENV1.
C) DENV3/1 chimeras contain DENV1-specific, DENV3-specific and cross-reactive epitopes. EC50 values of Vero-81 cell FRNT of DENV3-specific hmAb 5J7, cross-reactive hmAb EDEI C8 and DENV1-specific hmAbs 1F4 and 14c10.
D) Panel of 15 hmAbs were sorted into 3 groups by EC50 values of Vero-81 cell FRNT against chimeric DENV3/1 viruses. Group 1: Ten hmAbs do not neutralize any members of the chimeric dengue 3/1 panel. Group 2: hmAbs DENV-115, DENV-290 and −419 neutralize all chimeric DENV3/1 viruses (red circles). Group 3: hmAbs DENV-66 and −144 neutralize only DENV3/1 EDI-A and-B (blue arrows).
Table 1.
Identification of Six New Dengue Virus Serotype 3 Specific Human Neutralizing Antigenic Sites CELL-HOST-MICROBE-D-19–01064R2
| A Name | Recombinant Virus Backbone | DENV3 5J7 Neut | DENV1 1F4 Neut | AA changes | Epitope Transplant | Neutralized by hmAb |
|---|---|---|---|---|---|---|
| DENV4/3 M16 | DENV4 Baric genotype I | +++ | - | 36 aa | 5J7 DENV3 | 5J7 |
| DENV3/1 EDI A | DENV3 Baric genotype III | - | +++ | 23 aa | 1F4 DENV1 |
66, 115, 144, 290, 419 |
| DENV3/1 EDI B | DENV3 Baric genotype III | - | +++ | 23 aa | 1F4 + 14c10 DENV1 | 66, 115, 144, 290, 419 |
| DENV3/1 EDI/III C | DENV3 Baric genotype III | +++ | +++ | 35 aa | 1F4 + 14c10 DENV1 | 115, 290, 419, 5J7 |
| DENV3/1 EDI/III D | DENV3 Baric genotype III | +++ | 37 aa | 1F4 + 14c10 DENV1 | 115, 290, 419 | |
| DENV1/3 EDI A | DENV1 Baric | - | - | 18 aa | EDI surface aa DENV3 | 236, 286, 297, 298, 354, 404, 406, 437, 443, 144 |
| DENV1/3 EDI B | DENV1 Baric | - | - | 22 aa | EDI all aa DENV3 | 236, 286, 297, 298, 354, 404, 406, 437, 443, 144 |
| B Name | Recombinant Virus Backbone | Domain Swap | AA changes | DENV-236 | DENV-297 | DENV-415 | DENV-115 | DENV-419 | DENV-66 |
|---|---|---|---|---|---|---|---|---|---|
| DENV3 G-III | DENV3 baric genotype III Sri Lanka | None | 0 | +++ | +++ | +++ | +++ | +++ | +++ |
| DENV3 G-IV | DENV3 baric genotype IV Puerto Rico | None | 0 | - | - | + | - | - | - |
| DENV3 G-IV with G-III EDI | DENV3 baric genotype IV Puerto Rico | EDI | 9 aa | +++ | +++ | + | - | - | - |
| DENV3 G-IV with G-III EDII | DENV3 baric genotype IV Puerto Rico | EDII | 9 aa | - | - | + | +++ | +++ | - |
| DENV3 G-IV with G-III EDIII | DENV3 baric genotype IV Puerto Rico | EDIII | 6 aa | - | - | +++ | + | - | +++ |
| DENV3 G-with G-EDI | DENV3 baric genotype III Sri Lanka | EDI | 8 aa | - | - | ++ | +++ | +++ | +++ |
| DENV3 G-with G-EDII | DENV3 baric genotype III Sri Lanka | EDII | 10 aa | +++ | +++ | ++ | +++ | ||
| DENV3 G-with G-EDIII | DENV3 baric genotype III Sri Lanka | EDIII | 6 aa | +++ | +++ | +++ | +++ | +++ | - |
To demonstrate that appropriate epitope exchange, we investigated the ability of the DENV3 TS 5J7 hmAb and the DENV1 TS hmAbs 1F4 or 14c10 to neutralize the panel of wild-type or DENV3/1 chimeric viruses (Figure 2C). Whereas the DENV1 TS hmAb 1F4 neutralized all 4 of the DENV3/1 chimeras, only the DENV3/1 EDI/III-D (38 amino acids) chimera fully restored the DENV1 14c10 antibody neutralization phenotype, reflecting transplantation of the entire 14c10 epitope, which extends across EDI, EDIII and the EDI/II hinge region (Teoh et al., 2012). The other three chimeras partially restored the 14c10 neutralization phenotype. Residues Q52N, L53P and T55V in the EDI/II hinge region were critical for 14c10 neutralization in DENV1 chimeras (Figure 2C). Conversely, neutralization by hmAb 5J7 was retained in DENV3/1 EDI/III-C, but not in the DENV3/1 EDI/III-D recombinant virus, demonstrating the critical importance of these same three residues for hmAb 5J7 neutralization in DENV3 (Figure 2C). These data also support atomic structures showing that the hmAb 5J7 and 14c10 epitopes extend in opposite directions from an area of overlap within the EDI/II hinge in their respective serotypes. Even though the mAb 1F4 epitope overlapped the EDI/II hinge area (Fibriansah et al., 2014), variation in this area did not hinder its ability to neutralize viruses in the chimeric panel. As a control, the recombinant form of the CR hmAb EDE1 epitope-specific hmAb C8 also neutralized the panel of chimeric viruses.
We next tested the ability of each newly generated DENV3 hmAb to neutralize viruses in the chimeric DENV3/1 virus panel. The DENV3-specific hmAbs grouped into three classes (Figure 2D). Ten of the fifteen hmAbs (hmAbs DENV-236, −286, −297, −298, −354, −404, −406, −415, -437, or −443) did not neutralize the DENV3/1 chimeras, suggesting that loss of 23 core residues in the DENV3 EDI (that differ from DENV1 EDI) impacts binding and/or neutralization of this group (Figure 3D). These 10 hmAbs, designated as Group 1 hmAbs, likely target the core residues in the DENV3 EDI domain. The second cluster (designated Group 2) includes three hmAbs that efficiently neutralized wild-type DENV3 virus and the entire panel of DENV3/1 loss-of-function chimeric viruses (hmAbs DENV-115, −290 and −419). As the constructs with the largest transplanted regions include DENV1 residues from EDI and EDIII but leave EDII as found in DENV3, we predict that hmAbs DENV-115, −290 and −419 target residues in EDII and perhaps a small portion of EDIII of DENV3. The third group (hmAbs DENV-66 and DENV-144) neutralized only the chimeras with the smallest transplanted regions, DENV3/1 EDI-A and DENV3/1 EDI-B, suggesting that DENV3 residues outside of the EDI domain, perhaps in EDIII and/or a smaller footprint in EDI, likely contribute to binding and neutralization of the Group 3 hmAbs. Groups 1 and 3 contain a mixture of weakly to highly potent neutralizing hmAbs, whereas all three Group 2 hmAbs exhibited high neutralizing potency.
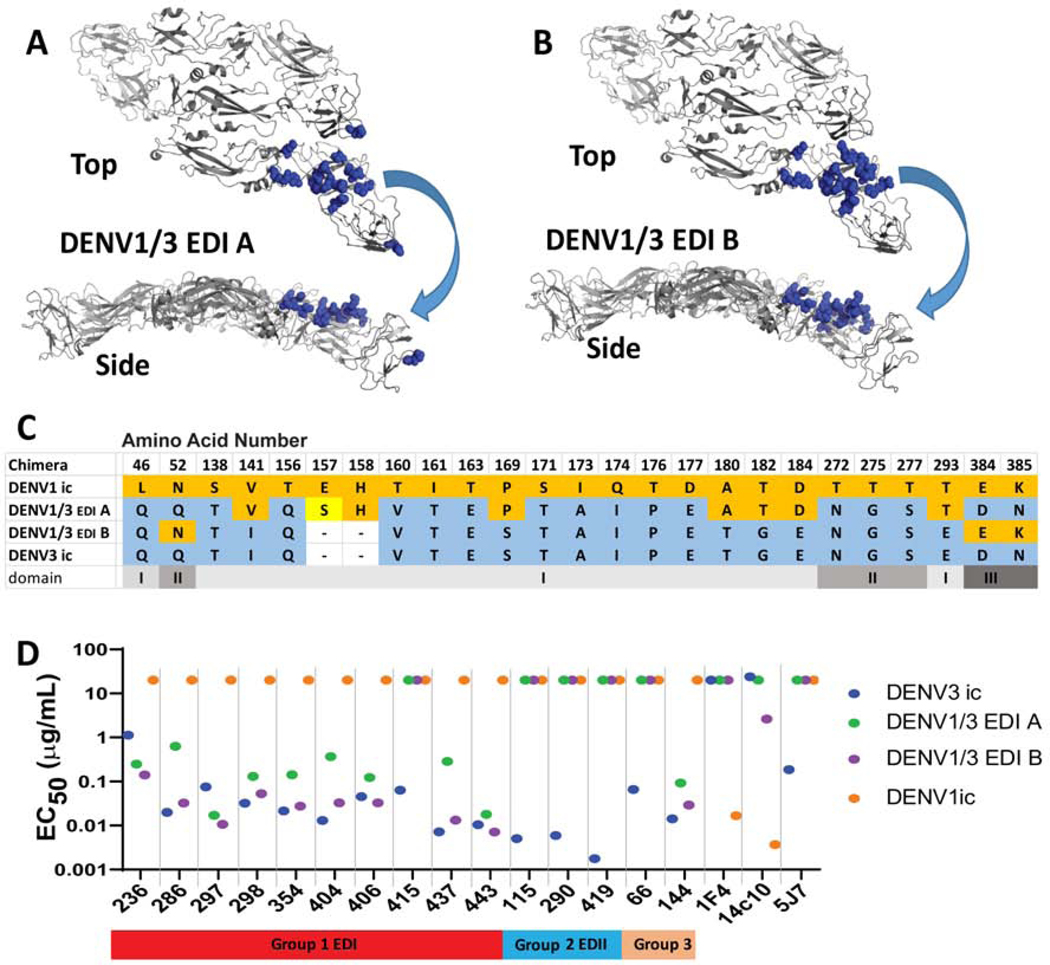
Figure 3. DENV1/3 EDI shows gain of function for 9 of 10 DENV3 hmAbs in group 1, mapping to EDI.
To interrogate gain of neutralization function, we constructed DENV1/3 EDI chimeric viruses with increased numbers of EDI residues from DENV3 introduced into the DENV1 backbone. PyMOL representations of changed residues in the DENV1/3 EDI-A chimera. Transplanted DENV1 residues are shown in orange spheres on a DENV3 backbone. Top and side views are shown.
A) DENV1/3 EDI-A chimera surface residues were changed.
B) DENV1/3 EDI-B chimera surface and interior residues were changed.
C) Amino acid alignment of changed residues in DENV1/3 EDI chimeras. DENV3 residues are shown in blue. DENV1 residues are shown in orange. A tissue culture adaptation in DENV3/1-A is shown in yellow. Blank spaces in DENV3 indicate residues not present in DENV1. All differences in EDI are represented except residues 29 and 47, which remained as DENV1 residues.
D) EC50 values of Vero-81 cell FRNT of hmAbs against chimeric DENV1/3 viruses. 9 of 10 group 1 hmAbs neutralized both chimeric DENV1/3 viruses. DENV1/3 EDI-B neutralization pattern is most similar to that of DENV3. As expected 1F4, 14c10 and 5J7 do not neutralize either DENV1/3 chimera.
Chimeric DENV1/3 gain-of-function (GOF) recombinant virus mapping.
Chimeric loss-of-function E glycoproteins may disrupt long-range protein-protein interactions and complicate interpretation of DENV3/1 antibody-epitope map locations. Therefore, we designed and recovered a panel of DENV1/3 gain-of-function (GOF) EDI mutant viruses (Figure 3A) to validate the predicted EDI map locations of the 10 Group 1 DENV3 hmAbs. Using a DENV1 molecular clone (Gallichotte et al., 2017), we introduced progressively larger portions of the EDI domain from DENV3 into DENV1 (Figure 3). In the DENV1/3-EDI-A recombinant virus, we replaced DENV1 surface contact residues for hmAbs 1F4 and 14c10 in EDI with the corresponding DENV3 residues. The virus expressed by this construct should not be neutralized by DENV1 TS hmAbs 1F4 or 14c10 nor the DENV3 TS mAb 5J7 (Figure 3A–C). In DENV1/3 EDI-B, we transplanted the entire EDI of DENV3 into DENV1, including both the previously described surface residues and variant interior residues (e.g., V141I, P169S, A180T, T182G, D184E and T293E). In addition, we deleted DENV1 residues E157 and H158, because these two amino acids do not exist in the DENV3 EDI domain. This construct probes the role of both the surface and interior residues in hmAb binding and neutralization. The two GOF chimeric viruses also replicated efficiently in Vero cells (Figure 3A–D, Figures S2, S3).
Consistent with the defined structural interaction domains of each antibody/epitope pair, both DENV1/3 EDI-A and DENV1/3 EDI-B chimeric viruses were not neutralized by the DENV1 TS hmAbs 1F4 and 14c10 nor the DENV3 TS hmAb 5J7 (Figure 3D). Upon testing the new panel of DENV1/3 EDI GOF chimeras against the ten Group 1 DENV3 hmAbs, we defined nine mAbs that efficiently neutralized both DENV1/3 chimeric viruses, but not DENV1. The DENV1/3 EDI-B chimera was neutralized more efficiently than the DENV 1/3 EDI-A chimera by seven of the nine hmAbs with similar neutralization potency compared to the DENV3 parental virus (e.g., Group 1a EDI hmAbs DENV-286, −298, 354, −404, −406, −437 and −443) (Figure 3D). Unexpectedly, two hmAbs (Group 1b EDI hmAbs DENV-236 and −297) neutralized both DENV1/3 EDI chimeras more efficiently than the parental DENV3 virus. These data suggest that the DENV3 Group 1 hmAbs engage the DENV3 EDI region using at least two different binding patterns that appear dictated, in part, by interior residues (e.g., V141I, P169S, A180T, T182G, D184E and T293E) in EDI of the chimeric viruses. A third binding pattern (Group 1c) is represented by the Group 1 hmAb DENV-415, which did not neutralize either of the DENV1/3 EDI GOF chimeras. The DENV1/3 EDI-A or DENV1/3 EDI-B chimeras did not alter the Group 2 antibody neutralization titers. In contrast, the Group 3 hmAbs had distinct patterns, with hmAb144 (Group 3b), but not hmAb66 (Group 3a), neutralizing DENV1/3 EDI-A and DENV1/3 EDI-B.
To determine if the DENV3-neutralizing hmAbs bound to epitopes contained within a single protomer or spanning the two protomers of the E homodimer, the hmAbs were tested for binding to DENV3 recombinant E (recE) monomer or stabilized recE dimers (Figure S4). Epitopes contained within a single E monomer or domain should be efficiently presented on recE monomers, whereas hmAbs with epitopes that spread across the dimer interface should bind preferentially to recE dimers (Dejnirattisai et al., 2015b; Metz et al., 2017). Indeed, hmAb DENV-415 did not bind to recE monomers but showed weak binding to recE dimers, suggesting it recognizes a quaternary epitope, perhaps requiring residues in neighboring E proteins in addition to EDI. Only Group 1b hmAbs DENV-236 and −297 showed no preference for recE dimers over monomers, suggesting that they recognize epitopes in the EDI of a single E protomer, whereas the remaining 12 hmAbs recognize quaternary epitopes that span the E homodimer similar to DENV-415.
Together, these data suggest the presence of functional neutralization epitopes on the surface of DENV3 E protein previously unidentified. Group 1 hmAbs DENV-236, −286, −297, −298, −354, −404, −406, −437 and −443, and to a lesser extent DENV-415, are predicted to recognize epitopes in EDI in at least 3 unique, yet perhaps overlapping, patterns from the previously described DENV3 neutralizing hmAb, 5J7. Group 2 (e.g., hmAbs DENV-115, −290 and −419) likely map in EDII. The group 3a DENV3 TS hmAb DENV-66 did not neutralize any of the DENV1/3 EDI GOF chimeric viruses, suggesting that its epitope overlaps with and/or resides outside of DENV3 EDI, perhaps in EDIII. Surprisingly, the Group 3b hmAb DENV-144 could neutralize both DENV1/3 EDI GOF chimeras (Figure 3D) and the DENV3/1 EDI LOF chimeras (Figure 2D). These data suggest the presence of another, likely complex, binding interface that depends on residues in DENV3 EDI and EDIII. However, it seems likely that the hmAb DENV-144 epitope site in EDI is resistant to the incorporation of extensive DENV1 variation across the region. This hypothesis is consistent with low-level cross neutralization by DENV-144 of the Nauru Westpac 1974 strain that encodes variation in EDI.
HmAb neutralization phenotypes for viruses of DENV3 genotypes I to IV.
To validate the location of the DENV3 epitopes, we determined if natural variation encoded within the DENV3 genotypic panel altered the neutralization profiles of hmAbs in the panel (Figure 4A and 4B). Viruses in the DENV3 recombinant genotype panel encode the E glycoproteins from genotypes I, II, III or IV (G-I to-IV) introduced into the DENV3 Sri Lanka G-III backbone (Messer et al., 2012). Although each genotype strain encodes distinct amino acid differences across EDI, EDII and EDIII, all DENV3 genotypes were highly sensitive to neutralization by hmAb 5J7. All Group 1a EDI hmAbs neutralized viruses from all 4 genotypes. In contrast, Group 1b hmAbs DENV-236 and DENV-297 did not neutralize the G-IV virus, which encoded seven unique amino acid substitutions in EDI (Figure 4C). These data further suggest Group 1b hmAbs DENV-236 and DENV-297 may use a different set of interaction residues in EDI as compared with Group 1a EDI hmAbs DENV-286, −298, −354, −404, −406, -437 and −443. Of note, Group 1c hmAb DENV-415 neutralized all of the viruses in the DENV3 genotype panel, although the EC50 values for neutralization varied by ~10-fold. G-III viruses were neutralized efficiently, whereas G-I, -II and-IV viruses were more resistant. Among the Group 2 hmAbs that may target EDII, hmAb DENV-290 efficiently neutralized viruses from all genotypes, whereas hmAbs DENV-115 and DENV-419 did not neutralize virus from G-IV (Figure 4C). G-IV contains nine unique amino acids substitutions in EDII (Figure 4A, 4B). Since hmAbs DENV-115, −290 and −419 neutralized all of the EDI/EDIII-exchanged DENV3/1 and DENV1/3 chimeras, these data support their recognition of at least two unique and/or partially overlapping epitopes in EDII. Group 3 hmAbs also demonstrated disparate neutralization phenotypes; Group 3b hmAb DENV-144 neutralized viruses from all four genotypes, whereas Group 3a hmAb DENV-66 neutralized only G-III strains. As EDIII contains amino acid substitutions in G-I, -II and-IV as compared to G-III, these data support mapping studies that DENV-66 recognizes EDIII (Figure 4C). For the hmAb DENV-144 epitope, we searched for areas that were not excluded by the DENV3/1 chimera panel and were conserved among the 4 genotypes. Although DENV-144 was the most complex functional epitope to define, our data suggest that it recognizes EDI perhaps at the ED I/III interface.
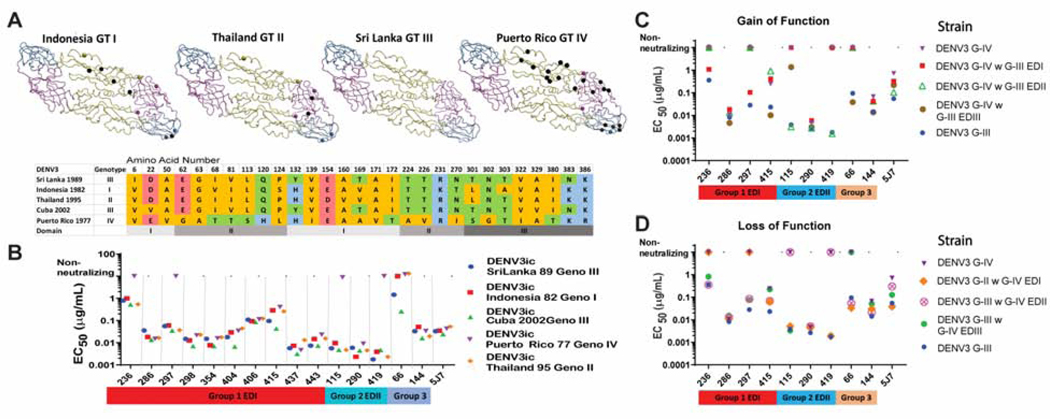
Figure 4. DENV3 genotype panel.
To identify natural variation that disrupts hmAb function, a panel of recombinant viruses were used that encoded E glycoproteins derived from the different DENV3 genotypes. Subsequently, DENV3 genotype chimeras were used to map hmAbs to EDI, EDII or EDIII in the E glycoprotein by gain-of-function or loss-of-function.
A) DENV3 E glycoprotein dimers for designated GI-IV are shown. Amino acid residues that differ from those in the Sri Lanka genotype III are shown as spheres. A black sphere indicates the residue is unique to that genotype and not shared between the genotypes. Colored spheres indicate residues seen in two or more genotypes.
B) Amino acid alignment. Amino acids with nonpolar side chains are colored orange, uncharged polar are green, acidic are red and basic are blue. Domains are indicated in gray bar at bottom.
C) Genotypic variation alters FRNT neutralization EC50 values for select DENV3 hmAbs. Genotype IV DENV3 escaped neutralization by some DENV3 hmAbs.
D) Gain-of-function genotype IV DENV3 chimeric viruses with EDI, EDII, or EDIII from genotype III DENV3 were used to map select hmAbs to specific domains of E glycoprotein (see Figure S5). EC50 values of Vero-81 cell FRNT for select hmAbs show gain-of-function for DENV-236, -297 and −415 when EDI is transplanted. DENV-115 and −419 show gain-of-function when EDII is transplanted. DENV-66 shows gain-of-function when EDIII is transplanted.
E) Loss-of-function genotype III DENV3 chimeric viruses with EDI, EDII, or EDIII from genotype IIV DENV3. EC50 values of Vero-81 cell FRNT for select hmAbs. DENV-236, −297 and −415 show loss-of-function when EDI is transplanted. DENV-115 and −419 show loss-of-function when EDII is transplanted. DENV-66 shows loss-of-function when EDIII is transplanted.
Fine mapping of epitopes using genotype differences.
As natural variation in DENV3 G-IV contains clustered variation in EDI, II and III that altered the neutralization profiles of Group 1–3 hmAbs, exchange of the ED regions between susceptible (e.g., G-III) and resistant (e.g., G-IV) genotypes of DENV3 should localize the epitope domain of selected Group 1, 2 and 3 hmAbs. We used reverse genetics to introduce either the EDI, II, or III regions from the resistant DENV3 G-IV E glycoprotein into the sensitive DENV3 G-III strain (Figure 4D–E, Figure S5) or vice versa, allowing us to map critical functional residues using both GOF and LOF studies. Focusing first on the Group 1b antibodies that efficiently neutralized DENV3 G-III but not G-IV viruses, we observed that hmAbs DENV-236 and −297 gained neutralizing activity against DENV3 G-IV mutant viruses encoding the EDI, but not the EDII or EDIII domains from G-III (Figure 4D). Additionally, hmAbs DENV-236 and −297 lost neutralizing activity against the sensitive DENV3 G-III mutant virus when EDI was replaced with G-IV EDI, but not EDII or EDIII (Figure 4E). Therefore, the epitopes for Group 1b hmAbs DENV-236 and-297 should reside in EDI. Group 1c hmAb DENV-415, which did not gain neutralizing activity against the DENV3/1 chimeras, had neutralization profiles that shifted nearly 10-fold when the G-III EDIII was present in this set of DENV3 genotype mutant viruses. HmAb DENV-415 gained neutralization potency (24-fold) of the DENV3 G-IV mutant when EDIII was converted to G-III, whereas converting EDI or EDII variation did not affect neutralization. Conversely, when EDIII of DENV3 G-III was replaced by EDIII of G-IV, we observed a 10-fold loss of neutralization potency. Based on these data, we hypothesize that the hmAb DENV-415 epitope region encompasses residues spanning across EDI-III, more so than the epitope targeted by other Group 1 hmAbs.
The Group 2 hmAbs DENV-115 and −419 demonstrated a gain-of-neutralization potential in the resistant DENV3 G-IV mutants when residues in EDII, but not EDI or EDIII, were converted to G-III residues (Figure 4D). Reciprocally, in the fully susceptible DENV3 G-III genetic backbone, neutralizing activity of these mAbs was lost when residues in EDII, but not EDI or EDIII, were changed to those in the DENV3 G-IV (Figure 4E). Of note, when the EDIII of DENV3 G-III was inserted into the resistant DENV3 G-IV backbone, hmAb DENV-115 gained the ability to neutralize this virus weakly, albeit >1,000-fold less potently than fully susceptible strains. These data further suggest that the epitope for hmAb DENV-115, but not DENV-419, is distinct and may extend from EDII into EDIII. Nonetheless, these data identify critical portions of the epitopes for DENV-115 and −419 in EDII. In agreement, passage of DENV3 G-III six times in the presence of high concentrations of DENV-115 resulted in a single, amino acid change, K93E, located in EDII. Finally, the group 3 mAb DENV-66 neutralized the resistant DENV3 G-IV backbone only when the variation in EDIII, but not EDI or EDII, was changed to residues in G-III. Similarly, hmAb DENV-66 failed to neutralize the susceptible DENV3 G-III virus when its EDIII was converted to that of G-IV EDIII. These results confirm our assignment of the epitope for hmAb DENV-66 in EDIII (Figure 4D–E).
In vivo protection studies.
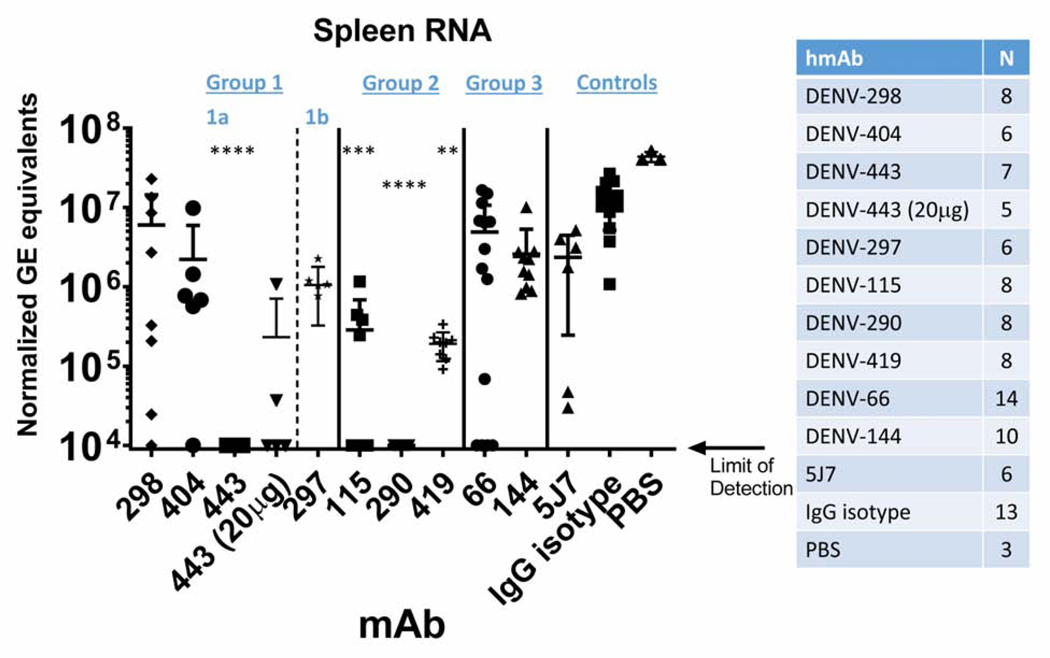
The in vitro neutralization potency of anti-flavivirus antibodies does not always correlate with in vivo activity, especially when antibodies target different epitopes or have distinct mechanisms of action (Mukherjee et al., 2014; Pierson and Diamond, 2015). The interferon α/β and γ receptor-deficient AG129 mouse strain is an established model for DENV replication and pathogenesis (Balsitis et al., 2010; Johnson and Roehrig, 1999; Messer et al., 2016; Raekiansyah et al., 2005; Sarathy et al., 2018; Shresta et al., 2004; Shresta et al., 2006). Using a DENV3 replication model, representative hmAbs from different individuals that targeted each of the four distinct interaction sites across EDI, II and III were evaluated for their ability to reduce viral load in the spleens of mice when administered as prophylaxis prior to inoculation with a wild-type DENV3 virus (DENV3 UNC3009, G-III) (Figure 5). The representative Group 1a hmAbs DENV-298, -404, and −443 along with Group 1c DENV-144, which recognize EDI and were isolated from three different individuals, showed mixed levels of capacity to reduce virus replication in vivo. The hmAbs DENV-298, −404 and −144 reduced virus titers by ~10-fold as compared with hmAb DENV-443, which reduced infection by >3 log10 genome equivalents (GE)/mg of GAPDH, below the limit of detection. Even 20 μg of hmAb DENV-443 reduced viral titers by 100-fold. These data were somewhat unexpected, as the in vitro neutralization potency of each Group 1a hmAb in culture was similar (e.g., IC50 values: DENV-443 = 4–7 ng/mL, DENV-404 = 8–18 ng/mL, DENV-298 = 15–23 ng/mL). The Group 2 EDII hmAbs DENV-115, −290 and −419, isolated from three different individuals who either experienced DENV3 as a primary or secondary infection, reduced virus titers by over ~2 to >3 log10 GE/μg of GAPDH. These DENV3 TS hmAbs also had high neutralizing potency in vitro (exhibiting IC50 values of 4–5 ng/mL), suggesting that EDII is an important site for antibody neutralization and antiviral activity in mice. In contrast, the Group 3 EDIII hmAb DENV-66 reduced virus titers by <10-fold in the spleen. For comparison, the previously described DENV3 neutralizing hmAb 5J7 reduced virus titers by an average of 10-fold as compared to the isotype control. Together, these data indicate that the hmAbs identified following DENV3 infections in children from Nicaragua are among the most potently neutralizing DENV3 hmAbs isolated to date, both in vitro and in vivo, and target diverse epitopes in EDI and EDII, and to a lesser extent in EDIII on DENV3.
Figure 5. Reduction of DENV3 viral burden in vivo by selected hmAbs.
AG129 mice were administered 50 mg hmAb (unless otherwise indicated) by intraperitoneal injection 24 hours prior to infection with 5×106 plaque forming units (PFU) of DENV3 UNC3009. Virus titers were assessed 72 hours post-infection using quantitative RT-PCR of RNA isolated from the spleens of infected mice and are expressed as genome equivalents (GE) normalized to mg of GAPDH. Group 1 DENV-443 and Group 2 DENV-115, −290, and −419 hmAbs reduced DENV3 viral load compared to IgG isotype antibody. The number of mice in each treatment group are indicated, comprising 5 independent experiments in total, with at least 2 independent experiments performed for each hmAb. The limit of detection is 104 GE/mg of GAPDH. Comparisons were performed using Kruskal-Wallis test with Dunn’s multiple comparisons (** p<0.005, *** p<0.0005, **** p<0.0001).
DISCUSSION
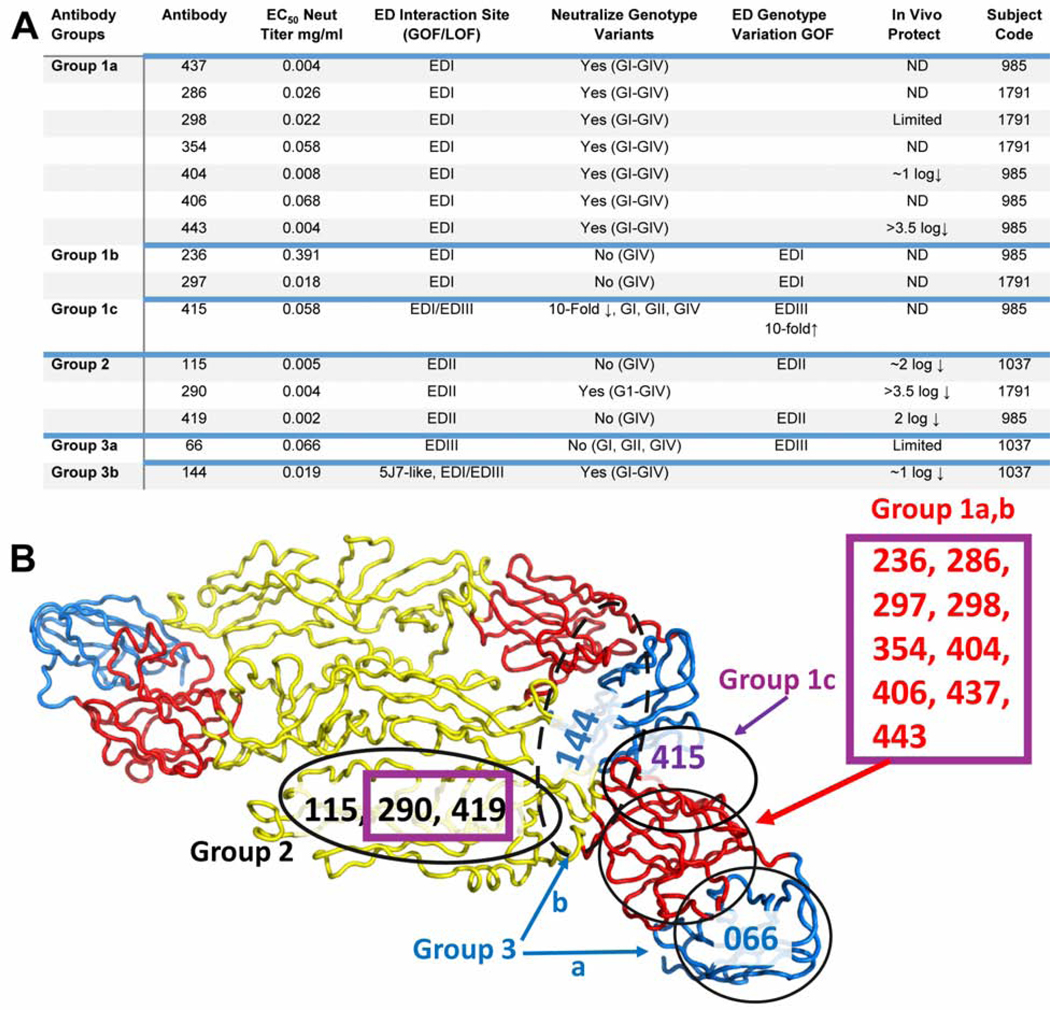
Dengue natural and vaccine-induced immunity relies on the development and maintenance of long-term protective antibody titers, and B and T cell memory responses. A tetravalent live attenuated dengue vaccine (Dengvaxia) sensitized rather than protected in DENV-naive individuals, whereas DENV-immune individuals were highly protected (Hadinegoro et al., 2015; Halstead, 2017, 2018a, b; Sridhar et al., 2018). The best studied correlate of protective immunity after DENV infection is the development of high serologic titers and high titers of serum neutralizing antibodies (Katzelnick et al., 2016; Sangkawibha et al., 1984). Moreover, recent studies with people exposed to natural DENV infections or live-attenuated vaccines indicate that the specificity (TS/epitope specificity) rather than total quantity of neutralizing antibodies correlates best with long-term protection (Gallichotte et al., 2018b; Henein et al., 2017; Moodie et al., 2018; Sridhar et al., 2018). Indeed, DENV4 genotype variation in the E glycoprotein strongly correlated with reduced neutralization titers after vaccination (Gallichotte et al., 2018b). These and other studies demonstrate a pressing need to identify improved correlates of antibody-mediated protective immunity. Here, we isolated 15 DENV3 TS neutralizing hmAbs from three Nicaraguan children who experienced DENV3 primary or secondary infection. Using both GOF-and LOF-epitope chimeric viruses, we identified three classes of neutralizing antibodies that likely reflect six to seven neutralizing antigenic sites in and across EDI, II and III of DENV3 (Figure 6A–B). All but two hmAbs in Group 1b bound to stabilized dimers of DENV3 E protein better than to monomeric E protein, supporting the hypothesis that most neutralizing anti-DENV hmAbs recognize quaternary epitopes (Magnani et al., 2017; Metz et al., 2017; Rouvinski et al., 2017). Importantly, these data suggest that the neutralizing antigenic repertoire of DENV3, and potentially other DENV serotypes, is more complex than previously recognized.
Figure 6. Six distinct neutralizing epitopes defined by panel of 15 DENV3 hmAbs.
We identified six distinct functional type specific neutralizing epitopes in DENV3. A) Summary of antibody properties, phenotypes and interaction sites using GOF DENV1/3 or LOF DENV3/1 chimeras, DENV3 genotype variants and DENV3 genotype chimeras.
B) Shown is a ribbon diagram of a DENV3 E trimer with EDI in red, EDII in yellow and EDIII in blue. Predicted functional epitope sites are shown as black circles. hmAbs in Group 1a interface with EDI differently from Group 1b, while Group 1c interacts in a more complex manner. The epitope for DENV-144 has the highest degree of uncertainty. HmAbs from secondary DENV3 infection are in purple squares.
Most of the durable human serum neutralizing response after primary infection is associated with TS antibodies that recognize a few well-defined epitopes centered within and spanning domains on the DENV E glycoprotein (de Alwis et al., 2012; Fibriansah et al., 2015b; Gallichotte et al., 2018a; Teoh et al., 2012). While our study has identified 15 hmAbs that map within and/or span DENV3 EDI, II and III, other less well characterized DENV3 hmAbs (DV74.4 and DV7.9.3) are predicted to bind in EDI/II (Beltramello et al., 2010). Another TS hmAb (P3D05) isolated after tetravalent live-attenuated vaccination remains unmapped (Magnani et al., 2017). The panels of hmAbs and chimeric viruses reported here will provide a useful resource to determine if uncharacterized mAbs recognize known or unique epitopes in DENV3. In contrast to murine DENV3 TS Abs, which principally target EDIII (Brien et al., 2010; Wahala et al., 2010), our studies support earlier work showing that epitopes entirely contained within EDIII of DENV are infrequently targeted by strongly neutralizing human antibodies.
Several groups have focused on defining the functional and structural properties of DENV CR antibodies elicited in adults after secondary infection (Dejnirattisai et al., 2015b; Li et al., 2018). However, the hmAbs elicited in pediatric subjects from DENV-endemic regions represent an understudied population who may differ in their capacity to recognize the number, location and complexity of DENV3 neutralizing epitopes in the E glycoprotein (Simon et al., 2015). Thus, analysis of the antibody repertoire in at-risk pediatric populations is important for evaluating vaccine performance. In subject 3243, who experienced a primary DENV3 infection followed by an inapparent secondary DENV1 infection, DENV3-specific neutralizing hmAbs (DENV-66, −115 and −144) targeted three distinct areas of the E glycoprotein. Whereas the epitope targeted by hmAb DENV-144 is complex and predicted to extend across a portion of EDI into EDIII (Figure 6A–B), mapping techniques suggest that hmAb DENV-115 recognizes a separate epitope in EDII that spans multiple protomers. Using similar approaches, we found that hmAb DENV-66 likely targets EDIII, analogous to DENV3 lateral-ridge epitope seen with murine mAbs (Brien et al., 2010). Thus, we have identified three spatially distinct DENV3-specific neutralizing epitopes in EDII and III induced by primary DENV3 infection in PBMCs collected after a secondary DENV1 infection. Unlike murine mAbs, the EDII-specific hmAb DENV-115, but not the EDII I-specific hmAb DENV-66, substantially reduced DENV3 viral load in the spleen in vivo.
The majority of our hmAbs were isolated from two children who experienced DENV3 secondary infections several years after a primary DENV2 infection. These two DENV3 secondary infections also induced TS antibodies that targeted EDII and EDI, which demonstrates the importance of these antigenic sites in TS DENV3 immunity. Together, these data support the idea that patient-specific polyclonal responses may target distinct neutralizing epitopes after infection. In these patients, ten of the twelve neutralizing hmAbs target EDI of the DENV3 E glycoprotein, and two of the hmAbs target EDII. This EDI epitope skewing was unexpected, given the expansive repertoire of epitopes targeted by antibodies from the first individual, 3243. DENV polyclonal neutralizing responses appear to target either an EDII/EDIII or an EDI epitope after a DENV2 primary infection (Gallichotte et al., 2018c) or a EDI/II hinge epitope following primary DENV1 and DENV3 infections (Andrade et al., 2017; Andrade et al., 2019). Using recombinant viruses and sera from many Nicaraguan pediatric subjects, substantial individual variation was noted in the proportion of DENV3 TS neutralizing antibody titers attributed to the 5J7 epitope (range, 0 to 100%), which further supports the notion of individual variation in epitope targeting (Andrade et al., 2017). This finding likely reflects inter-host immune variation associated with infection, differences in antibody germline gene frequency and use patterns, disparate flavivirus infection histories, and depth of epitope mapping analyses. In the children who experienced DENV2 primary infections in this study, it is possible that the antibody variable genes in pre-existing EDI DENV2-reactive B cells mutated during the DENV3 infection to convert clones from DENV2-reactive to DENV3-specific, leading to epitope skewing of the response. Immune responses between closely related flaviviruses are complex and may be impacted by pre-exposure histories that shape the antibody response to new strains or related viruses, as noted when Zika virus spread into DENV-endemic areas (Bhaumik et al., 2018; Grifoni et al., 2017; Stettler et al., 2016). Unfortunately, insufficient quantities of PBMCs prevented us from evaluating the antibody lineages identified in these children. In human recipients of the NIH tetravalent vaccine, both TS and CR neutralizing antibodies have been reported for DENV3, although it is unclear whether these antibodies map to the same or different epitopes as those reported after DENV3 infection (Magnani et al., 2017; Smith et al., 2013; Swanstrom et al., 2019).
Chimeric recombinant viruses provide a strategy for mapping epitope-specific responses of monoclonal or polyclonal antibodies (Gallichotte et al., 2018c; Gallichotte et al., 2017; Gallichotte et al., 2015; Widman et al., 2017). Functional characterization of the virus:antibody interface is also useful because the Group 1a hmAbs showed a 10-fold increase in their neutralization potency when the entire EDI from DENV3 was present, indicating that the surface and underlying residues in chimeric E glycoproteins are important. Group 1b antibodies are highly susceptible to natural variation in DENV3 genotypes, whereas Group 1a antibodies are not. When atomic-resolution structures of these epitopes become available, the functional epitopes will help delineate the critical interactions and mechanisms required for virus neutralization and DENV3 protective immunity.
All 15 hmAbs studied here efficiently neutralized DENV3 strains encoding a G-III E glycoprotein, which corresponds with the circulation of DENV3 G-III viruses throughout the Caribbean and Central America in 2004–11 (Gutierrez et al., 2011; OhAinle et al., 2011). However, natural variation altered the neutralization potency of a subset of antibodies targeting EDI (Group 1b hmAbs), EDII (hmAbs 115 and 419) and EDIII (hmAb 66) (Figure 6A). DENV3 genotypes G-I, -II, -III and-V circulate currently, whereas G-IV is derived from an ancestral lineage from the Caribbean (Puerto Rico:1963/77) that now is rare in human populations (King et al., 2008; Waman et al., 2017). Our DENV3 genotype panel includes E glycoproteins from G-I through-IV (Messer et al., 2012) and lacks some recently reported variation in this serotype. Historically, natural variation is thought to play a limited role in DENV immunologic escape from pre-existing immunity (Holmes and Twiddy, 2003). However, studies have demonstrated as much as 10-to 15-fold differences in DENV3 neutralization phenotypes across genotypes, using monoclonal and polyclonal antibodies collected following primary DENV3 infections (Messer et al., 2012; Sukupolvi-Petty et al., 2013; Wahala et al., 2010). Although speculative, these data suggest that DENV3 genotypic variation might contribute to breakthrough infections in rare individuals, especially those who developed limited polyclonal serum antibody responses that target one or a limited subset of neutralizing epitopes.
The in vivo potencies of the EDI antibodies were variable, and the panel of mAbs included both potent and weak inhibitors of virus replication in mice, as seen with the Group 1a hmAbs DENV-443 and −298, respectively. This phenotype may reflect subtle differences in epitope targeting and neutralization potency within EDI, an impact on antibody performance as a function of maturation status, or alternatively may reflect inherent differences in Fc effector functions encoded by these antibodies (Lee et al., 2013; Lofano et al., 2018). All three EDII antibodies tested potently reduced virus load in vivo, with DENV-290 being the most effective, suggesting the importance of EDII in protective immunity. Future studies are planned to evaluate the potency of a subset of these neutralizing human antibodies in a lethal DENV3 challenge mouse model.
The hmAbs, recombinant proteins and chimeric viruses will serve as key reagents for evaluating vaccine immunogenicity and for measuring epitope-and ED-specific responses associated with natural infections and/or vaccinations. As DENV-naive children receiving the Dengvaxia tetravalent vaccine appear at increased risk for severe DENV after infection (Sridhar et al., 2018), and TAK-003 showed reduced efficacy in seronegative populations against DENV3 (Biswal et al., 2019), there is a critical need for better correlates of protective immunity and improved vaccines in children. Our study demonstrates the importance of evaluating the TS neutralizing antibody responses in children experiencing primary or secondary infections with DENV. The DENV3 antibody neutralizing landscape is complex, with antibodies falling into multiple groups as described here and previously (Fibriansah et al., 2015b; Widman et al., 2017). As the most potent DENV3-specific hmAbs target EDI and EDII in vivo, it is possible that variation in the potency and epitope specificities of individual host responses after DENV3 infections or vaccination may result in complex patterns of neutralizing antibodies in polyclonal sera. Variation in the serological repertoire may correlate with protective immunity or susceptibility to repeat infection by the same or different DENV3 genotypes (Waggoner et al., 2016). The complexity of the DENV3 neutralizing antigenic landscape suggests that the diversity of neutralizing epitopes in other DENV serotypes also remains largely undiscovered. Given the hundreds of millions of DENV infections worldwide and the issues surrounding tetravalent vaccine outcomes, analysis of the antibody repertoire and epitope specificities elicited after vaccination may well determine efficacy and the likelihood of breakthrough infections leading to more severe disease in children and adults.
STAR METHODS
LEAD CONTACTS AND MATERIAL AVAILABILITY
Further information and requests for antibody reagents may be directed to and be fulfilled by the corresponding author: James E. Crowe, Jr. (james.crowe@vumc.org). Further information and requests for chimeric virus reagents may be directed to and be fulfilled by the corresponding author: Ralph S. Baric (rbaric@email.unc.edu). Requests for recombinant monomers and dimers should be directed to Aravinda deSilva (aravinda_desilva@med.unc.edu). Materials described in this paper are available for distribution under the Uniform Biological Material Transfer Agreement, a master agreement that was developed by the NIH to simplify transfers of biological research materials.
EXPERIMENTAL MODEL AND SUBJECT DETAILS
Human Subjects
The Pediatric Dengue Cohort Study is an ongoing prospective dengue cohort study that follows approximately 3,700 children ages 2–17 in District II of Managua, Nicaragua. The protocol for the Pediatric Dengue Cohort Study in Nicaragua was reviewed and approved by the Institutional Review Boards of the University of California, Berkeley, (#2010–09-2245) and the Nicaraguan Ministry of Health (NIC-MINSA/CNDR-CIRE-09/03/07–008.ver1). Parents or legal guardians of the subjects enrolled in the study provided written informed consent, and participants 6 years of age and older provided assent.
DENV infection and infection history.
A suspected dengue case was considered a symptomatic DENV infection when 1) DENV RNA was detected by reverse-transcriptase polymerase chain reaction (RT-PCR) (Balmaseda et al., 1999; Lanciotti et al., 1992), 2) DENV was isolated (Balmaseda et al., 1999), 3) seroconversion was observed in paired acute and convalescent phase sera by IgM capture ELISA (Balmaseda et al., 2003; Balmaseda et al., 1999), or 4) seroconversion and/or a ≥4-fold increase in total DENV-specific antibody titer in paired acute and convalescent sera was observed by Inhibition ELISA (Balmaseda et al., 2006; Fernandez and Vazquez, 1990). Inapparent DENV infections were identified through serological testing of paired annual blood draws from healthy subjects (Balmaseda et al., 2010; Kuan et al.,2008). Participants whose paired annual samples demonstrated seroconversion or a 4-fold or greater increase in total DENV-specific antibody titer by Inhibition ELISA (iELISA), but who had not experienced a documented febrile episode associated with acute DENV infection, were considered to have experienced an inapparent DENV infection (Balmaseda et al., 2010; Kuan et al., 2009)
One hundred and sixteen participants who entered the cohort DENV-naïve and had experienced at least two DENV infections as determined by the iELISA were selected, and neutralizing antibody titers (NT50) for all four DENV serotypes at each annual sample were determined using a flow cytometry-based assay with reporter viral particles (RVPs) representing the four serotypes and Raji cells expressing the DENV attachment factor DC-SIGN (Mattia et al., 2011; Montoya et al., 2013). PBMC samples collected after the second DENV infection for three such participants--985, 1791 and 3242--were used for isolation of DENV3-specific hmAbs. All individuals experienced one symptomatic RT-PCR-confirmed DENV3 infection. Individuals 985 and 1791 experienced primary inapparent DENV2 infection followed by secondary DENV3 infection, while individual 3242 experienced primary DENV3 infection followed by secondary inapparent DENV1 infection (Figure 1, Table S1).
PBMC preparation.
For PBMC preparation, blood samples were collected in Vacutainer tubes (Becton-Dickenson) with 5 mM EDTA as anticoagulant. Upon receipt at the Nicaraguan National Virology Laboratory, ~5 ml of blood was transferred into a Leucosep tube (Greiner Bio-One) containing 3 ml of Ficoll Histopaque (Sigma) and centrifuged at 500g for 20 minutes at room temperature. The PBMC fraction was collected and transferred to a tube containing 9 ml of PBS with 2% fetal bovine serum (FBS; Denville Scientific) and 1% penicillin/streptomycin (Sigma). Cells were washed and pelleted three times by centrifugation at 500g for 10 minutes and resuspended in RPMI 1640 complete medium (RPMI 1640, 10% FBS, 1% GlutaMAX™, 1% HEPES and 1% penicillin/streptomycin). Before the third wash, cells were counted using a hemocytometer (Sismex XS-1000i). After the third wash, cells were resuspended in cryovials at a concentration of 3 × 106 cells/ml in freezing medium (90% FBS, 10% dimethyl sulfoxide) and were placed in isopropanol containers (Mr. Frosty, Nalgene) at −80°C overnight and transferred to liquid nitrogen for storage (Michlmayr et al., 2017; Zompi et al., 2012).
Cell lines and viruses.
Vero-81 cells (ATCC# CCL-81) were maintained in Dulbecco’s modified Eagle’s/Ham’s F-12 50/50 Mix (DMEM/F-12 50/50) supplemented with non-essential amino acids (NEAA), glutamine and sodium bicarbonate (Vero cell medium) at 37°C. C6/36 cells (ATCC CRL-1660) were maintained in Gibco minimal essential medium (MEM) supplemented with 1% NEAA at 32°C. Both media were supplemented with 5% fetal bovine serum (FBS) and penicillin/streptomycin antibiotics. U937 cells expressing DC-SIGN (dendritic cell-specific intercellular adhesion molecule-3-grabbing nonintegrin), a known DENV attachment factor, were maintained as suspension cell cultures at 37°C with 5% CO2 in RPMI 1640 (Gibco) supplemented with 1% non-essential amino acids, 1% penicillin and streptomycin, and 5% fetal bovine serum (FBS; HyClone). The rDENV1 clone is based on DENV strain West Pac 74, the rDENV2 clone is based on DENV strain S16803, the rDENV3 clone is based on a Sri Lankan 1989 DENV strain and the DENV4 molecular clone was based on the sequence of Sri Lankan DENV strain 1992a, and have been previously described(Gallichotte et al., 2017; Gallichotte et al., 2015; Messer et al., 2012). The chimeric infectious clone rDENV4/3 M16 and the genotype panel of DENV3 also have been described previously(Widman et al., 2017).
AG129 Mice.
This study was performed in strict accordance with the recommendations in the Guide for the Care and Use of Laboratory Animals of the National Institutes of Health. All procedures were approved by the U.C. Berkeley Animal Care and Use Committee guidelines. AG129 mice were housed and bred in our biosafety level 1 animal facility at UC Berkeley under specific pathogen-free conditions.
METHOD DETAILS
Generation of DENV3-specific hMAbs.
Previously cryopreserved peripheral blood mononuclear cells (PBMCs) were thawed rapidly in a 37°C water bath and washed prior to transformation with Epstein-Barr virus (EBV) and incubated with CpG and additional supplements (Yu et al., 2008). Cultures were incubated at 37°C in 5% CO2 for 10 days prior to screening for DENV3-reactive cell lines with ELISA. The transformed B cell culture supernatants were screened by live virus capture ELISA for binding to a representative strain from each of the four DENV serotypes. The minimal frequency of DENV3-reactive B cells was estimated on the basis of the number of wells with DENV3-reactive supernatants as compared to the total number of lymphoblastoid cell line colonies in the transformation plates, as follows: [number of wells with DENV3-reactive supernatants]/[number of LCL colonies in the plate]. On the basis of the number of DENV positive wells and the number of transformed B cells tested (determined by average colony counts in transformed wells), the percentages of DENV E protein-reactive B cells in circulation were estimated to be 1.8, 1.1 and 1.6% of transformable B cells for subjects 3243, 985, and 1791, respectively, which were similar to B cell frequencies reported in earlier studies for DENV-immune adult subjects (Smith et al., 2014). Cells from wells with supernatants reacting in the DENV3 capture ELISA were subjected to cytofusion with HMMA2.5 nonsecreting myeloma cells, as previously described (Smith et al., 2013; Smith et al., 2012). Following cytofusion, hybridomas were selected for growth in HAT medium containing ouabain. Wells containing hybridomas producing DENV3-reactive antibodies were cloned biologically by limiting dilution plating followed by flow cytometric sorting for single cells using a FACSAria III cell sorter (BDBiosciences). Once clonal, the cell lines were used to produce mAb immunoglobulin G (IgG) in cell supernatants, using serum-free medium, followed by protein G column purification. One hybridoma line (DENV-419) secreted poorly, so we generated a recombinant form of the antibody and expressed it in mammalian ExpiCHO cells prior to protein G column purification. Here, in all three subjects, we focused on those antibodies that were TS against DENV3. This included 3 antibodies originating from subject 3243 (primary DENV3 infection; sample collected after secondary DENV infection) and 5 or 7 antibodies from subjects 1791 or 985, respectively, who had experienced DENV3 as a secondary DENV infection after primary DENV2.
Virus, rE and rEDIII ELISA.
To evaluate if the oligomeric state of the E protein influences the binding efficiency of the mAbs, we subjected the mAbs to an antigen-capture ELISA using DENV3 recombinant E (rE) proteins. DENV rE proteins exist in a concentration-and temperature-dependent monomer-to-dimer equilibrium (Kudlacek et al., 2018). At physiological conditions, rE is mainly present as a monomer (rEM). Stable DENV3 homodimers (rED) were generated by introducing a disulfide interaction at the EDII-dimer interface (A257C). Ni2+-coated ELISA plates (Pierce Thermo) were coated with 5 ng/pL DENV3 rEM or rED for 1 hour at 37°C. Next, the plates were blocked with TBS + 0.05% Tween-20 + 3% skim milk for 1 hour at 37°C. Plates subsequently were washed three times with TBS + 0.2% Tween-20 and incubated with serially diluted mAb (2–0.015 ng/pL) for 1 hour at 37°C. Next, plates were washed and incubated with 1:2,500 diluted alkaline-phosphatase (AP) conjugated anti-human IgG (Sigma) for 45 minutes at 37°C. After washing, wells were developed with AP substrate (Sigma) and absorbance was measured at 405 nm wavelength(Gallichotte et al., 2015).
Generation of the rDENV3/1and rDENV1/3 recombinant virus panels.
We previously described the generation of a DENV3/1 chimera, designated rDENV3/1 ED-1A (Messer et al., 2016). We use a four-component cDNA cloning system in which the DENV genome is divided into four segments that can be replicated separately as plasmids in Escherichia coli cells. Purified plasmids are cut with designated restriction enzymes to yield unique type IIS restriction endonuclease cleavage sites that can be ligated simultaneously to yield full-length DENV genome cDNA. A built-in T7 site is used to generate RNA, which is electroporated into C6/36 or Vero-81 cells to recover virus. Virus harvested from medium is subsequently passaged and sequence verified. To generate several additional chimeric rDENV3/1 viruses, we systematically increased the numbers and/or locations of amino acid residues from EDI and EDIII that were transplanted into DENV3 from DENV1. A closely matched derivative called DENV3/1 EDI-B (23 residues) was isolated, which extended the original DENV3/1 EDI-A transplanted region to include two residues in EDIII of the neighboring protomer (e.g., D384E and N385K), removed one DENV1 residue from the EDI/II hinge region (N52Q), removed one DENV1 residue from the interior of EDI (V141I) and corrected a tissue-culture-induced mutation at residue F46L. The design of the DENV3/1 EDI/III-C chimeric virus further reduced the number of transplanted residues in the EDI/II hinge region by 3 residues (V50A, P53L and V55T), but converted most of the DENV3 ED I and ED III domains to DENV1, thereby increasing the total number of transplanted residues to 35 amino acids (Table 1b). The final derivative, designated the DENV3/1 EDI/III-D chimera, builds upon the DENV3/1 EDI/III-C backbone by converting an additional 3 residues in the domain I/II hinge area of DENV3 to DENV1 (Q52N, L53P and T55V) resulting in a total of 38 residues transplanted into DENV3. The viruses were designed to gain DENV1 1F4 and 14c10 hmAb neutralizing epitopes, while differentially preserving the DENV3-specific hmAb 5J7 neutralizing epitope, allowing us to measure loss of neutralization with the new panel of DENV3 hmAb. As a result of our quadripartite infectious clone design, all changes were isolated to the A and B fragments of the DENV3 genome backbone. cDNAs encoding E proteins incorporating three increasing sizes of the DENV1 EDI/EDIII transplant were synthesized (BioBasic, Buffalo, NY) and incorporated into three different DENV3 fully assembled DNA genomes and transcribed. Then, the genome-length RNAs were electroporated into Vero-81 cells to generate a panel of viable recombinant rDENV3/1 viruses. Recombinant viruses were subjected to full-length sequencing to demonstrate the presence of appropriate subsets of mutations, as previously described(Gallichotte et al., 2018c; Gallichotte et al., 2017).
We also synthetically reconstructed two gain-of-function DENV1/3 recombinant chimeras. We replaced all of the varying surface residues in the EDI of our DENV1 ic with corresponding residues from DENV3 (DENV1/3 EDI-A). In parallel, we constructed a second chimera in which the varying residues in the surface and interior of the EDI of DENV1 were replaced with DENV3 residues (rDENV1/3 EDI-B). Both viruses were viable and sequenced confirmed, allowing for systematic measures of gain-of-function neutralization assays with the new panel of DENV hmAb.
Generation of DENV3 genotype III/IV domain exchange virus panel.
Recombinant DENV3 G-IV viruses encoding the G-III EDI, EDII or EDIII natural variation were recovered using reverse genetics Recombinant DENV3 G-III viruses encoding the G-IV EDI, G-IV EDII, or G-IV EDIII were isolated using reverse genetics. Briefly, we substituted residues in EDI, EDII or EDIII from our DENV3 Puerto Rico G-IV molecular clone into our Sri Lanka 89 G-III molecular clone or vice versa using the quadripartite system described above and electroporated into C6/36 or Vero cells. All six viruses were viable and sequence-confirmed to encode the appropriate ED specific natural variation from each genotype.
Vero cell titration and focus assays.
For viral titrations, viral stocks were diluted 10-fold serially in Vero cell medium supplemented with 2% heat-inactivated fetal bovine serum (HI-FBS; Hyclone Defined) and 1x antibiotic. The inoculum was added to Vero-81 cells that were seeded into a 96-well plate (2 × 104 cells/well) the previous day and incubated at 37°C for 1 hour, then overlaid with overlay medium (Opti-MEM I Grand Island, NY, with 1% methyl cellulose and 2% heat-inactivated FBS). Viral foci were detected at 44 to 48 h after infection, following fixation/permeabilization with 10% buffered formalin/0.01% saponin using primary murine mAbs 2H2 and 4G2 and secondary horseradish peroxidase (HRP)-conjugated goat anti-mouse IgG (Sigma), followed by TrueBlue substrate (KPL). Number and size of foci were analyzed with a CTL Immunospot instrument.
Vero cell neutralization assays.
Neutralization on Vero-81 cells has been described previously (Gallichotte et al., 2015). Briefly, monolayers of Vero-81 cells in 96-well plates were inoculated with a virus and antibody or serum mix that had been incubated for 1 h at 37°C to allow for Ab:virion binding. Following a 1 hr incubation on cells at 37°C for infection, cells and inoculum were overlaid with overlay medium (see above). Viral foci were detected at 44 to 48 h after infection, following fixation/permeabilization with 10% buffered formalin/0.01% saponin using primary mAbs 2H2 and 4G2 (Swanstrom et al., 2016) and secondary horseradish peroxidase (HRP)-conjugated goat anti-mouse IgG (Sigma), followed by TrueBlue substrate (KPL). Numbers of foci were analyzed with an Immunospot Analyzer instrument (Cellular Technology Limited). All hmAb neutralization assays were performed as eight point dilution curves done in duplicate with at least 2 independent experiments. Variable slope sigmoidal dose-response curves are calculated with top or bottom restraints of 100 or 0, respectively. EC50 is the concentration of antibody that neutralizes 50% of the virus being tested.
U937-DC-SIGN neutralization assay.
The neutralizing potency of the hmAbs was measured using a flow cytometry-based neutralization assay with the U937 human monocytic cell line stably transfected with DC-SIGN(Kraus et al., 2007). At an initial concentration of 15,000 ng/mL, hmAbs were serially diluted 3-fold 12 times in RPMI supplemented with 2% FBS. A dilution of virus that infects between 8–15% of the U937 cells (previously determined by virus titration) was added to the hmAb dilutions and incubated for 1 hour at 37°C. U937 cells then were added to each well and incubated for 2 hours at 37°C, following which unbound virus was removed by centrifugation at 252 x g for 5 minutes and resuspended in 100 μL RPMI medium. Cells were then incubated at 37°C for 24 hours. Next, cells were fixed in 4% paraformaldehyde, incubated for 10 minutes at room temperature, and centrifuged at 252 x g for 5 min. Following this, cells were blocked in permeabilization buffer (0.1% saponin, 5% bovine serum albumin in 1X phosphate-buffered saline [PBS]) for 30 minutes at room temperature. Then, cells were incubated with anti-E mAb 4G2 conjugated to Alexa 488, diluted in blocking buffer (0.5% bovine serum albumin and 0.02% sodium azide in 1X PBS) for 25 minutes at room temperature. Finally, cells were washed and resuspended in PBS. Acquisition of the infected cells was performed with a Guava flow cytometer (EMD Millipore) by gating Alexa Fluor 488 dye-positive cells. The data were analyzed using a nonlinear, 4-parameter dose-response regression analysis with Prism software (GraphPad). The NT50 was determined as the concentration of the hmAb dilution that achieved a 50% reduction of the infection compared to infection control. Data generated had to meet the quality control criteria, whereby the sigmoidal dose-response regression fit had to include an absolute sum of squares of <0.2 and a coefficient of determination (R2) of >0.9.
Antibody pressure escape study.
DENV3 Sri Lanka 89 ic was incubated with a >50% neutralizing concentration of mAb DENV3–115 of 0.01 mg/mL prior to inoculation of Vero-81 cell culture monolayers. Virus was grown for 4 days under an antibody pressure of 0.01 mg/mL, followed by a second passage in Vero-81 cells under similar conditions. The DENV3–115-selected virus was passaged 4 more times under increasing pressure to a final concentration of 0.1 mg/mL DENV3–115. RNA from the resulting DENV3–115-selected virus was isolated and sequenced. The only recovered non-silent mutation was A1211G, which resulted in a K93E mutation in EDII of the E glycoprotein.
Animal studies.
This study was performed in strict accordance with the recommendations in the Guide for the Care and Use of Laboratory Animals of the National Institutes of Health. All procedures were approved by the U.C. Berkeley Animal Care and Use Committee guidelines. AG129 mice were housed and bred in our biosafety level 1 animal facility at UC Berkeley under specific pathogen-free conditions. For infection experiments, mice 6–8 weeks of age, of both genders, were randomly assigned to experimental groups. Mice were transferred from our biosafety level 1 breeding facility to our biosafety level 2 infection facility 2–3 days before infection. Mice 6 to 8 weeks of age were administered 50 pg (unless otherwise indicated) of one of the newly isolated DENV3-specific hmAbs, DENV3 hmAb 5J7, or an isotype control antibody (IgG1) intraperitoneally (i.p.) in a total volume of 200 pL 24 hours prior to DENV inoculation. A sublethal dose (5 × 106 PFU) of rDENV3 strain UNC 3009, G-III, was administered intravenously (i.v.) in a total volume of 100 pL. Seventy-two hours post-infection, mice were sacrificed, spleens were harvested and placed in Trizol, and total RNA was extracted. Viral RNA burden and GAPDH levels in spleen were assessed by quantitative RT-PCR. Virus load in genome equivalents (GE) was normalized by dividing by ug of glutaraldehyde 3-phosphate dehydrogenase (GAPDH).
QUANTIFICATION AND STATISTICAL ANALYSIS
Statistical analysis was performed using Prism 5.0 (GraphPad, La Jolla, CA). Variable slope sigmoidal dose-response curves are calculated with top or bottom restraints of 100 or 0, respectively. EC50 is the concentration of antibody that neutralizes 50% of the virus being tested. Non-parametric Kruskal-Wallis test with Dunn’s multiple comparisons was calculated with multiplicity adjusted P-values. P-values are indicate by * symbol in plots; ** = p<0.005, *** = p<0.0005, **** = p<0.0001. Statistical details of experiments can be found in the figure legends.
Supplementary Material
KEY RESOURCES TABLE
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| DENV-66 (hybridoma-produced Ig) | This manuscript | N/A |
| DENV-115 (hybridoma-produced Ig) | This manuscript | N/A |
| DENV-144 (hybridoma-produced Ig) | This manuscript | N/A |
| DENV-236 (hybridoma-produced Ig) | This manuscript | N/A |
| DENV-297 (hybridoma-produced Ig) | This manuscript | N/A |
| DENV-286 (hybridoma-produced Ig) | This manuscript | N/A |
| DENV-290 (hybridoma-produced Ig) | This manuscript | N/A |
| DENV-298 (hybridoma-produced Ig) | This manuscript | N/A |
| DENV-254 (hybridoma-produced Ig) | This manuscript | N/A |
| DENV-404 (hybridoma-produced Ig) | This manuscript | N/A |
| DENV-406 (hybridoma-produced Ig) | This manuscript | N/A |
| DENV-415 (hybridoma-produced Ig) | This manuscript | N/A |
| DENV-419 (hybridoma-produced Ig) | This manuscript | N/A |
| DENV-437 (hybridoma-produced Ig) | This manuscript | N/A |
| DENV-443 (hybridoma-produced Ig) | This manuscript | N/A |
| DENV-5J7 (plasmid-produced Ig) | Beltramello, Williams et al. 2010 | N/A |
| DENV-14c10 (plasmid-produced Ig) | Teoh, Kukkaro et al. 2012 | N/A |
| DENV-1F4 (hybridoma-produced Ig) | Fibiansah, Tan et al. 2014 | N/A |
| EDE1-C8 (plasmid-produced Ig) | Rouivinski, Guardado-Calvo et al. 2015 | N/A |
| EDE1-C10 (plasmid-produced Ig) | Rouivinski, Guardado-Calvo et al. 2015 | N/A |
| DENV-2D22 (hybridoma-produced Ig) | Smith et al., 2015 | N/A |
| Goat anti-Human IgG (Fc)-AP | Meridian Life Science, Inc. | Cat# W99008A |
| DENV-4G2(hybridoma-produced Ig) | ATCC® HB-112™ | D1–4G2–4-15 |
| DENV-2H2(hybridoma-produced Ig) | ATCC® HB-114™ | D3–2H2–9-21 |
| Bacterial and Virus Strains | ||
| DENV1 Thailand/16007/1964 | Buddhari et al., 2014, PMC4199527 | N/A |
| DENV1 Nauru/West Pac/1974 | Puri et al., 2000, DOI: 10.1023/A:1008160123754 | N/A |
| DENV1 N1265–04 | Andrade et al., 2017 | N/A |
| DENV1ic West Pac 74 | Gallichotte et al., 2017 | N/A |
| DENV2 Thailand/16681/1964 | Buddhari et al., 2014, PMC4199527 | N/A |
| DENV2 Thailand/S16803/1974 | Kelly et al., 2011, DOI 10.1007/s11262–011-0602-z | N/A |
| DENV2 N172–06 | Andrade et al., 2017 | N/A |
| DENV2ic 16803 | Gallichotte et al., 2015 | N/A |
| DENV3 Philippines/16562/964 | Buddhari et al., 2014, PMC4199527 | N/A |
| DENV3 Thailand/CH53489/1973 | Wahala et al., 2010 | N/A |
| DENV3 N2845–09 | Hadjilaou et al., 2015, doi:10.4049/jimmunol.1500918 | N/A |
| DENV3ic SriLanka 89 GIII | Messer et al., 2012 | N/A |
| DENV3 UNC3009, G-III | Messer et al., 2016 | N/A |
| DENV4 Indonesia/1036/1976 | Buddhari et al., 2014, PMC4199527 | N/A |
| DENV4 Columbia/TVP-376/1982 | Sukupolve-Petty et al., 2013 | N/A |
| DENV4 N703–99 | Hadjilaou et al., 2015, doi:10.4049/jimmunol.1500918 | N/A |
| DENV4ic SriLanka 92 | Widman, Young et al. 2017 | N/A |
| DENV3/4 M16 | Widman, Young et al. 2017 | N/A |
| DENV3/1 EDI A | Messer, Yount et al. 2016 | N/A |
| DENV3/1 EDI B | This manuscript | N/A |
| DENV3/1 EDI/III C | This manuscript | N/A |
| DENV3/1 EDI/III D | This manuscript | N/A |
| DENV1/3 EDI A | This manuscript | N/A |
| DENV1/3 EDI B | This manuscript | N/A |
| DENV3 Indonesia 1982, genotype I | Messer, Yount et al. 2012 | N/A |
| DENV3 Thailand 1995, genotype II | Messer, Yount et al. 2012 | N/A |
| DENV3 Cuba 2002, genotype III | Messer, Yount et al. 2012 | N/A |
| DENV3 Puerto Rico 1977, genotype IV | Messer, Yount et al. 2012 | N/A |
| DENV3 GIV with GIII EDI | This manuscript | N/A |
| DENV3 GIV with GIII EDII | This manuscript | N/A |
| DENV3 GIV with GIII EDIII | This manuscript | N/A |
| DENV3 GIII with GIV EDI | This manuscript | N/A |
| DENV3 GIII with GIV EDII | This manuscript | N/A |
| DENV3 GIII with GIV EDII | This manuscript | N/A |
| Biological Samples | ||
| PBMCs from DENV infection survivors | This manuscript | Nicaraguan Pediatric Dengue Cohort Study Donor ID #3243, #985, and #1791 |
| Chemicals, Peptides, and Recombinant Proteins | ||
| 1-Step Ultra TMB-ELISA | Thermo Fisher Scientific | Cat# 34029 |
| Dulbecco’s Phosphate-Buffered Saline, 1X with calcium and magnesium | Corning Life Sciences | Cat# 21–030-CM |
| 50x HAT media supplement | Sigma-Aldrich | Cat# H0137 |
| Ouabain | Sigma-Aldrich | Cat# O3125 |
| HyClone insect cell culture medium | GE Healthcare Life Sciences | Cat# SH30280.03 |
| Fetal Bovine Serum, ultra-low IgG | Thermo Fisher Scientific | Cat# 16250078 |
| 100x Penicillin Streptomycin Glutamine | Thermo Fisher Scientific | Cat# 10378016 |
| ClonaCell-HY Medium E | Stem Cell Technologies | Cat# 03805 |
| ClonaCell-HY Medium A | Stem Cell Technologies | Cat# 03801 |
| DMEM, high glucose, GlutaMAX™ Supplement | Thermo Fisher Scientific | Cat# 10566–024 |
| Dulbecco’s modified Eagle’s/Ham’s F-12 50/50 Mix | Gibco | 10–092-CV |
| Opti-MEM I | Gibco | 31985–070 |
| GIBCO Hybridoma-SFM | Thermo Fisher Scientific | Cat# 12045076 |
| Recombinant DENV E protein | Kudlacek et al., 2018 | N/A |
| Recombinant DENV E protein stabilized dimers | Kudlacek et al., 2018 | N/A |
| CpG10103 (TCGTCGTTTTTCGGTCGTTTT) | Synthesized by Invitrogen | N/A |
| TrueBlue substrate | KPL | 0510–0050 |
| Cyclosporin A | Sigma-Aldrich | Cat# C1832 |
| Chk2 inhibitor | Sigma-Aldrich | Cat# C3742 |
| Non-fat dry milk | Bio Rad | Cat# 1706404 |
| Goat serum | Thermo Fisher Scientific | Cat# 16210072 |
| Critical Commercial Assays | ||
| Deposited Data | ||
| Experimental Models: Cell Lines | ||
| Mouse-human HMAA 2.5 myeloma cell line | Dr. Marshall Posner | N/A |
| DENV-66 hybridoma clone | This manuscript | N/A |
| DENV-115 hybridoma clone | This manuscript | N/A |
| DENV-144 hybridoma clone | This manuscript | N/A |
| DENV-236 hybridoma clone | This manuscript | N/A |
| DENV-297 hybridoma clone | This manuscript | N/A |
| DENV-298 hybridoma clone | This manuscript | N/A |
| DENV-286 hybridoma clone | This manuscript | N/A |
| DENV-290 hybridoma clone | This manuscript | N/A |
| DENV-254 hybridoma clone | This manuscript | N/A |
| DENV-404 hybridoma clone | This manuscript | N/A |
| DENV-406 hybridoma clone | This manuscript | N/A |
| DENV-415 hybridoma clone | This manuscript | N/A |
| DENV-419 hybridoma clone | This manuscript | N/A |
| DENV-437 hybridoma clone | This manuscript | N/A |
| DENV-443 hybridoma clone | This manuscript | N/A |
| C6/36 cells | ATCC | CRL-1660 |
| U937-DC-SIGN cells | ||
| Vero-81 cells | ATCC | CCL-81 |
| Experimental Models: Organisms/Strains | ||
| Mouse: AG129 strain | M. Aguet (Swiss Institute for Experimental Cancer Research, Epalinges, Switzerland) | N/A |
| Recombinant DNA | ||
| Primer: DENV1–4, D1: 5'-TCA ATA TGC TGA AAC GCGCGA GAA ACC G | Harris et al., 1998, PMID: 9705406 | N/A |
| Primer: DENV1–4, D2: 5’-TTGCACCAACAGTCAATGTCTTCAGGTTC | Harris et al., 1998, PMID: 9705406 | N/A |
| Primer: DENV1–4, TS1: 5'-CGT CTC AGT GAT CCG GGG G | Harris et al., 1998, PMID: 9705406 | N/A |
| Primer: DENV1–4, TS2: 5'-CGCCAC AAG GGC CAT GAA CAG | Harris et al., 1998, PMID: 9705406 | N/A |
| Primer: DENV1–4, TS3: 5'-TAA CAT CAT CAT GAG ACAGAG C | Harris et al., 1998, PMID: 9705406 | N/A |
| Primer: DENV1–4, DEN4: 5'-TGT TGT CTT AAA CAA GAG AGG TC | Harris et al., 1998, PMID: 9705406 | N/A |
| Primer: DENV UNC 3009 Forward: GGACTGGATACACGCACCCA | This manuscript | N/A |
| Primer: DENV UNC 3009 Reverse: CATGTCTCTACCTTCTCGACTTGTCT | This manuscript | N/A |
| DENV UNC 3009 Probe: ACCTGGATGTCGGCTGAAGGAGCTTG | This manuscript | N/A |
| TaqMan™ Rodent GAPDH Control Reagents | Applied Biosystems | Cat. 4308313 |
| Software and Algorithms | ||
| GraphPad Prism 7.2 | GraphPad Software, Inc. | https://www.graphpad.com |
| PyMOL | Schrödinger, LLC | https://www.pymol.org/ |
| Other | ||
| ÄKTA pure chromatography system | GE Healthcare Life Sciences | N/A |
| EL406 washer dispenser | BioTek | N/A |
| Biostack microplate stacker | BioTek | N/A |
| HiTrap Protein G High Performance | GE Healthcare Life Sciences | Cat# 17–0404-01 |
| Immunospot | CTL | N/A |
The identified human type-specific mAbs target diverse antigenic sites on DENV3 E
Chimeric DENV viruses encode functional DENV3 neutralizing epitopes
Functional neutralizing epitopes can be rapidly mapped using chimeric DENV viruses
Selected mAbs potently neutralize multiple genotype strains of DENV3 viruses
ACKNOWLEDGEMENTS
We thank members of the study teams at the Centro de Salud Sócrates Flores Vivas, the National Virology Laboratory in the Centro Nacional de Diagnóstico y Referencia, and the Sustainable Sciences Institute in Nicaragua for their dedication and high-quality work in conducting the study and in preparing the PBMCs. We are also grateful to the study participants and their families. We thank Rachel Sutton at VUMC for protein expression and purification work. This project received support from NIH grant P01 AI106695 to EH and AI107731 to AD. This work was also supported by the National Center for Research Resources, Grant UL1 RR024975–01 and the National Center for Advancing Translational Sciences, Grant 2 UL1 TR000445–06. The Nicaraguan Pediatric Cohort Study was also supported by NIH grant R01AI099631 to AB and grant VE-1 to EH from the Pediatric Dengue Vaccine Initiative. Flow cytometry experiments were performed in the VMC Flow Cytometry Shared Resource. The VMC Flow Cytometry Shared Resource is supported by the Vanderbilt Ingram Cancer Center (P30 CA68485) and the Vanderbilt Digestive Disease Research Center (DK058404). David R. Martinez is supported by an NIH NIAID T32 AI007151 and a Burroughs Wellcome Fund Postdoctoral Enrichment Program (BWF PDEP) Award. The content is solely the responsibility of the authors and does not necessarily represent the official views of the NIH.
Footnotes
DECLARATION OF INTERESTS. J.E.C. has served as a consultant for Takeda Vaccines, Sanofi Pasteur, Pfizer, and Novavax, is on the Scientific Advisory Boards of CompuVax and Meissa Vaccines, and is Founder of IDBiologics, Inc. M.S.D. is a consultant for Inbios and Emergent BioSolutions and on the Scientific Advisory Board of Moderna. R.S.B, A.d.S., and E.H. have served as consultants for Takeda Vaccines and Sanofi Pasteur. All other authors declare no conflict of interest. Vanderbilt University has applied for a patent related to the human antibodies. UNC has applied for a patent related to the chimeric viruses.
Data and Code Availability Statement.
The published article includes all datasets generated or analyzed during this study. This study did not generate code.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- Andrade DV, Katzelnick LC, Widman DG, Balmaseda A, de Silva AM, Baric RS, and Harris E. (2017). Analysis of Individuals from a Dengue-Endemic Region Helps Define the Footprint and Repertoire of Antibodies Targeting Dengue Virus 3 Type-Specific Epitopes. MBio 8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andrade DV, Warnes C, Young E, Katzelnick LC, Balmaseda A, de Silva AM, Baric RS, and Harris E. (2019). Tracking the polyclonal neutralizing antibody response to a dengue virus serotype 1 type-specific epitope across two populations in Asia and the Americas. Sci Rep 9, 16258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balmaseda A, Guzman MG, Hammond S, Robleto G, Flores C, Tellez Y, Videa E, Saborio S, Perez L, Sandoval E, et al. (2003). Diagnosis of dengue virus infection by detection of specific immunoglobulin M (IgM) and IgA antibodies in serum and saliva. Clin Diagn Lab Immunol 10, 317–322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balmaseda A, Hammond SN, Tellez Y, Imhoff L, Rodriguez Y, Saborio SI, Mercado JC, Perez L, Videa E, Almanza E, et al. (2006). High seroprevalence of antibodies against dengue virus in a prospective study of schoolchildren in Managua, Nicaragua. Trop Med Int Health 11, 935–942. [DOI] [PubMed] [Google Scholar]
- Balmaseda A, Sandoval E, Perez L, Gutierrez CM, and Harris E. (1999). Application of molecular typing techniques in the 1998 dengue epidemic in Nicaragua. Am J Trop Med Hyg 61, 893–897. [DOI] [PubMed] [Google Scholar]
- Balmaseda A, Standish K, Mercado JC, Matute JC, Tellez Y, Saborio S, Hammond SN, Nunez A, Aviles W, Henn MR, et al. (2010). Trends in patterns of dengue transmission over 4 years in a pediatric cohort study in Nicaragua. J Infect Dis 201, 5–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balsitis SJ, Williams KL, Lachica R, Flores D, Kyle JL, Mehlhop E, Johnson S, Diamond MS, Beatty PR, and Harris E. (2010). Lethal antibody enhancement of dengue disease in mice is prevented by Fc modification. PLoS Pathog 6, e1000790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beltramello M, Williams KL, Simmons CP, Macagno A, Simonelli L, Quyen NT, Sukupolvi-Petty S, Navarro-Sanchez E, Young PR, de Silva AM, et al. (2010). The human immune response to Dengue virus is dominated by highly cross-reactive antibodies endowed with neutralizing and enhancing activity. Cell Host Microbe 8, 271–283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhatt S, Gething PW, Brady OJ, Messina JP, Farlow AW, Moyes CL, Drake JM, Brownstein JS, Hoen AG, Sankoh O, et al. (2013). The global distribution and burden of dengue. Nature 496, 504–507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhaumik SK, Priyamvada L, Kauffman RC, Lai L, Natrajan MS, Cho A, Rouphael N, Suthar MS, Mulligan MJ, and Wrammert J. (2018). Pre-Existing Dengue Immunity Drives a DENV-Biased Plasmablast Response in ZIKV-Infected Patient. Viruses 11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biswal S, Reynales H, Saez-Llorens X, Lopez P, Borja-Tabora C, Kosalaraksa P, Sirivichayakul C, Watanaveeradej V, Rivera L, Espinoza F, et al. (2019). Efficacy of a Tetravalent Dengue Vaccine in Healthy Children and Adolescents. N Engl J Med 381, 2009–2019. [DOI] [PubMed] [Google Scholar]
- Brien JD, Austin SK, Sukupolvi-Petty S, O’Brien KM, Johnson S, Fremont DH, and Diamond MS (2010). Genotype-specific neutralization and protection by antibodies against dengue virus type 3. J Virol 84, 10630–10643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen R, and Vasilakis N. (2011). Dengue--quo tu et quo vadis? Viruses 3, 1562–1608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Alwis R, Smith SA, Olivarez NP, Messer WB, Huynh JP, Wahala WM, White LJ, Diamond MS, Baric RS, Crowe JE Jr., et al. (2012). Identification of human neutralizing antibodies that bind to complex epitopes on dengue virions. Proc Natl Acad Sci U S A 109, 7439–7444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Alwis R, Williams KL, Schmid MA, Lai CY, Patel B, Smith SA, Crowe JE, Wang WK, Harris E, and de Silva AM (2014). Dengue viruses are enhanced by distinct populations of serotype cross-reactive antibodies in human immune sera. PLoS Pathog 10, e1004386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dejnirattisai W, Wongwiwat W, Supasa S, Zhang X, Dai X, Rouvinski A, Jumnainsong A, Edwards C, Quyen NTH, Duangchinda T, et al. (2015a). A new class of highly potent, broadly neutralizing antibodies isolated from viremic patients infected with dengue virus. Nat Immunol 16, 170–177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dejnirattisai W, Wongwiwat W, Supasa S, Zhang X, Dai X, Rouvinsky A, Jumnainsong A, Edwards C, Quyen NT, Duangchinda T, et al. (2015b). Corrigendum: A new class of highly potent, broadly neutralizing antibodies isolated from viremic patients infected with dengue virus. Nat Immunol 16, 785. [DOI] [PubMed] [Google Scholar]
- Diamond MS, and Pierson TC (2015). Molecular Insight into Dengue Virus Pathogenesis and Its Implications for Disease Control. Cell 162, 488–492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferguson NM, Rodriguez-Barraquer I, Dorigatti I, Mier YT-RL, Laydon DJ, and Cummings DA(2016). Benefits and risks of the Sanofi-Pasteur dengue vaccine: Modeling optimal deployment. Science 353, 1033–1036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernandez RJ, and Vazquez S. (1990). Serological diagnosis of dengue by an ELISA inhibition method (EIM). Mem Inst Oswaldo Cruz 85, 347–351. [DOI] [PubMed] [Google Scholar]
- Fibriansah G, Ibarra KD, Ng TS, Smith SA, Tan JL, Lim XN, Ooi JS, Kostyuchenko VA, Wang J, de Silva AM, et al. (2015a). DENGUE VIRUS. Cryo-EM structure of an antibody that neutralizes dengue virus type 2 by locking E protein dimers. Science 349, 88–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fibriansah G, Tan JL, Smith SA, de Alwis AR, Ng TS, Kostyuchenko VA, Ibarra KD, Wang J, Harris E, de Silva A, et al. (2014). A potent anti-dengue human antibody preferentially recognizes the conformation of E protein monomers assembled on the virus surface. EMBO Mol Med 6, 358–371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fibriansah G, Tan JL, Smith SA, de Alwis R, Ng TS, Kostyuchenko VA, Jadi RS, Kukkaro P, de Silva AM, Crowe JE, et al. (2015b). A highly potent human antibody neutralizes dengue virus serotype 3 by binding across three surface proteins. Nat Commun 6, 6341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fleith RC, Lobo FP, Dos Santos PF, Rocha MM, Bordignon J, Strottmann DM, Patricio DO, Pavanelli WR, Lo Sarzi M, Santos CN, et al. (2016). Genome-wide analyses reveal a highly conserved Dengue virus envelope peptide which is critical for virus viability and antigenic in humans. Sci Rep 6, 36339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flipse J, and Smit JM (2015). The Complexity of a Dengue Vaccine: A Review of the Human Antibody Response. PLoS Negl Trop Dis 9, e0003749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallichotte EN, Baric RS, and de Silva AM (2018a). The Molecular Specificity of the Human Antibody Response to Dengue Virus Infections. Adv Exp Med Biol 1062, 63–76. [DOI] [PubMed] [Google Scholar]
- Gallichotte EN, Baric TJ, Nivarthi U, Delacruz MJ, Graham R, Widman DG, Yount BL, Durbin AP, Whitehead SS, de Silva AM, et al. (2018b). Genetic Variation between Dengue Virus Type 4 Strains Impacts Human Antibody Binding and Neutralization. Cell Rep 25, 1214–1224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallichotte EN, Baric TJ, Yount BL Jr., Widman DG, Durbin A, Whitehead S, Baric RS, and de Silva AM (2018c). Human dengue virus serotype 2 neutralizing antibodies target two distinct quaternary epitopes. PLoS Pathog 14, e1006934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallichotte EN, Menachery VD, Yount BL Jr., Widman DG, Dinnon KH 3rd, Hartman S, de Silva AM, and Baric RS (2017). Epitope Addition and Ablation via Manipulation of a Dengue Virus Serotype 1 Infectious Clone. mSphere 2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallichotte EN, Widman DG, Yount BL, Wahala WM, Durbin A, Whitehead S, Sariol CA, Crowe JE Jr., de Silva AM, and Baric RS (2015). A new quaternary structure epitope on dengue virus serotype 2 is the target of durable type-specific neutralizing antibodies. MBio 6, e01461–01415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grifoni A, Pham J, Sidney J, O’Rourke PH, Paul S, Peters B, Martini SR, de Silva AD, Ricciardi MJ, Magnani DM, et al. (2017). Prior Dengue Virus Exposure Shapes T Cell Immunity to Zika Virus in Humans. J Virol 91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gutierrez G, Standish K, Narvaez F, Perez MA, Saborio S, Elizondo D, Ortega O, Nunez A, Kuan G, Balmaseda A, et al. (2011). Unusual dengue virus 3 epidemic in Nicaragua, 2009. PLoS Negl Trop Dis 5, e1394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hadinegoro SR, Arredondo-Garcia JL, Capeding MR, Deseda C, Chotpitayasunondh T, Dietze R, Muhammad Ismail HI, Reynales H, Limkittikul K, Rivera-Medina DM, et al. (2015). Efficacy and Long-Term Safety of a Dengue Vaccine in Regions of Endemic Disease. N Engl J Med 373, 1195–1206. [DOI] [PubMed] [Google Scholar]
- Halstead SB (2015). Pathogenesis of Dengue: Dawn of a New Era. F1000Res 4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halstead SB (2017). Dengvaxia sensitizes seronegatives to vaccine enhanced disease regardless of age. Vaccine 35, 6355–6358. [DOI] [PubMed] [Google Scholar]
- Halstead SB (2018a). Safety issues from a Phase 3 clinical trial of a live-attenuated chimeric yellow fever tetravalent dengue vaccine. Hum Vaccin Immunother 14, 2158–2162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halstead SB (2018b). Which Dengue Vaccine Approach Is the Most Promising, and Should We Be Concerned about Enhanced Disease after Vaccination? There Is Only One True Winner. Cold Spring Harb Perspect Biol 10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halstead SB, Chow JS, and Marchette NJ (1973). Immunological enhancement of dengue virus replication. Nat New Biol 243, 24–26. [PubMed] [Google Scholar]
- Henein S, Swanstrom J, Byers AM, Moser JM, Shaik SF, Bonaparte M, Jackson N, Guy B, Baric R, and de Silva AM (2017). Dissecting Antibodies Induced by a Chimeric Yellow Fever-Dengue, Live-Attenuated, Tetravalent Dengue Vaccine (CYD-TDV) in Naive and Dengue-Exposed Individuals. J Infect Dis 215, 351–358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holmes EC, and Twiddy SS (2003). The origin, emergence and evolutionary genetics of dengue virus. Infect Genet Evol 3, 19–28. [DOI] [PubMed] [Google Scholar]
- Johnson AJ, and Roehrig JT (1999). New mouse model for dengue virus vaccine testing. J Virol 73, 783–786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katzelnick LC, Gresh L, Halloran ME, Mercado JC, Kuan G, Gordon A, Balmaseda A, and Harris E. (2017a). Antibody-dependent enhancement of severe dengue disease in humans. Science 358, 929–932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katzelnick LC, Harris E, and Participants in the Summit on Dengue Immune Correlates of, P. (2017b). Immune correlates of protection for dengue: State of the art and research agenda. Vaccine 35, 4659–4669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katzelnick LC, Montoya M, Gresh L, Balmaseda A, and Harris E. (2016). Neutralizing antibody titers against dengue virus correlate with protection from symptomatic infection in a longitudinal cohort. Proc Natl Acad Sci U S A 113, 728–733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- King CC, Chao DY, Chien LJ, Chang GJ, Lin TH, Wu YC, and Huang JH (2008). Comparative analysis of full genomic sequences among different genotypes of dengue virus type 3. Virol J 5, 63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kraus AA, Messer W, Haymore LB, and de Silva AM (2007). Comparison of plaque-and flow cytometry-based methods for measuring dengue virus neutralization. J Clin Microbiol 45, 3777–3780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuan G, Gordon A, Aviles W, Ortega O, Hammond SN, Elizondo D, Nunez A, Coloma J, Balmaseda A, and Harris E. (2009). The Nicaraguan pediatric dengue cohort study: study design, methods, use of information technology, and extension to other infectious diseases. Am J Epidemiol 170, 120–129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kudlacek ST, Premkumar L, Metz SW, Tripathy A, Bobkov AA, Payne AM, Graham S, Brackbill JA, Miley MJ, de Silva AM, et al. (2018). Physiological temperatures reduce dimerization of dengue and Zika virus recombinant envelope proteins. J Biol Chem 293, 8922–8933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuhn RJ, Dowd KA, Beth Post C, and Pierson TC (2015). Shake, rattle, and roll: Impact of the dynamics of flavivirus particles on their interactions with the host. Virology 479-–480, 508–517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lanciotti RS, Calisher CH, Gubler DJ, Chang GJ, and Vorndam AV (1992). Rapid detection and typing of dengue viruses from clinical samples by using reverse transcriptase-polymerase chain reaction. J Clin Microbiol 30, 545–551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee PD, Mukherjee S, Edeling MA, Dowd KA, Austin SK, Manhart CJ, Diamond MS, Fremont DH, and Pierson TC (2013). The Fc region of an antibody impacts the neutralization of West Nile viruses in different maturation states. J Virol 87, 13729–13740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J, Watterson D, Chang CW, Che XY, Li XQ, Ericsson DJ, Qiu LW, Cai JP, Chen J, Fry SR, et al. (2018). Structural and Functional Characterization of a Cross-Reactive Dengue Virus Neutralizing Antibody that Recognizes a Cryptic Epitope. Structure 26, 51–59 e54. [DOI] [PubMed] [Google Scholar]
- Lofano G, Gorman MJ, Yousif AS, Yu WH, Fox JM, Dugast AS, Ackerman ME, Suscovich TJ, Weiner J, Barouch Dv et al. (2018). Antigen-specific antibody Fc glycosylation enhances humoral immunity via the recruitment of complement. Sci Immunol 3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Magnani DM, Silveira CGT, Ricciardi MJ, Gonzalez-Nieto L, Pedreno-Lopez N, Bailey VK, Gutman MJ, Maxwell HS, Domingues A, Costa PR, et al. (2017). Potent Plasmablast-Derived Antibodies Elicited by the National Institutes of Health Dengue Vaccine. J Virol 91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mattia K, Puffer BA, Williams KL, Gonzalez R, Murray M, Sluzas E, Pagano D, Ajith S, Bower M, Berdougo E, et al. (2011). Dengue reporter virus particles for measuring neutralizing antibodies against each of the four dengue serotypes. PLoS One 6, e27252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Messer WB, Yount B, Hacker KE, Donaldson EF, Huynh JP, de Silva AM, and Baric RS (2012). Development and characterization of a reverse genetic system for studying dengue virus serotype 3 strain variation and neutralization. PLoS Negl Trop Dis 6, e1486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Messer WB, Yount BL, Royal SR, de Alwis R, Widman DG, Smith SA, Crowe JE Jr., Pfaff JM, Kahle KM, Doranz BJ, et al. (2016). Functional Transplant of a Dengue Virus Serotype 3 (DENV3)-Specific Human Monoclonal Antibody Epitope into DENV1. J Virol 90, 5090–5097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Metz SW, Gallichotte EN, Brackbill A, Premkumar L, Miley MJ, Baric R, and de Silva AM(2017). In Vitro Assembly and Stabilization of Dengue and Zika Virus Envelope Protein Homo-Dimers. Sci Rep 7, 4524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michlmayr D, Andrade P, Gonzalez K, Balmaseda A, and Harris E. (2017). CD14(+)CD16(+) monocytes are the main target of Zika virus infection in peripheral blood mononuclear cells in a paediatric study in Nicaragua. Nat Microbiol 2, 1462–1470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montoya M, Gresh L, Mercado JC, Williams KL, Vargas MJ, Gutierrez G, Kuan G, Gordon A, Balmaseda A, and Harris E. (2013). Symptomatic versus inapparent outcome in repeat dengue virus infections is influenced by the time interval between infections and study year. PLoS Negl Trop Dis 7, e2357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moodie Z, Juraska M, Huang Y, Zhuang Y, Fong Y, Carpp LN, Self SG, Chambonneau L, Small R, Jackson N, et al. (2018). Neutralizing Antibody Correlates Analysis of Tetravalent Dengue Vaccine Efficacy Trials in Asia and Latin America. J Infect Dis 217, 742–753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mukherjee S, Dowd KA, Manhart CJ, Ledgerwood JE, Durbin AP, Whitehead SS, and Pierson TC (2014). Mechanism and significance of cell type-dependent neutralization of flaviviruses. J Virol 88, 7210–7220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murphy BR, and Whitehead SS (2011). Immune response to dengue virus and prospects for a vaccine. Annu Rev Immunol 29, 587–619. [DOI] [PubMed] [Google Scholar]
- Nivarthi UK, Kose N, Sapparapu G, Widman D, Gallichotte E, Pfaff JM, Doranz BJ, Weiskopf D, Sette A, Durbin AP, et al. (2017). Mapping the Human Memory B Cell and Serum Neutralizing Antibody Responses to Dengue Virus Serotype 4 Infection and Vaccination. J Virol 91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- OhAinle M, Balmaseda A, Macalalad AR, Tellez Y, Zody MC, Saborio S, Nunez A, Lennon NJ, Birren BW, Gordon A, et al. (2011). Dynamics of dengue disease severity determined by the interplay between viral genetics and serotype-specific immunity. Sci Transl Med 3, 114ra128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patel B, Longo P, Miley MJ, Montoya M, Harris E, and de Silva AM (2017). Dissecting the human serum antibody response to secondary dengue virus infections. PLoS Negl Trop Dis 11, e0005554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pierson TC, and Diamond MS (2008). Molecular mechanisms of antibody-mediated neutralisation of flavivirus infection. Expert Rev Mol Med 10, e12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pierson TC, and Diamond MS (2015). A game of numbers: the stoichiometry of antibody-mediated neutralization of flavivirus infection. Prog Mol Biol Transl Sci 129, 141–166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raekiansyah M, Pramesyanti A, Bela B, Kosasih H, Ma’roef CN, Tobing SY, Rudiman PI, Alisjahbana B, Endi TP, Green S, et al. (2005). Genetic variations and relationship among dengue virus type 3 strains isolated from patients with mild or severe form of dengue disease in Indonesia and Thailand. Southeast Asian J Trop Med Public Health 36, 1187–1197. [PubMed] [Google Scholar]
- Rouvinski A, Dejnirattisai W, Guardado-Calvo P, Vaney MC, Sharma A, Duquerroy S, Supasa P, Wongwiwat W, Haouz A, Barba-Spaeth G, et al. (2017). Covalently linked dengue virus envelope glycoprotein dimers reduce exposure of the immunodominant fusion loop epitope. Nat Commun 8, 15411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salje H, Cummings DAT, Rodriguez-Barraquer I, Katzelnick LC, Lessler J, Klungthong C, Thaisomboonsuk B, Nisalak A, Weg A, Ellison D, et al. (2018). Reconstruction of antibody dynamics and infection histories to evaluate dengue risk. Nature 557, 719–723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sangkawibha N, Rojanasuphot S, Ahandrik S, Viriyapongse S, Jatanasen S, Salitul V, Phanthumachinda B, and Halstead SB (1984). Risk factors in dengue shock syndrome: a prospective epidemiologic study in Rayong, Thailand. I. The 1980 outbreak. Am J Epidemiol 120, 653–669. [DOI] [PubMed] [Google Scholar]
- Sarathy VV, White M, Li L, Kaiser JA, Campbell GA, Milligan GN, Bourne N, and Barrett ADT.(2018) Characterization of a murine model of non-lethal, symptomatic dengue virus infection. Sci Rep 8, 4900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shresta S, Kyle JL, Snider HM, Basavapatna M, Beatty PR, and Harris E. (2004). Interferon-dependent immunity is essential for resistance to primary dengue virus infection in mice, whereas T and B-cell-dependent immunity are less critical. J Virol 78, 2701–2710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shresta S, Sharar KL, Prigozhin DM, Beatty PR, and Harris E. (2006). Murine model for dengue virus-induced lethal disease with increased vascular permeability. J Virol 80, 10208–10217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simon AK, Hollander GA, and McMichael A. (2015). Evolution of the immune system in humans from infancy to old age. Proc Biol Sci 282, 20143085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith SA, de Alwis AR, Kose N, Jadi RS, de Silva AM, and Crowe JE Jr. (2014). Isolation of dengue virus-specific memory B cells with live virus antigen from human subjects following natural infection reveals the presence of diverse novel functional groups of antibody clones. J Virol 88, 12233–12241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith SA, de Alwis R, Kose N, Durbin AP, Whitehead SS, de Silva AM, and Crowe JE Jr. (2013). Human monoclonal antibodies derived from memory B cells following live attenuated dengue virus vaccination or natural infection exhibit similar characteristics. J Infect Dis 207, 1898–1908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith SA, Zhou Y, Olivarez NP, Broadwater AH, de Silva AM, and Crowe JE Jr. (2012). Persistence of circulating memory B cell clones with potential for dengue virus disease enhancement for decades following infection. J Virol 86, 2665–2675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sridhar S, Luedtke A, Langevin E, Zhu M, Bonaparte M, Machabert T, Savarino S, Zambrano B, Moureau A, Khromava A, et al. (2018). Effect of Dengue Serostatus on Dengue Vaccine Safety and Efficacy. N Engl J Med 379, 327–340. [DOI] [PubMed] [Google Scholar]
- Stettler K, Beltramello M, Espinosa DA, Graham V, Cassotta A, Bianchi S, Vanzetta F, Minola A, Jaconi S, Mele F, et al. (2016). Specificity, cross-reactivity, and function of antibodies elicited by Zika virus infection. Science 353, 823–826. [DOI] [PubMed] [Google Scholar]
- Sukupolvi-Petty S, Brien JD, Austin SK, Shrestha B, Swayne S, Kahle K, Doranz BJ, Johnson S, Pierson TC, Fremont DH, et al. (2013). Functional analysis of antibodies against dengue virus type 4 reveals strain-dependent epitope exposure that impacts neutralization and protection. J Virol 87, 8826–8842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swanstrom JA, Henein S, Plante JA, Yount BL, Widman DG, Gallichotte EN, Dean HJ, Osorio JE, Partidos CD, de Silva AM, et al. (2018). Analyzing the Human Serum Antibody Responses to a Live Attenuated Tetravalent Dengue Vaccine Candidate. J Infect Dis 217, 1932–1941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swanstrom JA, Nivarthi UK, Patel B, Delacruz MJ, Yount B, Widman DG, Durbin AP, Whitehead SS, De Silva AM, and Baric RS (2019). Beyond Neutralizing Antibody Levels: The Epitope Specificity of Antibodies Induced by National Institutes of Health Monovalent Dengue Virus Vaccines. J Infect Dis 220, 219–227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swanstrom JA, Plante JA, Plante KS, Young EF, McGowan E, Gallichotte EN, Widman DG, Heise MT, de Silva AM, and Baric RS (2016). Dengue Virus Envelope Dimer Epitope Monoclonal Antibodies Isolated from Dengue Patients Are Protective against Zika Virus. MBio 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teoh EP, Kukkaro P, Teo EW, Lim AP, Tan TT, Yip A, Schul W, Aung M, Kostyuchenko VA, Leo YS, et al. (2012). The structural basis for serotype-specific neutralization of dengue virus by a human antibody. Sci Transl Med 4, 139ra183. [DOI] [PubMed] [Google Scholar]
- Waggoner JJ, Balmaseda A, Gresh L, Sahoo MK, Montoya M, Wang C, Abeynayake J, Kuan G, Pinsky BA, and Harris E. (2016). Homotypic Dengue Virus Reinfections in Nicaraguan Children. J Infect Dis 214, 986–993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wahala WM, Donaldson EF, de Alwis R, Accavitti-Loper MA, Baric RS, and de Silva AM (2010). Natural strain variation and antibody neutralization of dengue serotype 3 viruses. PLoS Pathog 6, e1000821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waman VP, Kale MM, and Kulkarni-Kale U. (2017). Genetic diversity and evolution of dengue virus serotype 3: A comparative genomics study. Infect Genet Evol 49, 234–240. [DOI] [PubMed] [Google Scholar]
- Widman DG, Young E, Nivarthi U, Swanstrom JA, Royal SR, Yount BL, Debbink K, Begley M, Marcet S, Durbin A, et al. (2017). Transplantation of a quaternary structure neutralizing antibody epitope from dengue virus serotype 3 into serotype 4. Sci Rep 7, 17169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu X, McGraw PA, House FS, and Crowe JE Jr. (2008). An optimized electrofusion-based protocol for generating virus-specific human monoclonal antibodies. J Immunol Methods 336, 142–151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zompi S, Montoya M, Pohl MO, Balmaseda A, and Harris E. (2012). Dominant cross-reactive B cell response during secondary acute dengue virus infection in humans. PLoS Negl Trop Dis 6, e1568. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.