The outer membrane (OM) of Gram-negative bacteria is an asymmetric lipid bilayer that consists of inner leaflet phospholipids and outer leaflet lipopolysaccharides (LPS). The asymmetric character and unique biochemistry of LPS molecules contribute to the OM’s ability to function as a molecular permeability barrier that protects the bacterium against hazards in the environment. Assembly and regulation of the OM have been extensively studied for understanding mechanisms of antibiotic resistance and bacterial defense against host immunity; however, there is little knowledge on how Gram-negative bacteria release their OMs into their environment to manipulate their hosts.
KEYWORDS: outer membrane vesicles, OMV, glycerophospholipid, asymmetry, lipid A, endotoxin, cardiolipin, lipopolysaccharide, lipooligosaccharides, bleb, mla, lpxC, ftsH, pbgA, yejM, lapB/yciM, Toll-like receptor, TLR4, myeloid differentiation factor 2, MD-2, inflammation, immunity, inflammasome, microbial associated molecular patterns, pattern recognition receptor, pyroptosis, endocytosis, gasdermin, lipid rafts, caspases 11, 4, and 5, constriction, peptidoglycan, Tol-Pal, ompA, pldA, antibiotics, antimicrobial peptides, antibiotic resistance, permeability barrier, secretion systems, pagP, pagL, lpp
ABSTRACT
The outer membrane (OM) of Gram-negative bacteria is an asymmetric lipid bilayer that consists of inner leaflet phospholipids and outer leaflet lipopolysaccharides (LPS). The asymmetric character and unique biochemistry of LPS molecules contribute to the OM’s ability to function as a molecular permeability barrier that protects the bacterium against hazards in the environment. Assembly and regulation of the OM have been extensively studied for understanding mechanisms of antibiotic resistance and bacterial defense against host immunity; however, there is little knowledge on how Gram-negative bacteria release their OMs into their environment to manipulate their hosts. Discoveries in bacterial lipid trafficking, OM lipid homeostasis, and host recognition of microbial patterns have shed new light on how microbes secrete OM vesicles (OMVs) to influence inflammation, cell death, and disease pathogenesis. Pathogens release OMVs that contain phospholipids, like cardiolipins, and components of LPS molecules, like lipid A endotoxins. These multiacylated lipid amphiphiles are molecular patterns that are differentially detected by host receptors like the Toll-like receptor 4/myeloid differentiation factor 2 complex (TLR4/MD-2), mouse caspase-11, and human caspases 4 and 5. We discuss how lipid ligands on OMVs engage these pattern recognition receptors on the membranes and in the cytosol of mammalian cells. We then detail how bacteria regulate OM lipid asymmetry, negative membrane curvature, and the phospholipid-to-LPS ratio to control OMV formation. The goal is to highlight intersections between OM lipid regulation and host immunity and to provide working models for how bacterial lipids influence vesicle formation.
INTRODUCTION
Gram-negative bacteria enshroud themselves in dual membranes to survive in diverse environments. Their outer membrane (OM) is separated from their inner membrane (IM) by an aqueous periplasm and a thin peptidoglycan cell wall (Fig. 1). Bacteria construct the lipids for their OM in the cytosol, assemble them upon the IM, transport them across the periplasm, and insert them into the OM. This culminates in the assembly of the OM into an asymmetric bilayer of inner leaflet phospholipids and outer leaflet lipopolysaccharides (LPS), or lipooligosaccharides (LOS), collectively referred to as LPS molecules here (1). Bacteria maintain OM lipid asymmetry and balance the phospholipid-to-glycolipid ratio of their OMs to preserve the barrier that protects them from the environment (2, 3).
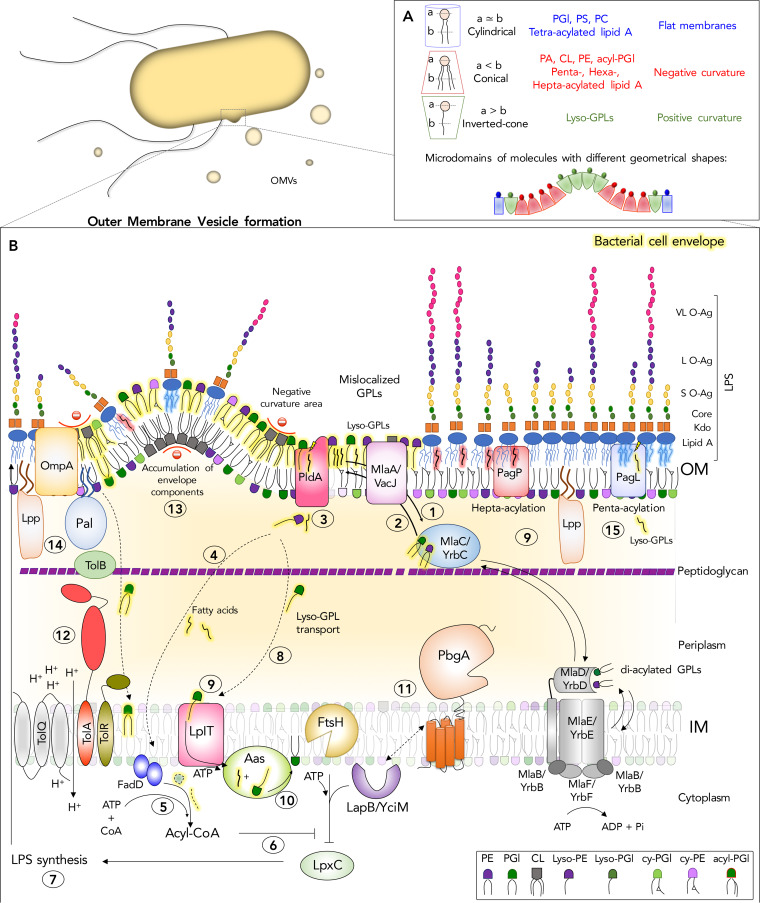
FIG 1.
Bacterial lipid trafficking, remodeling, and homeostasis systems involved in outer membrane vesicle (OMV) biogenesis. Gram-negative bacteria are diderm organisms whose cell envelope consists of an inner membrane (IM) and an outer membrane (OM) that are separated by a periplasmic space and a peptidoglycan layer. Lipid A is the amphipathic constituent of lipopolysaccharide (LPS) and forms the outer leaflet of the OM, while the inner leaflet consists of glycerophospholipids (GPL), making it an asymmetric bilayer. Gram-negative bacteria secrete lipids from their OMs in the form of OMVs and a variety of protein, lipid, and glycolipid components of the cell envelope. The process involves increasing the levels of amphipathic molecules in the outer leaflet of the OM, while not correspondingly increasing the levels in the inner leaflet. Depending upon the shape of lipids that accumulate in these microdomains, the bilayer can curve and pinch off from the surface. (A) Based upon a cross-sectional analysis of the head groups versus the acyl chains, GPLs and lipid A molecules adopt different geometrical shapes, such as conical, inverted conical, or cylindrical, which facilitates negative, positive, or lack of membrane curvature, respectively. (B) Gram-negative bacteria harbor multiple protein systems that work to maintain OM lipid asymmetry. (Step 1) The OM lipoprotein MlaA/VacJ prevents diffusion of GPLs into the OM outer leaflet by adopting an integral membrane conformation. (Step 2) The model of retrograde trafficking for Mla posits that MlaA donates the mislocalized GPLs to MlaC, and MlaC is an acceptor of phosphatidyglycerols (PGl) and phosphatidylethanolamines (PE). MlaC would bind to the MlaFEDB/YrbFEDB complex in the IM and transfer the GPLs to MlaD for their reintegration into the IM. The anterograde model posits that MlaC donates GPLs to MlaA, which is an acceptor in the OM. An anterograde mechanism would involve MlaC receiving GPLs from the IM complex and transporting them outwardly across the periplasm to the OM. In the OM, GPLs flip into the outer leaflet at a low frequency during growth and a greater frequency during OM damage. (Step 3) OM outer leaflet mislocalized GPLs are substrates for PldA, a beta-barrel phospholipase that generates lysophospholipids and fatty acids in the outer leaflet to maintain asymmetry (Step 4). Fatty acids diffuse back across the periplasm, cross the IM, and are converted by FadD to acyl-CoA (Step 5). Acyl-CoA, or another product originated from phospholipid metabolism in the OM, might act as a second messenger to modulate LpxC degradation (Step 6). This would allow bacteria to pause LpxC degradation and increase LPS biosynthesis (Step 7), thereby restoring balance to the level of GPLs and glycolipids that comprise the OM. PldA, and the OM beta barrel enzyme PagP, generate lysophospholipids (lyso-GPLs) from inverted GPLs. (Step 8) Lysophospholipids can act as detergents that disrupt the bilayer and bacteria presumably traffic these molecules back to the IM; however, this mechanism is not known. (Step 9) On their return to the IM, lysophospholipids are flipped across the membrane by the flippase, LplT. (Step 10) On the cytoplasmic surface, the acyltransferase Aas restores the lysophospholipids to their diacyl form. (Step 11) PbgA/YejM is an essential lipid-binding protein involved in regulating LPS biosynthesis. The model posits that PbgA-mediated LPS regulation works through LapB/YciM and LapB’s ability to modulate FtsH-mediated protein digestion, especially for LpxC. (Step 12) The Tol-Pal system forms a trans-envelope protein complex important for constricting the OM by forming energized interactions between the OM lipoprotein Pal and peptidoglycan, promoting-negative membrane curvature, and maintaining OM integrity. Loss of Tol-Pal causes GPLs to accumulate in the OM and results in OMV formation. (Step 13) Peptidoglycan fragments and misfolded proteins can also increase in the periplasm and cause changes in turgor pressure between the peptidoglycan layer and OM that might cause OMVs to form. (Step 14) In addition, major OM proteins like OmpA and the OM lipoprotein Lpp form periplasmic contacts with peptidoglycan, and alterations in these interactions have been shown to result in OMV release. (Step 15) Lipid A modifications by the PagP palmitoyltransferase and the PagL demyristoylase result in hyper- or hypoacylated forms of lipid A for LPS molecules, respectively. These changes in the lipid A acylation state impact the shape of the molecules in the outer leaflet and perhaps influence the local curvature of the bilayer. Abbreviations: CL, cardiolipin; PA, phosphatidic acid; PS, phosphatidylserine; acyl-PGL, acyl-phosphatidylglycerol; PC, phosphorylcholine; S O-Ag, short O-antigen; L O-Ag, long O-antigen; VL O-Ag, very long O-antigen; ATP, adenosine triphosphate.
Lipid A molecules are tetra-, penta-, hexa-, or hepta-acylated disaccharolipids that form the amphipathic base structure of the LPS supermolecule, and they are potent endotoxins that activate the immune system. The lipid A components of LPS molecules are microbe-associated molecular patterns (MAMPs) and immune ligands for eukaryotic pattern recognition receptors (PRRs). PRRs control inflammation, cell death, and host immunity in response to interactions with Gram-negative bacteria (4–8). The Toll-like receptor 4/myeloid differentiation factor-2 complex (TLR4/MD-2) and the mouse caspase-11, human caspase-4 and caspase-5, noncanonical inflammasome receptor bind and are activated by lipid A molecules on the surface and cytoplasm of host cells, respectively (Fig. 2) (5, 7–9). Specific interactions between the acyl chains of lipid A endotoxins and host receptors dictate the immune response to pathogens and even commensals. Tetra-acylated lipid A molecules can antagonize human TLR4/MD-2 signaling but provoke mouse signaling (10, 11). Furthermore, tetra-acylated lipid A molecules activate human caspase-4 but antagonize mouse caspase-11 (12). Gram-negative bacteria also produce cardiolipins (CLs) in their OMs, which are tetra-acylated diphosphatidylglycerols that engage TLR4/MD-2 and are agonists or antagonists depending upon the saturation state of their acyl chains (13–16).
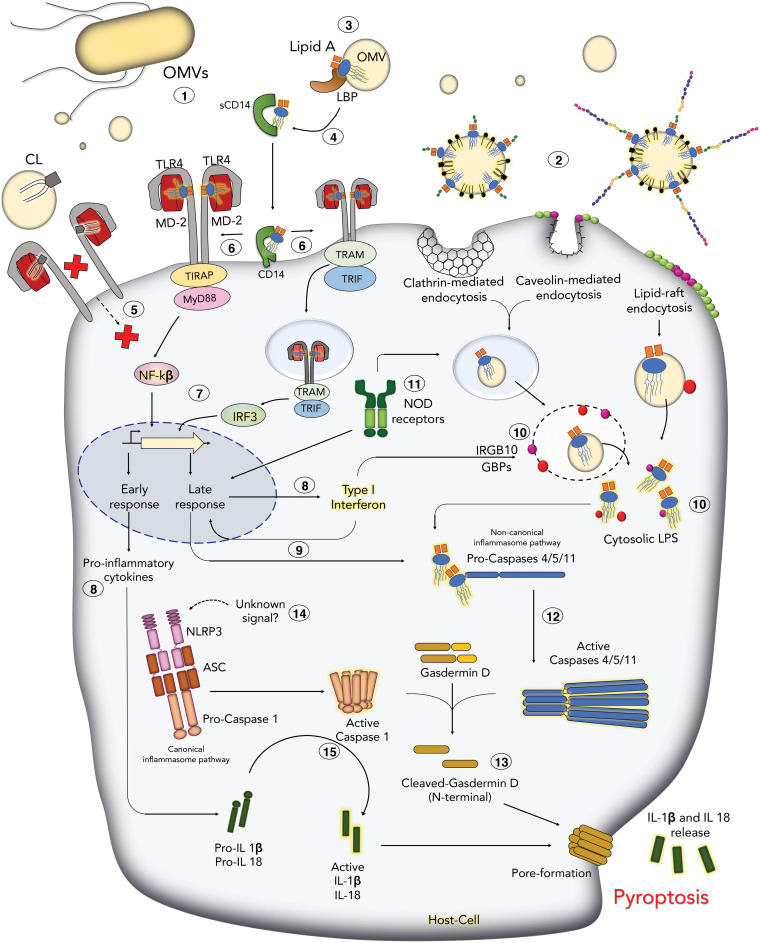
FIG 2.
Innate immune pattern recognition of bacterial lipids on OMVs leads to inflammation and death. Lipid A and cardiolipin (CL) are constituents of OMVs that engage pattern recognition receptors on membranes and in the cytosol of host cells. The LPS molecules carried by OMVs interact with a variety of proteins and systems. Lipid A components of these LPS molecules activate proinflammatory cytokine production and trigger an inflammatory cell death known as pyroptosis. Once released, OMVs (Step 1) can enter host cells through endocytosis (Step 2). In the bloodstream, OMVs interact with the circulating plasma protein, LPS-binding protein, or LBP (Step 3), which extracts a single LPS molecule from the micelle. LBP binds to soluble CD14 (sCD14) and membrane-anchored CD14 molecules. Interactions with LBP, CD14, and LPS result in the remodeling and presentation of the lipid A disaccharolipid component of LPS to MD-2 (Step 4). MD-2 interacts with lipid A by a sandwich-like mechanism, which influences TLR4 receptor dimerization and activation. CL can also interact with the TLR4/MD-2 complex on the surface (Step 5) and can act as an agonist or antagonist. Dimerization of TLR4/MD-2 (Step 6) recruits different adaptor proteins that will activate transcription factors such as nuclear factor κβ (NFκβ) and interferon regulatory factor 3 (IRF3) (Step 7), which will ultimately result in production of proinflammatory cytokines (inactive pro-IL-1β and pro-IL-18) and type I interferon (IFN-β) (Step 8). Type-I IFN also induces the expression of caspase-11 (Step 9). The production of type I IFN after TLR4 activation enhances the expression of cytoplasmic proteins, such as Interferon-related GTPase-10 (IRGB10) and guanylate-binding proteins (GBPs), which bind pathogen vacuoles and LPS molecules on cytosolic bacteria and OMVs (Step 10), and might facilitate endosome lysis or release of LPS molecules, and ultimately lipid A endotoxins, for detection by caspases-4/5/11. NOD receptors bind peptidoglycan fragments on OMVs and activate late inflammatory responses to pathogens (Step 11). Binding of the lipid A disaccharolipid by caspases-4/5/11 promotes autoprocessing and activation (Step 12). Activated caspase-4/5/11 cleaves gasdermin D, which causes gasdermin D to oligomerize and form large pores in the plasma membrane that alter ion homeostasis and cause cell lysis (Step 13). The NLRP3 noncanonical inflammasome is activated by an unknown signal that is downstream caspase-4/5/11 activation by lipid A molecules (Step 14). Signals received by NLRP3 activate autoprocessing of caspase-1 and activation of the canonical inflammasome. Active caspase-1 cleaves IL-1β and IL-18, as well as gasdermin D, which result in robust proinflammatory cell death, or pyroptosis (Step 15). The dual activation pathway combines the canonical and noncanonical inflammasomes and is specific to innate immune cells, which we have attempted to depict here.
Microbes release CL and LPS molecules as components of outer membrane vesicles (OMVs). OMVs are spheroidal lipid bilayers that emerge from the bacterial surface as asymmetric blebs, or vesicles, during division, shifts in nutrients and osmolarity, and encounters with antibiotics, reactive oxygen species (ROS), cationic antimicrobial peptides (CAMPs), and detergents (17). These extracellular lipid interfaces can contain the lipids, proteins, and sugars typically associated with the OM (Fig. 1B and 2). Concrete understanding of how OMVs are formed and secreted has not been attained, but advances in the field of bacterial lipid regulation and trafficking suggest that we are closer to possible mechanisms. Gram-negative bacteria secrete OMVs to enhance their virulence. In this minireview, we highlight how Gram-negative bacteria regulate OMV formation to influence immunity by altering OM lipid asymmetry, remodeling lipid shape, and controlling the phospholipid-to-glycolipid ratio in the OM.
LIPID A MOLECULES AND CARDIOLIPINS BIND TLR4 ON HOST CELL MEMBRANES TO CONTROL INFLAMMATION AND IMMUNITY
The innate immune system defends against pathogens by detecting MAMPs using PRRs. Since OMVs are vehicles for highly concentrated MAMPs, the lipid ligands on OMVs engage multiple receptors and initiate production of cytokines, chemokines, and antimicrobial molecules (18). In mammals, high concentrations of OMVs in circulation can induce massive inflammatory responses and endotoxic shock (19–23). Our focus is on the phospholipid and glycolipid constituents of OMVs that influence host immunity.
The lipid A endotoxin is a MAMP that is detected by the TLR4/MD-2 complex on host membranes.
Bacterial OMVs can act at a distance from the microbe and adhere to membranes (Fig. 2). Toll-like receptors (TLRs) are integral membrane receptors that detect microbial patterns (24). The lipid A component of LPS activates Toll-like receptor 4 (TLR4) through a biochemical interaction between lipid A and myeloid differentiation factor 2 (MD-2) (5, 25, 26). MD-2 accommodates the hydrophobic acyl chains of the lipid A moiety in a sandwich-like structure and then binds TLR4 to induce dimerization (27). Lipid A acyl chains are inserted deeply inside the MD-2 pocket, while phosphates are oriented outside and form hydrogen bonds with charged residues of MD-2 and TLR4 (Fig. 2) (27). The spatial configuration of lipid A molecules in the MD-2 sandwich is critical, as modifying the phosphorylation and acylation state of lipid A molecules impacts MD-2 binding and TLR4 signaling (4, 28–35).
The amphipathic nature of LPS molecules causes them to aggregate in aqueous media and form micelles (28). Since MD-2 engages a distinct moiety of the greater LPS molecule, the lipid A disaccharolipids must be liberated from the LPS micelles, or from the external leaflet of OMVs, by analogy. LPS-binding protein (LBP) and CD14 (cluster of differentiation 14) protein act upstream of TLR4/MD-2 to engage and dissociate lipid A structures from LPS aggregates and OMVs, which is necessary for TLR4/MD-2 activation (Fig. 2) (36, 37). LBP is an acute-phase plasma protein that extracts and transfers lipid A molecules to CD14, which is present on the membranes of myelomonocytic cells as a glycosylphosphatidylinositol (GPI)-anchored protein or as a soluble form in sera (sCD14) (38–40). Lipid A binding to CD14 enhances activation of innate immune effector cells like monocytes, macrophages, and polymorphonuclear cells (38, 41, 42). CD14 extracts a single lipid A molecule and dissociates from the micelle using electrostatic repulsion and residues on LBP (36, 43). LBP recruits subsequent apo-CD14 molecules to the micelle, which allows holo-CD14 to shuttle and deliver lipid A molecules to MD-2 (36). LBP protects humans against Gram-negative bacteria, and a polymorphism that affects LBP’s capacity to bind LPS molecules results in reduced levels of proinflammatory cytokines and worsened pneumonia and sepsis (44).
Upon binding the lipid A/MD-2 complex, TLR4 can signal the activation of two different pathways (Fig. 2). Surface dimerization of TLR4/MD-2 recruits the TIRAP (Toll-interleukin 1 receptor [TIR] domain containing adaptor protein) and myeloid differentiation primary response 88 (MyD88) adaptor proteins, resulting in an “early” activation of the NF-κB transcription factor and the production of proinflammatory cytokines (6). Alternatively, the dimerized receptor complex can be internalized, where it then recruits a different set of endosomal adaptor proteins, TRAP (transmembrane adaptor protein) and TRIF (TIR domain-containing adaptor protein inducing beta interferon), which promote “delayed” activation of NF-ĸB and production of type I interferons, through the IRF3 (interferon regulatory protein 3) transcription factor (Fig. 2) (45, 46).
Hexa-acylated, diphosphorylated, lipid A molecules are potent agonists of mouse and human TLR4/MD-2 that induce high levels of proinflammatory cytokines (47, 48). Chemical alterations to the lipid A molecule can impact TLR4/MD-2 signaling by generating immune-silent, or less-active, endotoxins (4, 34). Bacteria modify the number of acyl chains and phosphates on lipid A. This can result in inefficient MD-2 binding and decreased TLR4/MD2 activation (28–33). Five out of the six acyl chains of lipid A fit inside the MD-2 pocket, while the sixth is partially exposed and oriented toward TLR4, which favors dimerization (49). The intrinsic conformation of the hexa-acylated lipid A molecule favors hydrophilic interactions between the anionic phosphates on lipid A and the cationic residues on TLR4 (49). Tetra- or penta-acylated lipid A molecules have lower, if not antagonistic, effects on TLR4/MD-2 signaling (28, 50–56). Therefore, the spatial conformation of lipid A within MD-2 is critical for its interaction with MD-2 and subsequent binding and dimerization of TLR4.
Salmonella enterica serovar Typhimurium produce hepta-acylated lipid A molecules that cause decreased TLR4/MD-2 dimerization and reduced levels of proinflammatory cytokines (32). The absence of one or both phosphate groups in Helicobacter pylori, Porphyromonas gingivalis, Francisella novicida, and Leptospira interrogans results in a less toxic molecule with a reduced affinity for TLR4 compared to the diphosphorylated precursors (57–61). Gram-negative bacteria produce other tetra-acylated lipid molecules in their OMs, like cardiolipins, which concentrate at negatively curved regions of the bilayers, are components of OMVs, and interact with TLR4 to influence receptor activation by lipid A endotoxins (34, 62).
Bacterial cardiolipins are MAMPs that engage the TLR4/MD-2 complex.
Cardiolipins (CLs) are acidic diphosphatidylglycerols that adopt conical shapes and are concentrated at negatively curved poles and septa of bacterial cells (14, 63–66). CL is critical for mitochondria to function as energy-generating organelles, and oxidized CL molecules are damage signals (67, 68). During cellular stress, mitochondrial IM CL molecules are oxidized by cytochrome c and trafficked to the OM of the organelle. The oxidized CLs are externalized and then dock with the proapoptotic protein, Bid, as well as the canonical inflammasome by means of interactions with NLRP3 (nucleotide-binding oligomerization domain [NOD]-, leucine-rich repeat [LRR]- and pyrin domain-containing protein 3)/Acs/procaspase 1 (69–71). Moreover, CL interacts with Beclin1 and recruits autophagic machinery through its interaction with LC3 (72, 73).
Recent studies demonstrated that interactions between bacterial CL molecules and TLR4 might dampen inflammatory responses to endotoxin, presumably by blocking MD-2 binding to lipid A molecules (Fig. 2) (74). Compound 406 is a structural homologue of lipid IVA. Like CL, lipid IVA is a tetra-acylated amphiphile that antagonizes TLR4 signaling (75). Bacterial CL molecules can act as agonists or antagonists of TLR4/MD-2 signaling depending upon the saturation state of the acyl side chains (16). Unsaturated CLs are antagonists and compete with lipid A for TLR4/MD2 binding, thereby preventing receptor activation, while saturated CLs are TLR4/MD2 agonists (16). Moreover, the length of the CL acyl chain is related to the potency of the antagonistic effect. Specifically, the shorter the acyl chain (C14:1), the more anti-inflammatory the effect. CLs with C14 chain length behave as antagonists for human TLR4 (hTLR4), but not mouse TRL4 (mTLR4). This biochemical variation appears to be independent of the saturation state of the acyl chains (16). Similar results were observed for lipid IVA, suggesting that the conformation of C14-CL molecules within the MD2 pocket might somehow impede TLR4 dimerization and signaling (56, 76, 77). Gram-negative members of the intestinal microbiota might utilize CL to modulate inflammation, but how pathogens use CL to control TLR4 activity during infection is not fully understood (62).
ENDOTOXIN SENSING IN THE HOST CYTOSOL CAUSES PYROPTOSIS
The lipid antigens of extracellular and vacuolar pathogens are not restricted by host membranes (78). Eukaryotes also detect and discriminate lipid A endotoxins in their cell cytosol using PRRs. Bacteria release LPS molecules as products of antimicrobial damage, cell lysis, and OMV formation. In the cytosol, the lipid A endotoxins on LPS molecules are then somehow released from the core and O-antigen moieties before being detected by inflammatory caspases, caspase-4, -5, and -11 (79–81). Exact mechanisms of LPS entry into the cytosol via OMVs are not understood, and how lipid A molecules are liberated for detection from LPS superstructures on bacterial cells and as part of OMVs is an area of current focus.
OMV entry into the cytosol.
OMVs enter nonphagocytic cells from the extracellular environment through a variety of mechanisms. The best understood involve endocytosis and interactions with lipid rafts (46, 82–86). The endocytic route is mediated by either clathrin or caveolin depending upon the protein that oligomerizes to form the vesicle (Fig. 2) (82, 87). Clathrin assembles around membrane pits and forms polygonal lattices that result in large vesicles (200 nm in diameter) (88). OMVs released by H. pylori, enterohemorrhagic Escherichia coli (EHEC), enteroaggregative E. coli, and Brucella abortus enter the cell through this mechanism (87, 89–95). Evidence indicates that OMVs from Haemophilus influenzae, H. pylori, and Trichomonas vaginalis can enter through caveolae (87, 96, 97). Caveoleae are glycolipid rafts enriched in cholesterol, sphingolipids, and caveolins, which cause membrane invaginations of a smaller size (60 to 80 nm in diameter) (82, 86, 98–101). Perhaps this mechanism of entry enables bacteria to evade the immune response or avoid OMVs fusing with lysosomes (102–105). OMVs also use lipid rafts to enter (83–86). Lipid rafts are smaller (40 to 50 nm in diameter), mainly composed of (glyco-)sphingolipids and cholesterol, and diffuse along the plasma membrane (106–109). Lipid raft-mediated OMV entry was observed using infection models for Campylobacter jejuni, P. gingivalis, H. influenzae, and Pseudomonas aeruginosa (83, 96, 110, 111). OMV fusion with lipid rafts might facilitate delivery of toxins and MAMPs into host cells to induce immune activation or cell death, but the chemical-physical mechanism of entry through rafts is not understood (84, 85, 110, 112–115).
Guanylate-binding proteins bind to LPS molecules on bacteria and OMVs and can aid in caspase-11 detection of lipid A molecules in the cytosol.
Upon entering the cell, LPS molecules must somehow be processed and released by the host as lipid A endotoxins from their core and O-antigen components before their detection by caspase-4, -5, and -11. Extracellularly, these activities are contributed by LBP and CD14. In the cytoplasm, interferon-regulated genes (IRGs) and guanylate-binding proteins (GBPs) might mediate this activity.
Once inside the cell, OMVs might fuse with endosomal membranes or alternatively, lyse the endosome or host vesicle (81, 116). Host GBPs and IRGB10 (b10, a member of the family of immunity-related GTPases) are activated by lipid A binding to TLR4 on the surface (Fig. 2) (117–119). GBPs bind cytosolic OMVs by direct protein-LPS interactions and enhance caspase-11 activation and pyroptosis (119, 120). GBPs bind LPS molecules on the surfaces of Gram-negative pathogens that have entered the cytosol, and likely work with other cytosolic and membrane-associated host proteins to mediate vacuole lysis and LPS dissociation during infections with intravacuolar pathogens (IRGB10, galectin-3, and others) (121–126). Perhaps GBPs somehow alter the structure of the greater LPS supermolecule to promote specific interactions between lipid A molecules and the inflammatory caspase receptors (12, 127, 128). In this regard, GBP1 colocalizes with Shigella flexneri that produce smooth-type LPS greater than with bacteria that produce rough-type LPS, which lack the O antigen (129). We predict that additional lipid- and glycan-binding proteins engage lipid A endotoxins in the cytosol and influence the activity of the noncanonical inflammasome.
Caspase-4, -5, and -11 bind lipid A endotoxins in the cytoplasm and cleave gasdermin D to activate cell death.
The presence of endotoxin in the cytosol is detected by the caspase-11 PRR in mice and the caspase-4 and caspase-5 PRRs in humans (Fig. 2) (7, 130, 131). Anionic lipid A disaccharolipids bind to the caspase activation and recruitment domain (CARD) from caspase-4, caspase-5, and caspase-11 (caspases-4/5/11) through high-affinity electrostatic interactions with cationic amino acid residues (7). These activated proteases autoprocess and the active caspase-4, -5, and -11 fragments bind and cleave gasdermin D, which oligomerizes and forms large pores in the plasma membrane (132). Pore formation alters ion transport and homeostasis and ultimately causes lysis and an inflammatory cell death known as pyroptosis (133–136). Epithelial cells possess this LPS-induced pyroptotic pathway. Extracellular pathogens, like enterohemorrhagic E. coli, secrete OMVs that enter the cytoplasm of host-epithelial cells and induce caspase-11-driven cell death in this manner (81).
Dendritic cells and macrophages combine noncanonical inflammasome detection of LPS in the cytosol, with activation of the canonical inflammasome. The canonical inflammasome is comprised of Nlr3p, Acs, and pro-caspase-1 (127). Nlr3p/Acs/pro-caspase-1 activation results in caspase-1 autoprocessing into its active form, which then cleaves interleukin 1beta (IL-1β), interleukin 18 (IL-18), and gasdermin D, the latter through a binding and catalysis mechanism that is identical to caspase-4,-5, and -11 (Fig. 2) (132). Cleaved gasdermin D forms pores, and the cell releases IL-1β, which initiates a potent proinflammatory cell death in these phagocytes compared to epithelia (137–139).
The current model predicts that LPS activation of caspase-4, -5, and -11 results in gasdermin D-mediated pyroptosis, which is independent of Nlr3p/Acs/pro-caspase-1 activation (132). Changes in cation homeostasis as a consequence of cell pores might mediate the activation of Nlrp3/Acs/pro-caspase-1, resulting in production of the active form of IL-1β, IL-18, and ultimately, pyroptosis (140–142). Many pathogens that secrete OMVs initiate this pathway, such as EHEC O:157H7, S. Typhimurium, Citrobacter rodentium, Vibrio cholerae, Legionella pneumophila, P. gingivalis, Treponema denticola, and others (81, 126, 131, 137, 139, 143, 144).
The lipid A endotoxin can be extensively derivatized and enzymatically remodeled to influence interactions with TLR4/MD-2 (28–33, 145, 146). However, the particular modifications that influence the binding between lipid A and caspases are less well understood. The tetra-acylated lipid IVa molecule binds caspases-4 and -11, but not -5, and did not induce oligomerization of either protease, thereby preventing catalysis (7). F. novicida enters the cytosol and produces tetra-acylated lipid A molecules that activate the caspase-4 complex (12, 130, 147). Caspase-11 did not recognize tetra-acylated lipid A molecules, but was activated by penta-acylated molecules (130). Chlamydia trachomatis harbors endotoxins that completely avoid detection by the noncanonical inflammasome (148). The impact of lipid A hydroxylation and phosphorylation in caspase recognition has not been investigated.
In addition to caspases, eukaryotic host cells have nucleotide-binding oligomerization domain (NOD)-like receptors (NLRs), which are cytoplasmic PRRs (149). Each contains a central NOD region, a C-terminal leucine-rich repeat (LRR) region, and an N-terminal effector domain (149, 150). OMVs from H. pylori, V. cholerae, Neisseria meningitidis, and P. aeruginosa initiate proinflammatory cascades in a NOD-dependent manner, resulting in NF-ĸB and mitogen-activated protein kinase (MAPK) activation (84, 151–153). NOD1 and NOD2 are receptors that detect bacterial peptidoglycan fragments in the cytosol from OMVs produced from H. pylori and P. aeruginosa (84, 154, 155). NOD activation by the cytosolic pathogen S. flexneri induces autophagy and subsequent bacterial clearance (156). NOD detection of peptidoglycan fragments on OMVs is likely important for a variety of infections.
OUTER MEMBRANE VESICLE BIOGENESIS
Gram-negative bacteria produce OMVs as heterogeneous spheroidal particles that are extracellular lipid bilayers (10 to 300 nm in diameter) (Fig. 1B). OMVs are a subclass of microbial extracellular vesicles that are also produced by Gram-positive bacteria, mycoplasma, and fungi (157–163). Gram-negative bacteria produce OMVs as part of growth and survival, as well as from antimicrobial damage and bacterial cell lysis. The latter mechanisms potentially lead to mixing of bacterial compartments like the cytosol and plasma membrane. OMVs have been shown to carry IM lipids, nucleic acids, and other cytoplasmic molecules, peptidoglycan fragments, toxins, proteins, and small signaling molecules (164). This has led to many hypotheses and experimental examples of how OMVs are formed and the demonstration of their role in a multitude of bacterial processes, including cell physiology and bilayer homeostasis, molecular pathogenesis, horizontal gene transfer, quorum sensing, interspecies communication, toxin secretion, immunomodulation, and others (165).
Bacteria form vesicles in broth culture, on solid media, in biofilms, and during intracellular and extracellular survival as part of pathogenic and mutualistic relationships with eukaryotes (166–168). Certain conditions favor blebbing, such as elevated temperature, interactions with quorum-sensing quinolones, antibiotic exposure, encounters with bacteriophage endolysins, and nutrient deprivation (169–174). Their existence was described half of a century ago, yet we still do not firmly grasp how Gram-negative bacteria control OMV formation and regulate the secretion of lipids into their environment.
Multiple models have been proposed. For instance, as part of cell elongation and division, peptidoglycan must be turned over and low-molecular-weight muramyl peptide excision products can accumulate in the periplasm (164, 175, 176). Peptidoglycan fragment accumulation might increase the turgor pressure between the cell wall and OM, resulting in protrusion and blebbing (Fig. 1B) (177). The OM is connected to the cell wall by lipoproteins. For members of the family Enterobacteriaceae, Braun’s lipoprotein, or Lpp, is the predominant linker (178). Altering Lpp-peptidoglycan contacts might allow E. coli and S. Typhimurium to modulate vesicle production (179, 180). OMV biogenesis is impacted by the major OM beta-barrel protein, OmpA, in Acinetobacter baumannii and V. cholerae, which is an integral OM protein with a soluble peptidoglycan-interacting domain (Fig. 1B) (170, 181, 182).
Most Gram-negative bacteria possess the envelope-spanning Tol-Pal apparatus, which is a system of proteins that uses proton motive force across the IM to drive the OM lipoprotein, Pal, to bind peptidoglycan (Fig. 1B). Bacteria use this system to produce negative membrane curvature and regulate OM invagination at the division septum for fission (183–185). Genetic removal of the tol-pal system results in phospholipid accumulation within the OM and robust OMV formation (176, 186–189). Therefore, loss of interactions between peptidoglycan and the OM, and increases in OM phospholipids are associated with OMV formation. There is also evidence supporting a role for lipid shape in OMV formation.
Bacteria alter the structure of lipid A molecules within the outer leaflet to control OMV formation.
Bacteria regulate and remodel LPS structures by controlling the length and content of the O polysaccharides, or O antigens, derivatizing anionic phosphates on lipid A molecules with cationic aminoarabinose and phosphoethanolamine moieties, and by altering the number of phosphates and acyl chains on the lipid A disaccharolipid (4, 190). Multiple studies of different bacterial genera have shown that differentially acylated lipid A molecules adopt distinct shapes and configurations, which are driven by the number, length, and positioning of the hydrophobic moieties (191). In three dimensions, hexa-acylated lipid A is conical. Lipid A molecules with fewer numbers of acyl chains tend to acquire more cylindrical or inverted-conical structures, altering the membrane curvature (Fig. 1A) (191, 192). The lipid A acylation state, and thus the shape of molecules in the outer leaflet, affects the release of vesicles.
In particular, penta-acylated lipid A molecules are enriched in S. Typhimurium OMVs when the lipid A deacylase PagL is overexpressed (193). An independent study showed that hepta-acylated lipid A molecules are the predominant endotoxins loaded into OMVs when S. Typhimurium was exposed to acidic pH (5.5) and limiting Mg2+ concentrations (80). Recent evidence indicates that covalent modifications, such as the addition of phosphoethanolamine (pEtN) to lipid A can decrease OMV biogenesis in Citrobacter rodentium (194). These findings highlight how regulating lipid A structure might be a strategy to control vesicle release and to produce vesicles with altered biochemical properties.
THE BILAYER COUPLE AND LIPID SHAPE MODEL OF OMV BIOGENESIS
A universal mechanism of OMV production for Gram-negative bacteria does not exist. The “bilayer-couple” hypothesis supposes that an increase in amphipathic molecules within the outer leaflet causes the outer leaflet to expand relative to the inner leaflet. Outer leaflet insertion and expansion increase bilayer curvature and result in vesicle formation (195). Gram-negative bacteria share a system of envelope proteins known as the Mla system (maintenance of outer membrane lipid asymmetry), which is necessary for trafficking diacylated phospholipids across the periplasm, and for preventing diacylated phospholipids from accumulating in the outer leaflet (Fig. 1) (196–200). Bacterial downregulation of Mla increases the levels of phospholipids that are present in the outer leaflet and induces OMV formation (167, 170, 201).
Maintaining OM lipid asymmetry and forming OMVs.
Gram-negative bacteria synthesize phospholipids in the cytoplasm and transport them to the OM by a variety of energy-dependent and -independent mechanisms (Fig. 1B) (2, 3). Unlike LPS transport, phospholipid trafficking is bidirectional. Of the known protein systems, Mla is the best characterized (Fig. 1) (202). Mla proteins are conserved throughout Gram-negative bacteria and plant chloroplasts (203). The chloroplast system conducts retrograde phospholipid transport, which is necessary for the organelle to import essential phospholipids from the endoplasmic reticulum (204). Unlike chloroplasts, Gram-negative bacteria can synthesize all of their essential phospholipids, so the biochemical need to traffic intact phospholipids from the OM to the IM must involve a remodeling, recycling, or signaling mechanism at the IM (205–207). The bacterial Mla system is multifunctional. The OM lipoprotein component, MlaA/VacJ, adopts an integral membrane conformation that allows it to maintain OM lipid asymmetry, which it does by holding or maintaining phospholipids in their inner leaflet orientation (Fig. 1B) (196). Given the dual role of the system in promoting OM lipid asymmetry and phospholipid trafficking, the biochemical phenotypes of mla mutants have provided inconclusive data regarding the directionality of Mla transport across the periplasm (2, 3, 197, 198, 208).
MlaC is a soluble periplasmic protein that binds and ferries phospholipids across the periplasm. MlaC interacts with both MlaA, the OM lipoprotein, and MlaD, the IM phospholipid-binding and transfer protein (Fig. 1B) (197, 199). The retrograde model posits that MlaA binds and transfers mislocalized phospholipids to MlaC for ferrying across the periplasm back to the IM. The MlaD components of the IM complex, MlaBDEF, accept the phospholipids from MlaC and reinsert them into the IM (Fig. 1). In the anterograde model, MlaD of the MlaBDEF complex extracts phospholipids from the IM and transfers them to MlaC for export to the OM. MlaC subsequently transfers the phospholipids to MlaA, which orients the molecules in the inner leaflet of the OM and prevents their inversion into the outer leaflet (198, 208). The existing data support that the system likely works bidirectionally depending upon the environment and the use of the ATPase activity of MlaF (Fig. 1B) (198, 208).
In addition to MlaA, Gram-negative bacteria carry a gene that encodes the OM phospholipase beta-barrel enzyme, PldA, which degrades outer leaflet phospholipids and maintains OM lipid asymmetry (Fig. 1B) (209, 210). PldA hydrolyzes phospholipids that have become inverted into the outer leaflet into lysophospholipids and free fatty acids, which are then trafficked back to the IM by poorly understood mechanisms (Fig. 1B) (209, 211, 212). Lysophospholipids adopt inverted conical shapes in bilayers and act as detergents, so bacteria degrade or transport these molecules back to the inner leaflet of the IM (Fig. 1B) (213). The duration that lysophospholipids exist in the outer leaflet is unknown, but probably brief. mla mutants exhibit phenotypes similar to those of pldA mutants, and a double mutant (mla pldA) is more defective for barrier function, antimicrobial resistance, and maintaining lipid asymmetry than either single mutant (202). Bacterial strains with mutations in Mla proteins commonly accumulate phospholipids in the OM outer leaflet and produce OMVs (170, 201, 214). mla-null mutant phenotypes are suppressed by PldA upregulation (202). Conversely, a mutation in MlaA that allows constitutive phospholipid externalization, mlaA*, is suppressed by mutations that inactivate PldA (215). The consensus model is that Mla controls the access of phospholipids to the outer leaflet and PldA processes molecules that mistakenly flip across the bilayer.
H. influenzae, V. cholerae, E. coli, and Neisseria gonorrhoeae use the analogous system, VacJ/Yrb, to control OMV formation (170, 214). The previously mentioned pathogens respond to iron limitation by repressing the vacJ and yrb genes, which increase phospholipid externalization and OMV formation (170, 214). Recent evidence has shown that V. cholerae bacteria hypervesiculate to accelerate their adaptation to the host intestinal environment (167). The direct correlation between increasing phospholipids in the outer leaflet and forming OMVs fulfills the bilayer couple model; however, the contribution of the shapes of the particular phospholipids and lipid A molecules that result from phospholipid inversion must be also considered.
Phospholipid shape influences bilayer curvature and OMV formation.
Deformation of the bacterial OM is necessary for bacterial production of OMVs. Curvature can be modulated by controlling the level and structure of the individual lipid A and phospholipid molecules within the individual leaflets (Fig. 1) (216). The three-dimensional shape of lipids influences membrane curvature. Phospholipid shape depends upon the size of the polar head group and the number of acyl side chains. Diacylated bacterial phospholipids, such as phosphatidylserine (PS) or phosphatidylglycerol (PGl), have a cylindrical conformation and a flat surface area (Fig. 1A) (216–218) In contrast, CLs, phosphatidylethanolamines (PE), phosphatidic acids (PAs), and acyl-phosphatidylglycerols (acyl-PGl) adopt a conical configuration and generate negative membrane curvature when clustered (219). Bacteria concentrate CL at sites along the membranes that are negatively curved, such as at poles and division septa (64). Some monoacylated lysophospholipids, on account of their large head group and single fatty acid side chain, adopt an inverted conical shape, which favors formation of positively curved membranes (219, 220). It is generally thought that the introduction of negative curvature by concentrating conically shaped lipid molecules at particular sites along the cell surface results in vesiculation and OM lipid release from the bacterium.
Bacteria remodel lipid A molecules and phospholipids in the outer leaflet to influence membrane curvature and OMV formation.
During intracellular survival, S. Typhimurium releases OMVs that are trafficked throughout host cells, but the mechanism of vesicle formation is not fully resolved (78, 180). S. Typhimurium constitutively inverts phospholipids into the OM outer leaflet as a function of the PhoPQ two-component virulence regulators, whose signaling also activates OMV formation and is stimulated in the phagolysosome of macrophages by acidic pH, low divalent cation concentrations, and cationic antimicrobial peptides (80, 221). Activation increases the levels of triacylated acyl-PGl and hepta-acylated lipid A molecules, which contain palmitate. Synthesis of these molecules occurs in response to phospholipids becoming inverted into the outer leaflet by unknown mechanisms (222). PagP utilizes inverted phospholipids as the substrates for deacylation and acylation reactions that form lysophospholipids, acyl-PGls, and hepta-acylated lipid A molecules in the OM outer leaflet (Fig. 1B) (190, 221, 222). Each of these molecules adopts a shape that has been implicated in the ability of bacteria to establish membrane curvature.
The exact half-life of diacylated phospholipids in the OM outer leaflet is unknown, but like monoacylated lysophospholipids, it is probably short on account of PagP and PldA activity, or other analogous phospholipases in the OMs of nonenterobacterial organisms. Increased outer leaflet phospholipids and their enzymatic remodeling might lead to an increase in the abundance of curvature-generating molecules. It is interesting in this regard that S. Typhimurium bacteria increase the levels of CL molecules within their OMs in response to PhoPQ activation as a function of the PbgA transmembrane protein (190, 221, 222). We reason that a consensus model for how Gram-negative bacteria regulate OMV formation involves bilayer coupling and lipid shape.
Phospholipid recycling and lipid signaling across the periplasm.
The retrograde mechanism of lysophospholipid transport back to the IM is not understood (Fig. 1B). However, it is well established that E. coli uses the integral IM protein, lysophospholipid transporter (LplT), to move molecules across the IM. LplT hydrolyzes cytosolic ATP molecules and energizes lysophospholipid flipping from the periplasmic leaflet to the cytosolic leaflet of the IM (Fig. 1B) (223, 224). LplT works with the cytosolic Aas enzyme (acyl-acyl carrier protein [ACP] synthetase/lysophosopholipid acyltransferase). The LplT Aas enzymes cooperate to move lysophosphatidylethanolamines, lyso-PGls, and PLA2-hydrolyzed CLs across the IM and catalyze their reacylation (224). This recycling pathway might also provide an ill-defined signal transduction mechanism, which allows bacteria to sense and respond to changes on the surface (209).
The ability to couple OM homeostasis with phospholipid and LPS metabolism ensures that bacteria appropriately allot fatty acid and sugar resources during periods of limitation, damage, and repair. Unlike phospholipids, which are bidirectionally transported across the periplasm, LPS molecules are unidirectionally trafficked outward. Therefore, releasing OMVs might offer a mechanism for bacteria to secrete excess LPS glycolipids from their surface.
Regulating the level of phospholipids and LPS molecules in the OM bilayer.
The synthesis pathways for LPS and phospholipids compete for fatty acid resources in the cytosol (225, 226). Combined with the need to assemble an asymmetric bilayer, bacteria must appropriately regulate the relative rate of LPS and phospholipid production during feast or famine. The most well conserved and understood mechanism involves regulating the degradation of LpxC, the rate-limiting enzyme that catalyzes the first committed step for lipid A and therefore LPS formation (227, 228). LpxC is proteolyzed in a regulated manner by FtsH, an integral IM protease. Enterobacteriaceae increase the levels of the IM-associated tetratricopeptide repeat- and rubredoxin domain-containing protein, LapB/YciM, during stress, which interacts with FtsH and LpxC (229). In the working model, LapB binds FtsH and prompts FtsH to degrade LpxC (Fig. 1B) (230, 231). LapB is a key negative regulator of LPS biosynthesis for Enterobacteriaceae (229).
Recent work with S. Typhimurium suggests that the conserved enterobacterial IM protein PbgA/YejM uses its periplasmic domain to participate in LapB-mediated LPS regulation and phospholipid homeostasis (Fig. 1B) (206). Proteins that regulate LPS biosynthesis are essential for enterobacterial viability, including LpxC, FtsH, LapB, and PbgA (231–234). However, FtsH is dispensable when FabZ is hyperactive (230). FabZ is the dehydroxylase that catalyzes the first committed step in phospholipid biosynthesis by reducing the common C14 fatty acid precursor for the two pathways (230–235). Therefore, FabZ-driven phospholipid overproduction might serve to compensate for the high levels of LPS conferred by the inability to degrade LpxC. This hypothesis is consistent with the notion that Gram-negative bacteria balance the phospholipid and glycolipid composition of the OM to achieve homeostasis. The findings also support the possibility that regulated proteolysis of LpxC and other key regulators and biosynthesis proteins might allow Gram-negative bacteria to adjust LPS levels in the outer leaflet. It will be interesting to determine whether particular types of phospholipids with specific shapes are concomitantly regulated with LPS molecules.
Fatty acids are synthesized by the type II fatty acid synthesis (FASII) pathway, which has an initiation and elongation stage, followed by subsequent steps of the Kennedy pathway of glycerophospholipid biosynthesis and Raetz pathway of lipid A biosynthesis, to generate the bilayer-forming lipid components of the OM (236). The elongation intermediate, C14-OH, or β-hydroxymyristoyl-ACP (acyl carrier protein), can be utilized by either FabZ for phospholipid biogenesis, or LpxA for lipid A production (225, 235, 237). Pathway cross talk was demonstrated in E. coli when lpxA suppressor genotypes were shown to encode mutations in FabZ and when chemical inhibition of LpxC resulted in the selection of Klebsiella pneumoniae mutants with nucleotide polymorphisms encoding amino acid substitutions in both FabZ and LpxC (238, 239). Mutations in fabZ result in increased LPS levels, since synthesis is shifted toward lipid A biogenesis. Overactive FabZ enzymes suppress some phenotypes for bacteria that overproduce LPS molecules, likely by shifting synthesis away from LPS and toward phospholipids (225, 229, 238–242). Furthermore, interactome studies of LpxC and FtsH at various temperatures revealed binding between LpxC and phospholipid and fatty acid biosynthetic proteins (226).
Therefore, it is conceivable that LPS upregulation without concomitant increases in phospholipid biosynthesis could result in an overall increase in the LPS molecules that are present in the outer leaflet. Consistent with the bilayer-couple hypothesis, this increase in outer leaflet content without a coordinate increase in inner leaflet content, could result in expansion of the surface. Recently, a lipid-mediated signaling pathway involving MlaA, PldA, and LapB was identified (Fig. 1B) (209). This transenvelope signal transduction mechanism might allow Gram-negative bacteria to control the rate of LPS biosynthesis in response to phospholipids becoming inverted into the outer leaflet. The dominant allele of MlaA that results in constitutive phospholipid externalization, mlaA*, causes E. coli to increase the levels of LPS molecules on their surface (215). The mechanism of increasing LPS levels involves phospholipid hydrolysis in the outer leaflet by PldA and lipid signaling across the periplasm back to the IM (Fig. 1B) (209, 211–213). The model posits that fatty acids are lipid signals from the bacterial surface that move back across the IM and are converted to acyl coenzyme A (acyl-CoA), which somehow interacts with LapB and prevents the ability of LapB to downregulate LPS biosynthesis (206, 209). We envision that Gram-negative bacteria regulate the relative abundance of phospholipids and LPS molecules within their OM to orchestrate vesiculation and that this involves pathway cross talk between the protein systems that maintain asymmetry and those that control biosynthesis.
CONCLUDING REMARKS
The asymmetric character of the OM enhances its ability to function as a dynamic bacterial organelle. We have attempted to highlight the molecular complexity of the OM and its use as a secretory system for these microbes. Gram-negative bacteria are not all the same. Key differences include the presence of LOS molecules in some microbes and LPS molecules in others. LOS molecules are structurally similar but lack extended O polysaccharides (221). In some LOS-producing pathogens, complete loss of LOS glycolipids, and therefore lipid A endotoxins, is tolerated under certain conditions, like as a mechanism to resist killing by colistins (222, 223). The evidence suggests that the phospholipid and lipoprotein constituents of the OM provide a sufficient barrier for these pathogens to survive and replicate in the absence of LOS and lipid A molecules under select circumstances. In this regard, it is remarkable to now understand that the OM works as a major load-bearing element for Gram-negative bacteria, a role that had been exclusively attributed to peptidoglycan (243). Commensal species like Bacteroides fragilis secrete immunoregulatory OMVs to maintain tight junctions in the gut epithelium (34). This has important implications for antibiotic resistance and disease pathogenesis. In summary, OMVs provide both defense mechanisms and delivery approaches for pathogens and commensals to resist and modulate host immunity.
ACKNOWLEDGEMENT
This work was supported by R01AI139248 from the NIAID, which was awarded to Zachary D. Dalebroux.
Biographies

Nicole P. Giordano is a Ph.D. candidate in the Department of Microbiology and Immunology at the University of Oklahoma Health Sciences Center. She was born in Bridgewater, New Jersey, and completed her undergraduate work at High Point University. After receiving dual degrees (B.S.) in Biochemistry and Exercise Science, she completed a two-year postbaccalaureate fellowship at the U.S. Food & Drug Administration, studying Clostridium difficile response to oxygen. Afterwards, she moved to Oklahoma City and joined Dr. Dalebroux’s laboratory where she is working to understand how Salmonella enterica controls the lipid composition of the outer membrane, its interface with the host, using PbgA, a conserved enterobacterial protein. Understanding PbgA-mediated lipid regulation has aided in defining mechanisms of S. Typhimurium immune evasion and antimicrobial resistance.

Melina B. Cian, Ph.D., is a postdoctoral fellow in the Department of Microbiology and Immunology at the University of Oklahoma Health Sciences Center. She is originally from Reconquista, Santa Fe, Argentina. She received her B.S. in Clinical Biochemistry at the National University of Cordoba, Argentina. For her graduate studies, she joined the laboratory of Dr. Jose Echenique to conduct research on bacterial mechanisms of intracellular survival using the Gram-positive pathogen Streptococcus pneumoniae. During her Ph.D. work, she studied the synergistic mechanisms underlying S. pneumoniae and influenza A virus infections. For her postdoctoral work, she joined Dr. Dalebroux’s lab, where she is currently studying how S. Typhimurium regulates the lipids and glycolipids for their cell envelope to alter the host immune response at a cellular and systemic level.

Zachary D. Dalebroux, Ph.D., is an Assistant Professor in the Department of Microbiology and Immunology at the University of Oklahoma Health Sciences Center. Originally from Wisconsin, he received his B.Sc. in Bacteriology from the University of Wisconsin. He joined the laboratory of Dr. Michele Swanson at the University of Michigan for his Ph.D. in Microbiology and Immunology. During his Ph.D. work, he studied the role of guanosine tetraphosphate (ppGpp) and DksA in controlling Legionella pneumophila virulence gene regulation and host-to-host transmission in macrophages. He performed his postdoctoral studies at the University of Washington in Seattle in the laboratory of Dr. Samuel Miller. During his postdoc work, Dr. Dalebroux showed that S. Typhimurium used the PhoPQ virulence regulators to control the acidic glycerophospholipid content of the outer membrane to enhance their survival in their mammalian hosts. Since 2015, Dr. Dalebroux has been working as a principal investigator deciphering salmonella mechanisms of lipid regulation, which enhance bacterial immune evasion and promote antimicrobial resistance.
REFERENCES
- 1.Silhavy TJ, Kahne D, Walker S. 2010. The bacterial cell envelope. Cold Spring Harb Perspect Biol 2:a000414. doi: 10.1101/cshperspect.a000414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Powers MJ, Trent MS. 2019. Intermembrane transport: glycerophospholipid homeostasis of the Gram-negative cell envelope. Proc Natl Acad Sci U S A 116:17147–17155. doi: 10.1073/pnas.1902026116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Shrivastava R, Chng SS. 2019. Lipid trafficking across the Gram-negative cell envelope. J Biol Chem 294:14175–14184. doi: 10.1074/jbc.AW119.008139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Simpson BW, Trent MS. 2019. Pushing the envelope: LPS modifications and their consequences. Nat Rev Microbiol 17:403–416. doi: 10.1038/s41579-019-0201-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gioannini TL, Teghanemt A, Zhang D, Coussens NP, Dockstader W, Ramaswamy S, Weiss JP. 2004. Isolation of an endotoxin-MD-2 complex that produces Toll-like receptor 4-dependent cell activation at picomolar concentrations. Proc Natl Acad Sci U S A 101:4186–4191. doi: 10.1073/pnas.0306906101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Takeuchi O, Akira S. 2010. Pattern recognition receptors and inflammation. Cell 140:805–820. doi: 10.1016/j.cell.2010.01.022. [DOI] [PubMed] [Google Scholar]
- 7.Shi J, Zhao Y, Wang Y, Gao W, Ding J, Li P, Hu L, Shao F. 2014. Inflammatory caspases are innate immune receptors for intracellular LPS. Nature 514:187–192. doi: 10.1038/nature13683. [DOI] [PubMed] [Google Scholar]
- 8.Loppnow H, Brade H, Durrbaum I, Dinarello CA, Kusumoto S, Rietschel ET, Flad HD. 1989. IL-1 induction-capacity of defined lipopolysaccharide partial structures. J Immunol 142:3229–3238. [PubMed] [Google Scholar]
- 9.Kayagaki N, Wong MT, Stowe IB, Ramani SR, Gonzalez LC, Akashi-Takamura S, Miyake K, Zhang J, Lee WP, Muszyński A, Forsberg LS, Carlson RW, Dixit VM. 2013. Noncanonical inflammasome activation by intracellular LPS independent of TLR4. Science 341:1246–1249. doi: 10.1126/science.1240248. [DOI] [PubMed] [Google Scholar]
- 10.Golenbock DT, Hampton RY, Qureshi N, Takayama K, Raetz CR. 1991. Lipid A-like molecules that antagonize the effects of endotoxins on human monocytes. J Biol Chem 266:19490–19498. [PubMed] [Google Scholar]
- 11.Mishra V, Pathak C. 2019. Human Toll-like receptor 4 (hTLR4): structural and functional dynamics in cancer. Int J Biol Macromol 122:425–451. doi: 10.1016/j.ijbiomac.2018.10.142. [DOI] [PubMed] [Google Scholar]
- 12.Lagrange B, Benaoudia S, Wallet P, Magnotti F, Provost A, Michal F, Martin A, Di Lorenzo F, Py BF, Molinaro A, Henry T. 2018. Human caspase-4 detects tetra-acylated LPS and cytosolic Francisella and functions differently from murine caspase-11. Nat Commun 9:242. doi: 10.1038/s41467-017-02682-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Pineau B, Bourge M, Marion J, Mauve C, Gilard F, Maneta-Peyret L, Moreau P, Satiat-Jeunemaître B, Brown SC, De Paepe R, Danon A. 2013. The importance of cardiolipin synthase for mitochondrial ultrastructure, respiratory function, plant development, and stress responses in Arabidopsis. Plant Cell 25:4195–4208. doi: 10.1105/tpc.113.118018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mileykovskaya E, Dowhan W. 2000. Visualization of phospholipid domains in Escherichia coli by using the cardiolipin-specific fluorescent dye 10-N-nonyl acridine orange. J Bacteriol 182:1172–1175. doi: 10.1128/jb.182.4.1172-1175.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mileykovskaya E, Dowhan W. 2009. Cardiolipin membrane domains in prokaryotes and eukaryotes. Biochim Biophys Acta 1788:2084–2091. doi: 10.1016/j.bbamem.2009.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Pizzuto M, Lonez C, Baroja-Mazo A, Martinez-Banaclocha H, Tourlomousis P, Gangloff M, Pelegrin P, Ruysschaert JM, Gay NJ, Bryant CE. 2019. Saturation of acyl chains converts cardiolipin from an antagonist to an activator of Toll-like receptor-4. Cell Mol Life Sci 76:3667–3678. doi: 10.1007/s00018-019-03113-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Schwechheimer C, Kuehn MJ. 2015. Outer-membrane vesicles from Gram-negative bacteria: biogenesis and functions. Nat Rev Microbiol 13:605–619. doi: 10.1038/nrmicro3525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chandler CE, Ernst RK. 2017. Bacterial lipids: powerful modifiers of the innate immune response. F1000Res 6:1334. doi: 10.12688/f1000research.11388.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Park KS, Choi KH, Kim YS, Hong BS, Kim OY, Kim JH, Yoon CM, Koh GY, Kim YK, Gho YS. 2010. Outer membrane vesicles derived from Escherichia coli induce systemic inflammatory response syndrome. PLoS One 5:e11334. doi: 10.1371/journal.pone.0011334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Shah B, Sullivan CJ, Lonergan NE, Stanley S, Soult MC, Britt LD. 2012. Circulating bacterial membrane vesicles cause sepsis in rats. Shock 37:621–628. doi: 10.1097/SHK.0b013e318250de5d. [DOI] [PubMed] [Google Scholar]
- 21.Park KS, Lee J, Lee C, Park HT, Kim JW, Kim OY, Kim SR, Radinger M, Jung HY, Park J, Lotvall J, Gho YS. 2018. Sepsis-like systemic inflammation induced by nano-sized extracellular vesicles from feces. Front Microbiol 9:1735. doi: 10.3389/fmicb.2018.01735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Raeven P, Zipperle J, Drechsler S. 2018. Extracellular vesicles as markers and mediators in sepsis. Theranostics 8:3348–3365. doi: 10.7150/thno.23453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Marion CR, Lee J, Sharma L, Park KS, Lee C, Liu W, Liu P, Feng J, Gho YS, Dela Cruz CS. 2019. Toll-like receptors 2 and 4 modulate pulmonary inflammation and host factors mediated by outer membrane vesicles derived from Acinetobacter baumannii. Infect Immun 87:e00243-19. doi: 10.1128/IAI.00243-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.De Nardo D. 2015. Toll-like receptors: activation, signalling and transcriptional modulation. Cytokine 74:181–189. doi: 10.1016/j.cyto.2015.02.025. [DOI] [PubMed] [Google Scholar]
- 25.Ohto U, Fukase K, Miyake K, Shimizu T. 2012. Structural basis of species-specific endotoxin sensing by innate immune receptor TLR4/MD-2. Proc Natl Acad Sci U S A 109:7421–7426. doi: 10.1073/pnas.1201193109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ohto U, Yamakawa N, Akashi-Takamura S, Miyake K, Shimizu T. 2012. Structural analyses of human Toll-like receptor 4 polymorphisms D299G and T399I. J Biol Chem 287:40611–40617. doi: 10.1074/jbc.M112.404608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Park BS, Lee JO. 2013. Recognition of lipopolysaccharide pattern by TLR4 complexes. Exp Mol Med 45:e66. doi: 10.1038/emm.2013.97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Steimle A, Autenrieth IB, Frick JS. 2016. Structure and function: lipid A modifications in commensals and pathogens. Int J Med Microbiol 306:290–301. doi: 10.1016/j.ijmm.2016.03.001. [DOI] [PubMed] [Google Scholar]
- 29.Kong Q, Six DA, Liu Q, Gu L, Wang S, Alamuri P, Raetz CR, Curtiss R III. 2012. Phosphate groups of lipid A are essential for Salmonella enterica serovar Typhimurium virulence and affect innate and adaptive immunity. Infect Immun 80:3215–3224. doi: 10.1128/IAI.00123-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Trent MS, Pabich W, Raetz CR, Miller SI. 2001. A PhoP/PhoQ-induced lipase (PagL) that catalyzes 3-O-deacylation of lipid A precursors in membranes of Salmonella typhimurium. J Biol Chem 276:9083–9092. doi: 10.1074/jbc.M010730200. [DOI] [PubMed] [Google Scholar]
- 31.Teghanemt A, Zhang D, Levis EN, Weiss JP, Gioannini TL. 2005. Molecular basis of reduced potency of underacylated endotoxins. J Immunol 175:4669–4676. doi: 10.4049/jimmunol.175.7.4669. [DOI] [PubMed] [Google Scholar]
- 32.Aldapa-Vega G, Moreno-Eutimio MA, Berlanga-Taylor AJ, Jiménez-Uribe AP, Nieto-Velazquez G, López-Ortega O, Mancilla-Herrera I, Cortés-Malagón EM, Gunn JS, Isibasi A, Wong-Baeza I, López-Macías C, Pastelin-Palacios R. 2019. Structural variants of Salmonella Typhimurium lipopolysaccharide induce less dimerization of TLR4/MD-2 and reduced pro-inflammatory cytokine production in human monocytes. Mol Immunol 111:43–52. doi: 10.1016/j.molimm.2019.03.003. [DOI] [PubMed] [Google Scholar]
- 33.Coats SR, Berezow AB, To TT, Jain S, Bainbridge BW, Banani KP, Darveau RP. 2011. The lipid A phosphate position determines differential host Toll-like receptor 4 responses to phylogenetically related symbiotic and pathogenic bacteria. Infect Immun 79:203–210. doi: 10.1128/IAI.00937-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Shen Y, Giardino Torchia ML, Lawson GW, Karp CL, Ashwell JD, Mazmanian SK. 2012. Outer membrane vesicles of a human commensal mediate immune regulation and disease protection. Cell Host Microbe 12:509–520. doi: 10.1016/j.chom.2012.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Li Y, Wang Z, Chen J, Ernst RK, Wang X. 2013. Influence of lipid A acylation pattern on membrane permeability and innate immune stimulation. Mar Drugs 11:3197–3208. doi: 10.3390/md11093197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kim SJ, Kim HM. 2017. Dynamic lipopolysaccharide transfer cascade to TLR4/MD2 complex via LBP and CD14. BMB Rep 50:55–57. doi: 10.5483/bmbrep.2017.50.2.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Cochet F, Peri F. 2017. The role of carbohydrates in the lipopolysaccharide (LPS)/Toll-like receptor 4 (TLR4) signalling. Int J Mol Sci 18:E2318. doi: 10.3390/ijms18112318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hailman E, Lichenstein HS, Wurfel MM, Miller DS, Johnson DA, Kelley M, Busse LA, Zukowski MM, Wright SD. 1994. Lipopolysaccharide (LPS)-binding protein accelerates the binding of LPS to CD14. J Exp Med 179:269–277. doi: 10.1084/jem.179.1.269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Vesy CJ, Kitchens RL, Wolfbauer G, Albers JJ, Munford RS. 2000. Lipopolysaccharide-binding protein and phospholipid transfer protein release lipopolysaccharides from Gram-negative bacterial membranes. Infect Immun 68:2410–2417. doi: 10.1128/iai.68.5.2410-2417.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wurfel MM, Wright SD. 1997. Lipopolysaccharide-binding protein and soluble CD14 transfer lipopolysaccharide to phospholipid bilayers: preferential interaction with particular classes of lipid. J Immunol 158:3925–3934. [PubMed] [Google Scholar]
- 41.Tobias PS, Ulevitch RJ. 1993. Lipopolysaccharide binding protein and CD14 in LPS dependent macrophage activation. Immunobiology 187:227–232. doi: 10.1016/S0171-2985(11)80341-4. [DOI] [PubMed] [Google Scholar]
- 42.Pugin J, Schurer-Maly CC, Leturcq D, Moriarty A, Ulevitch RJ, Tobias PS. 1993. Lipopolysaccharide activation of human endothelial and epithelial cells is mediated by lipopolysaccharide-binding protein and soluble CD14. Proc Natl Acad Sci U S A 90:2744–2748. doi: 10.1073/pnas.90.7.2744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ryu JK, Kim SJ, Rah SH, Kang JI, Jung HE, Lee D, Lee HK, Lee JO, Park BS, Yoon TY, Kim HM. 2017. Reconstruction of LPS transfer cascade reveals structural determinants within LBP, CD14, and TLR4-MD2 for efficient LPS recognition and transfer. Immunity 46:38–50. doi: 10.1016/j.immuni.2016.11.007. [DOI] [PubMed] [Google Scholar]
- 44.Eckert JK, Kim YJ, Kim JI, Gurtler K, Oh DY, Sur S, Lundvall L, Hamann L, van der Ploeg A, Pickkers P, Giamarellos-Bourboulis E, Kubarenko AV, Weber AN, Kabesch M, Kumpf O, An HJ, Lee JO, Schumann RR. 2013. The crystal structure of lipopolysaccharide binding protein reveals the location of a frequent mutation that impairs innate immunity. Immunity 39:647–660. doi: 10.1016/j.immuni.2013.09.005. [DOI] [PubMed] [Google Scholar]
- 45.Rosadini CV, Kagan JC. 2017. Early innate immune responses to bacterial LPS. Curr Opin Immunol 44:14–19. doi: 10.1016/j.coi.2016.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kaparakis-Liaskos M, Ferrero RL. 2015. Immune modulation by bacterial outer membrane vesicles. Nat Rev Immunol 15:375–387. doi: 10.1038/nri3837. [DOI] [PubMed] [Google Scholar]
- 47.Backhed F, Normark S, Schweda EK, Oscarson S, Richter-Dahlfors A. 2003. Structural requirements for TLR4-mediated LPS signalling: a biological role for LPS modifications. Microbes Infect 5:1057–1063. doi: 10.1016/s1286-4579(03)00207-7. [DOI] [PubMed] [Google Scholar]
- 48.Zhou X, Gao X, Broglie PM, Kebaier C, Anderson JE, Thom N, Apicella MA, Sempowski GD, Duncan JA. 2014. Hexa-acylated lipid A is required for host inflammatory response to Neisseria gonorrhoeae in experimental gonorrhea. Infect Immun 82:184–192. doi: 10.1128/IAI.00890-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Park BS, Song DH, Kim HM, Choi BS, Lee H, Lee JO. 2009. The structural basis of lipopolysaccharide recognition by the TLR4-MD-2 complex. Nature 458:1191–1195. doi: 10.1038/nature07830. [DOI] [PubMed] [Google Scholar]
- 50.Zimmer SM, Zughaier SM, Tzeng YL, Stephens DS. 2007. Human MD-2 discrimination of meningococcal lipid A structures and activation of TLR4. Glycobiology 17:847–856. doi: 10.1093/glycob/cwm057. [DOI] [PubMed] [Google Scholar]
- 51.Hellum M, Troseid AS, Berg JP, Brandtzaeg P, Ovstebo R, Henriksson CE. 2017. The Neisseria meningitidis lpxL1 mutant induces less tissue factor expression and activity in primary human monocytes and monocyte-derived microvesicles than the wild type meningococcus. Innate Immun 23:196–205. doi: 10.1177/1753425916684201. [DOI] [PubMed] [Google Scholar]
- 52.D’Hauteville H, Khan S, Maskell DJ, Kussak A, Weintraub A, Mathison J, Ulevitch RJ, Wuscher N, Parsot C, Sansonetti PJ. 2002. Two msbB genes encoding maximal acylation of lipid A are required for invasive Shigella flexneri to mediate inflammatory rupture and destruction of the intestinal epithelium. J Immunol 168:5240–5251. doi: 10.4049/jimmunol.168.10.5240. [DOI] [PubMed] [Google Scholar]
- 53.Kawasaki K, Ernst RK, Miller SI. 2004. Deacylation and palmitoylation of lipid A by Salmonellae outer membrane enzymes modulate host signaling through Toll-like receptor 4. J Endotoxin Res 10:439–444. doi: 10.1179/096805104225006264. [DOI] [PubMed] [Google Scholar]
- 54.Kawasaki K, Ernst RK, Miller SI. 2004. 3-O-deacylation of lipid A by PagL, a PhoP/PhoQ-regulated deacylase of Salmonella typhimurium, modulates signaling through Toll-like receptor 4. J Biol Chem 279:20044–20048. doi: 10.1074/jbc.M401275200. [DOI] [PubMed] [Google Scholar]
- 55.Kawano M, Manabe T, Kawasaki K. 2010. Salmonella enterica serovar Typhimurium lipopolysaccharide deacylation enhances its intracellular growth within macrophages. FEBS Lett 584:207–212. doi: 10.1016/j.febslet.2009.11.062. [DOI] [PubMed] [Google Scholar]
- 56.Kim HM, Park BS, Kim JI, Kim SE, Lee J, Oh SC, Enkhbayar P, Matsushima N, Lee H, Yoo OJ, Lee JO. 2007. Crystal structure of the TLR4-MD-2 complex with bound endotoxin antagonist Eritoran. Cell 130:906–917. doi: 10.1016/j.cell.2007.08.002. [DOI] [PubMed] [Google Scholar]
- 57.Cullen TW, Giles DK, Wolf LN, Ecobichon C, Boneca IG, Trent MS. 2011. Helicobacter pylori versus the host: remodeling of the bacterial outer membrane is required for survival in the gastric mucosa. PLoS Pathog 7:e1002454. doi: 10.1371/journal.ppat.1002454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Coats SR, Jones JW, Do CT, Braham PH, Bainbridge BW, To TT, Goodlett DR, Ernst RK, Darveau RP. 2009. Human Toll-like receptor 4 responses to P. gingivalis are regulated by lipid A 1- and 4’-phosphatase activities. Cell Microbiol 11:1587–1599. doi: 10.1111/j.1462-5822.2009.01349.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Coats SR, To TT, Jain S, Braham PH, Darveau RP. 2009. Porphyromonas gingivalis resistance to polymyxin B is determined by the lipid A 4’-phosphatase, PGN_0524. Int J Oral Sci 1:126–135. doi: 10.4248/IJOS.09062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Kanistanon D, Powell DA, Hajjar AM, Pelletier MR, Cohen IE, Way SS, Skerrett SJ, Wang X, Raetz CR, Ernst RK. 2012. Role of Francisella lipid A phosphate modification in virulence and long-term protective immune responses. Infect Immun 80:943–951. doi: 10.1128/IAI.06109-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Que-Gewirth NL, Ribeiro AA, Kalb SR, Cotter RJ, Bulach DM, Adler B, Girons IS, Werts C, Raetz CR. 2004. A methylated phosphate group and four amide-linked acyl chains in Leptospira interrogans lipid A. The membrane anchor of an unusual lipopolysaccharide that activates TLR2. J Biol Chem 279:25420–25429. doi: 10.1074/jbc.M400598200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Coats SR, Hashim A, Paramonov NA, To TT, Curtis MA, Darveau RP. 2016. Cardiolipins act as a selective barrier to Toll-like receptor 4 activation in the intestine. Appl Environ Microbiol 82:4264–4278. doi: 10.1128/AEM.00463-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Romantsov T, Helbig S, Culham DE, Gill C, Stalker L, Wood JM. 2007. Cardiolipin promotes polar localization of osmosensory transporter ProP in Escherichia coli. Mol Microbiol 64:1455–1465. doi: 10.1111/j.1365-2958.2007.05727.x. [DOI] [PubMed] [Google Scholar]
- 64.Renner LD, Weibel DB. 2011. Cardiolipin microdomains localize to negatively curved regions of Escherichia coli membranes. Proc Natl Acad Sci U S A 108:6264–6269. doi: 10.1073/pnas.1015757108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Elias-Wolff F, Linden M, Lyubartsev AP, Brandt EG. 2019. Curvature sensing by cardiolipin in simulated buckled membranes. Soft Matter 15:792–802. doi: 10.1039/c8sm02133c. [DOI] [PubMed] [Google Scholar]
- 66.Beltran-Heredia E, Tsai FC, Salinas-Almaguer S, Cao FJ, Bassereau P, Monroy F. 2019. Membrane curvature induces cardiolipin sorting. Commun Biol 2:225. doi: 10.1038/s42003-019-0471-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Ardail D, Privat JP, Egret-Charlier M, Levrat C, Lerme F, Louisot P. 1990. Mitochondrial contact sites. Lipid composition and dynamics. J Biol Chem 265:18797–18802. [PubMed] [Google Scholar]
- 68.Epand RF, Tokarska-Schlattner M, Schlattner U, Wallimann T, Epand RM. 2007. Cardiolipin clusters and membrane domain formation induced by mitochondrial proteins. J Mol Biol 365:968–980. doi: 10.1016/j.jmb.2006.10.028. [DOI] [PubMed] [Google Scholar]
- 69.Iyer SS, He Q, Janczy JR, Elliott EI, Zhong Z, Olivier AK, Sadler JJ, Knepper-Adrian V, Han R, Qiao L, Eisenbarth SC, Nauseef WM, Cassel SL, Sutterwala FS. 2013. Mitochondrial cardiolipin is required for Nlrp3 inflammasome activation. Immunity 39:311–323. doi: 10.1016/j.immuni.2013.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Kagan VE, Tyurin VA, Jiang J, Tyurina YY, Ritov VB, Amoscato AA, Osipov AN, Belikova NA, Kapralov AA, Kini V, Vlasova II, Zhao Q, Zou M, Di P, Svistunenko DA, Kurnikov IV, Borisenko GG. 2005. Cytochrome c acts as a cardiolipin oxygenase required for release of proapoptotic factors. Nat Chem Biol 1:223–232. doi: 10.1038/nchembio727. [DOI] [PubMed] [Google Scholar]
- 71.Buland JR, Wasserloos KJ, Tyurin VA, Tyurina YY, Amoscato AA, Mallampalli RK, Chen BB, Zhao J, Zhao Y, Ofori-Acquah S, Kagan VE, Pitt BR. 2016. Biosynthesis of oxidized lipid mediators via lipoprotein-associated phospholipase A2 hydrolysis of extracellular cardiolipin induces endothelial toxicity. Am J Physiol Lung Cell Mol Physiol 311:L303–L316. doi: 10.1152/ajplung.00038.2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Dudek J. 2017. Role of cardiolipin in mitochondrial signaling pathways. Front Cell Dev Biol 5:90. doi: 10.3389/fcell.2017.00090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Huang W, Choi W, Hu W, Mi N, Guo Q, Ma M, Liu M, Tian Y, Lu P, Wang FL, Deng H, Liu L, Gao N, Yu L, Shi Y. 2012. Crystal structure and biochemical analyses reveal Beclin 1 as a novel membrane binding protein. Cell Res 22:473–489. doi: 10.1038/cr.2012.24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Balasubramanian K, Maeda A, Lee JS, Mohammadyani D, Dar HH, Jiang JF, St Croix CM, Watkins S, Tyurin VA, Tyurina YY, Kloditz K, Polimova A, Kapralova VI, Xiong Z, Ray P, Klein-Seetharaman J, Mallampalli RK, Bayir H, Fadeel B, Kagan VE. 2015. Dichotomous roles for externalized cardiolipin in extracellular signaling: promotion of phagocytosis and attenuation of innate immunity. Sci Signal 8:ra95. doi: 10.1126/scisignal.aaa6179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Mueller M, Brandenburg K, Dedrick R, Schromm AB, Seydel U. 2005. Phospholipids inhibit lipopolysaccharide (LPS)-induced cell activation: a role for LPS-binding protein. J Immunol 174:1091–1096. doi: 10.4049/jimmunol.174.2.1091. [DOI] [PubMed] [Google Scholar]
- 76.Ohto U, Fukase K, Miyake K, Satow Y. 2007. Crystal structures of human MD-2 and its complex with antiendotoxic lipid IVa. Science 316:1632–1634. doi: 10.1126/science.1139111. [DOI] [PubMed] [Google Scholar]
- 77.Meng J, Drolet JR, Monks BG, Golenbock DT. 2010. MD-2 residues tyrosine 42, arginine 69, aspartic acid 122, and leucine 125 provide species specificity for lipid IVA. J Biol Chem 285:27935–27943. doi: 10.1074/jbc.M110.134668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Garcia-del Portillo F, Stein MA, Finlay BB. 1997. Release of lipopolysaccharide from intracellular compartments containing Salmonella typhimurium to vesicles of the host epithelial cell. Infect Immun 65:24–34. doi: 10.1128/IAI.65.1.24-34.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.van Langevelde P, Kwappenberg KM, Groeneveld PH, Mattie H, van Dissel JT. 1998. Antibiotic-induced lipopolysaccharide (LPS) release from Salmonella typhi: delay between killing by ceftazidime and imipenem and release of LPS. Antimicrob Agents Chemother 42:739–743. doi: 10.1128/AAC.42.4.739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Bonnington KE, Kuehn MJ. 2016. Outer membrane vesicle production facilitates LPS remodeling and outer membrane maintenance in Salmonella during environmental transitions. mBio 7:e01532-16. doi: 10.1128/mBio.01532-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Vanaja SK, Russo AJ, Behl B, Banerjee I, Yankova M, Deshmukh SD, Rathinam V. 2016. Bacterial outer membrane vesicles mediate cytosolic localization of LPS and caspase-11 activation. Cell 165:1106–1119. doi: 10.1016/j.cell.2016.04.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Doherty GJ, McMahon HT. 2009. Mechanisms of endocytosis. Annu Rev Biochem 78:857–902. doi: 10.1146/annurev.biochem.78.081307.110540. [DOI] [PubMed] [Google Scholar]
- 83.Furuta N, Tsuda K, Omori H, Yoshimori T, Yoshimura F, Amano A. 2009. Porphyromonas gingivalis outer membrane vesicles enter human epithelial cells via an endocytic pathway and are sorted to lysosomal compartments. Infect Immun 77:4187–4196. doi: 10.1128/IAI.00009-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Kaparakis M, Turnbull L, Carneiro L, Firth S, Coleman HA, Parkington HC, Le Bourhis L, Karrar A, Viala J, Mak J, Hutton ML, Davies JK, Crack PJ, Hertzog PJ, Philpott DJ, Girardin SE, Whitchurch CB, Ferrero RL. 2010. Bacterial membrane vesicles deliver peptidoglycan to NOD1 in epithelial cells. Cell Microbiol 12:372–385. doi: 10.1111/j.1462-5822.2009.01404.x. [DOI] [PubMed] [Google Scholar]
- 85.Tsuda K, Furuta N, Inaba H, Kawai S, Hanada K, Yoshimori T, Amano A. 2008. Functional analysis of alpha5beta1 integrin and lipid rafts in invasion of epithelial cells by Porphyromonas gingivalis using fluorescent beads coated with bacterial membrane vesicles. Cell Struct Funct 33:123–132. doi: 10.1247/csf.08012. [DOI] [PubMed] [Google Scholar]
- 86.Riethmuller J, Riehle A, Grassme H, Gulbins E. 2006. Membrane rafts in host-pathogen interactions. Biochim Biophys Acta 1758:2139–2147. doi: 10.1016/j.bbamem.2006.07.017. [DOI] [PubMed] [Google Scholar]
- 87.Turner L, Bitto NJ, Steer DL, Lo C, D’Costa K, Ramm G, Shambrook M, Hill AF, Ferrero RL, Kaparakis-Liaskos M. 2018. Helicobacter pylori outer membrane vesicle size determines their mechanisms of host cell entry and protein content. Front Immunol 9:1466. doi: 10.3389/fimmu.2018.01466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Conner SD, Schmid SL. 2003. Regulated portals of entry into the cell. Nature 422:37–44. doi: 10.1038/nature01451. [DOI] [PubMed] [Google Scholar]
- 89.Parker H, Chitcholtan K, Hampton MB, Keenan JI. 2010. Uptake of Helicobacter pylori outer membrane vesicles by gastric epithelial cells. Infect Immun 78:5054–5061. doi: 10.1128/IAI.00299-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Olofsson A, Nygard Skalman L, Obi I, Lundmark R, Arnqvist A. 2014. Uptake of Helicobacter pylori vesicles is facilitated by clathrin-dependent and clathrin-independent endocytic pathways. mBio 5:e00979-14. doi: 10.1128/mBio.00979-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Bielaszewska M, Ruter C, Kunsmann L, Greune L, Bauwens A, Zhang W, Kuczius T, Kim KS, Mellmann A, Schmidt MA, Karch H. 2013. Enterohemorrhagic Escherichia coli hemolysin employs outer membrane vesicles to target mitochondria and cause endothelial and epithelial apoptosis. PLoS Pathog 9:e1003797. doi: 10.1371/journal.ppat.1003797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Bielaszewska M, Ruter C, Bauwens A, Greune L, Jarosch KA, Steil D, Zhang W, He X, Lloubes R, Fruth A, Kim KS, Schmidt MA, Dobrindt U, Mellmann A, Karch H. 2017. Host cell interactions of outer membrane vesicle-associated virulence factors of enterohemorrhagic Escherichia coli O157: intracellular delivery, trafficking and mechanisms of cell injury. PLoS Pathog 13:e1006159. doi: 10.1371/journal.ppat.1006159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Kunsmann L, Ruter C, Bauwens A, Greune L, Gluder M, Kemper B, Fruth A, Wai SN, He X, Lloubes R, Schmidt MA, Dobrindt U, Mellmann A, Karch H, Bielaszewska M. 2015. Virulence from vesicles: novel mechanisms of host cell injury by Escherichia coli O104:H4 outbreak strain. Sci Rep 5:13252. doi: 10.1038/srep13252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Pollak CN, Delpino MV, Fossati CA, Baldi PC. 2012. Outer membrane vesicles from Brucella abortus promote bacterial internalization by human monocytes and modulate their innate immune response. PLoS One 7:e50214. doi: 10.1371/journal.pone.0050214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Lee JJ, Kim DG, Kim DH, Simborio HL, Min W, Lee HJ, Her M, Jung SC, Watarai M, Kim S. 2013. Interplay between clathrin and Rab5 controls the early phagocytic trafficking and intracellular survival of Brucella abortus within HeLa cells. J Biol Chem 288:28049–28057. doi: 10.1074/jbc.M113.491555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Sharpe SW, Kuehn MJ, Mason KM. 2011. Elicitation of epithelial cell-derived immune effectors by outer membrane vesicles of nontypeable Haemophilus influenzae. Infect Immun 79:4361–4369. doi: 10.1128/IAI.05332-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Rai AK, Johnson PJ. 2019. Trichomonas vaginalis extracellular vesicles are internalized by host cells using proteoglycans and caveolin-dependent endocytosis. Proc Natl Acad Sci U S A 116:21354–21360. doi: 10.1073/pnas.1912356116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Rothberg KG, Heuser JE, Donzell WC, Ying YS, Glenney JR, Anderson RG. 1992. Caveolin, a protein component of caveolae membrane coats. Cell 68:673–682. doi: 10.1016/0092-8674(92)90143-z. [DOI] [PubMed] [Google Scholar]
- 99.Anderson RG. 1998. The caveolae membrane system. Annu Rev Biochem 67:199–225. doi: 10.1146/annurev.biochem.67.1.199. [DOI] [PubMed] [Google Scholar]
- 100.Stan RV. 2005. Structure of caveolae. Biochim Biophys Acta 1746:334–348. doi: 10.1016/j.bbamcr.2005.08.008. [DOI] [PubMed] [Google Scholar]
- 101.Amano A, Takeuchi H, Furuta N. 2010. Outer membrane vesicles function as offensive weapons in host-parasite interactions. Microbes Infect 12:791–798. doi: 10.1016/j.micinf.2010.05.008. [DOI] [PubMed] [Google Scholar]
- 102.Lim JS, Shin M, Kim HJ, Kim KS, Choy HE, Cho KA. 2014. Caveolin-1 mediates Salmonella invasion via the regulation of SopE-dependent Rac1 activation and actin reorganization. J Infect Dis 210:793–802. doi: 10.1093/infdis/jiu152. [DOI] [PubMed] [Google Scholar]
- 103.Rohde M, Muller E, Chhatwal GS, Talay SR. 2003. Host cell caveolae act as an entry-port for group A streptococci. Cell Microbiol 5:323–342. doi: 10.1046/j.1462-5822.2003.00279.x. [DOI] [PubMed] [Google Scholar]
- 104.Norkin LC, Wolfrom SA, Stuart ES. 2001. Association of caveolin with Chlamydia trachomatis inclusions at early and late stages of infection. Exp Cell Res 266:229–238. doi: 10.1006/excr.2001.5202. [DOI] [PubMed] [Google Scholar]
- 105.Bathori G, Cervenak L, Karadi I. 2004. Caveolae–an alternative endocytotic pathway for targeted drug delivery. Crit Rev Ther Drug Carrier Syst 21:67–95. doi: 10.1615/critrevtherdrugcarriersyst.v21.i2.10. [DOI] [PubMed] [Google Scholar]
- 106.Harder T, Simons K. 1997. Caveolae, DIGs, and the dynamics of sphingolipid-cholesterol microdomains. Curr Opin Cell Biol 9:534–542. doi: 10.1016/s0955-0674(97)80030-0. [DOI] [PubMed] [Google Scholar]
- 107.Brown DA, London E. 1998. Functions of lipid rafts in biological membranes. Annu Rev Cell Dev Biol 14:111–136. doi: 10.1146/annurev.cellbio.14.1.111. [DOI] [PubMed] [Google Scholar]
- 108.Edidin M. 2001. Shrinking patches and slippery rafts: scales of domains in the plasma membrane. Trends Cell Biol 11:492–496. doi: 10.1016/s0962-8924(01)02139-0. [DOI] [PubMed] [Google Scholar]
- 109.Mulcahy LA, Pink RC, Carter DR. 2014. Routes and mechanisms of extracellular vesicle uptake. J Extracell Vesicles 3:24641. doi: 10.3402/jev.v3.24641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Elmi A, Watson E, Sandu P, Gundogdu O, Mills DC, Inglis NF, Manson E, Imrie L, Bajaj-Elliott M, Wren BW, Smith DG, Dorrell N. 2012. Campylobacter jejuni outer membrane vesicles play an important role in bacterial interactions with human intestinal epithelial cells. Infect Immun 80:4089–4098. doi: 10.1128/IAI.00161-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Bauman SJ, Kuehn MJ. 2009. Pseudomonas aeruginosa vesicles associate with and are internalized by human lung epithelial cells. BMC Microbiol 9:26. doi: 10.1186/1471-2180-9-26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Barnaby R, Koeppen K, Stanton BA. 2019. Cyclodextrins reduce the ability of Pseudomonas aeruginosa outer-membrane vesicles to reduce CFTR Cl− secretion. Am J Physiol Lung Cell Mol Physiol 316:L206–L215. doi: 10.1152/ajplung.00316.2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Jin JS, Kwon SO, Moon DC, Gurung M, Lee JH, Kim SI, Lee JC. 2011. Acinetobacter baumannii secretes cytotoxic outer membrane protein A via outer membrane vesicles. PLoS One 6:e17027. doi: 10.1371/journal.pone.0017027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Kim YR, Kim BU, Kim SY, Kim CM, Na HS, Koh JT, Choy HE, Rhee JH, Lee SE. 2010. Outer membrane vesicles of Vibrio vulnificus deliver cytolysin-hemolysin VvhA into epithelial cells to induce cytotoxicity. Biochem Biophys Res Commun 399:607–612. doi: 10.1016/j.bbrc.2010.07.122. [DOI] [PubMed] [Google Scholar]
- 115.Bomberger JM, Maceachran DP, Coutermarsh BA, Ye S, O’Toole GA, Stanton BA. 2009. Long-distance delivery of bacterial virulence factors by Pseudomonas aeruginosa outer membrane vesicles. PLoS Pathog 5:e1000382. doi: 10.1371/journal.ppat.1000382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Russo AJ, Behl B, Banerjee I, Rathinam V. 2018. Emerging insights into noncanonical inflammasome recognition of microbes. J Mol Biol 430:207–216. doi: 10.1016/j.jmb.2017.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Gu L, Meng R, Tang Y, Zhao K, Liang F, Zhang R, Xue Q, Chen F, Xiao X, Wang H, Wang H, Billiar TR, Lu B. 2019. Toll-like receptor 4 signaling licenses the cytosolic transport of lipopolysaccharide from bacterial outer membrane vesicles. Shock 51:256–265. doi: 10.1097/SHK.0000000000001129. [DOI] [PubMed] [Google Scholar]
- 118.Pilla DM, Hagar JA, Haldar AK, Mason AK, Degrandi D, Pfeffer K, Ernst RK, Yamamoto M, Miao EA, Coers J. 2014. Guanylate binding proteins promote caspase-11-dependent pyroptosis in response to cytoplasmic LPS. Proc Natl Acad Sci U S A 111:6046–6051. doi: 10.1073/pnas.1321700111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Santos JC, Dick MS, Lagrange B, Degrandi D, Pfeffer K, Yamamoto M, Meunier E, Pelczar P, Henry T, Broz P. 2018. LPS targets host guanylate-binding proteins to the bacterial outer membrane for non-canonical inflammasome activation. EMBO J 37:e98089. doi: 10.15252/embj.201798089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Finethy R, Luoma S, Orench-Rivera N, Feeley EM, Haldar AK, Yamamoto M, Kanneganti TD, Kuehn MJ, Coers J. 2017. Inflammasome activation by bacterial outer membrane vesicles requires guanylate binding proteins. mBio 8:e01188-17. doi: 10.1128/mBio.01188-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Man SM, Karki R, Sasai M, Place DE, Kesavardhana S, Temirov J, Frase S, Zhu Q, Malireddi RKS, Kuriakose T, Peters JL, Neale G, Brown SA, Yamamoto M, Kanneganti TD. 2016. IRGB10 liberates bacterial ligands for sensing by the AIM2 and caspase-11-NLRP3 inflammasomes. Cell 167:382–396 e17. doi: 10.1016/j.cell.2016.09.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Selleck EM, Fentress SJ, Beatty WL, Degrandi D, Pfeffer K, Virgin HW IV, MacMicking JD, Sibley LD. 2013. Guanylate-binding protein 1 (Gbp1) contributes to cell-autonomous immunity against Toxoplasma gondii. PLoS Pathog 9:e1003320. doi: 10.1371/journal.ppat.1003320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Meunier E, Dick MS, Dreier RF, Schurmann N, Kenzelmann Broz D, Warming S, Roose-Girma M, Bumann D, Kayagaki N, Takeda K, Yamamoto M, Broz P. 2014. Caspase-11 activation requires lysis of pathogen-containing vacuoles by IFN-induced GTPases. Nature 509:366–370. doi: 10.1038/nature13157. [DOI] [PubMed] [Google Scholar]
- 124.Meunier E, Wallet P, Dreier RF, Costanzo S, Anton L, Ruhl S, Dussurgey S, Dick MS, Kistner A, Rigard M, Degrandi D, Pfeffer K, Yamamoto M, Henry T, Broz P. 2015. Guanylate-binding proteins promote activation of the AIM2 inflammasome during infection with Francisella novicida. Nat Immunol 16:476–484. doi: 10.1038/ni.3119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Paz I, Sachse M, Dupont N, Mounier J, Cederfur C, Enninga J, Leffler H, Poirier F, Prevost MC, Lafont F, Sansonetti P. 2010. Galectin-3, a marker for vacuole lysis by invasive pathogens. Cell Microbiol 12:530–544. doi: 10.1111/j.1462-5822.2009.01415.x. [DOI] [PubMed] [Google Scholar]
- 126.Chen S, Yang D, Wen Y, Jiang Z, Zhang L, Jiang J, Chen Y, Hu T, Wang Q, Zhang Y, Liu Q. 2018. Dysregulated hemolysin liberates bacterial outer membrane vesicles for cytosolic lipopolysaccharide sensing. PLoS Pathog 14:e1007240. doi: 10.1371/journal.ppat.1007240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Rathinam VA, Fitzgerald KA. 2016. Inflammasome complexes: emerging mechanisms and effector functions. Cell 165:792–800. doi: 10.1016/j.cell.2016.03.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Santos JC, Broz P. 2018. Sensing of invading pathogens by GBPs: at the crossroads between cell-autonomous and innate immunity. J Leukoc Biol 104:729–735. doi: 10.1002/JLB.4MR0118-038R. [DOI] [PubMed] [Google Scholar]
- 129.Piro AS, Hernandez D, Luoma S, Feeley EM, Finethy R, Yirga A, Frickel EM, Lesser CF, Coers J. 2017. Detection of cytosolic Shigella flexneri via a C-terminal triple-arginine motif of GBP1 inhibits actin-based motility. mBio 8:e01979-17. doi: 10.1128/mBio.01979-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Hagar JA, Powell DA, Aachoui Y, Ernst RK, Miao EA. 2013. Cytoplasmic LPS activates caspase-11: implications in TLR4-independent endotoxic shock. Science 341:1250–1253. doi: 10.1126/science.1240988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Bitto NJ, Baker PJ, Dowling JK, Wray-McCann G, De Paoli A, Tran LS, Leung PL, Stacey KJ, Mansell A, Masters SL, Ferrero RL. 2018. Membrane vesicles from Pseudomonas aeruginosa activate the noncanonical inflammasome through caspase-5 in human monocytes. Immunol Cell Biol 96:1120–1130. doi: 10.1111/imcb.12190. [DOI] [PubMed] [Google Scholar]
- 132.Wang K, Sun Q, Zhong X, Zeng M, Zeng H, Shi X, Li Z, Wang Y, Zhao Q, Shao F, Ding J. 2020. Structural mechanism for GSDMD targeting by autoprocessed caspases in pyroptosis. Cell 180:941–955.e20. doi: 10.1016/j.cell.2020.02.002. [DOI] [PubMed] [Google Scholar]
- 133.Shi J, Zhao Y, Wang K, Shi X, Wang Y, Huang H, Zhuang Y, Cai T, Wang F, Shao F. 2015. Cleavage of GSDMD by inflammatory caspases determines pyroptotic cell death. Nature 526:660–665. doi: 10.1038/nature15514. [DOI] [PubMed] [Google Scholar]
- 134.Liu X, Zhang Z, Ruan J, Pan Y, Magupalli VG, Wu H, Lieberman J. 2016. Inflammasome-activated gasdermin D causes pyroptosis by forming membrane pores. Nature 535:153–158. doi: 10.1038/nature18629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Aglietti RA, Estevez A, Gupta A, Ramirez MG, Liu PS, Kayagaki N, Ciferri C, Dixit VM, Dueber EC. 2016. GsdmD p30 elicited by caspase-11 during pyroptosis forms pores in membranes. Proc Natl Acad Sci U S A 113:7858–7863. doi: 10.1073/pnas.1607769113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Ruan J. 2019. Structural insight of gasdermin family driving pyroptotic cell death. Adv Exp Med Biol 1172:189–205. doi: 10.1007/978-981-13-9367-9_9. [DOI] [PubMed] [Google Scholar]
- 137.Aachoui Y, Leaf IA, Hagar JA, Fontana MF, Campos CG, Zak DE, Tan MH, Cotter PA, Vance RE, Aderem A, Miao EA. 2013. Caspase-11 protects against bacteria that escape the vacuole. Science 339:975–978. doi: 10.1126/science.1230751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Broz P, Ruby T, Belhocine K, Bouley DM, Kayagaki N, Dixit VM, Monack DM. 2012. Caspase-11 increases susceptibility to Salmonella infection in the absence of caspase-1. Nature 490:288–291. doi: 10.1038/nature11419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Case CL, Kohler LJ, Lima JB, Strowig T, de Zoete MR, Flavell RA, Zamboni DS, Roy CR. 2013. Caspase-11 stimulates rapid flagellin-independent pyroptosis in response to Legionella pneumophila. Proc Natl Acad Sci U S A 110:1851–1856. doi: 10.1073/pnas.1211521110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Gutierrez KD, Davis MA, Daniels BP, Olsen TM, Ralli-Jain P, Tait SW, Gale M Jr, Oberst A. 2017. MLKL activation triggers NLRP3-mediated processing and release of IL-1beta independently of gasdermin-D. J Immunol 198:2156–2164. doi: 10.4049/jimmunol.1601757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Muñoz-Planillo R, Kuffa P, Martínez-Colón G, Smith BL, Rajendiran TM, Núñez G. 2013. K+ efflux is the common trigger of NLRP3 inflammasome activation by bacterial toxins and particulate matter. Immunity 38:1142–1153. doi: 10.1016/j.immuni.2013.05.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Ruhl S, Broz P. 2015. Caspase-11 activates a canonical NLRP3 inflammasome by promoting K+ efflux. Eur J Immunol 45:2927–2936. doi: 10.1002/eji.201545772. [DOI] [PubMed] [Google Scholar]
- 143.Gurung P, Malireddi RK, Anand PK, Demon D, Vande Walle L, Liu Z, Vogel P, Lamkanfi M, Kanneganti TD. 2012. Toll or interleukin-1 receptor (TIR) domain-containing adaptor inducing interferon-beta (TRIF)-mediated caspase-11 protease production integrates Toll-like receptor 4 (TLR4) protein- and Nlrp3 inflammasome-mediated host defense against enteropathogens. J Biol Chem 287:34474–34483. doi: 10.1074/jbc.M112.401406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Kayagaki N, Warming S, Lamkanfi M, Vande Walle L, Louie S, Dong J, Newton K, Qu Y, Liu J, Heldens S, Zhang J, Lee WP, Roose-Girma M, Dixit VM. 2011. Non-canonical inflammasome activation targets caspase-11. Nature 479:117–121. doi: 10.1038/nature10558. [DOI] [PubMed] [Google Scholar]
- 145.Trent MS, Ribeiro AA, Doerrler WT, Lin S, Cotter RJ, Raetz CR. 2001. Accumulation of a polyisoprene-linked amino sugar in polymyxin-resistant Salmonella typhimurium and Escherichia coli: structural characterization and transfer to lipid A in the periplasm. J Biol Chem 276:43132–43144. doi: 10.1074/jbc.M106962200. [DOI] [PubMed] [Google Scholar]
- 146.Trent MS, Ribeiro AA, Lin S, Cotter RJ, Raetz CR. 2001. An inner membrane enzyme in Salmonella and Escherichia coli that transfers 4-amino-4-deoxy-L-arabinose to lipid A: induction on polymyxin-resistant mutants and role of a novel lipid-linked donor. J Biol Chem 276:43122–43131. doi: 10.1074/jbc.M106961200. [DOI] [PubMed] [Google Scholar]
- 147.Vinogradov E, Perry MB, Conlan JW. 2002. Structural analysis of Francisella tularensis lipopolysaccharide. Eur J Biochem 269:6112–6118. doi: 10.1046/j.1432-1033.2002.03321.x. [DOI] [PubMed] [Google Scholar]
- 148.Yang C, Briones M, Chiou J, Lei L, Patton MJ, Ma L, McClarty G, Caldwell HD. 2019. Chlamydia trachomatis lipopolysaccharide evades the canonical and noncanonical inflammatory pathways to subvert innate immunity. mBio 10:e00595-19. doi: 10.1128/mBio.00595-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Keestra-Gounder AM, Tsolis RM. 2017. NOD1 and NOD2: beyond peptidoglycan sensing. Trends Immunol 38:758–767. doi: 10.1016/j.it.2017.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Kim YK, Shin JS, Nahm MH. 2016. NOD-like receptors in infection, immunity, and diseases. Yonsei Med J 57:5–14. doi: 10.3349/ymj.2016.57.1.5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Chatterjee D, Chaudhuri K. 2013. Vibrio cholerae O395 outer membrane vesicles modulate intestinal epithelial cells in a NOD1 protein-dependent manner and induce dendritic cell-mediated Th2/Th17 cell responses. J Biol Chem 288:4299–4309. doi: 10.1074/jbc.M112.408302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Bielig H, Rompikuntal PK, Dongre M, Zurek B, Lindmark B, Ramstedt M, Wai SN, Kufer TA. 2011. NOD-like receptor activation by outer membrane vesicles from Vibrio cholerae non-O1 non-O139 strains is modulated by the quorum-sensing regulator HapR. Infect Immun 79:1418–1427. doi: 10.1128/IAI.00754-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Mukherjee T, Hovingh ES, Foerster EG, Abdel-Nour M, Philpott DJ, Girardin SE. 2019. NOD1 and NOD2 in inflammation, immunity and disease. Arch Biochem Biophys 670:69–81. doi: 10.1016/j.abb.2018.12.022. [DOI] [PubMed] [Google Scholar]
- 154.Canas MA, Fabrega MJ, Gimenez R, Badia J, Baldoma L. 2018. Outer membrane vesicles from probiotic and commensal Escherichia coli activate NOD1-mediated immune responses in intestinal epithelial cells. Front Microbiol 9:498. doi: 10.3389/fmicb.2018.00498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Irving AT, Mimuro H, Kufer TA, Lo C, Wheeler R, Turner LJ, Thomas BJ, Malosse C, Gantier MP, Casillas LN, Votta BJ, Bertin J, Boneca IG, Sasakawa C, Philpott DJ, Ferrero RL, Kaparakis-Liaskos M. 2014. The immune receptor NOD1 and kinase RIP2 interact with bacterial peptidoglycan on early endosomes to promote autophagy and inflammatory signaling. Cell Host Microbe 15:623–635. doi: 10.1016/j.chom.2014.04.001. [DOI] [PubMed] [Google Scholar]
- 156.Travassos LH, Carneiro LA, Ramjeet M, Hussey S, Kim YG, Magalhaes JG, Yuan L, Soares F, Chea E, Le Bourhis L, Boneca IG, Allaoui A, Jones NL, Nunez G, Girardin SE, Philpott DJ. 2010. Nod1 and Nod2 direct autophagy by recruiting ATG16L1 to the plasma membrane at the site of bacterial entry. Nat Immunol 11:55–62. doi: 10.1038/ni.1823. [DOI] [PubMed] [Google Scholar]
- 157.Lee EY, Choi DY, Kim DK, Kim JW, Park JO, Kim S, Kim SH, Desiderio DM, Kim YK, Kim KP, Gho YS. 2009. Gram-positive bacteria produce membrane vesicles: proteomics-based characterization of Staphylococcus aureus-derived membrane vesicles. Proteomics 9:5425–5436. doi: 10.1002/pmic.200900338. [DOI] [PubMed] [Google Scholar]
- 158.Hong SW, Kim MR, Lee EY, Kim JH, Kim YS, Jeon SG, Yang JM, Lee BJ, Pyun BY, Gho YS, Kim YK. 2011. Extracellular vesicles derived from Staphylococcus aureus induce atopic dermatitis-like skin inflammation. Allergy 66:351–359. doi: 10.1111/j.1398-9995.2010.02483.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Monnappa AK, Bari W, Seo JK, Mitchell RJ. 2018. The cytotoxic necrotizing factor of Yersinia pseudotuberculosis (CNFy) is carried on extracellular membrane vesicles to host cells. Sci Rep 8:14186. doi: 10.1038/s41598-018-32530-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.van Bergenhenegouwen J, Kraneveld AD, Rutten L, Kettelarij N, Garssen J, Vos AP. 2014. Extracellular vesicles modulate host-microbe responses by altering TLR2 activity and phagocytosis. PLoS One 9:e89121. doi: 10.1371/journal.pone.0089121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Chernov VM, Chernova OA, Mouzykantov AA, Efimova IR, Shaymardanova GF, Medvedeva ES, Trushin MV. 2011. Extracellular vesicles derived from Acholeplasma laidlawii PG8. ScientificWorldJournal 11:1120–1130. doi: 10.1100/tsw.2011.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Rodrigues ML, Nakayasu ES, Oliveira DL, Nimrichter L, Nosanchuk JD, Almeida IC, Casadevall A. 2008. Extracellular vesicles produced by Cryptococcus neoformans contain protein components associated with virulence. Eukaryot Cell 7:58–67. doi: 10.1128/EC.00370-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Vallejo MC, Matsuo AL, Ganiko L, Medeiros LC, Miranda K, Silva LS, Freymuller-Haapalainen E, Sinigaglia-Coimbra R, Almeida IC, Puccia R. 2011. The pathogenic fungus Paracoccidioides brasiliensis exports extracellular vesicles containing highly immunogenic alpha-galactosyl epitopes. Eukaryot Cell 10:343–351. doi: 10.1128/EC.00227-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Toyofuku M, Nomura N, Eberl L. 2019. Types and origins of bacterial membrane vesicles. Nat Rev Microbiol 17:13–24. doi: 10.1038/s41579-018-0112-2. [DOI] [PubMed] [Google Scholar]
- 165.Ellis TN, Kuehn MJ. 2010. Virulence and immunomodulatory roles of bacterial outer membrane vesicles. Microbiol Mol Biol Rev 74:81–94. doi: 10.1128/MMBR.00031-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Beveridge TJ. 1999. Structures of Gram-negative cell walls and their derived membrane vesicles. J Bacteriol 181:4725–4733. doi: 10.1128/JB.181.16.4725-4733.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Zingl FG, Kohl P, Cakar F, Leitner DR, Mitterer F, Bonnington KE, Rechberger GN, Kuehn MJ, Guan Z, Reidl J, Schild S. 2020. Outer membrane vesiculation facilitates surface exchange and in vivo adaptation of Vibrio cholerae. Cell Host Microbe 27:225–237.e8. doi: 10.1016/j.chom.2019.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Wang W, Chanda W, Zhong M. 2015. The relationship between biofilm and outer membrane vesicles: a novel therapy overview. FEMS Microbiol Lett 362:fnv117. doi: 10.1093/femsle/fnv117. [DOI] [PubMed] [Google Scholar]
- 169.Florez C, Raab JE, Cooke AC, Schertzer JW. 2017. Membrane distribution of the Pseudomonas quinolone signal modulates outer membrane vesicle production in Pseudomonas aeruginosa. mBio 8:e01034-17. doi: 10.1128/mBio.01034-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Roier S, Zingl FG, Cakar F, Durakovic S, Kohl P, Eichmann TO, Klug L, Gadermaier B, Weinzerl K, Prassl R, Lass A, Daum G, Reidl J, Feldman MF, Schild S. 2016. A novel mechanism for the biogenesis of outer membrane vesicles in Gram-negative bacteria. Nat Commun 7:10515. doi: 10.1038/ncomms10515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Gerritzen MJH, Martens DE, Uittenbogaard JP, Wijffels RH, Stork M. 2019. Sulfate depletion triggers overproduction of phospholipids and the release of outer membrane vesicles by Neisseria meningitidis. Sci Rep 9:4716. doi: 10.1038/s41598-019-41233-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Mashburn LM, Whiteley M. 2005. Membrane vesicles traffic signals and facilitate group activities in a prokaryote. Nature 437:422–425. doi: 10.1038/nature03925. [DOI] [PubMed] [Google Scholar]
- 173.Jasim R, Han ML, Zhu Y, Hu X, Hussein MH, Lin YW, Zhou QT, Dong CYD, Li J, Velkov T. 2018. Lipidomic analysis of the outer membrane vesicles from paired polymyxin-susceptible and -resistant Klebsiella pneumoniae clinical isolates. Int J Mol Sci 19:E2356. doi: 10.3390/ijms19082356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Kadurugamuwa JL, Clarke AJ, Beveridge TJ. 1993. Surface action of gentamicin on Pseudomonas aeruginosa. J Bacteriol 175:5798–5805. doi: 10.1128/jb.175.18.5798-5805.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.McBroom AJ, Johnson AP, Vemulapalli S, Kuehn MJ. 2006. Outer membrane vesicle production by Escherichia coli is independent of membrane instability. J Bacteriol 188:5385–5392. doi: 10.1128/JB.00498-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Bernadac A, Gavioli M, Lazzaroni JC, Raina S, Lloubes R. 1998. Escherichia coli tol-pal mutants form outer membrane vesicles. J Bacteriol 180:4872–4878. doi: 10.1128/JB.180.18.4872-4878.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Zhou L, Srisatjaluk R, Justus DE, Doyle RJ. 1998. On the origin of membrane vesicles in gram-negative bacteria. FEMS Microbiol Lett 163:223–228. doi: 10.1111/j.1574-6968.1998.tb13049.x. [DOI] [PubMed] [Google Scholar]
- 178.Braun V, Hantke K. 2019. Lipoproteins: structure, function, biosynthesis. Subcell Biochem 92:39–77. doi: 10.1007/978-3-030-18768-2_3. [DOI] [PubMed] [Google Scholar]
- 179.Schwechheimer C, Kulp A, Kuehn MJ. 2014. Modulation of bacterial outer membrane vesicle production by envelope structure and content. BMC Microbiol 14:324. doi: 10.1186/s12866-014-0324-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Deatherage BL, Lara JC, Bergsbaken T, Rassoulian Barrett SL, Lara S, Cookson BT. 2009. Biogenesis of bacterial membrane vesicles. Mol Microbiol 72:1395–1407. doi: 10.1111/j.1365-2958.2009.06731.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Valeru SP, Shanan S, Alossimi H, Saeed A, Sandstrom G, Abd H. 2014. Lack of outer membrane protein A enhances the release of outer membrane vesicles and survival of Vibrio cholerae and suppresses viability of Acanthamoeba castellanii. Int J Microbiol 2014:610190. doi: 10.1155/2014/610190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Moon DC, Choi CH, Lee JH, Choi CW, Kim HY, Park JS, Kim SI, Lee JC. 2012. Acinetobacter baumannii outer membrane protein A modulates the biogenesis of outer membrane vesicles. J Microbiol 50:155–160. doi: 10.1007/s12275-012-1589-4. [DOI] [PubMed] [Google Scholar]
- 183.Egan A. 2018. Bacterial outer membrane constriction. Mol Microbiol 107:676–687. doi: 10.1111/mmi.13908. [DOI] [PubMed] [Google Scholar]
- 184.Gerding MA, Ogata Y, Pecora ND, Niki H, de Boer PA. 2007. The trans-envelope Tol-Pal complex is part of the cell division machinery and required for proper outer-membrane invagination during cell constriction in E. coli. Mol Microbiol 63:1008–1025. doi: 10.1111/j.1365-2958.2006.05571.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Petiti M, Serrano B, Faure L, Lloubes R, Mignot T, Duche D. 2019. Tol energy-driven localization of Pal and anchoring to the peptidoglycan promote outer-membrane constriction. J Mol Biol 431:3275–3288. doi: 10.1016/j.jmb.2019.05.039. [DOI] [PubMed] [Google Scholar]
- 186.Masilamani R, Cian MB, Dalebroux ZD. 2018. Salmonella Tol-Pal reduces outer membrane glycerophospholipid levels for envelope homeostasis and survival during bacteremia. Infect Immun 86:e00173-18. doi: 10.1128/IAI.00173-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Shrivastava R, Jiang X, Chng SS. 2017. Outer membrane lipid homeostasis via retrograde phospholipid transport in Escherichia coli. Mol Microbiol 106:395–408. doi: 10.1111/mmi.13772. [DOI] [PubMed] [Google Scholar]
- 188.Cascales E, Bernadac A, Gavioli M, Lazzaroni JC, Lloubes R. 2002. Pal lipoprotein of Escherichia coli plays a major role in outer membrane integrity. J Bacteriol 184:754–759. doi: 10.1128/jb.184.3.754-759.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189.Turner L, Praszkier J, Hutton ML, Steer D, Ramm G, Kaparakis-Liaskos M, Ferrero RL. 2015. Increased outer membrane vesicle formation in a Helicobacter pylori tolB mutant. Helicobacter 20:269–283. doi: 10.1111/hel.12196. [DOI] [PubMed] [Google Scholar]
- 190.Needham BD, Trent MS. 2013. Fortifying the barrier: the impact of lipid A remodelling on bacterial pathogenesis. Nat Rev Microbiol 11:467–481. doi: 10.1038/nrmicro3047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191.Schromm AB, Brandenburg K, Loppnow H, Moran AP, Koch MH, Rietschel ET, Seydel U. 2000. Biological activities of lipopolysaccharides are determined by the shape of their lipid A portion. Eur J Biochem 267:2008–2013. doi: 10.1046/j.1432-1327.2000.01204.x. [DOI] [PubMed] [Google Scholar]
- 192.Seydel U, Schromm AB, Blunck R, Brandenburg K. 2000. Chemical structure, molecular conformation, and bioactivity of endotoxins. Chem Immunol 74:5–24. doi: 10.1159/000058754. [DOI] [PubMed] [Google Scholar]
- 193.Elhenawy W, Bording-Jorgensen M, Valguarnera E, Haurat MF, Wine E, Feldman MF. 2016. LPS remodeling triggers formation of outer membrane vesicles in Salmonella. mBio 7:e00940-16. doi: 10.1128/mBio.00940-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194.Sinha A, Nyongesa S, Viau C, Gruenheid S, Veyrier FJ, Le Moual H. 2019. PmrC (EptA) and CptA negatively affect outer membrane vesicle production in Citrobacter rodentium. J Bacteriol 201:e00454-18. doi: 10.1128/JB.00454-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 195.Schertzer JW, Whiteley M. 2012. A bilayer-couple model of bacterial outer membrane vesicle biogenesis. mBio 3:e00297-11. doi: 10.1128/mBio.00297-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196.Yeow J, Tan KW, Holdbrook DA, Chong ZS, Marzinek JK, Bond PJ, Chng SS. 2018. The architecture of the OmpC-MlaA complex sheds light on the maintenance of outer membrane lipid asymmetry in Escherichia coli. J Biol Chem 293:11325–11340. doi: 10.1074/jbc.RA118.002441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 197.Ercan B, Low WY, Liu X, Chng SS. 2019. Characterization of interactions and phospholipid transfer between substrate binding proteins of the OmpC-Mla system. Biochemistry 58:114–119. doi: 10.1021/acs.biochem.8b00897. [DOI] [PubMed] [Google Scholar]
- 198.Kamischke C, Fan J, Bergeron J, Kulasekara HD, Dalebroux ZD, Burrell A, Kollman JM, Miller SI. 2019. The Acinetobacter baumannii Mla system and glycerophospholipid transport to the outer membrane. Elife 8:e40171. doi: 10.7554/eLife.40171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 199.Ekiert DC, Bhabha G, Isom GL, Greenan G, Ovchinnikov S, Henderson IR, Cox JS, Vale RD. 2017. Architectures of lipid transport systems for the bacterial outer membrane. Cell 169:273–285.e17. doi: 10.1016/j.cell.2017.03.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 200.Chong ZS, Woo WF, Chng SS. 2015. Osmoporin OmpC forms a complex with MlaA to maintain outer membrane lipid asymmetry in Escherichia coli. Mol Microbiol 98:1133–1146. doi: 10.1111/mmi.13202. [DOI] [PubMed] [Google Scholar]
- 201.Davies C, Taylor AJ, Elmi A, Winter J, Liaw J, Grabowska AD, Gundogdu O, Wren BW, Kelly DJ, Dorrell N. 2019. Sodium taurocholate stimulates Campylobacter jejuni outer membrane vesicle production via down-regulation of the maintenance of lipid asymmetry pathway. Front Cell Infect Microbiol 9:177. doi: 10.3389/fcimb.2019.00177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 202.Malinverni JC, Silhavy TJ. 2009. An ABC transport system that maintains lipid asymmetry in the gram-negative outer membrane. Proc Natl Acad Sci U S A 106:8009–8014. doi: 10.1073/pnas.0903229106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 203.Casali N, Riley LW. 2007. A phylogenomic analysis of the Actinomycetales mce operons. BMC Genomics 8:60. doi: 10.1186/1471-2164-8-60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 204.Benning C. 2009. Mechanisms of lipid transport involved in organelle biogenesis in plant cells. Annu Rev Cell Dev Biol 25:71–91. doi: 10.1146/annurev.cellbio.042308.113414. [DOI] [PubMed] [Google Scholar]
- 205.Grogan DW, Cronan JE Jr. 1997. Cyclopropane ring formation in membrane lipids of bacteria. Microbiol Mol Biol Rev 61:429–441. doi: 10.1128/.61.4.429-441.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 206.Cian MB, Giordano NP, Masilamani R, Minor KE, Dalebroux ZD. 2019. Salmonella enterica serovar Typhimurium uses PbgA/YejM to regulate lipopolysaccharide assembly during bacteremia. Infect Immun 88:e00758-19. doi: 10.1128/IAI.00758-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 207.Raetz CR, Dowhan W. 1990. Biosynthesis and function of phospholipids in Escherichia coli. J Biol Chem 265:1235–1238. [PubMed] [Google Scholar]
- 208.Hughes GW, Hall SCL, Laxton CS, Sridhar P, Mahadi AH, Hatton C, Piggot TJ, Wotherspoon PJ, Leney AC, Ward DG, Jamshad M, Spana V, Cadby IT, Harding C, Isom GL, Bryant JA, Parr RJ, Yakub Y, Jeeves M, Huber D, Henderson IR, Clifton LA, Lovering AL, Knowles TJ. 2019. Evidence for phospholipid export from the bacterial inner membrane by the Mla ABC transport system. Nat Microbiol 4:1692–1705. doi: 10.1038/s41564-019-0481-y. [DOI] [PubMed] [Google Scholar]
- 209.May KL, Silhavy TJ. 2018. The Escherichia coli phospholipase PldA regulates outer membrane homeostasis via lipid signaling. mBio 9:e00379-18. doi: 10.1128/mBio.00379-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 210.Powers MJ, Trent MS. 2018. Phospholipid retention in the absence of asymmetry strengthens the outer membrane permeability barrier to last-resort antibiotics. Proc Natl Acad Sci U S A 115:E8518–E8527. doi: 10.1073/pnas.1806714115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 211.Bishop RE. 2008. Structural biology of membrane-intrinsic beta-barrel enzymes: sentinels of the bacterial outer membrane. Biochim Biophys Acta 1778:1881–1896. doi: 10.1016/j.bbamem.2007.07.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 212.Dekker N. 2000. Outer-membrane phospholipase A: known structure, unknown biological function. Mol Microbiol 35:711–717. doi: 10.1046/j.1365-2958.2000.01775.x. [DOI] [PubMed] [Google Scholar]
- 213.Zheng L, Lin Y, Lu S, Zhang J, Bogdanov M. 2017. Biogenesis, transport and remodeling of lysophospholipids in Gram-negative bacteria. Biochim Biophys Acta Mol Cell Biol Lipids 1862:1404–1413. doi: 10.1016/j.bbalip.2016.11.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 214.Baarda BI, Zielke RA, Le Van A, Jerse AE, Sikora AE. 2019. Neisseria gonorrhoeae MlaA influences gonococcal virulence and membrane vesicle production. PLoS Pathog 15:e1007385. doi: 10.1371/journal.ppat.1007385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 215.Sutterlin HA, Shi H, May KL, Miguel A, Khare S, Huang KC, Silhavy TJ. 2016. Disruption of lipid homeostasis in the Gram-negative cell envelope activates a novel cell death pathway. Proc Natl Acad Sci U S A 113:E1565–E1574. doi: 10.1073/pnas.1601375113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 216.McMahon HT, Gallop JL. 2005. Membrane curvature and mechanisms of dynamic cell membrane remodelling. Nature 438:590–596. doi: 10.1038/nature04396. [DOI] [PubMed] [Google Scholar]
- 217.Frolov VA, Shnyrova AV, Zimmerberg J. 2011. Lipid polymorphisms and membrane shape. Cold Spring Harb Perspect Biol 3:a004747. doi: 10.1101/cshperspect.a004747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 218.Furse S. 2017. Is phosphatidylglycerol essential for terrestrial life? J Chem Biol 10:1–9. doi: 10.1007/s12154-016-0159-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 219.Cooke IR, Deserno M. 2006. Coupling between lipid shape and membrane curvature. Biophys J 91:487–495. doi: 10.1529/biophysj.105.078683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 220.Graham TR, Kozlov MM. 2010. Interplay of proteins and lipids in generating membrane curvature. Curr Opin Cell Biol 22:430–436. doi: 10.1016/j.ceb.2010.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 221.Dalebroux ZD, Miller SI. 2014. Salmonellae PhoPQ regulation of the outer membrane to resist innate immunity. Curr Opin Microbiol 17:106–113. doi: 10.1016/j.mib.2013.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 222.Dalebroux ZD, Matamouros S, Whittington D, Bishop RE, Miller SI. 2014. PhoPQ regulates acidic glycerophospholipid content of the Salmonella Typhimurium outer membrane. Proc Natl Acad Sci U S A 111:1963–1968. doi: 10.1073/pnas.1316901111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 223.Harvat EM, Zhang YM, Tran CV, Zhang Z, Frank MW, Rock CO, Saier MH Jr. 2005. Lysophospholipid flipping across the Escherichia coli inner membrane catalyzed by a transporter (LplT) belonging to the major facilitator superfamily. J Biol Chem 280:12028–12034. doi: 10.1074/jbc.M414368200. [DOI] [PubMed] [Google Scholar]
- 224.Lin Y, Bogdanov M, Tong S, Guan Z, Zheng L. 2016. Substrate selectivity of lysophospholipid transporter LplT involved in membrane phospholipid remodeling in Escherichia coli. J Biol Chem 291:2136–2149. doi: 10.1074/jbc.M115.700419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 225.Emiola A, Andrews SS, Heller C, George J. 2016. Crosstalk between the lipopolysaccharide and phospholipid pathways during outer membrane biogenesis in Escherichia coli. Proc Natl Acad Sci U S A 113:3108–3113. doi: 10.1073/pnas.1521168113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 226.Thomanek N, Arends J, Lindemann C, Barkovits K, Meyer HE, Marcus K, Narberhaus F. 2018. Intricate crosstalk between lipopolysaccharide, phospholipid and fatty acid metabolism in Escherichia coli modulates proteolysis of LpxC. Front Microbiol 9:3285. doi: 10.3389/fmicb.2018.03285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 227.Anderson MS, Robertson AD, Macher I, Raetz CR. 1988. Biosynthesis of lipid A in Escherichia coli: identification of UDP-3-O-[(R)-3-hydroxymyristoyl]-alpha-D-glucosamine as a precursor of UDP-N2,O3-bis[(R)-3-hydroxymyristoyl]-alpha-D-glucosamine. Biochemistry 27:1908–1917. doi: 10.1021/bi00406a017. [DOI] [PubMed] [Google Scholar]
- 228.Young K, Silver LL, Bramhill D, Cameron P, Eveland SS, Raetz CR, Hyland SA, Anderson MS. 1995. The envA permeability/cell division gene of Escherichia coli encodes the second enzyme of lipid A biosynthesis. UDP-3-O-(R-3-hydroxymyristoyl)-N-acetylglucosamine deacetylase. J Biol Chem 270:30384–30391. doi: 10.1074/jbc.270.51.30384. [DOI] [PubMed] [Google Scholar]
- 229.Klein G, Kobylak N, Lindner B, Stupak A, Raina S. 2014. Assembly of lipopolysaccharide in Escherichia coli requires the essential LapB heat shock protein. J Biol Chem 289:14829–14853. doi: 10.1074/jbc.M113.539494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 230.Ogura T, Inoue K, Tatsuta T, Suzaki T, Karata K, Young K, Su LH, Fierke CA, Jackman JE, Raetz CR, Coleman J, Tomoyasu T, Matsuzawa H. 1999. Balanced biosynthesis of major membrane components through regulated degradation of the committed enzyme of lipid A biosynthesis by the AAA protease FtsH (HflB) in Escherichia coli. Mol Microbiol 31:833–844. doi: 10.1046/j.1365-2958.1999.01221.x. [DOI] [PubMed] [Google Scholar]
- 231.Mahalakshmi S, Sunayana MR, SaiSree L, Reddy M. 2014. yciM is an essential gene required for regulation of lipopolysaccharide synthesis in Escherichia coli. Mol Microbiol 91:145–157. doi: 10.1111/mmi.12452. [DOI] [PubMed] [Google Scholar]
- 232.Klein G, Raina S. 2019. Regulated assembly of LPS, its structural alterations and cellular response to LPS defects. Int J Mol Sci 20:E356. doi: 10.3390/ijms20020356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 233.Dalebroux ZD, Edrozo MB, Pfuetzner RA, Ressl S, Kulasekara BR, Blanc MP, Miller SI. 2015. Delivery of cardiolipins to the Salmonella outer membrane is necessary for survival within host tissues and virulence. Cell Host Microbe 17:441–451. doi: 10.1016/j.chom.2015.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 234.Rossi RM, Yum L, Agaisse H, Payne SM. 2017. Cardiolipin synthesis and outer membrane localization are required for Shigella flexneri virulence. mBio 8:e01199-17. doi: 10.1128/mBio.01199-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 235.Heath RJ, Rock CO. 1996. Roles of the FabA and FabZ beta-hydroxyacyl-acyl carrier protein dehydratases in Escherichia coli fatty acid biosynthesis. J Biol Chem 271:27795–27801. doi: 10.1074/jbc.271.44.27795. [DOI] [PubMed] [Google Scholar]
- 236.Parsons JB, Rock CO. 2013. Bacterial lipids: metabolism and membrane homeostasis. Prog Lipid Res 52:249–276. doi: 10.1016/j.plipres.2013.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 237.Anderson MS, Raetz CR. 1987. Biosynthesis of lipid A precursors in Escherichia coli. A cytoplasmic acyltransferase that converts UDP-N-acetylglucosamine to UDP-3-O-(R-3-hydroxymyristoyl)-N-acetylglucosamine. J Biol Chem 262:5159–5169. [PubMed] [Google Scholar]
- 238.Mostafavi M, Wang L, Xie L, Takeoka KT, Richie DL, Casey F, Ruzin A, Sawyer WS, Rath CM, Wei JR, Dean CR. 2018. Interplay of Klebsiella pneumoniae fabZ and lpxC mutations leads to LpxC inhibitor-dependent growth resulting from loss of membrane homeostasis. mSphere 3:e00508-18. doi: 10.1128/mSphere.00508-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 239.Mohan S, Kelly TM, Eveland SS, Raetz CR, Anderson MS. 1994. An Escherichia coli gene (FabZ) encoding (3R)-hydroxymyristoyl acyl carrier protein dehydrase. Relation to fabA and suppression of mutations in lipid A biosynthesis. J Biol Chem 269:32896–32903. [PubMed] [Google Scholar]
- 240.Clements JM, Coignard F, Johnson I, Chandler S, Palan S, Waller A, Wijkmans J, Hunter MG. 2002. Antibacterial activities and characterization of novel inhibitors of LpxC. Antimicrob Agents Chemother 46:1793–1799. doi: 10.1128/aac.46.6.1793-1799.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 241.Zeng D, Zhao J, Chung HS, Guan Z, Raetz CR, Zhou P. 2013. Mutants resistant to LpxC inhibitors by rebalancing cellular homeostasis. J Biol Chem 288:5475–5486. doi: 10.1074/jbc.M112.447607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 242.Emiola A, George J, Andrews SS. 2014. A complete pathway model for lipid A biosynthesis in Escherichia coli. PLoS One 10:e0121216. doi: 10.1371/journal.pone.0121216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 243.Rojas ER, Billings G, Odermatt PD, Auer GK, Zhu L, Miguel A, Chang F, Weibel DB, Theriot JA, Huang KC. 2018. The outer membrane is an essential load-bearing element in Gram-negative bacteria. Nature 559:617–621. doi: 10.1038/s41586-018-0344-3. [DOI] [PMC free article] [PubMed] [Google Scholar]