Abstract
Tandem mass spectrometry is an indispensable tool in proteomics used for protein sequencing and quantitation. On the basis of the sequential fragments usually generated from peptide ions via collision-induced dissociation, electron-transfer dissociation, or a combination of the two, probabilistic database search engines could be used for the identification of the peptides. The correct localization of posttranslational modifications (PTMs) poses a more challenging problem than the general identification of proteins. Histones are involved in the regulation of DNA transcription via the wealth of PTMs on their N-terminal tail. In this study, we analyzed the histone H4 peptide SGRGK incorporating four different posttranslational modifications: citrullination, acetylation, phosphorylation, and arginine methylation at various positions. The pentapeptides model the enzymatic cleavage of the N-terminal tail of human histone H4 protein by LysC protease. Fragmentation of the peptides was investigated using higher-energy collisional dissociation (HCD), electron-transfer dissociation (ETD), and electron-transfer higher-energy collisional dissociation (EThcD) on an ultrahigh resolution and mass accuracy instrument. We found that while all three techniques have their unique characteristics, advantages, and pitfalls, EThcD generated the most fragment ion-rich spectra. Despite potential ambiguities regarding exact fragment identities, full sequence coverage and PTM mapping may also be achievable. We also found novel neutral losses from the charge-reduced precursors characteristic to citrullination in ETD and EThcD which may be used in proteomic applications. N-Terminal acetylation and arginine methylation could also be confirmed by their characteristic neutral losses from the charge-reduced precursors.
Keywords: citrullination, acetylation, arginine methylation, posttranslational modifications, histone code, collision-induced dissociation, electron-transfer dissociation, tandem mass spectrometry
Introduction
Sequencing of peptides and detailed characterization of their posttranslational modifications (PTMs) could usually be carried out by tandem mass spectrometry using collisional activation (CID), activation by electron-transfer (ETD), or activation by electron transfer followed by collisional activation (EThcD).
Collisional activation (CID, or HCD in Orbitrap instruments) is an activation method that uses collisions with inert gases to impart enough energy to the precursors for fragmentation.1 It is the best option when peptides are relatively small (<20 amino acids), not highly charged (z ≤ 3), and devoid of labile PTMs (e.g., phosphorylation). Product ions mostly include y and b type ions2 originating from amide bond cleavages as well as neutral losses of small molecules that may or may not be characteristic to modifications present on the peptides. Despite some of its disadvantages, CID is still widely used due to the well-understood mechanism of the processes, reliability, and good sequence coverage of the peptides.
ETD was developed by Syka et al.3 and involves the transfer of electron(s) to multiply charged peptide species to initiate fragmentation. Being only efficient for peptides with higher charge densities, it is usually used for larger peptides or even proteins. Sequence coverage may also be higher than that of CID. ETD is applied as a complementary method to CID as it yields c and z type ions originating from N–Cα bond cleavages. Neutral losses are usually less frequent in ETD spectra. PTMs are preferentially preserved in ETD as opposed to CID. There is no fragmentation at the N-terminus of proline residues in ETD.
EThcD was first introduced by Frese et al.4 In EThcD, the transfer of electrons is followed by the collisional activation of all the products generated in the first step. The overall process yields b, y, c, and z ions and numerous neutral losses. It has been reported as the best method of choice for the analysis of phosphopeptides and glycopeptides5 due to the wealth of fragments generated, which considerably facilitate the localization of PTMs.
Histone-mediated gene regulation is one of the main fields where PTMs are of high importance.6 Acetylation of the N-terminus7 or lysine residues,8 methylation of arginine9 and lysine residues,10 and phosphorylation of serine residues11 have been known a long time along with the citrullination site at Arg-312 of the histone H4 N-terminal tail. While acetylation on the side chains in CID results in a mass increment of the corresponding fragments carrying the modification and may result in characteristic iminium ions, an N-terminal acetylation may give rise to b1 ions which are not stable in their original forms.13 No characteristic side chain loss attributed to acetylation has been previously described. On the other hand, neutral losses of methylamine, N-methylcarbodiimide, and N-methylguanidine as well as some iminium ion related fragments are reported to be characteristic to methylated arginine residues in CID MS/MS.14 Phosphorylated residues are highly prone to lose phosphoric acid—or even worse, transfer it to other unmodified hydroxyl side chains upon CID.15,16 In ETD, these side chain modifications remain preferentially intact.17 The presence of fragments corresponding to the neutral loss of isocyanic acid is selective for (homo)citrullinated residues in CID,18,19 although no distinctive side chain loss has been reported for citrullination in ETD. Moreover, we previously reported a cleavage preference at the C-termini of citrulline residues in the collision-induced tandem mass spectra of citrullinated peptides (citrulline effect).20,21
Histone modifications have been extensively studied in the past decade. Bottom-up, middle-down, and top-down approaches using CID/HCD and ETD techniques have all been applied to investigate the histone code.22−28 However, little work has explored the detailed and systematic fragmentation of small model peptides containing citrulline residues as well. Furthermore, we hypothesized that there should be additional uncharacterized neutral loss ions that may be useful for PTM localization or confirmation.
In this study, we synthesized the human histone H4 N-terminal pentapeptide SGRGK and its combinatorial variants bearing various PTMs: Ac-SGRGK, pSGRGK, Ac-pSGRGK, SGXGK, Ac-SGXGK, pSGXGK, Ac-pSGXGK, SGR(Me)GK, Ac-SGR(Me)GK, pSGR(Me)GK, Ac-pSGR(Me)GK, where X stands for the one-letter abbreviation for citrulline residues. Utilizing the current, commercially available activation techniques—namely, HCD, ETD, and EThcD—our aim was to examine the possible differences of the various tandem mass spectra. We took care to observe the neutral losses that could be used for improving PTM localization.
Experimental Procedures
Model pentapeptides were manually synthesized using the standard Fmoc/tBu strategy of solid phase peptide synthesis on Wang-resin resulting in peptides with free carboxy termini. PTMs—except for acetylation—were introduced by incorporating modified residues (Fmoc-Arg(Me, Pbf)-OH and Fmoc-Ser(PO(OBzl)OH)-OH)) into the sequence during synthesis. Acetylation was performed with acetic anhydride in basic conditions. All amino acids were purchased from Iris Biotech GmbH (Marktredwitz, Germany). After cleavage by trifluoroacetic acid in the presence of phenol, anisole, triisopropylsilane, and distilled water as scavengers, the resulting peptides were purified by HPLC-UV and were freeze-dried.
Prior to MS analysis, the freeze-dried samples were dissolved in acetonitrile–water (1:1, v/v), containing 0.1 v/v% formic acid. Final concentration of the peptides was 10 μM. These solutions were directly injected to the electrospray source of an Orbitrap Fusion Tribrid instrument (Thermo Scientific, Waltham, MA, USA) at a flow rate of 5 μL/min. Resolution was set to 120 000. An isolation width of 2 m/z was applied for MS/MS. For ETD and EThcD experiments, the ion activation time was set to 50 ms. Normalized collision energies (NCE) were set between 15 and 35% for HCD and EThcD. Collision energy dependence studies were carried out on a Q-Exactive Focus Hybrid Quadrupole-Orbitrap instrument (Thermo Scientific, Bremen, Germany) in the range of 10–48 eV with 2 eV steps at a resolution of 70 000. Raw data were visualized and annotated by mMass.29
Results
HCD Experiments
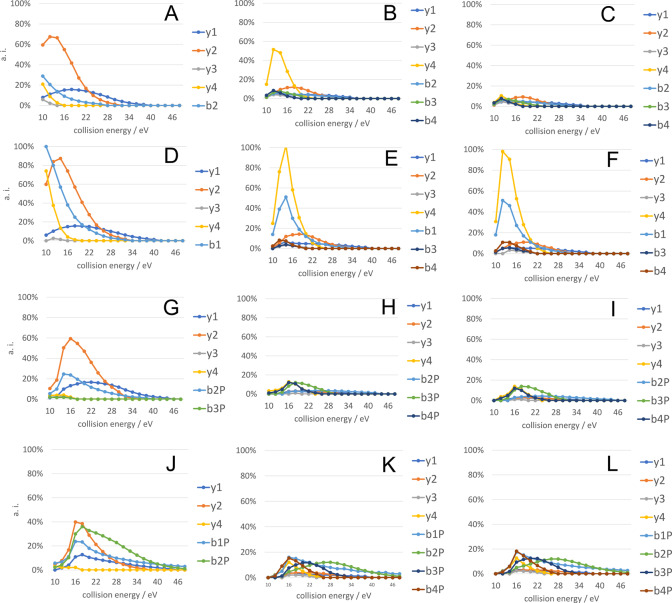
We previously demonstrated that the citrulline effect may substantially alter fragment ion distributions in CID.20,21 Therefore, in these experiments, we acquired tandem spectra at different collision energies to examine the effect of the various PTMs. Single stage MS data are summarized in Table 1. The results of collision energy dependence studies are summarized for doubly protonated precursor ions in Figure 1 and Supplementary Figures S1a–l in the case of all the 12 histone peptides. For ease of interpretation, only b and y type ions are depicted, other noncharacteristic fragments (peaks corresponding to an ammonia or water loss) are omitted. Singly and doubly protonated fragments of the same type are cumulated. Intact b ions could not be detected in the case of phosphopeptides, only fragments with a phosphoric acid loss. These are shown as biP in the figures.
Table 1. List of the Twelve Pentapeptide Sequences Originated from Human H4 Histone Protein.
| peptide sequence | measured [M + H]+ | calculated [M + H]+ | measured [M + 2H]2+ | calculated [M + 2H]2+ |
|---|---|---|---|---|
| SGRGK | 504.2889 | 504.2889 | 252.6480 | 252.6481 |
| Ac-SGRGK | 546.2997 | 546.2994 | 273.6530 | 273.6534 |
| pSGRGK | 584.2552 | 584.2552 | 292.6307 | 292.6312 |
| Ac-pSGRGK | 626.2653 | 626.2658 | 313.6362 | 313.6365 |
| SGR(Me)GK | 518.3042 | 518.3045 | 259.6557 | 259.6559 |
| Ac-SGR(Me)GK | 560.3147 | 560.3151 | 280.6608 | 280.6612 |
| pSGR(Me)GK | 598.2703 | 598.2709 | 299.6385 | 299.6391 |
| Ac-pSGR(Me)GK | 640.2808 | 640.2814 | 320.6437 | 320.6443 |
| SGXGK | 505.2723 | 505.2729 | 253.1394 | 253.1401 |
| Ac-SGXGK | 547.2831 | 547.2835 | 274.1447 | 274.1454 |
| pSGXGK | 585.2383 | 585.2392 | 293.1231 | 293.1232 |
| Ac-pSGXGK | 627.2493 | 627.2498 | 314.1282 | 314.1285 |
Figure 1.
HCD curves of appearance for the fragments originating from doubly protonated precursors of citrulline containing peptides (first column), NG-methylarginine containing peptides (second column) and arginine containing reference peptides (third column). SGZGK (A–C), Ac-SGZGK (D–F), pSGZGK (G–I), Ac-pSGZGK (J–L). Z = X (first column), Z = R(Me) (second column), and Z = R (third column). BiP fragments denote orthophosphoric acid loss from the corresponding bi ions.
Our results show that the y2 ion intensity is significantly higher for citrulline containing peptides compared to the arginine containing variants. This phenomenon can be explained by the citrulline effect. In addition, y2 intensity is usually higher than or as intense as that of other sequential fragments in its own spectrum. Conversely, we could not find any remarkable difference in the cleavage preference between NG-methylarginine and arginine containing histone peptides besides a more intense y4 ion for SGR(Me)GK vs SGRGK (Figure 1B–C).
For acetylated peptides, we observed b1 ions which are otherwise unstable if the N-terminus is unmodified. NG-Methylarginine containing peptides usually displayed at least two out of three previously described neutral losses14 at the same time, namely, the loss of methylamine (31.042 Da), N-methylguanidine (74.071 Da) and N-methylcarbodiimide (57.045). These could be used for confirmation of arginine methylation. By using these characteristic ions, false positive identification rates may also be reduced.
ETD Experiments
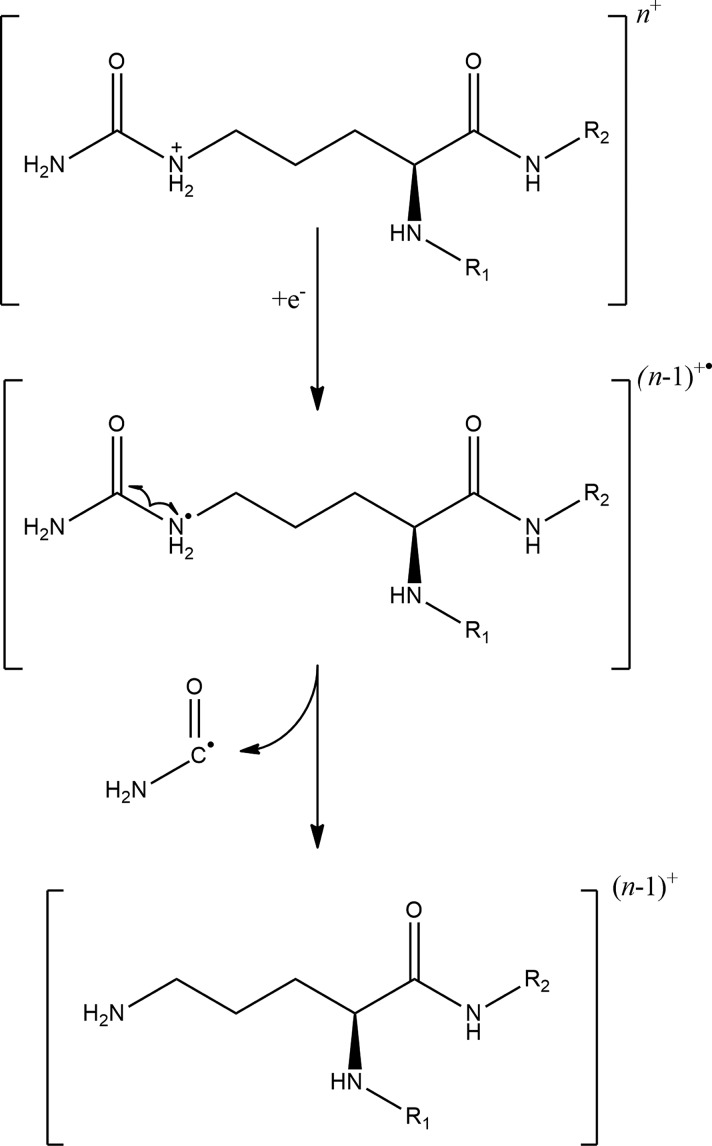
Electron-transfer dissociation is usually used for larger or highly charged peptides. For low charges, CID or HCD outperforms ETD in terms of sequence coverage.30 In our experiments, doubly charged ions were selected for fragmentation using ETD. In these cases, charge-reduced precursors could not be detected; their characteristic ammonia and other previously described losses could be observed. As it is expected, however, the most intense peaks in the ETD spectra correspond to precursor ions in the spectra due to the low efficiency of electron transfer to low-charged species. In these experiments, we mostly observed c and ż type fragments with a relatively lower occurrence of y and ȧ ions (Supplementary Figure S2(a–l)). The latter ones denote the radical type a ion that contains an extra hydrogen atom. Interestingly, we detected a neutral loss of 44.01 Da in the ETD MS/MS spectrum of the citrullinated SGXGK peptide possibly originating from the undetected charge-reduced precursor (Figure 2A). In the ETD spectrum of the unmodified SGRGK peptide, we obtained a similar loss that is 43.03 Da (Figure 2B). The difference between the two losses is the same as the increment for citrullinated versus unmodified arginine residues suggesting that these neutral losses can be associated with citrulline and arginine side chains. Arginine side chains may produce relatively strong fragment ion peaks corresponding to neutral losses in ETD. One of these possible reactions is the above-mentioned 43.03 Da loss which is suggested by Hunt et al.31 to be carbamimidoyl radical (NH=•C-NH2). We propose that the loss of 44.01 Da could be thus identified as the carbamoyl radical (O=•C-NH2) [Scheme 1]. Zolg et al. investigated the neutral losses characteristic to citrullination in HCD and ETD and found none for the latter technique.32 Our results show that the carbamoyl radical loss could, however, be selective for citrulline-containing peptides. The loss of CO2 which is frequently observed in ETD spectra has a very similar transition (43.990 Da). CO2 and carbamoyl loss differentiation therefore may be hard for higher charge-state precursors or instruments with lower resolution and mass accuracy. All citrulline-containing histone peptides in this study showed this fragmentation route as opposed to their arginine-containing variants. Therefore, we screened ETD spectra for additional selective losses and found that although being much less intense, the elimination of urea (H2N-CO-NH2) could also be indicative of citrullination which is analogous to the arginine-selective carbamidine loss (H2N–C(=•NH)-NH2) that was previously described only with a formula of CH5N3.33 However, urea loss is less frequently observed for citrullinated peptides. As expected, phosphate groups remained mostly intact in our ETD experiments. Almost all acetylated peptides displayed a relatively strong loss of 59.04 Da which could be attributed to the elimination of H3C–C(-OH)=•NH (ethanimidic acid) from the N-terminus (Figure 3). This could be elucidated by the original fragmentation mechanism proposed for electron-transfer dissociation by Syka et al.,3 if it is applied for the amide group of the acetylated N-terminus. A neutral loss with the same formula has already been reported for asparagine and glutamine side chains33 as well. NG-methylarginine residues also produced selective elimination products including CH3–NH2 (31.042 Da) or HN=•C-NH–CH3 (57.045 Da) (Figure 4),34 which are analogous to previously reported losses of NH3 (17.027 Da) and HN=•C-NH2 (43.030 Da) characteristic to arginine residues.
Figure 2.
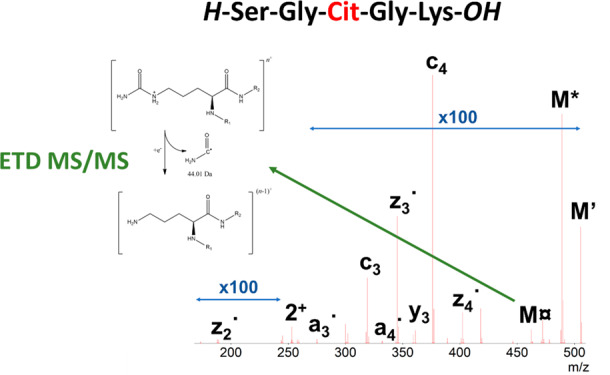
ETD tandem mass spectrum of doubly protonated SGXGK (A) and SGRGK (B). Precursor ions are denoted as “2+”, and the undetected charge-reduced precursors are only signed as M. Hydrogen atom loss is depicted as ’. Neutral losses of ammonia (*), CO (#), and the citrulline-selective carbamoyl radical (currency sign) and urea loss (u) are also shown for SGXGK along with arginine-selective carbamimidoyl radical (×) and carbamidine (Euro sign) for SGRGK. For ease of interpretation, precursor intensities are reduced to 1/100 of their original value.
Scheme 1. Suggested Mechanism for the Elimination of Carbamoyl Radical from Citrulline Side Chains in ETD Fragmentation.
Figure 3.
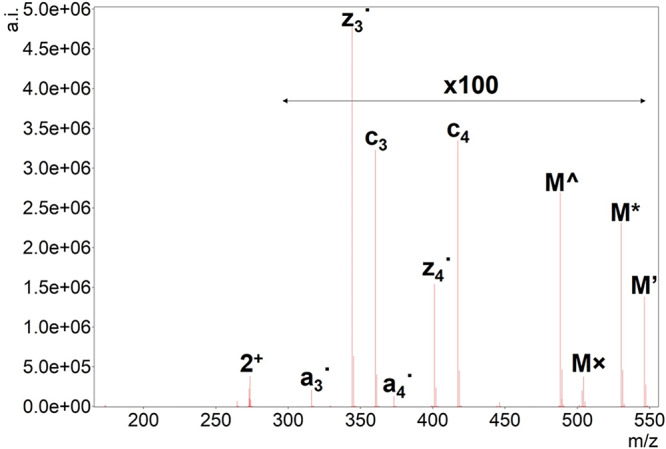
ETD tandem mass spectrum of doubly protonated Ac-SGRGK. Precursor ion is denoted as “2+”, the undetected charge-reduced precursor is only signed as M. Hydrogen atom loss is depicted as ’. Neutral losses of ammonia (*) and the arginine-selective carbamimidoyl radical (×) are also shown along with ethanimidic acid (^) loss characteristic to N-terminally acetylated peptides. For ease of interpretation, precursor intensity is reduced to 1/100 of its original value.
Figure 4.
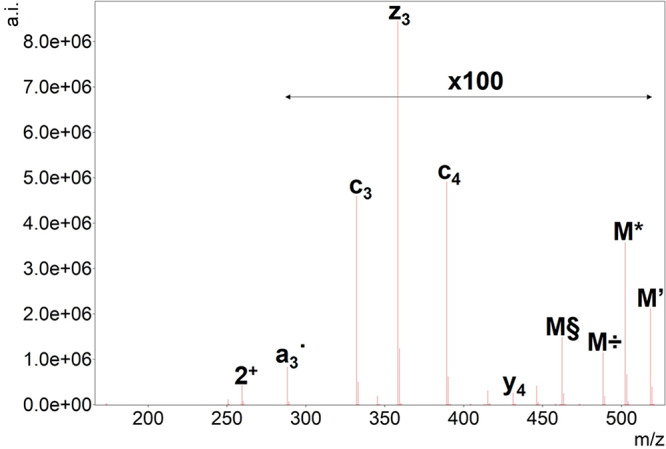
ETD tandem mass spectrum of doubly protonated SGR(Me)GK. Precursor ion is denoted as “2+”, and the undetected charge-reduced precursor is only signed as M. Hydrogen atom loss is depicted as ’. Neutral losses of ammonia (*) and the NG-methylarginine-selective methylamine (÷) and N-methylcarbodiimide losses (§) are also shown. For ease of interpretation, precursor intensity is reduced to 1/100 of its original value.
EThcD Experiments
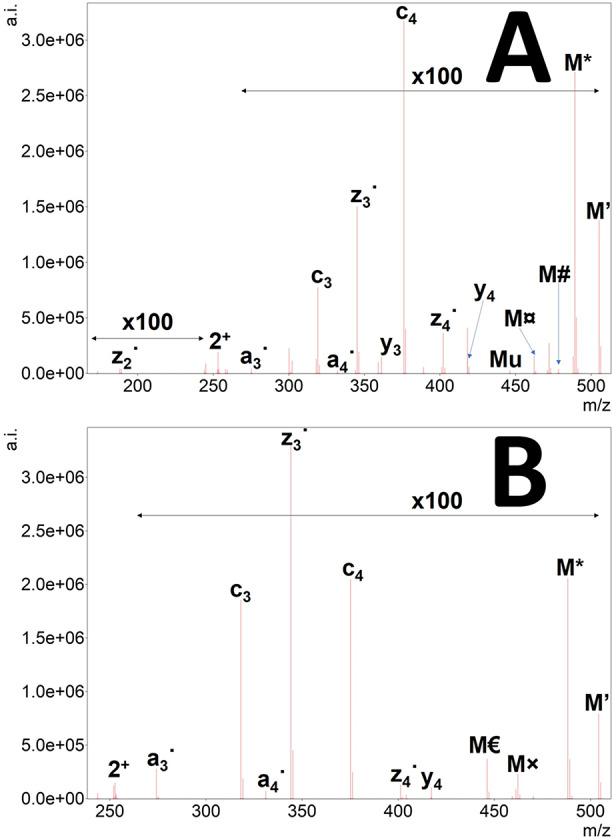
EThcD tandem mass spectra obtained for the histone H4 pentapeptides very much resemble HCD tandem mass spectra of singly charged precursors at high energies (Supplementary Figure S3(a–l)). In these cases, however, fragmentation efficiency seems much higher, and the whole m/z region is populated more evenly by fragment ions compared to HCD. Neutral losses are prevalent both for precursors and their fragments. Another feature is that the higher the proton mobility of a given precursor, the higher the similarity of a given EThcD spectrum to its HCD counterpart. On the other hand, the lower the proton mobility, the higher is the similarity of the given EThcD spectra to its ETD counterpart. In the case of EThcD spectra, however, differentiation between two fragments is often ambiguous. For example, the mass of a bi ion and a ci-NH3 are exactly the same, or the loss of a HNCO molecule from a [M + H]+ type precursor is the same as that of a O=•C-NH2 from a singly charged, charged-reduced precursor [M + 2H]+•. In general, several NH3 and even H2O losses could be observed for fragments of which the latter is less frequently seen in HCD generated spectra. The mass difference between NH3 and even H2O losses is 0.9840 Da, which is identical to the mass difference for deamidation and also citrullination. As examples, EThcD spectra of the doubly charged precursor of SGXGK and SGRGK peptides at 30% NCE are depicted in Figure 5. As NCE only compensates for mass difference and charge state differences, the lower efficacy of SGRGK fragmentation may be attributed to superior basicity of arginine residues, thus decreasing proton mobility.35 The citrulline effect is very pronounced as the peak corresponding to the y2 ion is the second highest in the spectrum of the citrullinated SGXGK peptide. In the upper m/z region, the ETD-selective losses could also be identified from the charge-reduced precursor (which is in itself not detectable) including NH3, carbamoyl radical, and urea along with a hydroxyl radical loss possibly from the serine side chain. Some of these losses could also be originated from the [M + H]+, which is formed by hydrogen atom abstraction from the charge-reduced precursor ([M + 2H]+˙). The presence of both processes could be inferred from the unusual “isotope” distributions (e.g., the peak of NH3 loss, see Figure S4). Another complicating feature in the spectra worth mentioning is the proper identification of z and ż ions which only differ by an H• radical (1.0078). Differentiation of hydrogen abstraction and isotope peaks may be hard. Very high resolution, sophisticated software annotation and high mass accuracy are needed, especially when low-mass posttranslational modifications (citrullination, deamidation) are also present. Usually, all fragments bearing citrulline residues display the loss of HNCO which is rarely seen in simple HCD experiments (Figure S5). This wealth of fragments may also be a disadvantage as it may increase the number of false positive identifications. Therefore, great care must be taken, and manual inspection is highly recommended during evaluation when fragments other than the intact fragment ions (b, y, c) are considered. However, the selective losses in the higher m/z region that can also be found in ETD spectra are reliable—at least in cases where only either citrulline or arginine is in the peptide sequence. For peptides containing one or more arginine and citrulline, the difference between the above-mentioned characteristic peaks is 0.9840 Da, which may also indicate the presence of citrullination beside the HNCO/carbamoyl radical loss.
Figure 5.
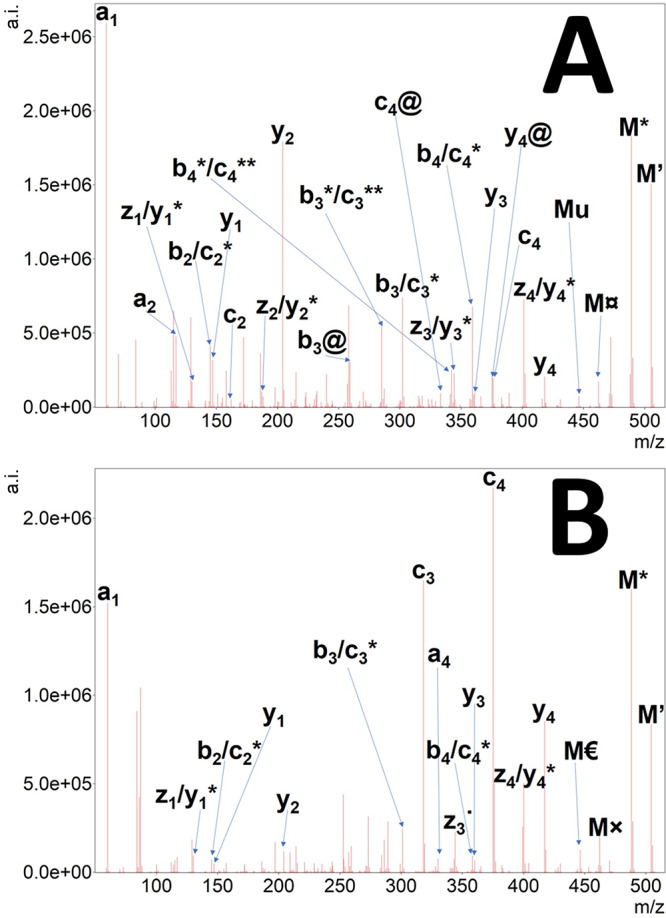
EThcD tandem mass spectrum of doubly protonated SGXGK (A) and SGRGK (B) histone peptides. Precursor ion is denoted as “2+”, the undetected charge-reduced precursor is only signed as M. Hydrogen atom loss is depicted as ’. Neutral losses of ammonia (*), water (deg), the citrulline-selective carbamoyl radical (currency sign) or isocyanic acid (@) and urea (u), and the arginine-selective carbamimidoyl radical (×) and carbamidine (Euro sign) eliminations are also shown.
In the case of Ac-SGXGK peptide, the highest sequential peak was attributed to the citrulline effect, while the second one—comparable to this—was the y4 ion (Figure 6). Cleavage preference at the N-terminus of glycine residues in mobile and partially mobile sequences has been reported earlier for CID spectra.36 Characteristic, modification-selective neutral losses in the upper m/z region are present indicating citrullination and acetylation in this histone peptide. The cleavage preference C-terminal to citrulline is also pronounced for the pSGXGK peptide containing a phosphorylated serine residue, while y2 intensity is much lower for pSGR(Me)GK (Figure 7). “False HNCO losses” from fragments still appeared for peptides that do not comprise citrulline residues (see Figure S6). The peaks corresponding to these fragments were mostly associated with b3 peaks of methyl-arginine and arginine-containing peptides. We also found a false positive b2-HNCO peak for the peptide Ac-SGXGK but none for other Cit-peptides. On the other hand, HNCO loss from precursor ions was only observed for citrulline-containing peptides being faithful reporters of this modification. ETD is often mentioned as a truly complementary technique of HCD. Even in the case of EThcD, it can be seen that a cleavage C-terminal to arginine or glycine—which are normally disfavored upon HCD—is prevalent for Arg-containing peptides (e.g., pSGRGK, Ac-SGRGK). In the case of Cit-containing peptides, EThcD fragmentation was still highly influenced by the citrulline effect. The MS/MS spectra of Ac-pSGXGK contains mainly iminium ions, suggesting much greater proton mobility. Yet, the citrulline effect and characteristic neutral losses are substantial in this case as well.
Figure 6.
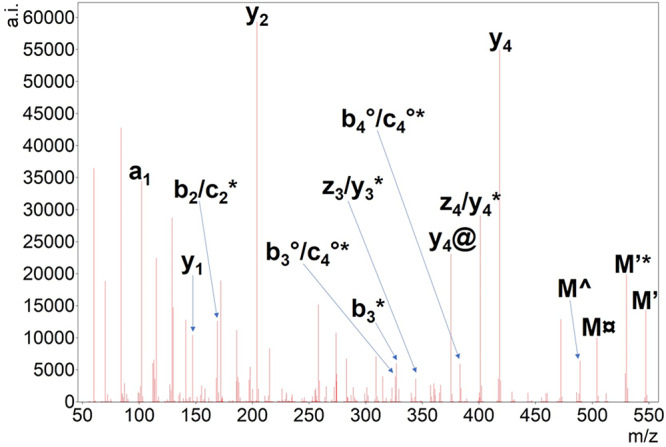
EThcD tandem mass spectrum of doubly protonated Ac-SGXGK histone peptide. The undetected charge-reduced precursor is only signed as M. Hydrogen atom loss is depicted as ’. Neutral losses of ammonia (*), water (deg), the citrulline-selective carbamoyl radical (currency sign) or isocyanic acid (@) are also shown. Note that the spectrum is very similar to an HCD one; radical losses could be explained by even-electron fragmentation from M’ as well.
Figure 7.
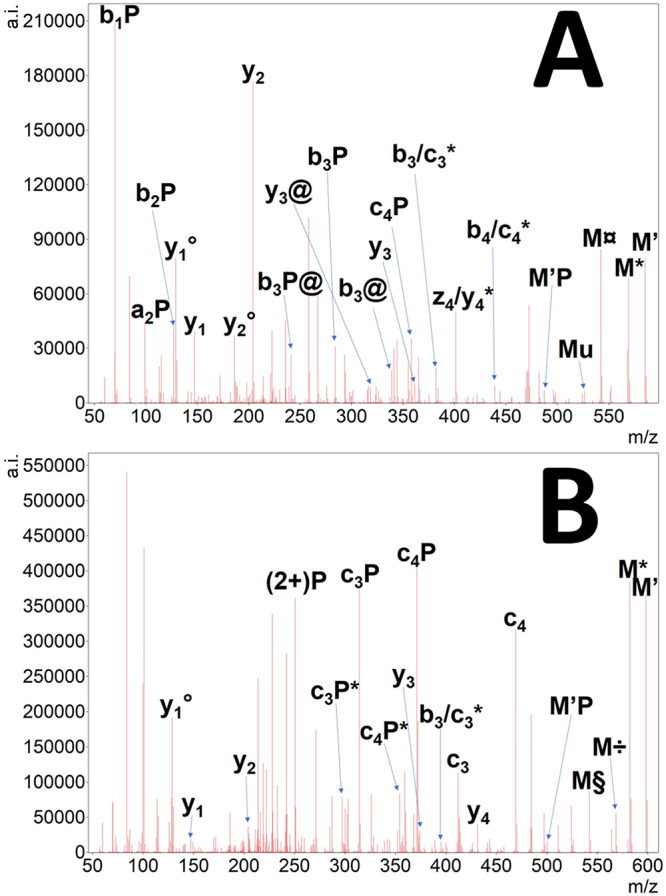
EThcD tandem mass spectrum of doubly protonated pSGXGK (A) and pSGR(Me)GK (B) histone peptides. The undetected charge-reduced precursor is only signed as M. Hydrogen atom loss is depicted as ’. Neutral losses of ammonia (*), water (deg), the citrulline-selective carbamoyl radical (currency sign), or isocyanic acid (@) as well as the NG-methylarginine-selective methylamine (÷) and N-methylcarbodiimide losses (§) are also shown. Note that all bi related fragments may represent ci* ones, and all zi related fragments may represent yi* as well.
Discussion
In this study, the effect of the presence of multiple posttranslational modifications (N-terminal acetylation, serine phosphorylation, and arginine citrullination/methylation) on the tandem mass spectra of human H4 histone peptides was examined by different activation methods including higher-energy collisional dissociation (HCD) with collision energy dependence, electron-transfer dissociation (ETD), and electron-transfer higher-energy collisional dissociation (EThcD).
In HCD spectra, the citrulline effect—i.e., the cleavage preference at the C-terminus of citrulline residues—was observed for Cit-peptides in a relatively wide collision energy range. Compared to the arginine-containing reference peptides, peaks corresponding to these fragments were significantly more intense. Isocyanic acid (HNCO) loss was also prominent from precursors (usually 10–20%, but up to 100% depending on proton mobility), but sometimes less prevalent and reliable for fragments (usually 1–30%).
ETD spectra of these doubly charged peptides provided some unexpected and characteristic high m/z peaks corresponding to radical losses from the charge-reduced precursors which could be utilized to confirm citrullination. The loss of a carbamoyl radical (•CO-NH2) from citrulline side chains—analogous to carbamimidoyl radical (HN=•C-NH2) elimination of arginine proposed by Hunt et al.—is described here for the first time along with carbamide loss. These products may not be intense when the charge state of the precursor is higher and may be suppressed by arginine side chain losses if arginine is also present in the sequence but are useful for confirmation of citrullination for lower-charged species. We also found the side chain losses associated with Arg-methylation (HN=•C-NH–CH3 and CH3–NH2).36 Acetylation could also be easily identified as the loss of C2H5NO (more precisely H3C–C(-OH)=NH in this case) can only originate from acetylation in the absence of Asn and Gln residues. Being a backbone cleavage, this peak usually tends to be rather intense compared to other neutral losses.
EThcD spectra of these histone peptides yielded the most numerous fragments for citrullinated peptides including the (•CO-NH2)/HNCO losses from (charge-reduced) precursors, HNCO losses from almost all fragments that bears a citrulline residue and citrulline effect in the case of Cit-peptides. Methylamine and N-methylcarbodiimide losses were also prevalent from the precursors of Arg(Me) containing peptides. Phosphorylated and acetylated peptides displayed their characteristic elimination reactions, too. These neutral loss intensities were very variable being usually ∼1–20% but orthophosphoric acid loss in some cases reached ∼70–80%. EThcD not only improved neutral losses’ yield for all types of PTMs but sequential fragment yields as well. Some fragments could not be differentiated from each other in EThcD, but most of these products refer to the same sequential information, potentially improving identification. However, the overlapping of isotope peaks due to the immense amount of neutral losses may pose a concern for a software-based annotation. The technique has its own pitfalls as well due to the high probability of generating combined losses having the same formulas but inferring to structurally different ions. If this is the case for such simple systems, fragmenting larger sized peptides would increase spectral complexity and may lead to erroneous assignments especially for quantitative studies. Thus, while EThcD was found to be beneficial for the localization of larger and labile modifications4 (e.g., glycosylation and phosphorylation), care must be taken when applied to small modifications (e.g., citrullination) and large precursor charge states.
Nevertheless, we suggest using EThcD for analyzing these PTMs for low mass peptides by restricting the automatic annotation to b, y, and c sequential ions and the above-mentioned characteristic neutral losses from precursors only, followed by manual inspection of the remaining product ion neutral losses, if necessary. In this case, a complete sequence coverage as well as exact PTM site determination becomes possible without significantly increasing false identifications. Another option is to use HCD for confirmation of the modifications and subsequent ETD fragmentation for unambiguous sequence information as was used previously for citrullinated peptides.37
Acknowledgments
The research was supported by the MTA Premium Post-Doctorate Research Program of the Hungarian Academy of Sciences (HAS, MTA) and GINOP-2.3.3-15-2016-00020. This work was completed in the ELTE Institutional Excellence Program (783-3/2018/FEKUTSRAT) supported by the Hungarian Ministry of Human Capacities. The research within Project No. VEKOP-2.3.3-15-2017-00020 was supported by the European Union and the State of Hungary, cofinanced by the European Regional Development Fund. Project No. 2018-1.2.1-NKP-2018-00005 has been implemented with the support provided from the National Research, Development and Innovation Fund of Hungary, financed under the 2018-1.2.1-NKP funding scheme.
Glossary
Abbreviations
- B
one-letter code for an amino acid if not stated otherwise
- CID
collision-induced dissociation
- Cit
three-letter abbreviation for citrulline
- ETD
electron-transfer dissociation
- EThcD
electron-transfer higher energy collisional dissociation
- HCD
higher energy collisional dissociation
- MS/MS
tandem mass spectrometry
- NCE
normalized collision energy
- pS
phosphorylated serine residue
- PTM
posttranslational modification
- X
one-letter abbreviation for citrulline (in the absence of IUPAC recommendation)
Supporting Information Available
The Supporting Information is available free of charge at https://pubs.acs.org/doi/10.1021/jasms.9b00036.
Figure S1: Fragment distribution of the peptides (a) SGXGK, (b) SGR(Me)GK, (c) SGRGK, (d) Ac-SGXGK, (e) Ac-SGR(Me)GK, (f) Ac-SGRGK, (g) pSGXGK, (h) pSGR(Me)GK, (i) pSGRGK, (j) Ac-pSGXGK, (k) Ac-pSGR(Me)GK and (l) Ac-pSGRGK upon higher-energy collision-induced dissociation (HCD) as a function of collision energy. Figure S2: ETD tandem mass spectra of peptides (a) SGRGK, (b) Ac-SGRGK, (c) pSGRGK, (d) Ac-pSGRGK, (e) SGR(Me)GK, (f) Ac-SGR(Me)GK, (g) pSGR(Me)GK, (h) Ac-pSGR(Me)GK, (i) SGXGK, (j) Ac-SGXGK, (k) pSGXGK, and (l) Ac-pSGXGK. Figure S3: EThcD tandem mass spectra of peptides (a) SGRGK, (b) Ac-SGRGK, (c) pSGRGK, (d) Ac-pSGRGK, (e) SGR(Me)GK, (f) Ac-SGR(Me)GK, (g) pSGR(Me)GK, (h) Ac-pSGR(Me)GK, (i) SGXGK, (j) Ac-SGXGK, (k) pSGXGK, and (l) Ac-pSGXGK. Figure S4: Representation of “unusual isotope distributions” in the EThcD spectrum of the peptide pSGXGK. Figure S5: Representation of isocyanic acid loss (@) from all fragments that contain the citrulline residue in an EThcD tandem mass spectrum. Figure S6: Representation of a false neutral loss of isocyanic acid (@) for the peptide pSGR(Me)GK which lacks a citrulline residue (PDF)
The authors declare no competing financial interest.
Supplementary Material
References
- Wells J. M.; McLuckey S. A. Collision-Induced Dissociation (CID) of Peptides and Proteins. Methods Enzymol. 2005, 402, 148–185. 10.1016/S0076-6879(05)02005-7. [DOI] [PubMed] [Google Scholar]
- Biemann K. Contributions of mass spectrometry to peptide and protein structure. Biol. Mass Spectrom. 1988, 16, 99–111. 10.1002/bms.1200160119. [DOI] [PubMed] [Google Scholar]
- Syka J. E. P.; Coon J. J.; Schroeder M. J.; Shabanowitz J.; Hunt D. F. Peptide and protein sequence analysis by electron transfer dissociation mass spectrometry. Proc. Natl. Acad. Sci. U. S. A. 2004, 101, 9528–9533. 10.1073/pnas.0402700101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frese C. K.; Zhou H.; Taus T.; Altelaar A. F. M.; Mechtler K.; Heck A. J. R.; Mohammed S. Unambiguous Phosphosite Localization using Electron-Transfer/Higher-Energy Collision Dissociation (EThcD). J. Proteome Res. 2013, 12, 1520–1525. 10.1021/pr301130k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glover M. S.; Yu Q.; Chen Z.; Shi X.; Kent K. C.; Li L. Characterization of intact sialylated glycopeptides and phosphorylated glycopeptides from IMAC enriched samples by EThcD fragmentation: Toward combining phosphoproteomics and glycoproteomics. Int. J. Mass Spectrom. 2018, 427, 35–42. 10.1016/j.ijms.2017.09.002. [DOI] [Google Scholar]
- Carlberg C.; Molnár F.. Human Epigenomics, 1st ed.; Springer Nature Pte Ltd., 2018. [Google Scholar]
- Song O.; Wang X.; Waterborg J. H.; Sternglanz R. An Nα-Acetyltransferase Responsible for Acetylation of the N-terminal Residues of Histones H4 and H2A. J. Biol. Chem. 2003, 278, 38109–38112. 10.1074/jbc.C300355200. [DOI] [PubMed] [Google Scholar]
- Grunstein M. Histone acetylation in chromatin structure and transcription. Nature 1997, 389, 349–352. 10.1038/38664. [DOI] [PubMed] [Google Scholar]
- Wang H.; Huang Z.-Q.; Xia L.; Feng Q.; Erdjument-Bromage H.; Strahl B. D.; Briggs S. D.; Allis C. D.; Wong J.; Tempst P.; Zhang Y. Methylation of Histone H4 at Arginine 3 Facilitating Transcriptional Activation by Nuclear Hormone Receptor. Science 2001, 293, 853–857. 10.1126/science.1060781. [DOI] [PubMed] [Google Scholar]
- Martin C.; Zhang Y. The diverse functions of histone lysine methylation. Nat. Rev. Mol. Cell Biol. 2005, 6, 838–849. 10.1038/nrm1761. [DOI] [PubMed] [Google Scholar]
- Barber C. M.; Turner F. B.; Wang Y.; Hagstrom K.; Taverna S. D.; Mollah S.; Ueberheide B.; Meyer B. J.; Hunt D. F.; Cheung P.; Allis C. D. The enhancement of histone H4 and H2A serine 1 phosphorylation during mitosis and S-phase is evolutionarily conserved. Chromosoma 2004, 112, 360–371. 10.1007/s00412-004-0281-9. [DOI] [PubMed] [Google Scholar]
- Fuhrmann J.; Thompson P. R. Protein Arginine Methylation and Citrullination in Epigenetic Regulation. ACS Chem. Biol. 2016, 11, 654–668. 10.1021/acschembio.5b00942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yalcin T.; Khouw C.; Csizmadia I. G.; Peterson M. R.; Harrison A. G. Why Are B ions Stable Species in Peptide Spectra?. J. Am. Soc. Mass Spectrom. 1995, 6, 1165–1174. 10.1016/1044-0305(95)00569-2. [DOI] [PubMed] [Google Scholar]
- Gehrig P. M.; Hunziker P. E.; Zahariev S.; Pongor S. Fragmentation pathways of NG-methylated and unmodified arginine residues in peptides studied by ESI-MS/MS and MALDI-MS. J. Am. Soc. Mass Spectrom. 2004, 15, 142–149. 10.1016/j.jasms.2003.10.002. [DOI] [PubMed] [Google Scholar]
- Palumbo A. M.; Reid G. E. Evaluation of Gas-Phase Rearrangement and Competing Fragmentation Reactions on Protein Phosphorylation Site Assignment Using Collision Induced Dissociation-MS/MS and MS3. Anal. Chem. 2008, 80, 9735–9747. 10.1021/ac801768s. [DOI] [PubMed] [Google Scholar]
- Bailey L. S.; Alves M.; Galy N.; Patrick A. L.; Polfer N. C. Mechanistic insights into intramolecular phosphate group transfer during collision induced dissociation of phosphopeptides. J. Mass Spectrom. 2019, 54, 449–458. 10.1002/jms.4351. [DOI] [PubMed] [Google Scholar]
- Chi A.; Huttenhower C.; Geer L. Y.; Coon J. J.; Syka J. E. P.; Bai D. L.; Shabanowitz J.; Burke D. J.; Troyanskaya O. G.; Hunt D. F. Analysis of phosphorylation sites on proteins from Saccharomyces cerevisiae by electron transfer dissociation (ETD) mass spectrometry. Proc. Natl. Acad. Sci. U. S. A. 2007, 104, 2193–2198. 10.1073/pnas.0607084104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hao G.; Wang D.; Gu J.; Shen Q.; Gross S. S.; Wang Y. Neutral Loss of Isocyanic Acid in Peptide CID Spectra: A Novel Diagnostic Marker for Mass Spectrometric Identification of Protein Citrullination. J. Am. Soc. Mass Spectrom. 2009, 20, 723–727. 10.1016/j.jasms.2008.12.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee C.-Y.; Wang D.; Wilhelm M.; Zolg D. P.; Schmidt T.; Schnatbaum K.; Reimer U.; Pontén F.; Uhlén M.; Hahne H.; Kuster B. Mining the Human Tissue Proteome for Protein Citrullination. Mol. Cell. Proteomics 2018, 17, 1378–1391. 10.1074/mcp.RA118.000696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steckel A.; Uray K.; Turiák L.; Gömöry Á.; Drahos L.; Hudecz F.; Schlosser G. Mapping the tandem mass spectrometric characteristics of citrulline-containing peptides. Rapid Commun. Mass Spectrom. 2018, 32, 844–850. 10.1002/rcm.8105. [DOI] [PubMed] [Google Scholar]
- Steckel A.; Schlosser G. Citrulline effect is a characteristic feature of deiminated peptides in tandem mass spectrometry. J. Am. Soc. Mass Spectrom. 2019, 30, 1586–1591. 10.1007/s13361-019-02271-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young N. L.; DiMaggio P. A.; Plazas-Mayorca M. D.; Baliban R. C.; Floudas C. A.; Garcia B. A. High Throughput Characterization of Combinatorial Histone Codes. Mol. Cell. Proteomics 2009, 8, 2266–2284. 10.1074/mcp.M900238-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plazas-Mayorca M. D.; Zee B. M.; Young N. L.; Fingerman I. M.; LeRoy G.; Briggs S. D.; Garcia B. A. One-Pot Shotgun Quantitative Mass Spectrometry Characterization of Histones. J. Proteome Res. 2009, 8, 5367–5374. 10.1021/pr900777e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jung H. R.; Sidoli S.; Haldbo S.; Sprenger R. R.; Schwammle V.; Pasini D.; Helin K.; Jensen O. N. Precision mapping of coexisting modifications in histone H3 tails from embryonic stem cells by ETD-MS/MS. Anal. Chem. 2013, 85, 8232–8239. 10.1021/ac401299w. [DOI] [PubMed] [Google Scholar]
- Jiang T.; Hoover M. E.; Holt M. V.; Freitas M. A.; Marshall A. G.; Young N. L. Middle-Down Characterization of the Cell Cycle Dependence of Histone H4 Posttranslational Modifications and Proteoforms. Proteomics 2018, 18, 1700442 10.1002/pmic.201700442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pesavento J. J.; Bullock C. R.; LeDuc R. D.; Mizzen C. A.; Kelleher N. L. Combinatorial Modification of Human Histone H4 Quantitated by Two-dimensional Liquid Chromatography Coupled with Top Down Mass Spectrometry. J. Biol. Chem. 2008, 283, 14927–14937. 10.1074/jbc.M709796200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dang X.; Scotcher J.; Wu S.; Chu R. K.; Tolić N.; Ntai I.; Thomas P. M.; Fellers R. T.; Early B. P.; Zheng Y.; Durbin K. R.; Leduc R. D.; Wolff J. J.; Thompson C. J.; Pan J.; Han J.; Shaw J. B.; Salisbury J. P.; Easterling M.; Borchers C. H.; Brodbelt J. S.; Agar J. N.; Paša-Tolić L.; Kelleher N. L.; Young N. L. The first pilot project of the consortium for top-down proteomics: a status report. Proteomics 2014, 14, 1130–40. 10.1002/pmic.201300438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holt M. V.; Wang T.; Young N. L. One-Pot Quantitative Top- and Middle-Down Analysis of GluC-Digested Histone H4. J. Am. Soc. Mass Spectrom. 2019, 30, 2514. 10.1007/s13361-019-02219-1. [DOI] [PubMed] [Google Scholar]
- Strohalm M.; Hassman M.; Košata B.; Kodíček M. mMass data miner: an open source alternative for mass spectrometric data analysis. Rapid Commun. Mass Spectrom. 2008, 22, 905–908. 10.1002/rcm.3444. [DOI] [PubMed] [Google Scholar]
- Good D. M.; Wirtala M.; McAlister G. C.; Coon J. J. Performance Characteristics of Electron Transfer Dissociation Mass Spectrometry. Mol. Cell. Proteomics 2007, 6, 1942–1951. 10.1074/mcp.M700073-MCP200. [DOI] [PubMed] [Google Scholar]
- Hunt D. F.; Shabanowitz J.; Bai D. L. Peptide Sequence Analysis by Electron Transfer Dissociation Mass Spectrometry: A Web-Based Tutorial. J. Am. Soc. Mass Spectrom. 2015, 26, 1256–1258. 10.1007/s13361-015-1105-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zolg D. P.; Wilhelm M.; Schmidt T.; Médard G.; Zerweck J.; Knaute T.; Wenschuh H.; Reimer U.; Schnatbaum K.; Kuster B. ProteomeTools: Systematic Characterization of 21 Post-translational Protein Modifications by Liquid Chromatography Tandem Mass Spectrometry (LC-MS/MS) Using Synthetic Peptides. Mol. Cell. Proteomics 2018, 17, 1850–1863. 10.1074/mcp.TIR118.000783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xia Q.; Lee M. V.; Rose C. M.; Marsh A. J.; Hubler S. L.; Wenger C. D.; Coon J. J. Characterization and Diagnostic Value of Amino Acid Side Chain Neutral Losses Following Electron-Transfer Dissociation. J. Am. Soc. Mass Spectrom. 2011, 22, 255–264. 10.1007/s13361-010-0029-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snijders A. P. L.; Hung M.-L.; Wilson S. A.; Dickman M. J. Analysis of Arginine and Lysine Methylation Utilizing Peptide Separations at Neutral pH and Electron Transfer Dissociation Mass Spectrometry. J. Am. Soc. Mass Spectrom. 2010, 21, 88–96. 10.1016/j.jasms.2009.09.010. [DOI] [PubMed] [Google Scholar]
- Wysocki V. H.; Tsaprailis G.; Smith L. L.; Breci L. A. Mobile and localized protons: a framework for understanding peptide dissociation. J. Mass Spectrom. 2000, 35, 1399–1406. . [DOI] [PubMed] [Google Scholar]
- Tabb D. L.; Smith L. L.; Breci L. A.; Wysocki V. H.; Lin D.; Yates J. R. Statistical Characterization of Ion Trap Tandem Mass Spectra from Doubly Charged Tryptic Peptides. Anal. Chem. 2003, 75, 1155–1163. 10.1021/ac026122m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Creese A. J.; Grant M. M.; Chapple I. L. C.; Cooper H. J. On-line liquid chromatography neutral loss-triggered electron transfer dissociation mass spectrometry for the targeted analysis of citrullinated peptides. Anal. Methods 2011, 3, 259–266. 10.1039/C0AY00414F. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.