Neutrophils kill invading microbes and therefore represent the first line of defense of the innate immune response. Activated neutrophils assemble NADPH oxidase to convert substantial amounts of molecular oxygen into superoxide, which, after dismutation into peroxide, serves as the substrate for the generation of the potent antimicrobial hypochlorous acid (HOCl) in the phagosomal space. In this minireview, we explore the most recent insights into physiological consequences of HOCl stress. Not surprisingly, Gram-negative bacteria have evolved diverse posttranslational defense mechanisms to protect their proteins, the main targets of HOCl, from HOCl-mediated damage.
KEYWORDS: disulfide bond formation, posttranslational modifications, chaperedoxin, biofilm formation, motility, biofilm, hypochlorous acid, molecular chaperone, N-chlorination, neutrophilic oxidants, oxidative burst, protein aggregation, oxidative stress, polyphosphate
ABSTRACT
Neutrophils kill invading microbes and therefore represent the first line of defense of the innate immune response. Activated neutrophils assemble NADPH oxidase to convert substantial amounts of molecular oxygen into superoxide, which, after dismutation into peroxide, serves as the substrate for the generation of the potent antimicrobial hypochlorous acid (HOCl) in the phagosomal space. In this minireview, we explore the most recent insights into physiological consequences of HOCl stress. Not surprisingly, Gram-negative bacteria have evolved diverse posttranslational defense mechanisms to protect their proteins, the main targets of HOCl, from HOCl-mediated damage. We discuss the idea that oxidation of conserved cysteine residues and partial unfolding of its structure convert the heat shock protein Hsp33 into a highly active chaperone holdase that binds unfolded proteins and prevents their aggregation. We examine two novel members of the Escherichia coli chaperone holdase family, RidA and CnoX, whose thiol-independent activation mechanism differs from that of Hsp33 and requires N-chlorination of positively charged amino acids during HOCl exposure. Furthermore, we summarize the latest findings with respect to another bacterial defense strategy employed in response to HOCl stress, which involves the accumulation of the universally conserved biopolymer inorganic polyphosphate. We then discuss sophisticated adaptive strategies that bacteria have developed to enhance their survival during HOCl stress. Understanding bacterial defense and survival strategies against one of the most powerful neutrophilic oxidants may provide novel insights into treatment options that potentially compromise the ability of pathogens to resist HOCl stress and therefore may increase the efficacy of the innate immune response.
INTRODUCTION
One major challenge for all organisms living in an aerobic world is that of dealing with the consequences of oxidative stress, a situation that emerges from the accumulation of reactive oxygen species (ROS) in the cell. ROS such as superoxide, peroxide, and hydroxyl radicals are generated through the reduction of molecular oxygen, which can occur during electron transfer in the respiratory chain, by enzymes such as NADPH oxidases, and in organelles such as peroxisomes (1). Bacteria naturally encounter oxidative stress in different sources (recently reviewed in reference 122), which can represent a burden for their survival. One such scenario occurs when bacteria enter the host; as the first-line defense, phagocytic cells of the innate immune response, particularly neutrophils, phagocytize invading pathogens. Within the phagosome of neutrophils, bacteria are confronted with a complex mixture of highly antimicrobial compounds, including reactive oxygen and chlorine species (RO/CS) (2, 3). In this minireview, we first describe how phagocytic cells generate RO/CS as a powerful strategy to kill invading pathogens. We then focus on the most abundant and powerful neutrophil-derived RO/CS, hypochlorous acid (HOCl), and discuss the cellular targets that are most sensitive to HOCl. Finally, we examine redox-regulated mechanisms that bacteria utilize to protect their proteome against the harmful proteotoxic effects of HOCl.
NEUTROPHILS GENERATE RO/CS DURING OXIDATIVE BURST
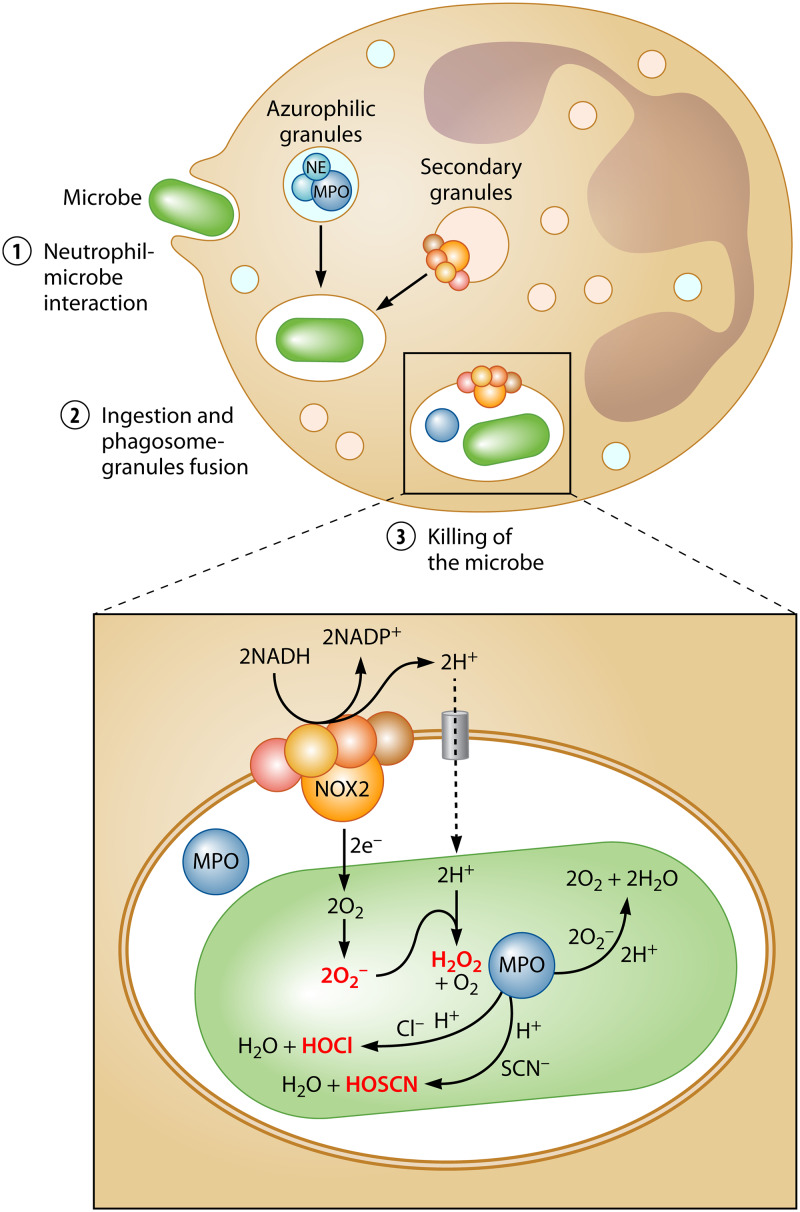
Neutrophils are recruited to the sites of infection to rapidly carry out their microbicidal activity, which relies on several mechanisms such as phagocytosis, degranulation of antimicrobial proteins, and the release of neutrophil extracellular traps (NETs) (4). During phagocytosis, neutrophils identify and engulf foreign cellular material by endocytosis in order to eliminate the ingested particles. Along with the invagination of the plasma membrane, specific azurophilic granules containing antimicrobial enzymes such as NADPH oxidase complex 2 (NOX2), myeloperoxidase (MPO), and digestive enzymes fuse with the phagosomal compartment and release their content (5). As part of the antimicrobial action, neutrophils activate a powerful oxidative burst by converting large amounts of oxygen into superoxide radicals (3, 6). This event is mediated through the activation of NOX2, a multicomponent electron-transfer complex (a more detailed review on NOX2 and its enzymology can be found in reference 6). Stimulated by proinflammatory mediators, microbes, soluble stimuli, or activation of pattern recognition receptors (PRRs), NOX2 assembles in the phagosomal membrane and catalyzes the reduction of molecular oxygen by transfer of a single electron (Fig. 1). Coming from NADPH, the electron is directionally transferred via the NOX2 complex to molecular oxygen in the phagosomal space, resulting in the formation of superoxide (O2*−) (7). Notably, NOX2 activation by phagocytosis is a rapid and vigorous event: the assembly of NOX2 takes only ∼45 s and results in a level of oxygen consumption that is up to 100-fold higher than the level seen with basal metabolic activity of neutrophils (8). Enzymatic characterization of NOX2 revealed that oxygen concentrations of as little as 20 μM are sufficient to maintain maximal activity, suggesting rapid superoxide production even at low oxygen concentrations (9). However, the directional NOX2-mediated electron transfer from cytoplasm to the phagosomal space has two consequences: a charge imbalance (which would ultimately result in membrane depolarization and NOX2 inhibition) and a proton gradient (due to increased proton release in the cytosol and a higher demand for protons in the phagosome caused by superoxide dismutation). Neutrophils overcome this limitation by activating the voltage-gated proton channel VSOP/HV1, which compensates for the discrepancies in close coordination with the NADPH oxidase complex (10). The heme-dependent peroxidase MPO, an enzyme that is released by phagosomal degranulation and constitutes 5% of the total protein expressed in neutrophils (11), rapidly dismutates superoxide into peroxide (H2O2). Peroxide, in turn, serves as the substrate for the oxidation of available (pseudo)halides such as chloride, bromide, and thiocyanate to produce hypochlorous acid (HOCl), hypobromous acid (HOBr), and hypothiocyanous acid (HOSCN), respectively (5, 6, 12) (Fig. 1). The outcome of the MPO-catalyzed reaction is determined not only by the differences in availability of the three anions but also by the distinct levels of selectivity of the enzyme for each of them: ∼70% to ∼80% of all peroxide consumed by MPO yields HOCl, and the remainder is accounted for by HOSCN, as the level of formation of HOBr is negligibly low due to the low concentration of available bromide (13). However, both HOCl and HOSCN are produced in the phagosomal space and likely react with phagosomal constituents before they reach the microbe. Indeed, it has been shown that tyrosine chlorination was more pronounced in phagosomal proteins than in proteins of ingested bacteria, suggesting that phagosomal proteins are exposed to a higher HOCl concentration (14). Besides methionine and thiol groups, HOCl shows high reactivity toward amino groups, resulting in the formation of chloramines (15). However, it is likely that protein chloramines decompose into smaller chloramines and/or aldehydes, which are more stable and readily penetrate into the bacterial cell. Despite having lower reactivities than HOCl, chloramines still show prolonged bactericidal activities. However, all oxidants have powerful antimicrobial activity and (at least) HOCl (and corresponding downstream products) and HOSCN are crucial for the immunological functions of neutrophils (6). In addition to the bacterial cell, only a minor part of the extracellular medium is taken up by phagocytosis (14). As a result, the space between the phagosomal membrane and the particle’s surface is small and extremely concentrated, with these oxidants making the phagosomal space well equipped for the efficient killing of the ingested microbe.
FIG 1.
Neutrophil-mediated RO/CS formation. Neutrophils represent the first line of defense of the innate immune system. To kill invading bacteria, they generate powerful antimicrobials in a multistep process. (Upper panel) First, neutrophils sense and bind to the invading microbe (step 1), which results in the ingestion of the microbe as well as in granule-phagosome fusion events (step 2). Subsequently, granular proteins and RO/CS are released into the phagosome (step 3), where myeloperoxidase catalyzes the production of RO/CS. (Lower panel) Superoxide (O2•−) is produced by activated NADPH oxidases (NOX2) located in the phagosome membrane. The charge is balanced through proton channels (gray). O2•− is mainly dismutated into peroxide (H2O2) by myeloperoxidase (MPO), an enzyme that is released from azurophil granules. Moreover, MPO catalyzes the reaction of H2O2 with physiological concentrations of chloride or thiocyanate to yield hypochlorous acid (HOCl) or hypothiocyanous acid (HOSCN), respectively. The rate of production of HOCl is determined by the availability of chloride, as superoxide competes with chloride for MPO and thus controls the rate of production of HOCl. All neutrophilic RO/CS are produced in the phagosomal space, and some readily penetrate into the bacterial cell.
PHYSIOLOGICAL CONSEQUENCES OF HOCl-MEDIATED OXIDATIVE STRESS
Neutrophilic oxidants such as HOCl, HOSCN, H2O2, and O2*− differ significantly in their concentrations and reactivities with cellular macromolecules. Both H2O2 and O2*− likely function only as intermediates for the production of HOCl rather than directly killing microbes during phagocytosis (16). With its limited reactivity and low ability to penetrate the bacterial cell, O2*− is immediately dismutated into H2O2. H2O2 is thiol specific; however, except for selected peroxidases, its reactivity with most cellular macromolecules is rather slow (∼101 M−1s−1) and does not cause significant protein aggregation. Even though H2O2 is bactericidal at sufficient concentrations, it likely is consumed by MPO rapidly after it is generated (3, 17). Both the physiological consequences and cellular defenses of superoxide-mediated and/or peroxide-mediated oxidative stress have recently been reviewed (18–20). On the other hand, RO/CS, i.e., HOCl/HOBr/HOSCN, are extremely reactive and bactericidal even at low micromolar levels (21, 22). One well-known target of HOCl is the amino acid cysteine (3, 6). HOCl or related chloramines can cause oxidative thiol modifications which are either reversible (i.e., sulfenic acids and disulfide bonds) or irreversible (i.e., sulfinic acid and sulfonic acid) (1). However, HOCl reacts equally well with methionine and, much more slowly, with amines. Chloramines, the products of reactions between amines and HOCl, show moderate reactivity with high selectivity for sulfur centers. HOSCN, in contrast, targets specifically thiol groups. Reversible thiol modifications often result in severe structural and functional consequences, while irreversible thiol modifications lead to protein aggregation and degradation (1, 23). Recent work revealed overlapping outcomes for treatments of Pseudomonas aeruginosa with HOCl and HOBr, as the two oxidants target nongrowing cells more efficiently than growing cells and elicit similar stress responses (24). Results of treatment with HOSCN are, however, distinctly different, affecting primarily actively growing cells and invoking different stress responses. However, all three oxidants cause extensive protein unfolding and aggregation (24, 25). RO/CS cause pleiotropic phenotypes in bacterial cells and can oxidize and damage virtually any cellular molecule, including selected amino acids, lipids, metal centers, and nucleic acids, leading to microbial death (26) and explaining why the absence of MPO in isolated neutrophils results in strongly impaired killing of a wide range of bacteria (3, 15). In vivo thiol-trapping experiments identified a clear increase in cysteine oxidation of proteins in phagocytized compared to nonphagocytized Escherichia coli (27). Recent work with genetically encoded redox probes beautifully illustrated the severity of oxidative stress experienced by E. coli when trapped in the phagosome of neutrophils (2). The same study provided strong evidence for identification of HOCl as the main component of the complex RO/CS mixture produced in neutrophils. Several other findings further support the idea that neutrophilic oxidants, particularly HOCl, fulfill a crucial function in killing invading microorganisms as follows. (i) NOX deficiency and lower NOX activity result in reduced oxidation of the proteome in phagocytized bacteria (2) and higher susceptibility to microbial infection in mice (28). (ii) Individuals with chronic granulomatous disease, a genetic disease with impaired NADPH oxidase activity, appear to be more prone to microbial infections (11). (iii) Compared to their free-living counterparts, bacteria experience the largest amount of oxidative stress when enclosed in the phagosome (2). (iv) Pretreatment of neutrophils with an MPO inhibitor resulted in a much lower level of oxidation of the bacterial proteins and reduced release of RO/CS (2). (v) Neutrophils show significantly impaired bacterial killing and reduced HOCl production when cultured in a low-chloride environment (6). Bacteria respond to sublethal HOCl stress by significant changes in gene expression, including activation of HOCl-specific transcriptional regulators (26, 29, 30). A significant disadvantage of stress-mediated transcriptional and/or translational responses is the lengthy time from the detection of the stress until the newly synthesized proteins take action, which can take up to 1 h in bacteria (31, 32). This is particularly pressing during exposure to HOCl, an extremely fast-acting oxidant that oxidatively modifies cysteine and methionine residues with reaction constants of ∼108 M−1s−1 so that damage occurs long before cells can respond (3, 31, 33, 34). It is well established that the generation of ATP is impaired in HOCl-stressed cells. The limited levels of ATP are a consequence of oxidative inactivation of redox-regulated enzymes in ATP-synthesizing pathways, including GAPDH (glyceraldehyde-3-phosphate dehydrogenase) and F0F1-ATP synthase, as well as of the rerouting of ATP into the chaperone holdase polyphosphate (polyP) (see below) (25, 35–37). The lack of ATP and the oxidative inactivation of proteins essential for translation likely help to explain why global protein synthesis is downregulated during HOCl stress, which, of course, reduces the risk of toxic protein aggregate formation (38). An additional consequence is the repression of the global transcription under these error-prone conditions. However, these changes are certainly not sufficient to successfully fight the threats of HOCl-derived oxidative stress.
CELLULAR DEFENSES AGAINST HOCl STRESS: RAPID ACTIVATION OF CHAPERONE HOLDASES HELPS HOCl-STRESSED CELLS COPE WITH HOCl-INDUCED PROTEIN AGGREGATION
HOCl-mediated protein modifications can damage proteins, induce the loss of their structure and function, and hence lead to excessive protein aggregation and cell death (1, 25). Therefore, it is not surprising that microbes have developed posttranslational mechanisms to reduce aggregation-sensitive folding intermediates, utilizing a powerful proteostasis network. Molecular chaperones and proteases fulfill these functions; they support de novo folding of newly translated polypeptides, prevent irreparable stress-induced misfolding of native proteins within the crowded and metastable cellular environment, and degrade proteins that are irreversibly damaged and/or refolding incompetent (38, 39). The chaperone network is complex and includes multichaperone machineries such as DnaK/DnaJ/GrpE and GroEL/ES, where cochaperones are needed for substrate binding, their foldase activity, and/or substrate release. However, the fact that most of them rely on ATP for their function and some are oxidation-sensitive disqualifies them from actively participating in the protein homeostasis under severe oxidative stress conditions (35, 40). Instead, the cell employs a stress-specific chaperone, holdase, that deals with this dilemma even when ATP availability is limited. In this minireview, we review the latest findings concerning four holdases that, despite distinct activation mechanisms, all have been shown to bind unfolded proteins during exposure to HOCl and prevent them from aggregation. When nonstress conditions are restored, ATP-dependent chaperones take over and mediate the refolding of the client proteins.
(i) Hsp33 holdase activity is activated by cysteine oxidation mediated partial unfolding.
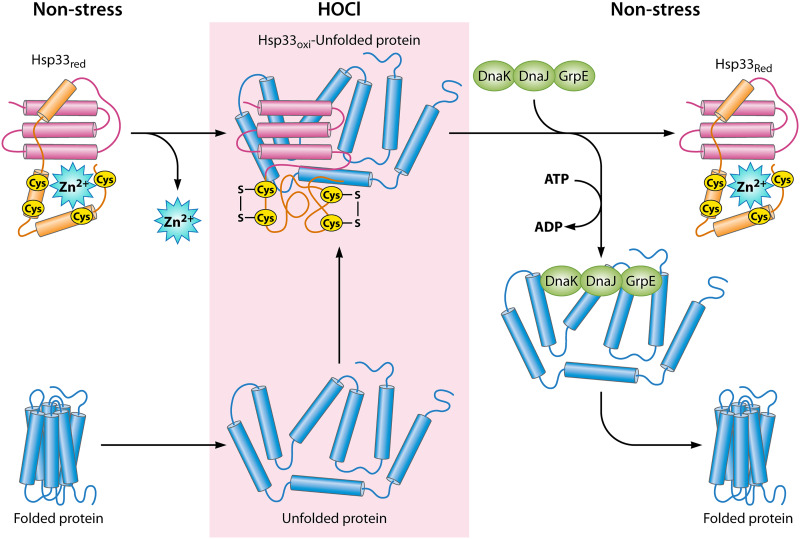
Hsp33, a highly conserved heat shock protein in bacteria, green algae, and several parasites such as Leishmania, was the first redox-regulated chaperone to be described and was identified almost 2 decades ago (41). As commonly seen for most molecular chaperones under the control of the heat shock response, expression of Hsp33 is induced when protein unfolding intermediates accumulate in the cell (42, 43). Given the many studies on Hsp33 (1, 38, 42, 44–47), its mode of action during oxidative stress is well understood. Hsp33 consists of a compactly folded N-terminal domain, a highly flexible ∼40-amino-acid (aa)-long linker region, and a C-terminal redox-sensing domain. Four highly conserved cysteine residues are located within the C-terminal domain which function as redox sensors. Under nonstress conditions, the cysteines are present in their reduced state and coordinate a Zn2+ ion with high affinity, resulting in a the formation of a compactly folded and chaperone-inactive Hsp33 monomer (48) (Fig. 2). The tetrahedral arrangement by which the cysteine residues coordinate the Zn2+ ion under nonstress conditions likely explains why cysteines bound to zinc display a higher reactivity with HOCl at a near-diffusion-limited rate than cysteine or methionine alone (49, 50). Thus, under HOCl-mediated protein unfolding conditions, all four cysteine residues are rapidly oxidized and Hsp33 forms two intramolecular disulfide bonds. The cysteine oxidation causes the release of the Zn2+ ion, resulting in widespread conformational rearrangements. These include partial unfolding of the central linker region as well as dimerization, and (potentially) even higher levels of oligomer formation of oxidized Hsp33 monomers follow, ultimately resulting in the activation of the chaperone function (Fig. 2) (25, 51, 52). The unfolded, hydrophilic linker region binds to unfolded client proteins via electrostatic interaction (51, 53). This effect is further stabilized through hydrophobic interactions between the client and the well-folded N-terminal domain of Hsp33 (53). Additional activators of Hsp33’s chaperone function have been identified over the years, including reactive bromine species (e.g., HOBr) (24), a combination of peroxide stress and heat stress (41, 48), bile salts (54), and, as recently noted, atmospheric-pressure plasma (55). All of those conditions cause oxidative protein unfolding and lead to the aggregation of many essential proteins in the absence of Hsp33 in vivo (24, 35, 54–56). Not surprisingly, bacteria lacking Hsp33 are significantly more sensitive under those stress conditions. Inactivation of Hsp33 requires both reducing and refolding conditions. Once reducing conditions are restored, oxidized Hsp33 dimers are converted to reduced Hsp33 dimers, which are still bound to their client proteins, likely until ATP-dependent foldase systems such as the DnaK/DnaJ/GrpE systems are fully functional for subsequent refolding of the client proteins. However, a recent study suggested that Hsp33 may be involved in the process of refolding of their client proteins (53).
FIG 2.
Reversible cysteine oxidation causes conditional disorder to activate Hsp33. One way for bacteria to fight the proteotoxic effects of HOCl stress is to employ the stress-specific molecular chaperone Hsp33, which is present as a chaperone-inactive monomer under nonstress conditions. Reduced Hsp33 is well folded due to the coordination of a Zn2+ ion by four highly conserved cysteine residues in the C-terminal domain. Upon exposure to protein-unfolding conditions, such as HOCl stress, the cysteines are oxidized, resulting in the formation of two disulfide bonds, zinc release, and massive structural rearrangements. Similarly to its client proteins, Hsp33 partially unfolds during HOCl stress; however, the disordered conformation is essential for the chaperone activity of Hsp33. The disulfide bonds are reduced upon return to nonstress conditions, and client proteins are transferred to the ATP-dependent DnaK/DnaJ/GrpE system for refolding.
(ii) Reversible N-chlorination as a novel mechanism for chaperone activation.
Recently, RidA was added to the list of holdases known to protect E. coli from HOCl-mediated protein aggregation (57). E. coli RidA belongs to the functionally highly diverse YjgF/YER057c/UK114 protein family and appears to be bifunctional. Under nonstress conditions, the protein is chaperone inactive, functions as an enamine/imine deaminase by accelerating the release of ammonia from enamine/imine intermediates generated by the threonine dehydratase IlvA, and even prevents the accumulation of toxic metabolites in the cell (57–59). In the presence of HOCl, however, RidA no longer accelerated the deaminase reaction but instead almost completely inhibited IlvA’s dehydratase activity (57). This observation indicated that RidA and IlvA form a tight complex that negatively affects the activity of IlvA. In vitro studies with unfolded IlvA and other model substrates revealed that HOCl-oxidized RidA rapidly turns into a powerful holdase and prevents the aggregation of its client proteins (Fig. 3). The RidA chaperone function is HOCl specific, as none of the other tested oxidizing agents (e.g., diamide and peroxide) activated RidA’s chaperone activity. In a previous study, RidA was identified as one of the very few proteins affected by a cysteine modification after treatment with the nitrosative stress-causing agent peroxynitrite (60). It was therefore obvious that the activation of RidA’s chaperone function during HOCl stress might be mediated via oxidation of its single conserved cysteine residue. Surprisingly, this was not the case as the cysteine-free RidA variant still gained full chaperone activity (57). Consistent with the 70% reduction in the number of free amino groups present in HOCl-treated RidA compared to its untreated counterpart, mass spectrometry (MS) analysis of HOCl-activated RidA identified the presence of multiple chlorinated RidA species. These findings suggested that HOCl activates RidA by N-chlorination of the ε-amino groups of lysines, guanidinium groups of arginines, and/or the terminal amino group. This type of chaperone activation was unexpected, as N-chlorination of proteins can lead to protein inactivation, unfolding, fragmentation, cross-linking, and, ultimately, protein aggregation (6, 61). Biophysical studies revealed that HOCl-mediated activation of RidA is accompanied by the formation of higher oligomers and substantially increased hydrophobicity, two features that facilitate the binding of unfolded proteins and therefore represent characteristic features of chaperones (39). Treatment with physiological redox systems such as Trx or glutathione (GSH) reversed the chlorination of RidA, caused the inactivation of RidA’s chaperone function, and reactivated its enamine/imine deaminase activity in vitro (57). However, it remains to be elucidated whether DnaK/DnaJ/GrpE and/or GroEL/ES can serve as a foldase system to refold the client proteins. The RidA-deficient cells were substantially more susceptible to HOCl stress than the wild-type cells, indicating that RidA represents a remarkable HOCl-specific bacterial defense strategy that effectively protects E. coli against severe HOCl stress in vivo. Pulldown assays using cell lysates identified highly abundant cytosolic proteins of the primary metabolism as potential in vivo substrates and therefore pointed to a rather unspecific RidA-client interaction as well as to broad substrate specificity. Since HOCl chlorinates primary amines in the phagosome, resulting in chloramines (31, 34, 62), RidA may very well play a role as a bacterial defense strategy to fight phagocytosis. However, it remains to be seen whether the HOCl levels produced during the oxidative burst are sufficient to efficiently activate RidA’s chaperone function during phagocytosis. RidA homologues seem to be functionally diverse, as HOCl-mediated chlorination turned the S. aureus RidA homologue into a potent RNase that did not exhibit any detectable chaperone activity (63). It is therefore necessary to develop a better understanding of the identity and/or number of the amino acids with free amino groups that are needed for activation and inactivation of RidA’s chaperone function.
FIG 3.
N-chlorination turns RidA and CnoX into powerful chaperone holdases. Similarly to Hsp33, CnoX (upper panel) and RidA (lower panel) serve as powerful chaperone holdases in E. coli that prevent unfolded proteins from aggregation during HOCl stress. HOCl-mediated N-chlorination increases the surface hydrophobicity of both proteins and is required for activation of their chaperone function. N-chlorination is reversible, and at least RidA forms oligomers when present in its chlorinated form (lower panel). Once nonstress conditions are restored, CnoX (upper panel) transfers the substrate proteins to ATP-dependent chaperone foldase systems such as DnaK/DnaJ/GrpE or GroEL/ES and initiates refolding of the substrate proteins. Moreover, CnoX protects the substrate from overoxidation through the formation of a mixed disulfide. While reduced Trx-SH or GSH can reverse the chlorination of RidA (lower panel), the foldase system(s) required for refolding of the client proteins has not yet been identified.
Soon after, CnoX was identified as another member of the HOCl stress defense system in E. coli. It is mechanistically very similar to RidA (57); while it is chaperone inactive under nonstress conditions, N-chlorination of CnoX resulted in increased hydrophobicity and turned the protein into a powerful holdase (Fig. 3) (64). The locations of the chlorinated residues were identified in four atypical tetratricopeptide repeats (TPR) at the C terminus, representing motifs that mediate protein-protein interactions and are prevalent in folding factors (65). N-chlorination of the C-terminal TPR domain was essential for the survival of E. coli, as a CnoX variant lacking the TPR domain was chaperone inactive and unable to complement a cnoX-deficient strain in vivo, likely because the increase in hydrophobicity is a requirement for the interaction with unfolded substrates (64). Earlier studies had pointed toward a potential protein-protective role of CnoX, as cnoX-deficient cells were highly sensitive to elevated temperature (66). Independent studies further confirmed its role as a chaperone, as it was shown to form complexes with unfolded substrate proteins, to cooperate directly with DnaK, and to affect cell division and DNA replication by assisting in folding DNA polymerase 3 (66–68). Therefore, mechanisms other than N-chlorination must exist that lead to the activation of E. coli CnoX’s chaperone function. Interestingly, HOCl-mediated N-chlorination was not required for activation of the holdase activity of Caulobacter crescentus CnoX, most likely due to its overall surface hydrophobicity being higher than that seen with its E. coli homologue (69). Therefore, constitutively active CnoX might be an inherent part of the proteostasis network in C. crescentus even in the absence of HOCl exposure, a stress situation that this nonpathogenic species most likely would not experience in its natural aquatic habitat. However, the two homologues share the ability to transfer the substrate proteins to foldase systems such as DnaK/DnaJ/GrpE, which then initiate the refolding process in an ATP-dependent manner (Fig. 3) (64, 69). Both homologues were also shown to efficiently cooperate in vitro and in vivo with GroEL/ES, another ATP-dependent foldase system that assists numerous proteins to regain their native structure, activity, and ability to perform their specific functions (64, 69). Intriguingly, it was the very first time that substrate transfer from a holdase to GroEL/ES was shown in vitro. This unique feature was also supported by two additional observations: (i) a direct interaction between CnoX and GroEL was previously reported (66, 70) and (ii) CnoX also protected various client proteins from aggregation that accept only GroEL/ES as a refolding system (64). However, in addition to its holdase activity, CnoX was shown to confer redox protection to its client proteins. Thiol groups are present in low-molecular-weight thiols such as the antioxidant GSH, in free cysteines, and in side chains of cysteine residues of polypeptides (reviewed in reference 1) and are, along with methionine residues, the most reactive targets of HOCl. Therefore, another consequence of HOCl stress is the formation of intra- or intermolecular disulfide bonds in proteins, which can result in protein misfolding and aggregation, because ATP-dependent chaperone systems do not act as reducing agents and are not capable of refolding oxidized proteins. Instead, independent oxidoreductases, i.e., members of the thioredoxin (Trx) family or glutaredoxin (Grx) family, are required to first reduce the disulfide bonds via direct thiol/disulfide exchange before DnaK/DnaJ/GrpE or GroEL/GroES can initiate the refolding process (71, 72). This view has now been corrected, as C. crescentus CnoX was previously shown to combine its holdase with thioredoxin activity (69). The latter is mediated by two surface-exposed cysteine residues in the conserved WCGPC motif of the N-terminal Trx fold. Thus, CnoX is the first bacterial protein identified as having the ability to prevent aggregation and reduce nonnative disulfide bonds in substrate proteins, thereby providing a powerful alternative to independent Trx/Grx systems. E. coli CnoX, on the other hand, most likely lacks oxidoreductase activity due to a mutation in one of the conserved cysteines of the Trx fold (64, 73), even though a dissenting report exists (68). The protein contains an additional surface-exposed cysteine residue; however, the great distance between the two cysteine residues excludes the possibility of disulfide bond formation. Intriguingly, a CnoX variant lacking this cysteine residue was unable to complement the phenotype of a cnoX-deficient strain, suggesting a crucial role during HOCl stress (64). In fact, this cysteine residue was responsible for the formation of mixed disulfide bonds with cysteine residues in substrate proteins in vivo and therefore represents an elegant way for CnoX to protect the substrate proteins from irreversible overoxidation (Fig. 3). Efficient reduction of the mixed disulfide complex in vivo required the presence of reduced glutathione, which itself is a prominent target of HOCl, suggesting that substrate transfer to the ATP-dependent folding machineries can occur only when nonstress conditions are restored. However, both the thioredoxin activity of C. crescentus CnoX and the ability of E. coli CnoX to form mixed disulfide are redox functions that, in combination with the chaperone activity, are unique properties of the newly founded chaperedoxin protein family. Overall, N-chlorination represents a reversible thiol-independent chaperone activation mechanism that fully depends on exposure to HOCl. Chaperone activation mediated by the oxidation of cysteine residues, such as occurs in Hsp33, is rather specific and almost exclusively affects highly conserved cysteines (25, 74). N-chlorination, on the other hand, is much more unspecific. Therefore, it is not unlikely that more chaperones exist that utilize this type of activation mechanism. Indeed, additional holdases activated by N-chlorination, including blood plasma proteins, were discovered over the last 2 years (75).
(iii) Conversion of ATP into the protein scaffold polyphosphate.
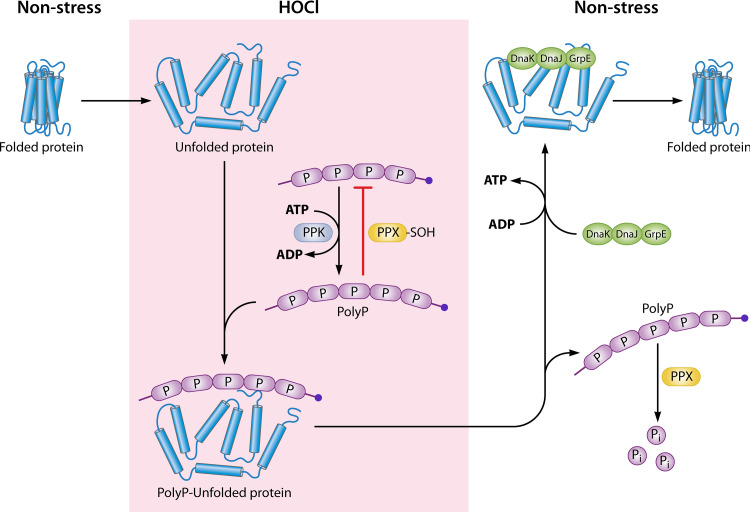
Utilization of protein-based chaperones is only one way by which Gram-negative bacteria protect their proteome against HOCl stress. One of the most efficient posttranslational stress response systems that bacteria employ to survive extended periods of HOCl is the conversion of ATP into the biopolymer polyP (37). PolyP is highly conserved, present in prokaryotic and eukaryotic species, and structurally extremely simple, as it consists of a linear arrangement of phosphate molecules (up to ∼1,000 Pi/chain) covalently linked via high-energy phosphoanhydride bonds (76). Much of the relevant early work was done by the late Arthur Kornberg, who identified the enzymatic systems responsible for synthesis and degradation of polyP. In bacteria, generation of polyP(n+1) is reversibly catalyzed by nonessential polyphosphate kinase 1 (PPK1), which transfers a terminal phosphate of ATP to a growing chain of polyP(n) (Fig. 4) (77, 78). Several cues, such as nutrient shift, exposure to osmotic changes, acidic pH, and high temperatures, have been identified that trigger the cellular accumulation of polyP (24, 37, 79–82). Degradation of polyP into monomeric Pi molecules is performed by an exopolyphosphatase (PPX) (83). The ability to genetically manipulate polyP levels in vivo allowed detailed phenotypic characterization and provided insights into the various roles of polyP, e.g., those involving increased susceptibility to elevated temperatures, heavy metals, oxidants, and starvation (24, 37, 76, 79, 82, 84). Moreover, cells lacking the ability to produce polyP are defective in biofilm formation, quorum sensing, virulence, and motility (85–90). However, the mechanism by which polyP achieves these seemingly unrelated functions remained unclear. Many excellent reviews exist that discuss the manifold roles of polyP in prokaryotes and eukaryotes (76, 91–94). Therefore, we focus here exclusively on the recent discovery that polyP serves as a physiologically relevant chemical chaperone during HOCl stress. Various Gram-negative bacteria, including E. coli, Vibrio cholerae, and Pseudomonas aeruginosa, accumulate large amounts of polyP in response to HOCl or related hypohalous acids (24, 37). HOCl-treated E. coli cells were previously shown to experience a 50% drop in their ATP levels and showed substantial upregulation of several phosphate-starvation genes (37, 95). PolyP-deficient cells were substantially more susceptible to neutrophilic oxidants than wild-type cells, suggesting that polyP counteracts the proteotoxic effects of HOCl (24, 37). The first connection to the role of polyP as a protein-stabilizing scaffold was made with the observation that the accumulation of aggregated proteins in vivo under high-temperature conditions or in the presence of hypohalous acids was much more pronounced in the absence of polyP (24, 37, 79). Consistently, expression of heat shock response genes in polyP-deficient cells was upregulated upon stress treatment, indicating the cell’s need for molecular chaperones. In vitro studies confirmed that polyP binds a broad set of oxidatively damaged and otherwise unfolded proteins in a chain-length and concentration-dependent manner and prevents their aggregation by maintaining them in a refolding-competent conformation (37, 79). Once nonstress conditions were restored, client release was triggered by PPX-mediated polyP hydrolysis or PPK-catalyzed reconversion into ATP (Fig. 4). However, the release of the client proteins requires the presence of ATP-dependent foldase systems such as DnaK/DnaJ/GrpE (37, 79). Conversion of ATP into polyP offers a great deal for the cell; polyP does not depend on transcription or translation, is inert to oxidative stress, and does not require ATP for its chaperone function. Moreover, it can be reconverted to ATP by PPK if nonstress conditions are restored. Once refueled, the ATP pool can then be utilized by ATP-dependent foldases to promote protein refolding, making the conversion of ATP into the highly effective protein scaffold polyP an ideal alternative for the cell to combat the detrimental consequences of HOCl stress (Fig. 4). Most strikingly, unfolded client proteins remained fully soluble even at near-boiling temperatures in the presence of physiological concentrations of polyP (37, 79). Structural analyses of the complexes revealed the formation of β-sheet structures in these typically α-helical proteins (79). The fact that no interactions were observed between polyP and native proteins suggests that polyP interacts with emerging β-sheet structures that form during stress-induced protein unfolding. However, additional structural analyses performed at high resolution will be necessary to characterize the precise nature of the polyP-client protein interaction in more detail as well as proteomic studies to identify polyP substrates in vivo. Several open issues remain, particularly concerning the regulation of polyP synthesis/degradation in vivo. PolyP synthesis appears to be primarily regulated on posttranslational levels, as ppk transcript levels were unaffected during nutrient starvation and HOCl stress (37, 95, 96). Moreover, the absence of PPX does not result in significant changes in polyP levels under nonstress conditions. These observations suggest that polyP homeostasis must be controlled either directly or through stress-mediated regulators, which energetically makes sense given that polyP generation directly affects the ATP pool of the cell. A recent study revealed that the RNA polymerase secondary channel binding transcriptional regulator DksA affects polyP synthesis in vivo (97). In addition, one redox-regulated mechanism has been identified that contributes to the accumulation of polyP under severe conditions of HOCl stress: HOCl oxidizes a cysteine residue located in close proximity to the polyP-binding site of PPX, resulting in the transient inhibition of its exopolyphosphatase activity until nonstress conditions are restored (37). Even though this explanation goes a long way toward enabling full understanding of the diverse and pleiotropic phenotypes associated with polyP, it became clear over the past years that, given all the phenotypes of polyP-deficient cells, the bacterium-specific PPK1 represents an ideal drug target. Recently, mesalamine, the gold standard in treating patients with mild to moderate ulcerative colitis, was identified as a potent inhibitor of bacterial polyP production in a wide range of Gram-negative bacteria and in members of the gut microbiome (87). By mediating decreases in the polyP levels in the cell, mesalamine mimics the phenotypes reported previously for polyP-deficient cells (24, 87) and, therefore, may have a much broader application than originally anticipated. Additional studies have suggested that some probiotic bacteria potentially secrete polyP to modulate host inflammatory responses in the gut (98). Given polyP’s role as a signal molecule for the control of inflammation in mammals (99, 100) and the better understanding of polyP’s molecular mechanism developed over the past few years, the door is now open for more precisely directed investigations of, e.g., whether the observed effects on the immune cells are based on polyP’s protein scaffolding function, potentially revealing broader implications for this simple but powerful molecule.
FIG 4.
Conversion of ATP into protein-scaffold polyphosphate. Under HOCl stress conditions, bacteria convert significant amounts of ATP into long chains of polyphosphate (polyP), a reaction catalyzed by the bacterium-specific enzyme polyP kinase (PPK). The reversible oxidative inactivation of the polyP-degrading enzyme exopolyphosphatase (PPX) further contributes to the accumulation of polyP in the cell. PolyP functions as a protein-stabilizing scaffold that binds protein unfolding intermediates, stabilizes them in a soluble β-sheet-rich conformation, and effectively prevents protein aggregation both in vitro and in vivo. Once nonstress conditions are restored, chaperone foldases such as the DnaK/DnaJ/GrpE system take over to refold polyP-bound client proteins.
HOCl STIMULATES ADAPTIVE SURVIVAL STRATEGIES IN PATHOGENIC BACTERIA
Among the most important strategies enabling bacterial pathogens to persist in the host is their ability to perform chemotaxis and form biofilms. The cohesive extracellular matrix of biofilm bacteria provides numerous survival benefits, including protection against attacks by immune cells, reduced metabolic activity, and up to 1,000-fold decreased susceptibility to antimicrobial treatment compared to their planktonic counterparts (101, 102). The change from a planktonic and motile lifestyle to sessile biofilms requires the cells to attach to biotic or abiotic surfaces, a process that is regulated by a complex regulatory network of different signaling pathways (103, 104). As a result, substantial changes in gene expression occur which cause loss of motility and increase expression of fimbriae and pili (105–107). Moreover, the level of production of components of the extracellular matrix such as exopolysaccharides and extracellular DNA is substantially elevated. Many of these changes are attributed to the accumulation of the second messenger cyclic dimeric GMP (c-di-GMP), a key biofilm regulator in numerous pathogenic bacteria (108–111). So far, only a few environmental stimuli affecting c-di-GMP homeostasis have been identified. One of them is sublethal HOCl stress, which was shown to trigger the ability of P. aeruginosa to attach to abiotic surfaces and form biofilms by activating c-di-GMP synthesis (Fig. 5) (112). This effect appears to be mediated through induced expression of the P. aeruginosa 3177 (PA3177) gene, which encodes the diguanylate cyclase (DGC) PA3177, an enzyme that catalyzes the condensation of two GTP molecules to form c-di-GMP (108). The same DGC was also found to contribute to antibiotic and ROS tolerance in P. aeruginosa biofilm cells (113). Induced PA3177 gene expression appeared to be HOCl specific and was unaffected during exposure to other oxidants such as peroxide and paraquat (112). Overexpression of PA3177 had a strong stimulating impact on autoaggregation and biofilm formation, indicating that changes in the expression level of this protein are indeed responsible for the switch from the planktonic to the biofilm lifestyle. HOCl-stimulated biofilm cells displayed substantially higher cell surface hydrophobicity, a result of c-di-GMP-mediated upregulation of exopolysaccharide expression and a potential reason for the increased surface attachment. Notably, HOCl is not the only stimulator of biofilm formation, as several studies have presented evidence that bacteria increase their tendency to form protective biofilms in response to sublethal concentrations of detergents, antibiotics, or biocides (114, 115). c-di-GMP appears to be the mediator in many cases (116). In fact, the E. coli DGC YdeH was shown to play an important role in surface attachment not only under nonstress conditions but also particularly for aminoglycoside-mediated biofilm induction (116); however, the mechanism is still unknown. Given that aminoglycosides can cause oxidative stress, it is tempting to speculate whether YdeH activation occurs via redox sensing. Despite recent attempts to explain potential mechanisms behind biofilm stimulation (117), the precise mechanism behind PA3177’s activation during HOCl stress remains enigmatic. Future studies will be needed to elucidate whether PA3177 expression is controlled by a redox-controlled transcriptional or translational regulator. Alternatively, it is also possible that the elevated level of c-di-GMP production is a result of posttranslational regulation.
FIG 5.
HOCl sensing affects biofilm formation and chemotaxis in pathogenic bacteria. (Upper panel) Sublethal concentrations of HOCl turn planktonic P. aeruginosa into sessile biofilms. Upon exposure to HOCl, transcription of the P. aeruginosa PA3177 gene encoding a diguanylate cyclase is induced via an as-yet-unknown regulatory pathway. This results in elevated c-di-GMP levels (black arrow) and subsequent induction of exopolysaccharide Pel and Psl production (dotted arrow). The increased hydrophobicity triggers the surface attachment and ultimately results in the switch from planktonic growth to nonmotile biofilm formation. (Lower panel) Cytosolic transducer-like protein D (TlpD) serves as a redox sensor in H. pylori to translate changes in HOCl concentrations. In the absence of HOCl, TlpD’s conserved cysteine in the chemoreceptor zinc-binding (CZB) domain is reduced and coordinates a Zn2+ ion. In its reduced state, TlpD induces the autophosphorylation of chemotaxis phosphorelation system CheA/CheY. CheY-Pi directly interacts with the flagellar rotor (labeled yellow), resulting in a temporary reversal of flagellar rotation (as indicated by arrows). In the presence of HOCl, the conserved cysteine is oxidized, Zn2+ is released, and the signaling is reversibly inactivated, resulting in a smooth swimming behavior (chemoattraction).
One such HOCl-mediated redox-sensing adaptive survival strategy has recently been identified in Helicobacter pylori, a persistent colonizer of the stomach (50). H. pylori employs a strong antioxidant network that includes four highly expressed peroxidredoxins and catalases and is therefore ideally suited to resist infiltrating neutrophils and inflammation, both of which lead to elevated HOCl concentrations in the stomach (118). In fact, H. pylori not only tolerates but actively senses HOCl by modifying its swimming behavior to move toward even higher HOCl concentrations. The pathogen employs a cytoplasmic chemoreceptor, TlpD, that senses micromolar concentrations of HOCl by reversibly oxidizing a conserved cysteine located in the chemoreceptor zinc-binding domain (CZB) (119). In its reduced state, the cysteine coordinates a Zn2+ ion and allows TlpD to promote the autophosphorylation of the histidine kinase CheA, which subsequently phosphorylates the response regulator CheY to induce chemorepulsion (Fig. 5). However, in the presence of micromolar concentrations of HOCl, TlpD’s conserved cysteine is reversibly oxidized, impairing the stimulatory function of TlpD. The inactivation of TlpD is based on oxidation-mediated conformational changes and effectively decreases the availability of phosphorylated CheA/CheY (Fig. 5) (50). As a result, H. pylori no longer performs chemorepulsion but instead performs chemoattraction. tlpD-deficient H. pylori strains are defective in colonization of the stomach antrum, a region that is associated with high inflammation and is therefore rich in HOCl (120). Overall, these recent discoveries suggest that biofilm formation and chemoattraction potentially are specific redox-regulated mechanisms that allow bacteria to thrive during HOCl stress in the host.
CONCLUDING REMARKS
The production of RO/CS, such as the powerful oxidant HOCl, is successfully utilized by cells of the innate immune response to combat invading pathogens. The mechanisms by which HOCl contributes to bacterial killing in the phagosome are still far from being fully understood, although much progress has been made to increase our understanding of how bacteria respond to and defend against HOCl stress. We have discussed some of the major strategies that Gram-negative bacteria employ to protect their proteome under HOCl-mediated oxidative stress conditions. Activated within minutes, ATP-independent holdases are ideally suited to the protection of cells against conditions that lead to sudden HOCl-mediated protein unfolding and aggregation even in the ATP-depleted cellular environment during HOCl stress. However, more research is needed to elucidate how many more of the enzymes, transcriptional regulators, or structural proteins that are present under nonstress conditions turn into effective holdases during HOCl stress. Intriguingly, many of the strategies that bacteria have evolved in order to fight HOCl stress are bacterium specific. Hsp33 has recently been used as a DNA vaccine candidate to protect flounders from Vibrio anguillarum infections by eliciting local and systemic immune responses (121). Therefore, HOCl-specific defense mechanisms have the great potential to serve as suitable targets for novel antimicrobial therapies given the important antimicrobial role that HOCl plays during innate immunity and host defense.
ACKNOWLEDGMENTS
We thank both reviewers for their positive comments and helpful feedback.
The Department of Biological Sciences is acknowledged for supporting the attendance of J.-U.D. at the 2019 Microbial Adhesion and Signal Transduction Gordon Research Conference (GRC).
Figures 2 to 5 were created with BioRender. Patrick Lane is acknowledged for enhancing all figures.
Biographies

Sadia Sultana is currently a first-year Ph.D. student in the Molecular and Cellular Biology graduate program at Illinois State University, Normal, IL, USA. She obtained her B.Sc. degree in 2016 and her M.Sc. degree in microbiology in 2017 from the University of Dhaka, Bangladesh. Sadia aims to better understand how pathogenic bacteria defend antimicrobial oxidants produced by the host immune system. Her long-term career goal is to establish herself as an academic scientist with a research focus in bacterial pathogenesis finding answers to unique and medically relevant questions.

Alessandro Foti obtained his B.Sc. from Palermo University, Palermo, Italy, and his M.Sc. degree in cell biology from the University of Rome, Rome, Italy. He received his Ph.D. training in enzymology under the supervision of Silke Leimkuehler at the University of Potsdam, Potsdam, Germany, where he studied the mechanisms of superoxide production by human aldehyde oxidase. In 2017, he joined the group of Arturo Zychlinsky as a postdoctoral research fellow at the Max Planck Institute of Infection Biology in Berlin, Germany, where he studies the physiological mechanisms of ROS production in neutrophils. He focuses on the processes by which neutrophils produce ROS as an antimicrobial strategy but also as intracellular signaling molecules to regulate immunological functions of neutrophils.

Jan-Ulrik Dahl began his academic training as a Diplom (M.Sc. equivalent) and Ph.D. student in the laboratory of Silke Leimkuehler at the University of Potsdam, Potsdam, Germany, specializing in the field of protein biochemistry. He then completed his postdoctoral research in the laboratory of Ursula Jakob at the University of Michigan, which was funded by a research fellowship from the German Research Foundation (DFG). In 2019, he started his independent career as an Assistant Professor of Microbiology at Illinois State University, Normal, IL, USA. Dr. Dahl is interested in bacterial responses to stress, particularly in defense systems of Gram-negative bacteria in response to neutrophilic oxidants.
REFERENCES
- 1.Dahl J-U, Gray MJ, Jakob U. 2015. Protein quality control under oxidative stress conditions. J Mol Biol 427:1549–1563. doi: 10.1016/j.jmb.2015.02.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Degrossoli A, Müller A, Xie K, Schneider JF, Bader V, Winklhofer KF, Meyer AJ, Leichert LI. 2018. Neutrophil-generated HOCl leads to non-specific thiol oxidation in phagocytized bacteria. Elife 7:e32288. doi: 10.7554/eLife.32288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Winterbourn CC, Kettle AJ. 2013. Redox reactions and microbial killing in the neutrophil phagosome. Antioxid Redox Signal 18:642–660. doi: 10.1089/ars.2012.4827. [DOI] [PubMed] [Google Scholar]
- 4.Amulic B, Cazalet C, Hayes GL, Metzler KD, Zychlinsky A. 2012. Neutrophil function: from mechanisms to disease. Annu Rev Immunol 30:459–489. doi: 10.1146/annurev-immunol-020711-074942. [DOI] [PubMed] [Google Scholar]
- 5.Hurst JK. 2012. What really happens in the neutrophil phagosome? Free Radic Biol Med 53:508–520. doi: 10.1016/j.freeradbiomed.2012.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Winterbourn CC, Kettle AJ, Hampton MB. 2016. Reactive oxygen species and neutrophil function. Annu Rev Biochem 85:765–792. doi: 10.1146/annurev-biochem-060815-014442. [DOI] [PubMed] [Google Scholar]
- 7.Nguyen GT, Green ER, Mecsas J. 2017. Neutrophils to the ROScue: mechanisms of NADPH oxidase activation and bacterial resistance. Front Cell Infect Microbiol 7:373. doi: 10.3389/fcimb.2017.00373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Nunes P, Demaurex N, Dinauer MC. 2013. Regulation of the NADPH oxidase and associated ion fluxes during phagocytosis. Traffic 14:1118–1131. doi: 10.1111/tra.12115. [DOI] [PubMed] [Google Scholar]
- 9.Gabig TG, Bearman SI, Babior BM. 1979. Effects of oxygen tension and pH on the respiratory burst of human neutrophils. Blood 53:1133–1139. doi: 10.1182/blood.V53.6.1133.1133. [DOI] [PubMed] [Google Scholar]
- 10.DeCoursey TE. 2010. Voltage-gated proton channels find their dream job managing the respiratory burst in phagocytes. Physiology (Bethesda) 25:27–40. doi: 10.1152/physiol.00039.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hampton MB, Kettle AJ, Winterbourn CC. 1998. Inside the neutrophil phagosome: oxidants, myeloperoxidase, and bacterial killing. Blood 92:3007–3017. doi: 10.1182/blood.V92.9.3007. [DOI] [PubMed] [Google Scholar]
- 12.Davies MJ. 2011. Myeloperoxidase-derived oxidation: mechanisms of biological damage and its prevention. J Clin Biochem Nutr 48:8–19. doi: 10.3164/jcbn.11-006FR. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Meotti FC, Jameson GNL, Turner R, Harwood DT, Stockwell S, Rees MD, Thomas SR, Kettle AJ. 2011. Urate as a physiological substrate for myeloperoxidase implications for hyperuricemia and inflammation. J Biol Chem 286:12901–12911. doi: 10.1074/jbc.M110.172460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Green JN, Kettle AJ, Winterbourn CC. 2014. Protein chlorination in neutrophil phagosomes and correlation with bacterial killing. Free Radic Biol Med 77:49–56. doi: 10.1016/j.freeradbiomed.2014.08.013. [DOI] [PubMed] [Google Scholar]
- 15.Klebanoff SJ, Kettle AJ, Rosen H, Winterbourn CC, Nauseef WM. 2013. Myeloperoxidase: a front-line defender against phagocytosed microorganisms. J Leukoc Biol 93:185–198. doi: 10.1189/jlb.0712349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Weiss SJ. 1989. Tissue destruction by neutrophils. N Engl J Med 320:365–376. doi: 10.1056/NEJM198902093200606. [DOI] [PubMed] [Google Scholar]
- 17.Winterbourn CC, Hampton MB, Livesey JH, Kettle AJ. 2006. Modeling the reactions of superoxide and myeloperoxidase in the neutrophil phagosome: implications for microbial killing. J Biol Chem 281:39860–39869. doi: 10.1074/jbc.M605898200. [DOI] [PubMed] [Google Scholar]
- 18.Imlay JA. 2008. Cellular defenses against superoxide and hydrogen peroxide. Annu Rev Biochem 77:755–776. doi: 10.1146/annurev.biochem.77.061606.161055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Imlay JA. 2015. Transcription factors that defend bacteria against reactive oxygen species. Annu Rev Microbiol 69:93–108. doi: 10.1146/annurev-micro-091014-104322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Imlay JA. 2013. The molecular mechanisms and physiological consequences of oxidative stress: lessons from a model bacterium. Nat Rev Microbiol 11:443–454. doi: 10.1038/nrmicro3032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Nagl M, Hess MW, Pfaller K, Hengster P, Gottardi W. 2000. Bactericidal activity of micromolar N-chlorotaurine: evidence for its antimicrobial function in the human defense system. Antimicrob Agents Chemother 44:2507–2513. doi: 10.1128/aac.44.9.2507-2513.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Love DT, Barrett TJ, White MY, Cordwell SJ, Davies MJ, Hawkins CL. 2016. Cellular targets of the myeloperoxidase-derived oxidant hypothiocyanous acid (HOSCN) and its role in the inhibition of glycolysis in macrophages. Free Radic Biol Med 94:88–98. doi: 10.1016/j.freeradbiomed.2016.02.016. [DOI] [PubMed] [Google Scholar]
- 23.Cremers CM, Jakob U. 2013. Oxidant sensing by reversible disulfide bond formation. J Biol Chem 288:26489–26496. doi: 10.1074/jbc.R113.462929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Groitl B, Dahl J-U, Schroeder JW, Jakob U. 2017. Pseudomonas aeruginosa defense systems against microbicidal oxidants. Mol Microbiol 106:335–350. doi: 10.1111/mmi.13768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Winter J, Ilbert M, Graf PCF, Ozcelik D, Jakob U. 2008. Bleach activates a redox-regulated chaperone by oxidative protein unfolding. Cell 135:691–701. doi: 10.1016/j.cell.2008.09.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gray MJ, Wholey W-Y, Jakob U. 2013. Bacterial responses to reactive chlorine species. Annu Rev Microbiol 67:141–160. doi: 10.1146/annurev-micro-102912-142520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Xie K, Bunse C, Marcus K, Leichert LI. 2019. Quantifying changes in the bacterial thiol redox proteome during host-pathogen interaction. Redox Biol 21:101087. doi: 10.1016/j.redox.2018.101087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Vethanayagam RR, Almyroudis NG, Grimm MJ, Lewandowski DC, Pham CTN, Blackwell TS, Petraitiene R, Petraitis V, Walsh TJ, Urban CF, Segal BH. 2011. Role of NADPH oxidase versus neutrophil proteases in antimicrobial host defense. PLoS One 6:e28149. doi: 10.1371/journal.pone.0028149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Parker BW, Schwessinger EA, Jakob U, Gray MJ. 2013. The RclR protein is a reactive chlorine-specific transcription factor in Escherichia coli. J Biol Chem 288:32574–32584. doi: 10.1074/jbc.M113.503516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gebendorfer KM, Drazic A, Le Y, Gundlach J, Bepperling A, Kastenmüller A, Ganzinger KA, Braun N, Franzmann TM, Winter J. 2012. Identification of a hypochlorite-specific transcription factor from Escherichia coli. J Biol Chem 287:6892–6903. doi: 10.1074/jbc.M111.287219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pattison DI, Davies MJ. 2001. Absolute rate constants for the reaction of hypochlorous acid with protein side chains and peptide bonds. Chem Res Toxicol 14:1453–1464. doi: 10.1021/tx0155451. [DOI] [PubMed] [Google Scholar]
- 32.Dukan S, Dadon S, Smulski DR, Belkin S. 1996. Hypochlorous acid activates the heat shock and soxRS systems of Escherichia coli. Appl Environ Microbiol 62:4003–4008. doi: 10.1128/AEM.62.11.4003-4008.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Storkey C, Davies MJ, Pattison DI. 2014. Reevaluation of the rate constants for the reaction of hypochlorous acid (HOCl) with cysteine, methionine, and peptide derivatives using a new competition kinetic approach. Free Radic Biol Med 73:60–66. doi: 10.1016/j.freeradbiomed.2014.04.024. [DOI] [PubMed] [Google Scholar]
- 34.Hawkins CL, Pattison DI, Davies MJ. 2003. Hypochlorite-induced oxidation of amino acids, peptides and proteins. Amino Acids 25:259–274. doi: 10.1007/s00726-003-0016-x. [DOI] [PubMed] [Google Scholar]
- 35.Winter J, Linke K, Jatzek A, Jakob U. 2005. Severe oxidative stress causes inactivation of DnaK and activation of the redox-regulated chaperone Hsp33. Mol Cell 17:381–392. doi: 10.1016/j.molcel.2004.12.027. [DOI] [PubMed] [Google Scholar]
- 36.Barrette WC, Hannum DM, Wheeler WD, Hurst JK. 1989. General mechanism for the bacterial toxicity of hypochlorous acid: abolition of ATP production. Biochemistry 28:9172–9178. doi: 10.1021/bi00449a032. [DOI] [PubMed] [Google Scholar]
- 37.Gray MJ, Wholey WY, Wagner NO, Cremers CM, Mueller-Schickert A, Hock NT, Krieger AG, Smith EM, Bender RA, Bardwell JCA, Jakob U. 2014. Polyphosphate is a primordial chaperone. Mol Cell 53:689–699. doi: 10.1016/j.molcel.2014.01.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Reichmann D, Voth W, Jakob U. 2018. Maintaining a healthy proteome during oxidative stress. Mol Cell 69:203–213. doi: 10.1016/j.molcel.2017.12.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Frydman J. 2001. Folding of newly translated proteins in vivo: the role of molecular chaperones. Annu Rev Biochem 70:603–647. doi: 10.1146/annurev.biochem.70.1.603. [DOI] [PubMed] [Google Scholar]
- 40.Khor HK, Fisher MT, Schöneich C. 2004. Potential role of methionine sulfoxide in the inactivation of the chaperone GroEL by hypochlorous acid (HOCl) and peroxynitrite (ONOO-). J Biol Chem 279:19486–19493. doi: 10.1074/jbc.M310045200. [DOI] [PubMed] [Google Scholar]
- 41.Jakob U, Muse W, Eser M, Bardwell JC. 1999. Chaperone activity with a redox switch. Cell 96:341–352. doi: 10.1016/s0092-8674(00)80547-4. [DOI] [PubMed] [Google Scholar]
- 42.Voth W, Jakob U. 2017. Stress-activated chaperones: a first line of defense. Trends Biochem Sci 42:899–913. doi: 10.1016/j.tibs.2017.08.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Verghese J, Abrams J, Wang Y, Morano KA. 2012. Biology of the heat shock response and protein chaperones: budding yeast (Saccharomyces cerevisiae) as a model system. Microbiol Mol Biol Rev 76:115–158. doi: 10.1128/MMBR.05018-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Groitl B, Jakob U. 2014. Thiol-based redox switches. Biochim Biophys Acta 1844:1335–1343. doi: 10.1016/j.bbapap.2014.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kumsta C, Jakob U. 2009. Redox-regulated chaperones. Biochemistry 48:4666–4676. doi: 10.1021/bi9003556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Bardwell JCA, Jakob U. 2012. Conditional disorder in chaperone action. Trends Biochem Sci 37:517–525. doi: 10.1016/j.tibs.2012.08.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Reichmann D, Jakob U. 2013. The roles of conditional disorder in redox proteins. Curr Opin Struct Biol 23:436–442. doi: 10.1016/j.sbi.2013.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Graumann J, Lilie H, Tang X, Tucker KA, Hoffmann JH, Vijayalakshmi J, Saper M, Bardwell JC, Jakob U. 2001. Activation of the redox-regulated molecular chaperone Hsp33–a two-step mechanism. Structure 9:377–387. doi: 10.1016/s0969-2126(01)00599-8. [DOI] [PubMed] [Google Scholar]
- 49.Lebrun V, Ravanat J-L, Latour J-M, Sénèque O. 2016. Near diffusion-controlled reaction of a Zn(Cys)4 zinc finger with hypochlorous acid. Chem Sci 7:5508–5516. doi: 10.1039/c6sc00974c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Perkins A, Tudorica DA, Amieva MR, Remington SJ, Guillemin K. 2019. Helicobacter pylori senses bleach (HOCl) as a chemoattractant using a cytosolic chemoreceptor. PLoS Biol 17:e3000395. doi: 10.1371/journal.pbio.3000395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Reichmann D, Xu Y, Cremers CM, Ilbert M, Mittelman R, Fitzgerald MC, Jakob U. 2012. Order out of disorder: working cycle of an intrinsically unfolded chaperone. Cell 148:947–957. doi: 10.1016/j.cell.2012.01.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ilbert M, Horst J, Ahrens S, Winter J, Graf PCF, Lilie H, Jakob U. 2007. The redox-switch domain of Hsp33 functions as dual stress sensor. Nat Struct Mol Biol 14:556–563. doi: 10.1038/nsmb1244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Moayed F, Bezrukavnikov S, Naqvi MM, Groitl B, Cremers CM, Kramer G, Ghosh K, Jakob U, Tans SJ. 2020. The anti-aggregation holdase Hsp33 promotes the formation of folded protein structures. Biophys J 118:85–95. doi: 10.1016/j.bpj.2019.10.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Cremers CM, Knoefler D, Vitvitsky V, Banerjee R, Jakob U. 2014. Bile salts act as effective protein-unfolding agents and instigators of disulfide stress in vivo. Proc Natl Acad Sci U S A 111:E1610–E1619. doi: 10.1073/pnas.1401941111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Krewing M, Stepanek JJ, Cremers C, Lackmann J-W, Schubert B, Müller A, Awakowicz P, Leichert LIO, Jakob U, Bandow JE. 2019. The molecular chaperone Hsp33 is activated by atmospheric-pressure plasma protecting proteins from aggregation. J R Soc Interface 16:20180966. doi: 10.1098/rsif.2018.0966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Wholey W-Y, Jakob U. 2012. Hsp33 confers bleach resistance by protecting elongation factor Tu against oxidative degradation in Vibrio cholerae. Mol Microbiol 83:981–991. doi: 10.1111/j.1365-2958.2012.07982.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Müller A, Langklotz S, Lupilova N, Kuhlmann K, Bandow JE, Leichert L. 2014. Activation of RidA chaperone function by N-chlorination. Nat Commun 5:5804. doi: 10.1038/ncomms6804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Lambrecht JA, Schmitz GE, Downs DM. 2013. RidA proteins prevent metabolic damage inflicted by PLP-dependent dehydratases in all domains of life. mBio 4:e00033-13. doi: 10.1128/mBio.00033-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Lambrecht JA, Flynn JM, Downs DM. 2012. Conserved YjgF protein family deaminates reactive enamine/imine intermediates of pyridoxal 5’-phosphate (PLP)-dependent enzyme reactions. J Biol Chem 287:3454–3461. doi: 10.1074/jbc.M111.304477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Lindemann C, Lupilova N, Müller A, Warscheid B, Meyer HE, Kuhlmann K, Eisenacher M, Leichert LI. 2013. Redox proteomics uncovers peroxynitrite-sensitive proteins that help Escherichia coli to overcome nitrosative stress. J Biol Chem 288:19698–19714. doi: 10.1074/jbc.M113.457556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Hawkins CL, Davies MJ. 1998. Hypochlorite-induced damage to proteins: formation of nitrogen-centred radicals from lysine residues and their role in protein fragmentation. Biochem J 332:617–625. doi: 10.1042/bj3320617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Grisham MB, Jefferson MM, Melton DF, Thomas EL. 1984. Chlorination of endogenous amines by isolated neutrophils. Ammonia-dependent bactericidal, cytotoxic, and cytolytic activities of the chloramines. J Biol Chem 259:10404–10413. [PubMed] [Google Scholar]
- 63.Kim HJ, Kwon A-R, Lee B-J. 2018. A novel chlorination-induced ribonuclease YabJ from Staphylococcus aureus. Biosci Rep 38:BSR20180768. doi: 10.1042/BSR20180768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Goemans CV, Vertommen D, Agrebi R, Collet JF. 2018. CnoX is a chaperedoxin: a holdase that protects its substrates from irreversible oxidation. Mol Cell 70:614–627.e7. doi: 10.1016/j.molcel.2018.04.002. [DOI] [PubMed] [Google Scholar]
- 65.Kenneth Allan R, Ratajczak T. 2011. Versatile TPR domains accommodate different modes of target protein recognition and function. Cell Stress Chaperones 16:353–367. doi: 10.1007/s12192-010-0248-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Kthiri F, Le H-T, Tagourti J, Kern R, Malki A, Caldas T, Abdallah J, Landoulsi A, Richarme G. 2008. The thioredoxin homolog YbbN functions as a chaperone rather than as an oxidoreductase. Biochem Biophys Res Commun 374:668–672. doi: 10.1016/j.bbrc.2008.07.080. [DOI] [PubMed] [Google Scholar]
- 67.Le H-T, Gautier V, Kthiri F, Kohiyama M, Katayama T, Richarme G. 2011. DNA replication defects in a mutant deficient in the thioredoxin homolog YbbN. Biochem Biophys Res Commun 405:52–57. doi: 10.1016/j.bbrc.2010.12.122. [DOI] [PubMed] [Google Scholar]
- 68.Caldas T, Malki A, Kern R, Abdallah J, Richarme G. 2006. The Escherichia coli thioredoxin homolog YbbN/Trxsc is a chaperone and a weak protein oxidoreductase. Biochem Biophys Res Commun 343:780–786. doi: 10.1016/j.bbrc.2006.03.028. [DOI] [PubMed] [Google Scholar]
- 69.Goemans CV, Beaufay F, Arts IS, Agrebi R, Vertommen D, Collet JF. 2018. The chaperone and redox properties of CnoX chaperedoxins are tailored to the proteostatic needs of bacterial species. mBio 9:e01541-18. doi: 10.1128/mBio.01541-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Lin J, Wilson MA. 2011. Escherichia coli thioredoxin-like protein YbbN contains an atypical tetratricopeptide repeat motif and is a negative regulator of GroEL. J Biol Chem 286:19459–19469. doi: 10.1074/jbc.M111.238741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Berndt C, Lillig CH, Holmgren A. 2008. Thioredoxins and glutaredoxins as facilitators of protein folding. Biochim Biophys Acta 1783:641–650. doi: 10.1016/j.bbamcr.2008.02.003. [DOI] [PubMed] [Google Scholar]
- 72.Holmgren A, Johansson C, Berndt C, Lönn ME, Hudemann C, Lillig CH. 2005. Thiol redox control via thioredoxin and glutaredoxin systems. Biochem Soc Trans 33:1375–1377. doi: 10.1042/BST20051375. [DOI] [PubMed] [Google Scholar]
- 73.Pan JL, Bardwell J. 2006. The origami of thioredoxin-like folds. Protein Sci 15:2217–2227. doi: 10.1110/ps.062268106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Barbirz S, Jakob U, Glocker MO. 2000. Mass spectrometry unravels disulfide bond formation as the mechanism that activates a molecular chaperone. J Biol Chem 275:18759–18766. doi: 10.1074/jbc.M001089200. [DOI] [PubMed] [Google Scholar]
- 75.Ulfig A, Schulz AV, Müller A, Lupilov N, Leichert LI. 2019. N-chlorination mediates protective and immunomodulatory effects of oxidized human plasma proteins. Elife 8:e47395. doi: 10.7554/eLife.47395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Rao NN, Gómez-García MR, Kornberg A. 2009. Inorganic polyphosphate: essential for growth and survival. Annu Rev Biochem 78:605–647. doi: 10.1146/annurev.biochem.77.083007.093039. [DOI] [PubMed] [Google Scholar]
- 77.Ahn K, Kornberg A. 1990. Polyphosphate kinase from Escherichia coli. Purification and demonstration of a phosphoenzyme intermediate. J Biol Chem 265:11734–11739. [PubMed] [Google Scholar]
- 78.Akiyama M, Crooke E, Kornberg A. 1992. The polyphosphate kinase gene of Escherichia coli. Isolation and sequence of the ppk gene and membrane location of the protein. J Biol Chem 267:22556–22561. [PubMed] [Google Scholar]
- 79.Yoo NG, Dogra S, Meinen BA, Tse E, Haefliger J, Southworth DR, Gray MJ, Dahl J-U, Jakob U. 2018. Polyphosphate stabilizes protein unfolding intermediates as soluble amyloid-like oligomers. J Mol Biol 430:4195–4208. doi: 10.1016/j.jmb.2018.08.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Mullan A, Quinn JP, McGrath JW. 2002. Enhanced phosphate uptake and polyphosphate accumulation in Burkholderia cepacia grown under low pH conditions. Microb Ecol 44:69–77. doi: 10.1007/s00248-002-3004-x. [DOI] [PubMed] [Google Scholar]
- 81.Ault-Riche D, Fraley CD, Tzeng C-M, Kornberg A. 1998. Novel assay reveals multiple pathways regulating stress-induced accumulations of inorganic polyphosphate in Escherichia coli. J Bacteriol 180:1841–1847. doi: 10.1128/JB.180.7.1841-1847.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Shiba T, Tsutsumi K, Yano H, Ihara Y, Kameda A, Tanaka K, Takahashi H, Munekata M, Rao NN, Kornberg A. 1997. Inorganic polyphosphate and the induction of rpoS expression. Proc Natl Acad Sci U S A 94:11210–11215. doi: 10.1073/pnas.94.21.11210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Akiyama M, Crooke E, Kornberg A. 1993. An exopolyphosphatase of Escherichia coli. The enzyme and its ppx gene in a polyphosphate operon. J Biol Chem 268:633–639. [PubMed] [Google Scholar]
- 84.Crooke E, Akiyama M, Rao NN, Kornberg A. 1994. Genetically altered levels of inorganic polyphosphate in Escherichia coli. J Biol Chem 269:6290–6295. [PubMed] [Google Scholar]
- 85.Rashid MH, Kornberg A. 2000. Inorganic polyphosphate is needed for swimming, swarming, and twitching motilities of Pseudomonas aeruginosa. Proc Natl Acad Sci U S A 97:4885–4890. doi: 10.1073/pnas.060030097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Rashid MH, Rao NN, Kornberg A. 2000. Inorganic polyphosphate is required for motility of bacterial pathogens. J Bacteriol 182:225–227. doi: 10.1128/jb.182.1.225-227.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Dahl J-U, Gray MJ, Bazopoulou D, Beaufay F, Lempart J, Koenigsknecht MJ, Wang Y, Baker JR, Hasler WL, Young VB, Sun D, Jakob U. 2017. The anti-inflammatory drug mesalamine targets bacterial polyphosphate accumulation. Nat Microbiol 2:16267. doi: 10.1038/nmicrobiol.2016.267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Fraley CD, Rashid MH, Lee SSK, Gottschalk R, Harrison J, Wood PJ, Brown MRW, Kornberg A. 2007. A polyphosphate kinase 1 (ppk1) mutant of Pseudomonas aeruginosa exhibits multiple ultrastructural and functional defects. Proc Natl Acad Sci U S A 104:3526–3531. doi: 10.1073/pnas.0609733104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Kim K-S, Rao NN, Fraley CD, Kornberg A. 2002. Inorganic polyphosphate is essential for long-term survival and virulence factors in Shigella and Salmonella spp. Proc Natl Acad Sci U S A 99:7675–7680. doi: 10.1073/pnas.112210499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Tan S, Fraley CD, Zhang M, Dailidiene D, Kornberg A, Berg DE. 2005. Diverse phenotypes resulting from polyphosphate kinase gene (ppk1) inactivation in different strains of Helicobacter pylori. J Bacteriol 187:7687–7695. doi: 10.1128/JB.187.22.7687-7695.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Lempart J, Jakob U. 2019. Role of polyphosphate in amyloidogenic processes. Cold Spring Harb Perspect Biol 11:a034041. doi: 10.1101/cshperspect.a034041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Xie L, Jakob U. 2019. Inorganic polyphosphate, a multifunctional polyanionic protein scaffold. J Biol Chem 294:2180–2190. doi: 10.1074/jbc.REV118.002808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Gray MJ, Jakob U. 2015. Oxidative stress protection by polyphosphate—new roles for an old player. Curr Opin Microbiol 24:1–6. doi: 10.1016/j.mib.2014.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Kampinga HH. 2014. Chaperoned by prebiotic inorganic polyphosphate molecules: an ancient transcription-independent mechanism to restore protein homeostasis. Mol Cell 53:685–687. doi: 10.1016/j.molcel.2014.02.023. [DOI] [PubMed] [Google Scholar]
- 95.Gray MJ, Wholey W-Y, Parker BW, Kim M, Jakob U. 2013. NemR is a bleach-sensing transcription factor. J Biol Chem 288:13789–13798. doi: 10.1074/jbc.M113.454421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Rudat AK, Pokhrel A, Green TJ, Gray MJ. 2018. Mutations in Escherichia coli polyphosphate kinase that lead to dramatically increased in vivo polyphosphate levels. J Bacteriol 200:e00697-17. doi: 10.1128/JB.00697-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Gray MJ. 2019. Inorganic polyphosphate accumulation in Escherichia coli is regulated by DksA but not by (p)ppGpp. J Bacteriol 201:e00664-18. doi: 10.1128/JB.00664-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Segawa S, Fujiya M, Konishi H, Ueno N, Kobayashi N, Shigyo T, Kohgo Y. 2011. Probiotic-derived polyphosphate enhances the epithelial barrier function and maintains intestinal homeostasis through integrin-p38 MAPK pathway. PLoS One 6:e23278. doi: 10.1371/journal.pone.0023278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Baker CJ, Smith SA, Morrissey JH. 2019. Polyphosphate in thrombosis, hemostasis, and inflammation. Res Pract Thromb Haemost 3:18–25. doi: 10.1002/rth2.12162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Smith SA, Morrissey JH. 2014. Polyphosphate: a new player in the field of hemostasis. Curr Opin Hematol 21:388–394. doi: 10.1097/MOH.0000000000000069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Mah TF, O'Toole GA. 2001. Mechanisms of biofilm resistance to antimicrobial agents. Trends Microbiol 9:34–39. doi: 10.1016/S0966-842X(00)01913-2. [DOI] [PubMed] [Google Scholar]
- 102.Mah T-F, Pitts B, Pellock B, Walker GC, Stewart PS, O'Toole GA. 2003. A genetic basis for Pseudomonas aeruginosa biofilm antibiotic resistance. Nature 426:306–310. doi: 10.1038/nature02122. [DOI] [PubMed] [Google Scholar]
- 103.Hall-Stoodley L, Costerton JW, Stoodley P. 2004. Bacterial biofilms: from the natural environment to infectious diseases. Nat Rev Microbiol 2:95–108. doi: 10.1038/nrmicro821. [DOI] [PubMed] [Google Scholar]
- 104.O'Toole GA, Wong GC. 2016. Sensational biofilms: surface sensing in bacteria. Curr Opin Microbiol 30:139–146. doi: 10.1016/j.mib.2016.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Whiteley M, Bangera MG, Bumgarner RE, Parsek MR, Teitzel GM, Lory S, Greenberg EP. 2001. Gene expression in Pseudomonas aeruginosa biofilms. Nature 413:860–864. doi: 10.1038/35101627. [DOI] [PubMed] [Google Scholar]
- 106.Orazi G, O’Toole GA. 2020. “It takes a village”: mechanisms underlying antimicrobial recalcitrance of polymicrobial biofilms. J Bacteriol 202:e00530-19. doi: 10.1128/JB.00530-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Branda SS, Vik S, Friedman L, Kolter R. 2005. Biofilms: the matrix revisited. Trends Microbiol 13:20–26. doi: 10.1016/j.tim.2004.11.006. [DOI] [PubMed] [Google Scholar]
- 108.Hengge R, Gründling A, Jenal U, Ryan R, Yildiz F. 2016. Bacterial signal transduction by cyclic Di-GMP and other nucleotide second messengers. J Bacteriol 198:15–26. doi: 10.1128/JB.00331-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Boyd CD, O'Toole GA. 2012. Second messenger regulation of biofilm formation: breakthroughs in understanding c-di-GMP effector systems. Annu Rev Cell Dev Biol 28:439–462. doi: 10.1146/annurev-cellbio-101011-155705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Jenal U, Malone J. 2006. Mechanisms of cyclic-di-GMP signaling in bacteria. Annu Rev Genet 40:385–407. doi: 10.1146/annurev.genet.40.110405.090423. [DOI] [PubMed] [Google Scholar]
- 111.Povolotsky TL, Hengge R. 2012. “Life-style” control networks in Escherichia coli: signaling by the second messenger c-di-GMP. J Biotechnol 160:10–16. doi: 10.1016/j.jbiotec.2011.12.024. [DOI] [PubMed] [Google Scholar]
- 112.Strempel N, Nusser M, Neidig A, Brenner-Weiss G, Overhage J. 2017. The oxidative stress agent hypochlorite stimulates c-di-GMP synthesis and biofilm formation in Pseudomonas aeruginosa. Front Microbiol 8:2311. doi: 10.3389/fmicb.2017.02311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Poudyal B, Sauer K. 2018. The PA3177 gene encodes an active diguanylate cyclase that contributes to biofilm antimicrobial tolerance but not biofilm formation by Pseudomonas aeruginosa. Antimicrob Agents Chemother 62:e01049-18. doi: 10.1128/AAC.01049-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Klebensberger J, Birkenmaier A, Geffers R, Kjelleberg S, Philipp B. 2009. SiaA and SiaD are essential for inducing autoaggregation as a specific response to detergent stress in Pseudomonas aeruginosa. Environ Microbiol 11:3073–3086. doi: 10.1111/j.1462-2920.2009.02012.x. [DOI] [PubMed] [Google Scholar]
- 115.Hoffman LR, D'Argenio DA, MacCoss MJ, Zhang Z, Jones RA, Miller SI. 2005. Aminoglycoside antibiotics induce bacterial biofilm formation. Nature 436:1171–1175. doi: 10.1038/nature03912. [DOI] [PubMed] [Google Scholar]
- 116.Boehm A, Steiner S, Zaehringer F, Casanova A, Hamburger F, Ritz D, Keck W, Ackermann M, Schirmer T, Jenal U. 2009. Second messenger signalling governs Escherichia coli biofilm induction upon ribosomal stress. Mol Microbiol 72:1500–1516. doi: 10.1111/j.1365-2958.2009.06739.x. [DOI] [PubMed] [Google Scholar]
- 117.Ranieri MR, Whitchurch CB, Burrows LL. 2018. Mechanisms of biofilm stimulation by subinhibitory concentrations of antimicrobials. Curr Opin Microbiol 45:164–169. doi: 10.1016/j.mib.2018.07.006. [DOI] [PubMed] [Google Scholar]
- 118.Stent A, Every AL, Sutton P. 2012. Helicobacter pylori defense against oxidative attack. Am J Physiol Gastrointest Liver Physiol 302:G579–G587. doi: 10.1152/ajpgi.00495.2011. [DOI] [PubMed] [Google Scholar]
- 119.Draper J, Karplus K, Ottemann KM. 2011. Identification of a chemoreceptor zinc-binding domain common to cytoplasmic bacterial chemoreceptors. J Bacteriol 193:4338–4345. doi: 10.1128/JB.05140-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Rolig AS, Shanks J, Carter JE, Ottemann KM. 2012. Helicobacter pylori requires TlpD-driven chemotaxis to proliferate in the antrum. Infect Immun 80:3713–3720. doi: 10.1128/IAI.00407-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Xu H, Xing J, Tang X, Sheng X, Zhan W. 2019. Immune response and protective effect against Vibrio anguillarum induced by DNA vaccine encoding Hsp33 protein. Microb Pathog 137:103729. doi: 10.1016/j.micpath.2019.103729. [DOI] [PubMed] [Google Scholar]
- 122.Imlay JA. 2019. Where in the world do bacteria experience oxidative stress? Environ Microbiol 21:521–530. doi: 10.1111/1462-2920.14445. [DOI] [PMC free article] [PubMed] [Google Scholar]