Acinetobacter baumannii is a nosocomial pathogen capable of causing a range of diseases, including respiratory and urinary tract infections and bacteremia. Treatment options are limited due to the increasing rates of antibiotic resistance, underscoring the importance of identifying new targets for antimicrobial development. During infection, A. baumannii must acquire nutrients for replication and survival. These nutrients include carbon- and nitrogen-rich molecules that are needed for bacterial growth.
KEYWORDS: Acinetobacter, histidine, infection, pneumonia
ABSTRACT
Acinetobacter baumannii is a nosocomial pathogen capable of causing a range of diseases, including respiratory and urinary tract infections and bacteremia. Treatment options are limited due to the increasing rates of antibiotic resistance, underscoring the importance of identifying new targets for antimicrobial development. During infection, A. baumannii must acquire nutrients for replication and survival. These nutrients include carbon- and nitrogen-rich molecules that are needed for bacterial growth. One possible nutrient source within the host is amino acids, which can be utilized for protein synthesis or energy generation. Of these, the amino acid histidine is among the most energetically expensive for bacteria to synthesize; therefore, scavenging histidine from the environment is likely advantageous. We previously identified the A. baumannii histidine utilization (Hut) system as being linked to nutrient zinc homeostasis, but whether the Hut system is important for histidine-dependent energy generation or vertebrate colonization is unknown. Here, we demonstrate that the Hut system is conserved among pathogenic Acinetobacter and regulated by the transcriptional repressor HutC. In addition, the Hut system is required for energy generation using histidine as a carbon and nitrogen source. Histidine was also detected extracellularly in the murine lung, demonstrating that it is bioavailable during infection. Finally, the ammonia-releasing enzyme HutH is required for acquiring nitrogen from histidine in vitro, and strains inactivated for hutH are severely attenuated in a murine model of pneumonia. These results suggest that bioavailable histidine in the lung promotes Acinetobacter pathogenesis and that histidine serves as a crucial nitrogen source during infection.
INTRODUCTION
Amino acids are a fundamental component of life and, as such, their concentrations in biological systems are tightly regulated. While humans require several amino acids from dietary sources, many bacterial species are capable of fully synthesizing all 20 genetically encoded amino acids. Among these, histidine is the fourth most energetically expensive amino acid to synthesize (1). Histidine is derived from 5′-phosphoribosyl-1-pyrophosphate (PRPP), which also serves as a precursor for other critical pathways, including purine biosynthesis (2). However, when histidine is in excess, many bacteria possess mechanisms to catabolize this amino acid for energy generation via the histidine utilization (Hut) system. While some bacteria are capable of histidine carbon source utilization, this is not universal in all bacteria, with Escherichia coli being a notable model organism unable to catabolize histidine (3). The core Hut enzymes, which are comprised of HutH, HutU, HutI, and HutG, are conserved across domains and result in the production of ammonia, glutamate, and the one-carbon compound formate or formamide depending on the organism (3). Fundamental biological processes have been uncovered through investigations into the Hut system, including the discovery of catabolite repression and the identification of the master regulator CodY in Bacillus subtilis (4, 5). Expression of the Hut system is controlled through a variety of transcriptional regulatory systems, with the HutC repressor serving as one of the main regulators (6–8). Despite the widespread occurrence of the Hut system in bacteria, the contribution of histidine catabolism and the Hut system to bacterial pathogenesis is largely unknown (3).
One organism that encodes a Hut system is the opportunistic Gram-negative bacterial pathogen Acinetobacter baumannii (9). A. baumannii is a frequent cause of nosocomial infections, including wound and burn infections, urinary tract infections, and ventilator-associated pneumonia (10). The ability of A. baumannii to cause disease is at least partly driven by its metabolic and genetic plasticity (11). These features have led to the emergence of multidrug-resistant A. baumannii infections and prompted the World Health Organization (12) and the Centers for Disease Control and Prevention (13) to list A. baumannii as a top threat for which new therapeutics are needed (12, 13). Importantly, during A. baumannii infections, nutrients must come from the host, but nutritional sources within vertebrates are not well defined and likely complex (11).
We previously demonstrated that histidine is capable of forming complexes with the nutrient metal zinc (9). HutH is the first enzyme in the Hut system, and in A. baumannii HutH liberates zinc from zinc-histidine complexes through the catabolism of histidine to urocanate and ammonia (9, 14). However, HutH is also required for growth on histidine as the sole carbon source, which suggests this enzyme and the Hut system may perform broader physiological roles in A. baumannii. Here, we identified histidine and other amino acids within the extracellular space of the lungs, indicating that amino acids are bioavailable to A. baumannii within the vertebrate host. We dissected aspects of the A. baumannii Hut system and discovered that the system is present in other pathogenic Acinetobacter but absent in several environmental Acinetobacter species. In addition, we defined HutC as a transcriptional repressor of A. baumannii hut and showed that HutH serves as an important factor for acquiring nitrogen from histidine. Finally, we determined that HutH is critical for A. baumannii lung colonization and dissemination to the liver, which suggests that histidine serves as an important nitrogen source during A. baumannii pneumonia.
RESULTS
Histidine is bioavailable to A. baumannii in the vertebrate lung.
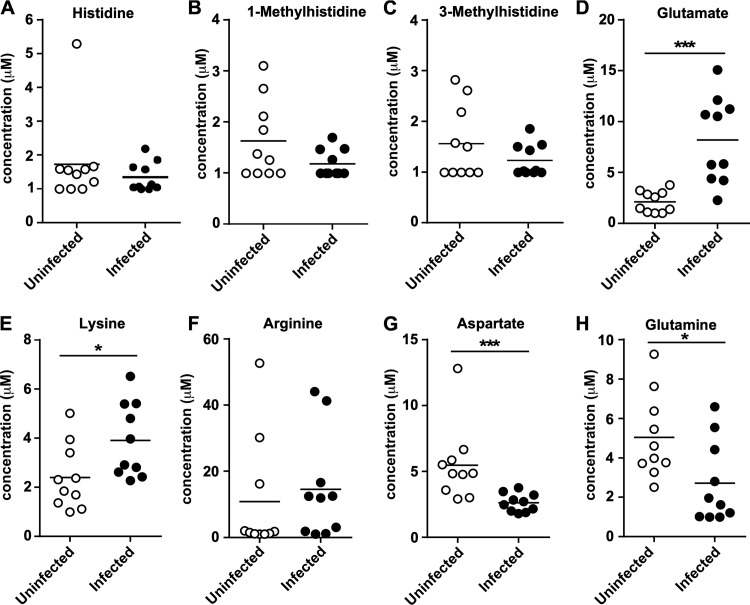
A. baumannii is a leading cause of ventilator-associated pneumonia, which implies that nutrient availability within the lung is sufficient to sustain A. baumannii during infection. Given that A. baumannii can utilize histidine as a carbon source (9), we hypothesized that histidine is bioavailable during infection. To determine whether histidine or other amino acids are available to A. baumannii during lung colonization, mice were intranasally inoculated with A. baumannii. At 36 h postinfection (hpi), bronchial alveolar lavage fluid (BALF) was collected from infected and uninfected mice. Immune cells were removed, and the samples were sterile filtered before being subjected to metabolite analysis to quantify amino acid and metabolite concentrations. This analysis revealed that histidine and its modified forms 1-methylhistidine and 3-methylhistidine, which are weak inducers of the Hut system in Klebsiella spp. (15), are detectable in both infected and uninfected BALF (Fig. 1A to C). In addition, glutamate, which is the end product of the Hut system, was significantly elevated in the BALF during infection (Fig. 1D). Aside from histidine, the other positively charged amino acids lysine and arginine were detectable within the BALF, with lysine levels being elevated during infection. Aspartate and glutamine were also detected, but both were significantly lower during infection (Fig. 1G and H). A variety of other metabolites and amino acids were detected (see Fig. S1 in the supplemental material), such as ethanolamine, which was significantly reduced with infection (Fig. S1B), and taurine, which was elevated in the BALF during infection (Fig. S1C). Taken together, these results establish histidine and other amino acids as being detectable within the extracellular space of the lungs during infection.
FIG 1.
Histidine is bioavailable within the murine lung. (A to H) BALF was collected at 36 hpi from mice and compared to uninfected counterparts after metabolite analysis for the indicated molecule. *, P < 0.05; ***, P < 0.001 (as determined by Mann-Whitney U test; n = 10 mice per group).
The Hut system is conserved in pathogenic Acinetobacter.
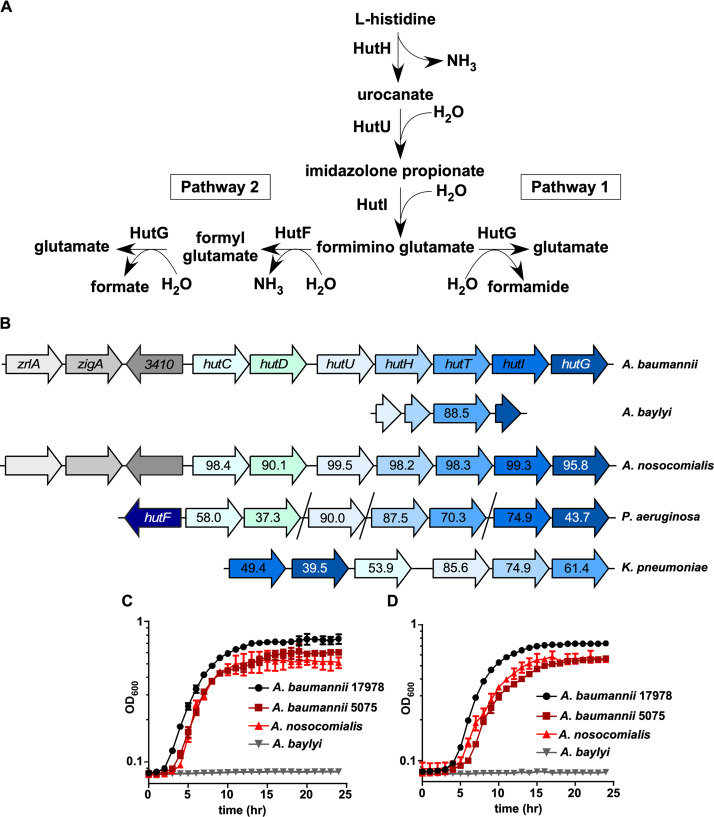
Since histidine was detectable within the BALF of mice, we investigated how the Hut system is organized in A. baumannii. In general, the Hut system is comprised of four core enzymes, HutHUIG, that facilitate the conversion of histidine to glutamate that can be further modified to generate α-ketoglutarate and enter the tricarboxylic acid cycle (Fig. 2A). In bacteria, there are two general pathways defined for Hut systems, termed pathway 1 and pathway 2 (Fig. 2A) (3). Both pathways yield glutamate and ammonia, but pathway 1 generates formamide, while pathway 2 generates a second molecule of ammonia and formate. To determine the relative conservation of the Hut system in Acinetobacter relative to other proteobacteria, genomic analyses and protein alignments were performed using EMBL Needle (16). A. baumannii ATCC 17978 was used as the basis for all alignments. Upon analyzing the genomes of pathogenic and nonpathogenic Acinetobacter species, it was revealed that the closely related pathogenic species Acinetobacter nosocomialis possesses an intact Hut system, with the core enzymes and putative regulators having >90% amino acid identity to A. baumannii (Fig. 2B). The A. nosocomialis Hut system also has the same genomic organization as A. baumannii, where it is encoded adjacent to the predicted zinc-regulated genes zigA and zrlA (9, 17). This conservation extends to other strains of A. baumannii, as well as less frequently isolated pathogenic species, including A. pittii and A. haemolyticus (18) (Table 1). Interestingly, the human skin commensal A. lwoffii, which is sometimes pathogenic in immunocompromised individuals (19), does not possess an intact Hut system (Table 1). In addition, the environmental species Acinetobacter baylyi does not possess a complete Hut system and is absent in several other environmental Acinetobacter species (Table 1). However, the histidine transporter HutT appears to be conserved with >88% amino acid identity in A. baylyi (Fig. 2B). The A. baylyi genome also possesses short genomic remnants of hutU, hutI, and hutG surrounding the gene encoding HutT (Fig. 2B). These findings predict that A. baylyi is unable to catabolize histidine for growth, while pathogenic strains such as A. baumannii and A. nosocomialis have maintained this metabolic attribute.
FIG 2.
The histidine utilization system is broadly conserved. (A) Model for the two defined pathways for Hut-mediated histidine catabolism (3). (B) Genomic organization of hut systems in A. baumannii ATCC 17978, A. baylyi ADP1, A. nosocomialis M2, Pseudomonas aeruginosa PA14, and Klebsiella pneumoniae ATCC 43816. Numbers within arrows denote the percent protein identity compared to A. baumannii ATCC 17978. (C) Representative growth curve monitored based on the OD600 over time of A. baumannii 17978, A. baumannii 5075, A. nosocomialis M2, and A. baylyi ADP1 in M9 minimal media with histidine as the sole carbon source. (D) Representative growth curve monitored based on the OD600 over time of A. baumannii 17978, A. baumannii 5075, A. nosocomialis M2, and A. baylyi ADP1 in M9 minimal media with histidine as the sole carbon and nitrogen source.
TABLE 1.
Percent protein identities of HutC and other Hut enzymes across bacterial strains and species relative to A. baumannii ATCC 17978
| Straina | Protein (% identity) |
||||
|---|---|---|---|---|---|
| HutC | HutH | HutU | HutI | HutG | |
| Acinetobacter baumannii 19606 | 99.2 | 100 | 100 | 98.8 | 97.7 |
| Acinetobacter baumannii 5075 | 99.2 | 100 | 99.8 | 99.3 | 98.4 |
| Acinetobacter baumannii MDR-ZJ06 | 99.2 | 100 | 100 | 99.3 | 98.0 |
| Acinetobacter calcoaceticus | 99.6 | 100 | 100 | 99.3 | 98.4 |
| Acinetobacter nosocomialis | 98.4 | 98.2 | 99.5 | 99.3 | 95.8 |
| Acinetobacter pittii | 99.2 | 98.0 | 98.7 | 98.5 | 76.8 |
| Acinetobacter haemolyticus | 90.4 | 94.7 | 95.3 | 83.0 | 68.6 |
| Pseudomonas aeruginosa | 58.0 | 87.5 | 90.0 | 74.9 | 43.7 |
| Klebsiella pneumoniae | 53.9 | 74.9 | 85.6 | 49.4 | 39.5 |
No Hut proteins were identified for A. albensis, A. baylyi, A. boissieri, A. indicus, or A. lwoffii.
Sequence analysis revealed that in A. baumannii, the four core Hut enzymes are more similar to Pseudomonas spp. that possess pathway 2 Hut systems, as opposed to Klebsiella spp. that possess pathway 1. Pathway 2 Hut systems also include hutD, which is a Hut-related gene that occurs downstream of the HutC repressor and whose protein function is not well defined (Fig. 2B) (3). We noted that A. baumannii encodes a HutD in the same genomic position as in Pseudomonas. Intriguingly, A. baumannii lacks hutF, which is required for pathway 2 Hut systems, and therefore possesses the four-enzyme system typical of pathway 1. These findings suggest that A. baumannii possesses a hybrid Hut system that includes aspects of both pathway 1 and pathway 2.
In organisms containing an intact Hut system, histidine can be utilized as the sole carbon and nitrogen sources. To test functional outcomes from sequence predictions, wild-type strains of the pathogens A. baumannii (strains ATCC 17978 and ABUW5075) and A. nosocomialis and the environmental species A. baylyi were grown in minimal media with l-histidine supplemented as the sole carbon and/or nitrogen source. As predicted, multiple strains of A. baumannii and A. nosocomialis can use histidine as the sole energy source, and A. baylyi is unable to grow under these conditions (Fig. 2C and D). In addition, A. baumannii requires an intact Hut system to support growth on histidine, since genetic inactivation of the first enzyme in the catabolic pathway hutH, or the histidine transporter hutT, prevents growth (Fig. S2A). As predicted based on sequence analysis (Table 1), this feature is conserved in the recent multidrug-resistant A. baumannii strain ABUW5075 (Fig. S2A and B). Taken together, these results establish Hut-mediated histidine catabolism as a feature of A. baumannii physiology.
HutC transcriptionally regulates the Hut system.
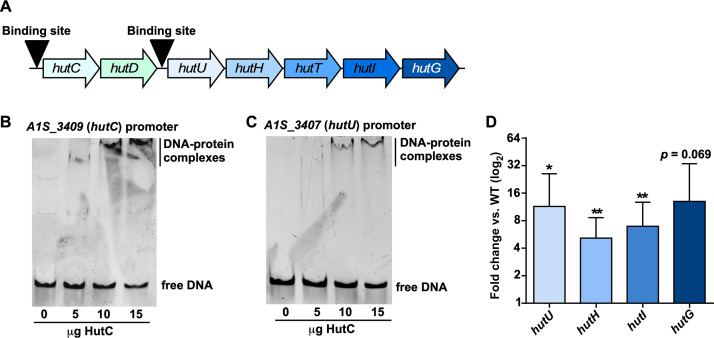
In both the pathway 1 and 2 Hut systems, the primary transcriptional regulation occurs via HutC-mediated repression of the genes encoding the core enzymes, although other regulators for the Hut system have been defined (7, 8, 20). Despite earlier-noted differences between the A. baumannii and P. aeruginosa Hut systems, the overall organization of the A. baumannii Hut system is similar to that of pseudomonads (Fig. 2B) (3). In P. putida and P. fluorescens, 14-nucleotide HutC binding sites were identified upstream of both the hutCD genes and genes encoding the Hut enzymes (8, 21). Based on these observations, we predicted that A. baumannii HutC binds the promoter region upstream of hutCD and hutUHTIG (Fig. 3A). Using these defined sequences, alignments identified putative HutC binding sites in the promoter regions of A. baumannii hutC and hutU (Fig. S3A). To experimentally validate HutC binding, recombinant HutC was purified (Fig. S3B) and subjected to electrophoretic mobility shift assays (EMSAs). Incubation of recombinant HutC with DNA containing the promoter region of hutC (Fig. 3B; Fig. S3D) or hutU (Fig. 3C; Fig. S3D) resulted in visible DNA-protein complexes that were absent with a nonspecific DNA probe (Fig. S3C and D). In order to determine the nature of HutC-mediated regulation of the core Hut enzymes, hutC was genetically inactivated, and expression of the Hut enzymes was monitored using quantitative reverse transcription-PCR (RT-PCR). This analysis revealed that inactivation of hutC resulted in significant upregulation of the genes encoding the Hut enzymes relative to wild-type (WT) A. baumannii (Fig. 3D), demonstrating that HutC likely functions as a transcriptional repressor of the Hut system in this organism.
FIG 3.
HutC transcriptionally regulates A. baumannii hut. (A) Model detailing predicted HutC binding locations upstream of hutC and hutU. (B) EMSA performed with recombinant HutC and a DNA probe containing the hutC promoter region. (C) EMSA performed with recombinant HutC and a DNA probe containing the hutU promoter region. (D) Quantitative RT-PCR analysis of genes encoding the Hut enzymes in a strain lacking hutC grown to mid-log and compared to expression in WT A. baumannii. *, P < 0.05; **, P < 0.01 (as determined by Student t test with a value of 1 from three independent experiments).
Using P. putida, P. fluorescens, and A. baumannii HutC-binding motifs (Fig. S3A), we next analyzed the A. baumannii genome for other possible HutC-regulated genes. This analysis revealed two genes with 14-nucleotide motifs containing high conservation to HutC binding sites, which encode a putative purine-cytosine permease (A1S_1504; CTTGTATATACATG) and a putative oxidoreductase (A1S_2459; AGTGTATATACAAA). These results are consistent with findings in Brucella abortus that HutC likely has regulatory targets outside the Hut system (22). Collectively, these data define HutC as a transcriptional regulator of the Hut system in A. baumannii and suggest HutC targets exist outside histidine catabolism.
HutH permits histidine nitrogen source utilization.
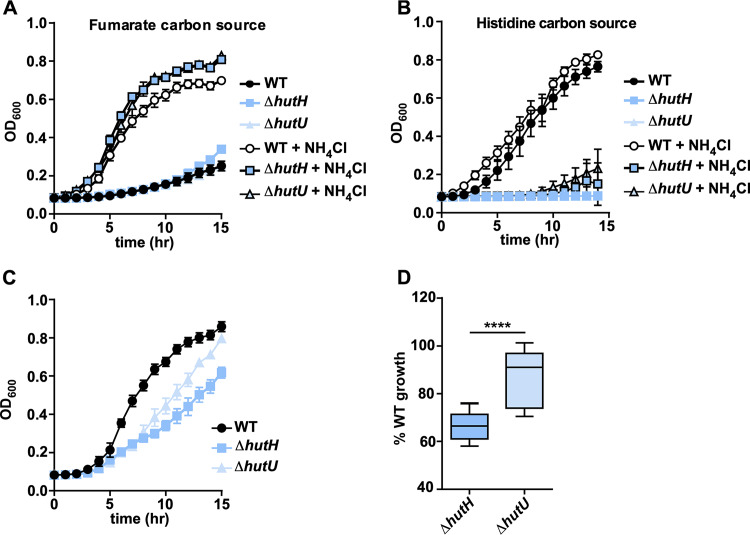
Hut-mediated histidine catabolism proceeds from l-histidine to the first intermediate urocanate via the histidine ammonia lyase HutH (Fig. 2A). We have previously confirmed the enzymatic activity of HutH and its requirement for histidine carbon source utilization (9). However, the first step of histidine catabolism mediated by HutH releases ammonia, which suggests that histidine may serve as a nitrogen source for A. baumannii in the presence of other carbon sources that provide no nitrogen. To assess the contribution of HutH-mediated ammonium release for growth, WT A. baumannii and strains genetically inactivated for hutH or hutU, the second enzyme in the Hut pathway (Fig. 2A), were grown in minimal media in the presence or absence of ammonium chloride. When fumarate is provided as the sole carbon source, ammonium chloride is required for growth of all three strains because fumarate metabolism does not provide any nitrogen (Fig. 4A). When histidine is provided as the sole carbon source, the WT strain is proficient for growth regardless of ammonium chloride supplementation, while the hutH and hutU mutants are both unable to grow (Fig. 4B). If fumarate is provided as the carbon source and supplemented with histidine as the sole nitrogen source, WT A. baumannii has normal growth kinetics, while the hutH mutant growth is inhibited (Fig. 4C). However, the hutU mutant is partially restored in its ability to grow under these conditions and grows significantly better than the hutH mutant (Fig. 4C and D). In addition, the hutH dependence on histidine for carbon and nitrogen source utilization can be complemented by expressing hutH in trans (Fig. S4A and B). These results are consistent with a model whereby A. baumannii utilizes histidine as a nitrogen source in combination with other, more energetically efficient carbon sources, such as fumarate, through the action of HutH.
FIG 4.
HutH promotes A. baumannii histidine nitrogen source utilization. (A) Growth monitored based on the OD600 of WT A. baumannii 17978 and ΔhutH and ΔhutU strains in M9 minimal media with fumarate as the sole carbon source with or without NH4Cl. (B) Growth monitored based on the OD600 of WT A. baumannii 17978 and ΔhutH and ΔhutU strains in M9 minimal media with histidine as the sole carbon source with or without NH4Cl. (C) Representative growth curve monitored based on the OD600 of WT A. baumannii 17978 and ΔhutH and ΔhutU strains in M9 minimal media with fumarate as the sole carbon source and histidine as the sole nitrogen source. (D) Percent growth relative to WT at 15 h for ΔhutH and ΔhutU strains with fumarate as the sole carbon source and histidine as the sole nitrogen source. ****, P < 0.0001 (as determined by Student t test and combined from three independent experiments).
HutH contributes to A. baumannii pathogenesis.
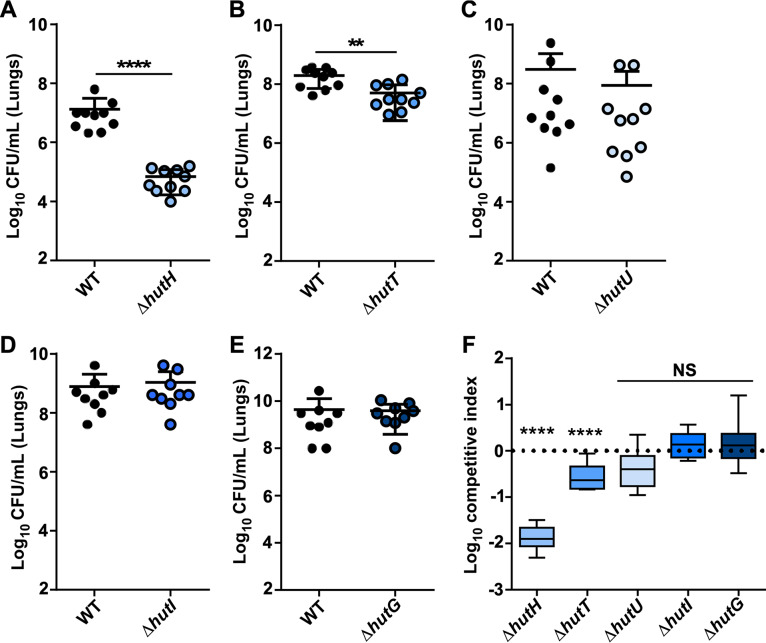
Given the conservation of the Hut system in pathogenic Acinetobacter and the detection of histidine within the BALF, we sought to determine whether components of this system are important for survival within the vertebrate host. Eight- to ten-week-old C57BL6/J mice were intranasally inoculated with WT A. baumannii in pairwise combinations with hut mutants, and the infection progressed for 36 h before mice were humanely euthanized and bacterial burdens enumerated. These experiments revealed that the hutH mutant was severely attenuated within the lungs and defective in dissemination to the liver (Fig. 5A; Fig. S5A), with a 100-fold decrease in its ability to compete against the WT strain (Fig. 5F). In addition, the hutT histidine transport mutant also displayed a colonization defect in the lungs and liver (Fig. 5B and F; see also Fig. S5B). However, no other hut mutants displayed any significant defect in the lungs or in the liver (Fig. 5C to F; see also Fig. S5C to E). We previously demonstrated that HutH-mediated histidine catabolism may release zinc from histidine-zinc complexes to promote survival in low zinc environments generated by the metal-binding properties of the host protein calprotectin (CP) (9). However, the hutH mutant remained defective in the lungs of CP−/− mice, and the hutU mutant was unaffected, suggesting that hutH contributes specifically to histidine metabolism (Fig. S5F to H). Collectively, these results establish HutH-mediated histidine catabolism as a key determinant of A. baumannii pathogenicity.
FIG 5.
Components of the Hut system are important for A. baumannii pathogenesis. Eight- to ten-week-old mice were intranasally infected with a 1:1 mixture of WT and mutant A. baumannii. (A) Bacterial burdens of WT and ΔhutH strains recovered at 36 hpi in the lungs. (B) Bacterial burdens of WT and ΔhutT strains recovered at 36 hpi in the lungs. (C) Bacterial burdens of WT and ΔhutU strains recovered at 36 hpi in the lungs. (D) Bacterial burdens of WT and ΔhutI strains recovered at 36 hpi in the lungs. (E) Bacterial burdens of WT and ΔhutG strains recovered at 36 hpi in the lungs. **, P < 0.01; ****, P < 0.0001 (as determined by Mann-Whitney U-test with Dunnett’s multiple comparisons; n = 9 to 10 mice per group). (F) Competitive index ([input mutant/WT]/[output mutant/WT]) for panels A to E for the various mutants in the lungs. ****, P < 0.0001 (as determined by Student t test against value of 1).
DISCUSSION
The ability of opportunistic bacterial pathogens to persist in diverse environments is likely a major contributor to their disease-causing potential. A. baumannii produces all required biological cofactors from simple starting materials, including biosynthesis of the full suite of amino acids. While amino acids serve as the building blocks of proteins, many bacteria can metabolize amino acids as nutrient sources. Histidine is one of the most energetically expensive amino acids to synthesize, so acquiring it from the environment is more efficient than biosynthesis. As such, both the import of histidine and its degradation are tightly regulated to maintain a cellular supply for protein synthesis (3). Here, we demonstrate that histidine and other amino acids are detectable within the vertebrate lung during infection. In addition, we describe the sole pathway for histidine nutrient utilization in A. baumannii and demonstrate that the catabolic enzymes are conserved among several pathogenic Acinetobacter species. Further, HutC serves as a transcriptional repressor of hut. The Hut system is required for A. baumannii to catabolize histidine as a carbon source, but it also permits histidine degradation for nitrogen source utilization. This nitrogen source utilization requires ammonia release by the HutH enzyme, and a strain lacking hutH is defective for histidine nitrogen source utilization and is attenuated in a mouse model of A. baumannii pneumonia. These findings suggest that histidine serves as an important nitrogen source for A. baumannii within the lung and that inhibiting A. baumannii HutH or limiting bioavailable histidine may be a viable strategy for combating A. baumannii pathogenesis.
While HutC is a major regulator of the Hut system, other transcriptional regulators may influence expression of hut gene expression in A. baumannii. In Pseudomonas additional regulators include the CbrAB and NtrBC two-component systems (3). In carbon-limiting conditions, CbrAB serves as a transcriptional activator of hut genes, and nitrogen-limiting conditions are sensed by NtrBC to activate hut (21). These two-component systems have functional overlap and are independently sufficient to activate hut genes (21). In addition, the function of HutD, which is encoded directly downstream of HutC in A. baumannii and in Pseudomonas species, is not well defined but may alter the expression of hut genes (23). Given the importance of fine-tuning histidine levels within the bacterial cell, the A. baumannii Hut system regulation is likely multifaceted and is an exciting area of future investigation.
Amino acid profiling of the extracellular lung contents revealed that histidine and other amino acids are detectable, and likely bioavailable, to A. baumannii and other bacterial pathogens. The levels of histidine and several amino acids did not significantly change with infection, which is consistent with metabolite analysis performed on BALF following Klebsiella pneumoniae infection (24). However, other amino acids did change with infections status, such as glutamine and glutamate, which are known modulators of macrophage activity (25). Since many amino acids and small molecules were detectable within the BALF, A. baumannii or other bacterial pathogens could occupy unique metabolic niches depending on their individual nutrient requirements. For example, citrulline levels were significantly elevated during infection, and citrulline catabolism is important for Streptococcus pyogenes pathogenesis (26). These findings suggest that targeting specific amino acid acquisition pathways could be effective at limiting bacterial pathogenesis more broadly.
The observation that HutH is critical for survival within the vertebrate host suggests that the Hut system and histidine utilization have broader impacts beyond simple energy generation. For example, A. baumannii encodes a histidine decarboxylase that converts histidine to histamine, which is an essential precursor for the siderophore acinetobactin (27, 28). We also previously demonstrated that HutH-mediated histidine catabolism plays a role in liberating Zn from histidine-Zn complexes during nutrient limitation (9). Further, A. baumannii grown in l-histidine as the sole carbon source has enhanced biofilm formation that is dependent on an intact Hut system (29). Histidine also stimulates the production of extracellular proteases in the marine opportunistic bacterial pathogen Vibrio alginolyticus, and this production is dependent on hutH (30). In B. abortus, the causative agent of brucellosis, the HutC repressor regulates not only hut expression but also serves as a coactivator of the type IV secretion system that is responsible for effector translocation to host cells (22). Consistent with this, we identified two possible HutC binding sites in the promoters of a putative nucleoside permease and oxidoreductase, and other HutC-regulated genes may exist as well. Collectively, these findings illustrate the diversity in which Hut-mediated histidine metabolism and sensing may impact bacterial pathogenesis and physiology more broadly. Our observation that several pathogenic Acinetobacter species maintain the Hut system supports a model whereby some bacteria use host-derived amino acids for survival during infection.
This study demonstrates that the Hut system, and specifically HutH, is important for the pathogenesis of A. baumannii, where histidine may serve as a nitrogen source alongside more energetically favorable carbon sources to promote A. baumannii lung colonization. These data highlight an exciting intersection between bacterial amino acid metabolism and host-pathogen interactions that may be targeted for therapeutic development.
MATERIALS AND METHODS
Bacterial strains and culture conditions.
Experiments were performed using A. baumannii strain ATCC 17978 and its derivatives unless otherwise noted. Transposon mutants in the A. baumannii ABUW5075 background were purchased from the University of Washington A. baumannii mutant library, and transposon insertion was confirmed by PCR (31). Acinetobacter nosocomialis M2 was kindly provided by Mario Feldman (Washington University, St. Louis, MO). Acinetobacter baylyi ADP1 was purchased from the American Type Culture Collection. Cloning was performed in E. coli DH5α, and protein expression was performed in E. coli BL21(DE3). All experiments involving liquid cultures were performed in lysogeny broth (LB) at 37°C with aeration unless otherwise stated.
Bacterial mutant generation.
The ΔhutH, ΔhutU, ΔhutT, ΔhutI, and ΔhutG strains were generated previously (9). To generate the hutC::Km (A1S_3409) strain (ΔhutC), approximately 1,000 bp of DNA in the 5′ and 3′ flanking regions surrounding the open reading frame were amplified from A. baumannii genomic DNA, and the kanamycin resistance gene aph was amplified from pUC18K1 (32). The three PCR products were joined together using Gibson assembly into pFLP2 and sequence verified (33). pFLP2 was then electroporated into A. baumannii, plated onto LB agar with kanamycin at 40 μg/ml (Km40), and grown overnight at 37°C. Transformants were patched to LB Km40 or LB agar with 10% sucrose to isolate Km-resistant (Kmr) and sucrose-sensitive merodiploids. Merodiploid strains were grown in LB overnight at 37°C to resolve the plasmid. Cultures were serially diluted, plated on LB agar with 10% sucrose, and incubated at 37°C overnight. The resulting Kmr sucrose-resistant strains were screened for the loss of hutC and replacement with aph by multiple PCRs. The relevant primers are listed in Table 2.
TABLE 2.
Oligonucleotides used in this study
| Oligonucleotide | Sequence (5′–3′) | Description |
|---|---|---|
| hutC mutant construction and confirmation | ||
| HutC FL1For | GGTTAAAAAGGATCGATCCTCTAGACATCAAAACTTGAAAAATAGTG | Cloning into pFLP2, 5′ flank, forward |
| HutC FL1 Rev | TAGTTAGTCACATATGAACAAATACTCAAAAAGGCAG | Cloning into pFLP2, 5′ flank, reverse |
| HutC Kan For | AGTATTTGTTCATATGTGACTAACTAGGAGGAATAAATG | Cloning into pFLP2, 3′ flank, forward |
| HutC Kan Rev | ATCAATTCAACATATGTCATTATTCCCTCCAGGTAC | Cloning into pFLP2, 3′ flank, reverse |
| HutC FL2 For | GGAATAATGACATATGTTGAATTGATCAGGGCCG | Cloning into pFLP2 for Km cassette from pUC18-k1, forward |
| HutC FL2 Rev | AAGTTCCTATTCTCTAGGGGGATCCTTCTGCAACGTCCGGATC | Cloning into pFLP2 for Km cassette from pUC18-k1, reverse |
| EMSA probes | ||
| hutC_EMSA_R | AAC AAA TAC TCA AAA AGG CAG CAG GCA | Upstream of hutC |
| hutC_EMSA_F1 | GGC ACT GGC TTA GCA GAA ATA ACT | Upstream of hutC |
| hutU_EMSA_R | ACG CAA TGG GGC TTC AGT TAA | Upstream of hutU |
| hutU_EMSA_F1 | TAC GAG CAA GCC TCT TCA CGC | Upstream of hutU |
| A1S_3412_F | CCA CGC GAG ATT TGG TCC AA | Internal to A1S_3412 |
| A1S_3412_R | GCT TGG CCT TTA GTC GCC C | Internal to A1S_3412 |
Growth in minimal media.
Overnight bacterial cultures were diluted 1:50 into 1× M9 salts with no nitrogen source added (33.7 mM Na2HPO4, 22 mM KH2PO4, 8.55 mM NaCl) for 1.5 h at 37°C and 180 rpm with shaking. Cultures were then diluted 1:100 in a 96-well plate containing M9 minimal media (1× M9 salts, 1 mM MgSO4, 0.3 mM CaCl2, 1× Vishniac’s trace metal mix with or without 9.35 mM NH4Cl with l-histidine and/or fumarate as the sole carbon and/or nitrogen source [0.5%, wt/vol]). Growth was monitored over time by monitoring optical density at 600 nm (OD600) on an EPOCH 2 microplate reader (BioTek) from at least three independent experiments unless otherwise noted.
HutC expression and purification.
The HutC open reading frame was cloned into pET15b with an N-terminal His tag (pET15bHutC) and transformed into E. coli BL21(DE3)/pREL. Overnight cultures of pET15bHutC were inoculated into terrific broth supplemented with carbenicillin (50 μg/ml) and chloramphenicol (34 μg/ml) and grown to an OD of 0.5. Cells were induced with 0.5 mM IPTG (isopropyl-β-d-thiogalactopyranoside) and grown for an additional 5 h. Cultures were pelleted, and pellets were frozen at –80°C until processing. To purify protein, pellets were thawed on ice in lysis buffer (50 mM NaH2PO4, 300 mM NaCl, 20 mM imidazole, 1 mg/ml lysozyme) and lysed using an Emulsiflex with five passes at 20,000 lb/in2. Insoluble material was removed by centrifuging lysates at 8,000 × g for 10 min, followed by ultracentrifugation at 100,000 × g for 1 h at 4°C. Supernatants were collected and applied to Ni-NTA columns (Qiagen) preequilibrated with lysis buffer. The protein loaded column was washed with wash buffer (50 mM NaH2PO4, 300 mM NaCl, 25 mM imidazole), and protein was eluted with an imidazole gradient ranging from 50 to 300 mM. Protein purity was confirmed by SDS-PAGE and Coomassie staining. Purified HutC was dialyzed overnight into buffer containing 20 mM Tris-HCl (pH 8) and 150 mM NaCl. After dialysis, the protein samples were mixed with glycerol to 10% and frozen at –80°C.
Complementation.
WT and ΔhutH A. baumannii strains were electroporated with the empty plasmid pWH1266 or a plasmid containing hutH under the A1S_2682 constitutive promoter as described previously (9), and the strains were grown overnight in 50 μg/ml carbenicillin. Strains were back-diluted to 1:50 into 1× M9 salts with no nitrogen source added (33.7 mM Na2HPO4, 22 mM KH2PO4, 8.55 mM NaCl) for 1.5 h at 37°C and 180 rpm with shaking. Cultures were then inoculated 1:100 into a 96-well plate with M9 minimal media (1× M9 salts, 1 mM MgSO4, 0.3 mM CaCl2, 1× Vishniac’s trace metal mix ± 9.35 mM NH4Cl with l-histidine and/or fumarate as the sole carbon and/or nitrogen source [0.5%, wt/vol]). Growth was monitored over time by monitoring the OD600 on an EPOCH 2 microplate reader.
Quantitative RT-PCR.
Overnight cultures of A. baumannii were diluted 1:5,000 in LB and grown to the mid-exponential phase. Bacterial cultures were then mixed with an equal volume of acetone-ethanol (1:1) prior to storage at –80°C until processing. For RNA extraction, cells were pelleted and resuspended in LETS buffer (0.1 M LiCl, 0.01 M Na2EDTA, 0.01 M Tris-HCl [pH 7.4], 0.2% sodium dodecyl sulfate) and lysed using Lysis Matrix B tubes (MP Biologicals) and a FastPrep-24 (MP) bead beater. Samples were heated to 55°C for 5 min and pelleted at 21,000 × g for 10 min. The top phase was combined with TRIzol and incubated at room temperature for 5 min. Chloroform was mixed with each sample, incubated for 3 min, and centrifuged for 15 min at 4°C at 21,000 × g. The top layer was collected, and RNA was precipitated with isopropyl alcohol for 10 min at room temperature. Samples were spun for 10 min at 21,000 × g at 4°C. Supernatant was removed, and pellets were washed with 70% ethanol. The pellet was dissolved in water, and DNA contamination was removed by adding RQ1 and RQ1 buffer (Promega) and RNase inhibitor (Thermo Fisher Scientific) for an incubation period of 2 h at 37°C. After DNase treatment, RNA was purified using the RNeasy minikit (Qiagen) according to the manufacturer’s protocol. RNA was quantified, and 2 μg of RNA was used for cDNA synthesis using M-MLV reverse transcriptase and buffer (Thermo Fisher Scientific) and mixing in random hexamers (Thermo Fisher Scientific) and deoxynucleoside triphosphate (Thermo Fisher Scientific). cDNA synthesis and purity were confirmed by PCR using 16S primers (r01) (34) with a “no RT” control. Synthesized cDNA was diluted 1:50 for quantitative RT-PCR using iQ SYBR green Supermix (Bio-Rad) on a Bio-Rad CFX96 real-time thermocycler. Relative expression was determined using the ΔΔCT value as previously described (34). Relevant primers were previously published (9) or are listed in Table 2.
Electrophoretic mobility shift assays.
EMSAs were performed generally as described previously (35). Briefly, DNA probes were designed to include predicted HutC binding sites in the hutC and hutU promoter regions and encompass 231 and 224 bp upstream of their respective initiation codons. A negative-control DNA probe was designed targeting the coding region of A1S_3412 (zrlA), which is a non-HutC-regulated gene. DNA probes were generated after amplification from genomic A. baumannii DNA using KAPA polymerase with appropriate primers (Table 2) and processed using a PCR cleanup kit (Thermo Fisher Scientific). Equimolar amounts of various DNA probes (300 fmol) were incubated with recombinant His-tagged HutC in binding buffer (20 mM Tris-HCl [pH 7.5], 500 mM NaCl, 2.5 mM MgCl2, 0.45 mM EDTA, 0.05% Nonidet P-40, 10% glycerol) for 30 min at 37°C. After incubation, samples were mixed with 6× EMSA loading dye (15% glycerol, 0.25% bromophenol blue, 0.25% xylene cyanol in 1× Tris-borate-EDTA [TBE]). Samples were loaded onto prerun 5% TBE gels (Bio-Rad) and run at 100 V until dye front reached the gel bottom. Gels were stained with SYBR green (Invitrogen) in 0.5× TBE for 20 min, washed twice with Millipore water, and imaged on a Bio-Rad ChemiDoc MP system with a SYBR green filter applied. Densitometry analysis was performed using the ImageJ gel analyzing tool, where the HutC protein-bound band intensity was compared to the unbound DNA probe for each protein concentration. Gel images are representative of at least three independent experiments.
Mouse model of A. baumannii pneumonia.
Eight- to ten-week-old male (ΔhutH and ΔhutU infections) and female (ΔhutI, ΔhutT, and ΔhutG infections) C57BL6/J mice were purchased from Jackson Laboratories, and male S100A9−/− (CP−/−) mice were bred in-house. Mice were housed with standard Vanderbilt University Medical Center (VUMC) facility chow and bedding with a 24-h light-dark cycle. Mice were inoculated 1:1 with a mixture of WT and mutant A. baumannii strains totaling 3 × 108 CFU in 40 μl of phosphate-buffered saline (PBS). At 36 hpi, mice were euthanized, and CFU were enumerated in various organs following tissue homogenization and dilution plating on LB and LB Km. The limit of detection of this assay is 100 CFU/ml. All animal experiments were approved by the VUMC Institutional Care and Use Committee and conform to the policies and guidelines established by VUMC, the Animal Welfare Act, the National Institutes of Health, and the American Veterinary Medical Association.
BALF metabolite profiling.
Eight-week-old male C57BL6/J mice were inoculated with A. baumannii for 36 h before euthanasia, at which time bronchial alveolar lavage fluid (BALF) was collected in 0.8 ml of PBS, as described previously (36). BALF was centrifuged for 5 min at 1,500 × g to remove immune cells and flash-frozen in liquid nitrogen. Upon thawing, samples were filtered through a 0.22-μm filter (Millipore) and subjected to amino acid analysis using a Biochrom 30 amino acid analyzer physiological system through the Vanderbilt University Hormone Assay and Analytical Services Core. This analysis utilizes a lithium-based ion exchange with post-column ninhydrin, which is then analyzed for quantitative results.
Sequence analysis.
A. baumannii ATCC 17978 protein sequences for components of the Hut system were obtained from the Kyoto Encyclopedia of Genes and Genomes database (37). These sequences were used as the input for protein Basic Local Alignment Search Tool (BLAST) to find homologs in the following organisms: A. baumannii ATCC 19606, A. baumannii 5075, A. baumannii MDR-JZ06, A. calcoaceticus TAXID 471, A. nosocomialis M2, A. pittii PHEA-2, A. haemolyticus ATCC 19194, A. baylyi ADP1, A. boissieri TAXID 1219383, A. indicus TAXID 756892, A. albensis TAXID 1673609, A. lwoffii TAXID 28090, Pseudomonas aeruginosa PA14, and Klebsiella pneumoniae ATCC 43816. These sequences were then aligned against A. baumannii ATCC 17978 using the EMBL pairwise sequence alignment tool, and the percent protein identity was recorded.
Quantification and statistical analyses.
Statistical analyses were performed using GraphPad Prism 7 and Microsoft Excel. The statistical tests used, the significance values, and the group sizes are indicated in the corresponding figure legends and method details.
Supplementary Material
ACKNOWLEDGMENTS
We thank members of the Skaar laboratory for critical review of the manuscript.
The work presented here was supported by National Institutes of Health (NIH) grant R01 AI101171 (E.P.S.). Z.R.L. was supported by NIH F31 AI136255, and L.D.P. was supported by NIH K99 HL143441. The VUMC Hormone Assay and Analytical Core was supported by NIH grants DK059637 and DK020593.
Footnotes
Supplemental material is available online only.
REFERENCES
- 1.Akashi H, Gojobori T. 2002. Metabolic efficiency and amino acid composition in the proteomes of Escherichia coli and Bacillus subtilis. Proc Natl Acad Sci U S A 99:3695–3700. doi: 10.1073/pnas.062526999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ingle RA. 2011. Histidine biosynthesis. Arabidopsis Book 9:e0141. doi: 10.1199/tab.0141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bender RA. 2012. Regulation of the histidine utilization (hut) system in bacteria. Microbiol Mol Biol Rev 76:565–584. doi: 10.1128/MMBR.00014-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Magasanik B. 1961. Catabolite repression. Cold Spring Harbor Symp Quant Biol 26:249–256. doi: 10.1101/sqb.1961.026.01.031. [DOI] [PubMed] [Google Scholar]
- 5.Sonenshein AL. 2007. Control of key metabolic intersections in Bacillus subtilis. Nat Rev Microbiol 5:917–927. doi: 10.1038/nrmicro1772. [DOI] [PubMed] [Google Scholar]
- 6.Brill WJ, Magasanik B. 1969. Genetic and metabolic control of histidase and urocanase in Salmonella Typhimurium, strain 15–59. J Biol Chem 244:5392–5402. [PubMed] [Google Scholar]
- 7.Schwacha A, Bender RA. 1990. Nucleotide sequence of the gene encoding the repressor for the histidine utilization genes of Klebsiella aerogenes. J Bacteriol 172:5477–5481. doi: 10.1128/jb.172.9.5477-5481.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Allison SL, Phillips AT. 1990. Nucleotide sequence of the gene encoding the repressor for the histidine utilization genes of Pseudomonas putida. J Bacteriol 172:5470–5476. doi: 10.1128/jb.172.9.5470-5476.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Nairn BL, Lonergan ZR, Wang J, Braymer JJ, Zhang Y, Calcutt MW, Lisher JP, Gilston BA, Chazin WJ, de Crecy-Lagard V, Giedroc DP, Skaar EP. 2016. The response of Acinetobacter baumannii to zinc starvation. Cell Host Microbe 19:826–836. doi: 10.1016/j.chom.2016.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Peleg AY, Seifert H, Paterson DL. 2008. Acinetobacter baumannii: emergence of a successful pathogen. Clin Microbiol Rev 21:538–582. doi: 10.1128/CMR.00058-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Harding CM, Hennon SW, Feldman MF. 2018. Uncovering the mechanisms of Acinetobacter baumannii virulence. Nat Rev Microbiol 16:91–102. doi: 10.1038/nrmicro.2017.148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.World Health Organization. 2017. Guidelines for the prevention and control of carbapenem-resistant Enterobacteriaceae, Acinetobacter baumannii, and Pseudomonas aeruginosa in health care facilities. World Health Organization, Geneva, Switzerland. [PubMed] [Google Scholar]
- 13.Centers for Disease Control and Prevention. 2019. Antibiotic resistance threats in the United States. Centers for Disease Control and Prevention, Atlanta, GA. [Google Scholar]
- 14.Lonergan ZR, Skaar EP. 2019. Nutrient zinc at the host-pathogen interface. Trends Biochem Sci 44:1041–1056. doi: 10.1016/j.tibs.2019.06.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Schlesinger S, Magasanik B. 1964. Effect of alpha-methylhistidine on the control of histidine synthesis. J Mol Biol 9:670–682. doi: 10.1016/s0022-2836(64)80174-1. [DOI] [PubMed] [Google Scholar]
- 16.Li W, Cowley A, Uludag M, Gur T, McWilliam H, Squizzato S, Park YM, Buso N, Lopez R. 2015. The EMBL-EBI bioinformatics web and programmatic tools framework. Nucleic Acids Res 43:W580–W584. doi: 10.1093/nar/gkv279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lonergan ZR, Nairn BL, Wang J, Hsu YP, Hesse LE, Beavers WN, Chazin WJ, Trinidad JC, VanNieuwenhze MS, Giedroc DP, Skaar EP. 2019. An Acinetobacter baumannii, zinc-regulated peptidase maintains cell wall integrity during immune-mediated nutrient sequestration. Cell Rep 26:2009–2018.e6. doi: 10.1016/j.celrep.2019.01.089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wong D, Nielsen TB, Bonomo RA, Pantapalangkoor P, Luna B, Spellberg B. 2017. Clinical and pathophysiological overview of Acinetobacter infections: a century of challenges. Clin Microbiol Rev 30:409–447. doi: 10.1128/CMR.00058-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Regalado NG, Martin G, Antony SJ. 2009. Acinetobacter lwoffii: bacteremia associated with acute gastroenteritis. Travel Med Infect Dis 7:316–317. doi: 10.1016/j.tmaid.2009.06.001. [DOI] [PubMed] [Google Scholar]
- 20.Hu L, Allison SL, Phillips AT. 1989. Identification of multiple repressor recognition sites in the hut system of Pseudomonas putida. J Bacteriol 171:4189–4195. doi: 10.1128/jb.171.8.4189-4195.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhang XX, Rainey PB. 2008. Dual involvement of CbrAB and NtrBC in the regulation of histidine utilization in Pseudomonas fluorescens SBW25. Genetics 178:185–195. doi: 10.1534/genetics.107.081984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sieira R, Arocena GM, Bukata L, Comerci DJ, Ugalde RA. 2010. Metabolic control of virulence genes in Brucella abortus: HutC coordinates virB expression and the histidine utilization pathway by direct binding to both promoters. J Bacteriol 192:217–224. doi: 10.1128/JB.01124-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhang XX, George A, Bailey MJ, Rainey PB. 2006. The histidine utilization (hut) genes of Pseudomonas fluorescens SBW25 are active on plant surfaces, but are not required for competitive colonization of sugar beet seedlings. Microbiology 152:1867–1875. doi: 10.1099/mic.0.28731-0. [DOI] [PubMed] [Google Scholar]
- 24.Silver RJ, Paczosa MK, McCabe AL, Balada-Llasat JM, Baleja JD, Mecsas J. 2019. Amino acid biosynthetic pathways are required for Klebsiella pneumoniae growth in immunocompromised lungs and are druggable targets during infection. Antimicrob Agents Chemother 63:e02674-18. doi: 10.1128/AAC.02674-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gras G, Porcheray F, Samah B, Leone C. 2006. The glutamate-glutamine cycle as an inducible, protective face of macrophage activation. J Leukoc Biol 80:1067–1075. doi: 10.1189/jlb.0306153. [DOI] [PubMed] [Google Scholar]
- 26.Cusumano ZT, Watson ME Jr, Caparon MG. 2014. Streptococcus pyogenes arginine and citrulline catabolism promotes infection and modulates innate immunity. Infect Immun 82:233–242. doi: 10.1128/IAI.00916-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Actis LA, Smoot JC, Barancin CE, Findlay RH. 1999. Comparison of differential plating media and two chromatography techniques for the detection of histamine production in bacteria. J Microbiol Methods 39:79–90. doi: 10.1016/s0167-7012(99)00099-8. [DOI] [PubMed] [Google Scholar]
- 28.Dorsey CW, Beglin MS, Actis LA. 2003. Detection and analysis of iron uptake components expressed by Acinetobacter baumannii clinical isolates. J Clin Microbiol 41:4188–4193. doi: 10.1128/jcm.41.9.4188-4193.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Cabral MP, Soares NC, Aranda J, Parreira JR, Rumbo C, Poza M, Valle J, Calamia V, Lasa I, Bou G. 2011. Proteomic and functional analyses reveal a unique lifestyle for Acinetobacter baumannii biofilms and a key role for histidine metabolism. J Proteome Res 10:3399–3417. doi: 10.1021/pr101299j. [DOI] [PubMed] [Google Scholar]
- 30.Bowden G, Mothibeli MA, Robb FT, Woods DR. 1982. Regulation of hut enzymes and intracellular protease activities in Vibrio alginolyticus hut mutants. J Gen Microbiol 128:2041–2045. doi: 10.1099/00221287-128-9-2041. [DOI] [PubMed] [Google Scholar]
- 31.Gallagher LA, Ramage E, Weiss EJ, Radey M, Hayden HS, Held KG, Huse HK, Zurawski DV, Brittnacher MJ, Manoil C. 2015. Resources for genetic and genomic analysis of emerging pathogen Acinetobacter baumannii. J Bacteriol 197:2027–2035. doi: 10.1128/JB.00131-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Menard R, Sansonetti PJ, Parsot C. 1993. Nonpolar mutagenesis of the ipa genes defines IpaB, IpaC, and IpaD as effectors of Shigella flexneri entry into epithelial cells. J Bacteriol 175:5899–5906. doi: 10.1128/jb.175.18.5899-5906.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hoang TT, Karkhoff-Schweizer RR, Kutchma AJ, Schweizer HP. 1998. A broad-host-range Flp-FRT recombination system for site-specific excision of chromosomally-located DNA sequences: application for isolation of unmarked Pseudomonas aeruginosa mutants. Gene 212:77–86. doi: 10.1016/s0378-1119(98)00130-9. [DOI] [PubMed] [Google Scholar]
- 34.Mortensen BL, Rathi S, Chazin WJ, Skaar EP. 2014. Acinetobacter baumannii response to host-mediated zinc limitation requires the transcriptional regulator Zur. J Bacteriol 196:2616–2626. doi: 10.1128/JB.01650-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Juttukonda LJ, Green ER, Lonergan ZR, Heffern MC, Chang CJ, Skaar EP. 2018. Acinetobacter baumannii OxyR regulates the transcriptional response to hydrogen peroxide. Infect Immun 87:e00413-18. doi: 10.1128/IAI.00413-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Palmer LD, Maloney KN, Boyd KL, Goleniewska AK, Toki S, Maxwell CN, Chazin WJ, Peebles RS Jr, Newcomb DC, Skaar EP. 2019. The innate immune protein S100A9 protects from T-helper cell type 2-mediated allergic airway inflammation. Am J Respir Cell Mol Biol 61:459–468. doi: 10.1165/rcmb.2018-0217OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kanehisa M, Goto S. 2000. KEGG: Kyoto encyclopedia of genes and genomes. Nucleic Acids Res 28:27–30. doi: 10.1093/nar/28.1.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.