Chronic bacterial infections are caused by pathogens that persist within their hosts and avoid clearance by the immune system. Treatment and/or detection of such pathogens is difficult, and the resulting pathologies are often deleterious or fatal. There is an urgent need to develop protective vaccines and host-directed therapies that synergize with antibiotics to prevent pathogen persistence and infection-associated pathologies. However, many persistent pathogens, such as Mycobacterium tuberculosis, actively target the very host pathways activated by vaccination.
KEYWORDS: immune evasion, intracellular pathogens, Mycobacterium tuberculosis, T cell, vaccine development
ABSTRACT
Chronic bacterial infections are caused by pathogens that persist within their hosts and avoid clearance by the immune system. Treatment and/or detection of such pathogens is difficult, and the resulting pathologies are often deleterious or fatal. There is an urgent need to develop protective vaccines and host-directed therapies that synergize with antibiotics to prevent pathogen persistence and infection-associated pathologies. However, many persistent pathogens, such as Mycobacterium tuberculosis, actively target the very host pathways activated by vaccination. These immune evasion tactics blunt the effectiveness of immunization strategies and are impeding progress to control these infections throughout the world. Therefore, it is essential that M. tuberculosis immune evasion-related pathogen virulence strategies are considered to maximize the effectiveness of potential new treatments. In this review, we focus on how Mycobacterium tuberculosis infects antigen-presenting cells and evades effective immune clearance by the adaptive response through (i) manipulating antigen presentation, (ii) repressing T cell-activating costimulatory molecules, and (iii) inducing ligands that drive T cell exhaustion. In this context, we will examine the challenges that bacterial virulence strategies pose to developing new vaccines. We will then discuss new approaches that will help dissect M. tuberculosis immune evasion mechanisms and devise strategies to bypass them to promote long-term protection and prevent disease progression.
INTRODUCTION
The adaptive immune response has evolved to detect and destroy invading bacterial pathogens while simultaneously protecting the tissues of the host from damage (1). During an acute infection, adaptive responses synergize with innate pathways to drive bacterial clearance (2). A subset of pathogens, like Mycobacterium tuberculosis, cause persistent infections by employing virulence mechanisms that evade immune detection and inhibit adaptive responses (3, 4). This results in M. tuberculosis infections causing the most deaths by infectious disease each year (5). To prevent the ongoing epidemic, it is essential to develop an effective vaccine that protects against lung disease. A major challenge to M. tuberculosis vaccine development, however, is accounting for bacterial immune evasion tactics. M. tuberculosis effectively modulates adaptive responses from within the intracellular niche in antigen-presenting cells (APCs) that prevent T cell responses from sterilizing the infection (3, 6). Current vaccination methods activate the very processes that M. tuberculosis targets. Thus, it is essential to understand how M. tuberculosis suppresses adaptive responses to develop new approaches that bypass M. tuberculosis-mediated immune evasion. In this review, we highlight a subset of mechanisms used by Mycobacterium spp. to inhibit T cell responses from within APCs and draw similarities with other persistent bacterial pathogens. We will then discuss new approaches that might allow a full understanding of the M. tuberculosis-mediated immune evasion that is needed to develop vaccines that overcome pathogen virulence and protect against M. tuberculosis.
OVERVIEW OF M. TUBERCULOSIS IMMUNE CELL INTERACTIONS
M. tuberculosis is a facultative intracellular pathogen that resides inside APCs, including a variety of macrophage and dendritic cell (DC) subsets (7, 8). Upon inhalation of M. tuberculosis-containing aerosol droplets, M. tuberculosis efficiently targets alveolar macrophages that line the alveoli (8). Ideally, the initial infection by M. tuberculosis would stimulate inflammation in alveolar macrophages to activate protective adaptive immune responses that quickly respond to the lung and eradicate the infection. However, alveolar macrophages do not robustly detect or respond to M. tuberculosis infection, which results in a blunted inflammatory response and delays adaptive immune activation over 2 weeks (8–10). This delay is unlike other lung infections such as those caused by influenza virus or respiratory syncytial virus (11). These viral infections develop a robust pathogen-specific T cell response within 1 week, suggesting that M. tuberculosis actively uses the alveolar macrophages to avoid rapid adaptive immune activation and detection. Eventually, M. tuberculosis-infected alveolar macrophages migrate from the alveoli into the interstitial space through the direct activity of the specialized type VII secretion system ESX-1 (8). In the interstitial space, inflammatory macrophages and dendritic cell populations are infected by M. tuberculosis, triggering robust inflammation that causes the onset of adaptive immunity. M. tuberculosis antigens are trafficked to the draining lymph nodes by dendritic cells, where they activate M. tuberculosis-specific T cells that are required to protect against disease (12, 13).
For T cells to be activated during an M. tuberculosis infection, they must receive two distinct signals in the lung draining lymph node (14). Signal one is dependent on the antigen specificity of the T cell receptor (TCR) which detects pathogen-derived peptides loaded into major histocompatibility complex class I or II (MHC-I or MHC-II, respectively) (14). These peptide-MHC complexes are then presented on the surface of APCs to naive T cells. The second signal, also known as costimulation, is delivered to the T cell through the ligation of inflammation-induced molecules such as CD80, CD86, or CD40 on the surface of the APC (15, 16). Binding of distinct costimulatory molecule by T cells can skew their function, enhancing or inhibiting M. tuberculosis control (6, 17). In addition to signals one and two, a third signal, driven by stimulatory cytokines, enhances the activation of T cells, in particular, CD8+ T cells (reviewed in reference 4). Following their activation in the lymph node, T cells then traffic to the lung environment in search of infected cells to eradicate (13). In the lungs, direct contact of both CD4+ T cells and CD8+ T cells with cells harboring M. tuberculosis can partially control disease, yet they are insufficient to sterilize the infection (4, 13, 18). The reasons T cells fail to fully control M. tuberculosis infection are complex. M. tuberculosis actively prevents effective detection by T cells and drives T cell exhaustion that limits the protective potential of T cells (3, 7). It is also possible that M. tuberculosis evolved to use T cell responses to help drive transmission. Unlike viruses like influenza virus, which actively evade immunity by mutating antigens to prevent detection, M. tuberculosis does not evolve rapidly, and T cell antigens are known to be hyperconserved, with few mutations across lineages (19, 20). This means that the antigens activating the M. tuberculosis T cell responses are very conserved across the human population. This has led some to hypothesize that M. tuberculosis actively stimulates robust T cell responses to drive tissue damage and subsequent transmission. Together, these data suggest that T cells are essential to protect against M. tuberculosis infection, yet their role in disease progression needs to be more carefully understood.
While T cells are required for protection against tuberculosis (TB), how T cells mechanistically contribute to protection remains unclear. T cells can protect by controlling antimicrobial resistance pathways which directly restrict bacterial growth or by regulating disease tolerance, the ability to withstand an infection and the subsequent tissue damage (1, 21). Given that T cells are unable to provide sterilizing immunity against M. tuberculosis, it is likely that many antimicrobial mechanisms activated by T cells are ineffective against M. tuberculosis or are actively inhibited by the pathogen. Evidence also suggests that dysregulated T cell responses, such as increased gamma interferon (IFN-γ) production or altered mitochondrial dynamics, can drive failed disease tolerance (22, 23). Together, these studies suggest that an important role of T cells during M. tuberculosis infection is to promote disease tolerance against infection. Given the modest protection provided by even the best new vaccine candidates, the question of whether a T cell-mediated vaccine could activate antimicrobial resistance mechanisms remains uncertain. Evidence from the nonhuman primate model suggests that an ongoing M. tuberculosis infection can eliminate subsequent infections with new M. tuberculosis strains, but the mechanisms of this protection and the harnessing of these mechanisms for a vaccine remain unknown (24). However, a recent study examining the protective response in nonhuman primates immunized with Mycobacterium bovis BCG by the intravenous route suggests that this is a possibility (25). In this study, the authors found sterilizing immunity that was driven by a unique CD4+ T cell subpopulation that must now be further investigated for their protective potential.
Many current vaccine strategies for M. tuberculosis are aimed at augmenting T cell responses that are required to control M. tuberculosis. Most of these protective pathways were identified because hosts lacking single immune genes were more susceptible to M. tuberculosis infection than are otherwise healthy hosts. However, in many healthy individuals infected with fully virulent M. tuberculosis, these responses are inadequate on their own (4, 7, 26). A possible reason that current approaches are not yielding the progress hoped is that they do not account for M. tuberculosis immune evasion tactics. M. tuberculosis has developed distinct mechanisms that delay, block, and distract host immune mechanisms that might be the best equipped to eradicate the disease. Without fully understanding how M. tuberculosis avoids normally effective immune mechanisms, the challenge to overcome M. tuberculosis virulence strategies and develop fully protective vaccines remains.
MECHANISMS OF M. TUBERCULOSIS IMMUNE EVASION
M. tuberculosis inhibits antigen processing and presentation to prevent T cell-mediated clearance.
The ability of T cells to recognize their cognate antigens presented in MHC molecules is essential to activate T cell responses that detect and destroy infected cells in the lung environment. However, M. tuberculosis infection delays the activation of antigen-specific T cell responses, skews the antigen specificity of T cells, and prevents the effective detection of infected cells (13, 27, 28) (Fig. 1). M. tuberculosis-infected mice elicit a muted antigen-specific CD4+ T cell response compared to that with BCG-infected mice as a result of suboptimal M. tuberculosis antigen presentation independent of antigen levels, suggesting that M. tuberculosis interferes with optimal immune activation (29). Additionally, M. tuberculosis is able to evade direct killing by T cells through metabolic functions such as de novo tryptophan synthesis that bypasses the induction of indoleamine 2,3 dioxygenase (IDO) and tryptophan restriction in host cells (30). Thus, M. tuberculosis effectively manipulates the activation and effector functions of M. tuberculosis-specific CD4 and CD8+ T cells by modulating antigen processing and presentation in MHC molecules that contribute to pathogen persistence in the lungs.
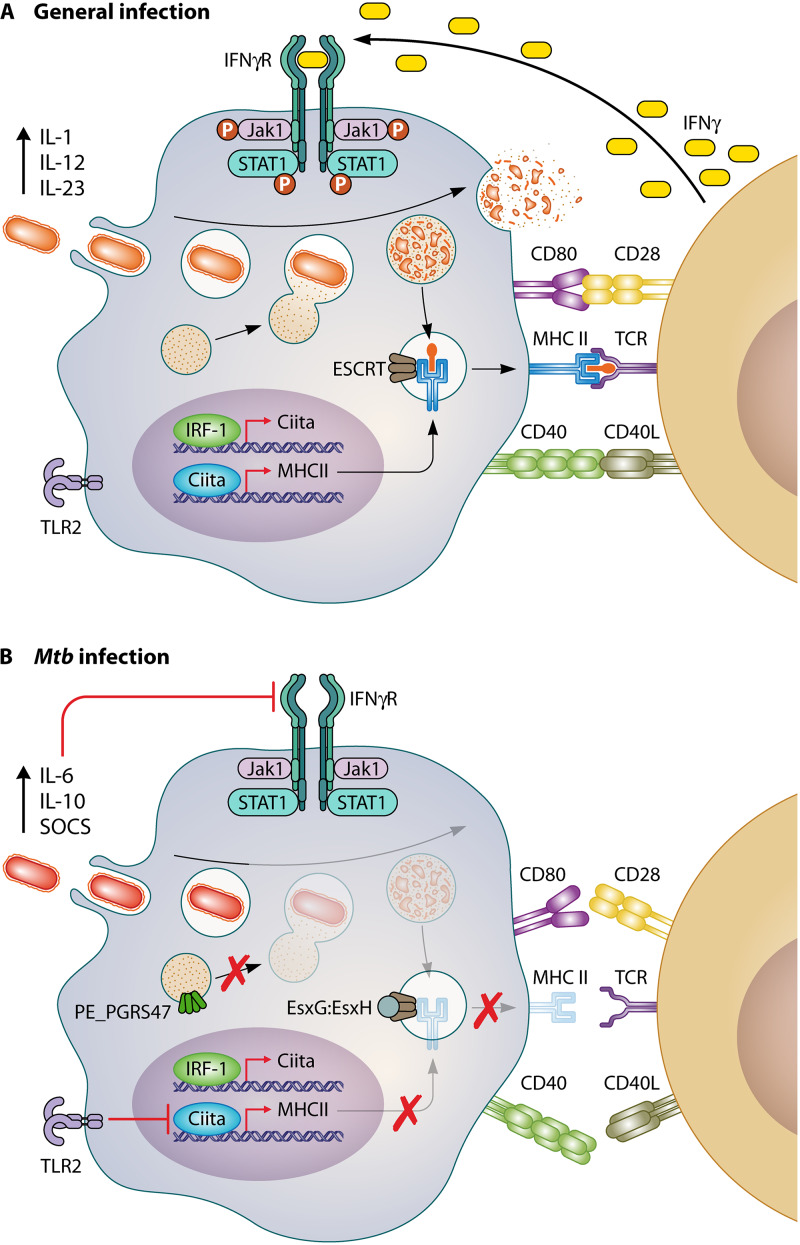
FIG 1.
M. tuberculosis inhibits effective antigen presentation. (A) Schematic of a normal response to phagocytosed bacteria. Antigen-presenting cells phagocytose a pathogen. Fusion of the phagolysosome causes degradation and production of pathogen-specific peptides. Pathogen-derived peptides then bind to the MHC-II complex and are trafficked to the surface of the cell. There, T cells recognize the presented antigen leading to increased immune cell recruitment, cytokine production, and antibody memory. (B) A schematic of M. tuberculosis (Mtb)-mediated evasion of antigen presentation. The APC engulfs M. tuberculosis, and the expression of PE_PGRS47 and EsxG-EsxH inhibits phagolysosome fusion by preventing the degradation of M. tuberculosis antigens. Innate immune detection of lipoproteins results in the activation of TLR2 by M. tuberculosis, which inhibits the induction of CIITA and MHC-II through unknown mechanisms. Together, these M. tuberculosis-mediated alterations prevent the effective expression of MHC molecules and prevents effective clearance by M. tuberculosis-specific T cells.
Inhibition of phagolysosome fusion and maturation.
M. tuberculosis-specific T cell responses are delayed in their activation, which allows uninhibited M. tuberculosis growth over the first weeks of infection (8, 13, 26). This delay is due to several factors, including M. tuberculosis-mediated inhibition of antigen processing and loading of antigens into the MHC. For all APCs, fusion of the phagocytosed particles in a phagosome with the lysosome is a general mechanism to kill the pathogen and provide pathogen-derived peptides that are then loaded into MHC-II molecules (31, 32). M. tuberculosis efficiently prevents lysosome fusion, growing instead in a modified phagosome compartment that is protected against bacterial degradation (33). This inhibition results in inefficient MHC-II loading of M. tuberculosis-derived peptides and prevents efficient CD4+ T cell detection. One way that M. tuberculosis blocks lysosome fusion is through the manipulation of the host endosomal sorting complex required for transport (ESCRT) (34–36). ESCRT sorts ubiquitin-labeled surface receptors into intraluminal vesicles of multivesicular bodies to be degraded in the lysosome and loaded into MHC-II molecules (36). M. tuberculosis secretes EsxH and EsxG via the ESX-3 type VII secretion system. These effectors then dimerize and inhibit phagosome maturation through the interactions with the ESCRT pathway (34, 35). EsxG-EsxH-dependent ESCRT inhibition limits the activation of M. tuberculosis-specific CD4+ T cells in both macrophages and dendritic cells, suggesting that these virulence components are key to the delay in T cell activation during infection. In the absence of EsxH, T cells are more effectively activated and better control infection. In contrast, M. tuberculosis strains overexpressing EsxG-EsxH further limit the activation of CD4+ T cells. Therefore, EsxH impairs phagolysosome fusion in an ESCRT-dependent manner, resulting in the suboptimal antigen processing and presentation of M. tuberculosis proteins that are needed to activate CD4+ T cells.
M. tuberculosis employs multiple effectors to modulate other host phagosomal fusion pathways. Maturation of the phagosome depends on the recruitment and dissociation of multiple membrane markers, including phosphatidylinositol-3-phosphate (PI3P), the acidifying proton pumps V-ATPase and Hv1, and multiple Rab-GTPases, each of which is disrupted by M. tuberculosis (37–42). The SecA2-dependent secreted factors SapM and PknG inhibit both phagosome and autophagosome maturation by inhibiting PI3P phosphorylation, Rab5 dissociation, and Rab7 recruitment (43). Additionally, the secreted protein tyrosine phosphatase PtpA specifically binds and inhibits V-ATPase trafficking to the phagosome (44). It is currently unknown how M. tuberculosis inhibits Hv1 localization to the phagosome. Another recent study identified that the M. tuberculosis protein PE_PGRS47 plays an important role in modulating antigen presentation by inhibiting lysosome function (45). Rather than directly preventing lysosome fusion with the phagosome, PE_PGRS47 manipulates the host autophagy pathway. Autophagy is a highly conserved mechanism used by cells to remove unnecessary, damaged components and plays a role in controlling intracellular pathogens (46). Fusion of autophagosomes with the lysosome allows foreign antigens to be efficiently loaded and presented on MHC-II molecules (47). PE_PGRS47 limits MHC-II antigen presentation by preventing effective autophagosome-lysosome fusion. PE_PGRS47 deletion mutants show increased autophagic vesicles, in addition to more acidified phagosomes and increased lysosome fusion (45). The inhibition of autophagosome-lysosome fusion by PE_PGRS47 has functional consequences on the activation of CD4+ T cells. Infection of mice with PE_PGRS47-deficient strains resulted in significantly more activated CD4+ T cells due to the increase in M. tuberculosis antigens loaded into MHC-II molecules. This directly shows that pathogen-mediated inhibition of lysosome fusion is an effective immune evasion tactic to avoid rapid CD4+ T cell responses. While M. tuberculosis possesses numerous pathways to inhibit host phagolysosome fusion/maturation, it also expresses at least one failsafe mechanism to neutralize acidification that the bacteria do experience. A comparative lipidomics study between M. tuberculosis and BCG identified an M. tuberculosis-exclusive robustly produced extracellular lipid, 1-tuberculosinyladenosine (1-TbAd), which was shown to possess acid-neutralizing properties, result in protected growth at low pH, and induce phagosomal swelling in infected human macrophages (48, 49).
When viewed through this lens, it is not surprising how prevalent the inhibition of phagolysosome maturation and lysosome fusion is among successful pathogens. For example, Chlamydia trachomatis, Salmonella enterica serovar Typhimurium, and Brucella abortus all inhibit lysosome fusion by producing specific virulence factors (50–52). While it remains to be directly tested for these pathogens, evading lysosome fusion might not only protect bacterial viability but also hinder CD4+ T cell activation, similar to what occurs during M. tuberculosis infection. In addition, given the importance of lysosome inhibition to virulence, several M. tuberculosis factors may contribute in parallel to ensure success. It is essential to identify these redundant mechanisms in M. tuberculosis to delineate strategies that overcome lysosome inhibition and drive a more rapid and robust activation of protective CD4+ T cells.
Inhibition of MHC surface expression.
Once M. tuberculosis-specific T cells are activated in the draining lymph node, they must then traffic to the lung and identify M. tuberculosis-infected cells to control pathogen growth and contain disease (13). For both CD4+ and CD8+ T cells, this is known to require direct contact with the MHC on infected cells within the lung environment (4, 18). On resting macrophages, the surface expression of MHC-II is moderate, which limits the ability of macrophages to stimulate CD4+ T cells directly (53). However, the addition of cytokines such as IFN-γ results in the robust upregulation of MHC-II by the class II transactivator (CIITA) (53, 54). Intracellular pathogens have evolved mechanisms to directly inhibit the induction of CIITA and prevent MHC-II upregulation. Chlamydia trachomatis, for example, directly degrades the transcription factors USF-1 and RFX-5 that are required for IFN-γ-dependent upregulation of MHC-II (55, 56). In contrast, M. tuberculosis infection does not appear to actively target MHC-II upregulation but rather uses innate immune detection of M. tuberculosis against the host. Several studies have found that mycobacterial lipoproteins and other cell envelope components inhibit MHC-II upregulation by acting as Toll-like receptor 2 (TLR2) agonists (3, 57). The 19-kDa lipoprotein of M. tuberculosis limits MHC-I expression and prevents IFN-γ-induced HLA-DR (an MHC-II surface receptor), the FcγR1 (a high-affinity IgG receptor), and CIITA expression (58–60). M. tuberculosis also inhibits macrophage responses to IFN-γ by inducing cytokines like interleukin 6 (IL-6) which can inhibit Th1 differentiation while inducing the suppressor of cytokine signaling (SOCS) (61, 62). SOCS inhibits STAT1 phosphorylation, thus limiting antigen presentation. These findings are similar to those observed during Brucella abortus infection, which also induces IL-6 secretion to inhibit IFN-γ-mediated induction of interferon regulatory factor 1 (IRF-1) and CIITA (63). By stimulating innate responses that are hard wired into APCs, M. tuberculosis effectively prevents the subsequent induction of MHC molecules that would help T cell-mediated clearance. Data suggest that M. tuberculosis stimulates TLR2, TLR4, and TLR9, yet how these innate receptors directly or indirectly prevent the upregulation of surface MHC molecules remains to be fully understood (64). Additionally, how to overcome the initial TLR activation following M. tuberculosis infection to strongly upregulate MHC molecules is an outstanding question that must be addressed in the future.
Inhibition of direct T cell detection.
Even if infected APCs induced robust surface expression of MHC molecules, the question remains of whether T cells are capable of detecting M. tuberculosis-infected cells. Work from several groups suggests that M. tuberculosis might use decoy antigens to drive dominant T cell responses against proteins that are subsequently downregulated in M. tuberculosis as persistence begins (27, 28, 65, 66). This allows M. tuberculosis to evade protective T cell responses by eliminating the expression of the antigens for which the majority of T cells are specific. This evasion strategy results in a T cell repertoire that is focused on nonprotective antigens that do not improve disease outcome. Similarly, recent studies from the Behar group suggest an inability of M. tuberculosis-specific CD8+ cells to directly detect infected macrophages, which may explain a limited role for CD8+ cells in vivo (28). It has been a conundrum as to why CD8+ T cells do not play a more central role in the protective response against M. tuberculosis. M. tuberculosis secretes antigens directly into the cytosol, and CD8+ T cells are well equipped to eliminate cells infected with intracellular pathogens, yet the loss of CD8+ T cells results in a minimal change in M. tuberculosis disease progression (4). Interestingly, this mirrors findings for other intracellular pathogens like Chlamydia trachomatis, where CD8+ T cells play a minimal role in protection but contribute greatly to the immunopathology that occurs during chronic infections (67, 68).
In recent studies, M. tuberculosis-infected macrophages were detected by CD4+ T cells specific for ESAT-6 and Ag85b and restricted M. tuberculosis growth (28). Yet, polyclonal CD4+ T cells isolated from the infected lungs of mice did not effectively detect infected macrophages (27). In contrast, CD8+ T cells specific for the immunodominant antigen TB10.4 could not detect M. tuberculosis in macrophages, while polyclonal CD8+ T cells could (27, 28). These findings raise the possibility that M. tuberculosis carefully controls what antigens are available for presentation in infected cells to dictate the T cell repertoire that is activated. Clearly, this virulence strategy allows for the effective evasion of protective T cell responses, yet how M. tuberculosis controls antigen availability and prevents effective detection by T cells on macrophages remains unknown. In these reports, there were no obvious problems with antigen processing or antigen presentation (32, 64). However, it was noted that infection with BCG results in a greater capacity of T cells to recognize infected macrophages, suggesting that the RD1 locus may play a role in masking infected macrophages. Any future vaccine must overcome the ability of M. tuberculosis to distract T cell responses away from protective antigens to allow the detection of infected cells.
M. tuberculosis modulates costimulatory molecules to skew T cell effector responses.
In addition to antigen-specific activation that is mediated by peptide-MHC complexes, effective T cell responses require efficient costimulation (16). Costimulation, or signal 2, occurs through the ligation of a variety of molecules on APCs with their cognate ligands on the surface of T cells (14, 16). While the interaction between CD28 on T cells and CD80/CD86 on APCs is most commonly studied, other molecules on APCs, including CD40 and OX40L, also play an important role. How each costimulatory molecule influences immune responses to distinct pathogens remains unclear, as costimulatory molecules can augment the MHC-TCR signal and influence the subsequent effector response. The absence of any costimulatory signal in the presence of antigen presentation results in T cell anergy, a state in which the T cell remains alive but is largely unresponsive to further stimulus (69). Given the importance of costimulation in shaping the subsequent host response, pathogens like M. tuberculosis directly manipulate these molecules to the benefit of the pathogen (Fig. 2). While our understanding of the mechanisms employed by M. tuberculosis to target distinct costimulatory pathways remains lagging, there is significant evidence that targeting distinct costimulation networks during immunization may improve M. tuberculosis infection outcomes.
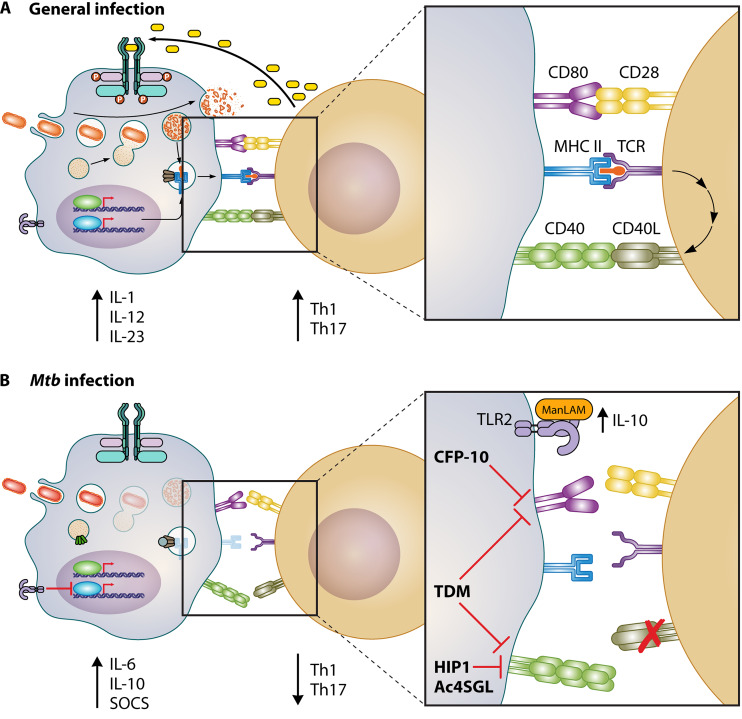
FIG 2.
M. tuberculosis uses multiple effectors to manipulate costimulatory molecule activity and CD4+ T cell polarization. (A) Schematic of the general response of costimulatory molecules in APCs during infection with an intracellular pathogen. APCs induce CD80/86:CD28 and/or CD40:CD40L binding, resulting in the proliferation of CD4+ T cells and Th-1/Th-17 polarizing cytokine secretion from the APC. (B) Schematic of M. tuberculosis-mediated evasion of costimulatory. Infection of APCs with M. tuberculosis results in inhibition of costimulatory molecule expression. ManLAM from M. tuberculosis interacts with TLR2 to induce IL-10 secretion, repressing Th1 polarization. TDM inhibits the induction of CD80/86 and CD40, while Hip1 and Ac4SGL block robust activation of CD40. These virulence traits change the overall cytokine response and prevent the protective capacity of M. tuberculosis-specific T cells from eradicating the infection.
A central aspect of costimulatory molecules is that their expression is increased on the surface of APCs following the activation of pathogen-associated molecular patterns and the subsequent cytokine response (15, 16). This regulation pattern ensures that T cell activation only occurs in the correct inflammatory context. Both type I and type II interferons as well as tumor necrosis factor alpha (TNF-α) feed back into APCs to drive the expression of CD40, OX40L, CD80, and CD86 to maximal levels (15, 70, 71). Thus, M. tuberculosis is well positioned to manipulate the induction of costimulatory molecules by carefully controlling the inflammatory response it induces during infection. Recent evidence suggests that while each costimulation marker plays an important role in driving a protective response against M. tuberculosis, these pathways are not fully functional during virulent M. tuberculosis infection (6).
Costimulation between CD80/CD86 and CD28 helps activate the Th1 effector responses during M. tuberculosis infection (72). Loss of CD28 results in muted Th1 induction and uncontrolled M. tuberculosis growth (72). In addition, the loss of CD40 results in severe susceptibility to M. tuberculosis infection due to the altered effector profile of CD4+ T cells, while the loss of OX40L prevents the efficient induction of Th1 vaccine responses against M. bovis BCG (73, 74). Therefore, each distinct costimulatory molecule may help influence the overall host responses against M. tuberculosis. This is similar to other persistent pathogens like Chlamydia trachomatis, where a combination of costimulatory molecules appears to be required for effective protective immunity (75, 76). This highlights a supportive role for all costimulatory molecules during persistent infections that must be better understood.
Because costimulatory molecules play a critical role in determining the effector T cell response during M. tuberculosis infection, these pathways are actively manipulated during infection. Several reports suggest that during the chronic phase of M. tuberculosis infection, CD80, CD86, and CD40 are all downregulated in the lung, limiting the capacity of responding T cells to find infected cells and escape anergy (77). One mechanism driving the inhibition of costimulatory molecules is the presence of the surface lipid trehalose 6,6′-dimycolate (TDM) (78). Distinct macrophage populations, including alveolar and peritoneal macrophages, that were infected with wild-type M. tuberculosis did not express high levels of CD40, CD80, or CD86, while strains that were delipidated for TDM strongly induced the expression of these molecules. This broad inhibition is similar to the effect of VacA from Helicobacter pylori, which induces the dendritic cell transcription factor E2F1 to suppress activation (79, 80). It remains to be known if TDM directly induces E2F1 in a similar manner and should be pursued in the future. In addition to TDM, the mycobacterial cell wall component mannosylated lipoarabinomannan (ManLAM) binds TLR2, resulting in increased IL-10 production which prevents the robust differentiation of Th1 cells (81). Altogether, the inflammatory makeup of M. tuberculosis surface lipids directly impacts the magnitude of costimulatory molecule induction, thereby skewing T cells away from protective effector responses.
How M. tuberculosis modulates the makeup of surface lipids to impact costimulatory signals remains unclear. One possible mechanism is mediated by the serine hydrolase Hip1. Hip1 was identified in a genome-wide transposon screen and subsequently found to be required for M. tuberculosis to persist long term in animals (82, 83). Later studies found that Hip1 is important for GroEL hydrolysis and that Hip1 inhibits the inflammatory response to M. tuberculosis (84–86). The loss of Hip1 results in the increased expression of costimulatory molecules, in particular, CD40, on infected APCs (17, 85). In addition, the loss of Hip1 increases the production of proinflammatory cytokines that drive both Th1 and Th17 responses against M. tuberculosis infection. This observation has important implications, as wild-type M. tuberculosis infections do not effectively activate Th17 responses, even though a balanced Th1/Th17 response provides improved overall protection against M. tuberculosis. To test this hypothesis directly, Rengarajan and colleagues examined how the presence or absence of Hip1 changes CD40 expression on M. tuberculosis-infected APCs to skew the effector T cell response (17). Their results clearly show that CD40 is required to activate a subset of CD4+ T cells to produce IL-17. Infecting cells with a Hip1 mutant or treating wild-type M. tuberculosis-infected dendritic cells with purified CD40L-trimer overcame Hip1-mediated CD40 repression and induced a more balanced Th1/Th17 CD4+ T cell response. This balanced effector response of CD4+ T cells improved the capacity of the host to restrict M. tuberculosis growth and prevent disease. These important studies not only show that M. tuberculosis modifies detection by the innate immune system to avoid robust costimulation but also provide a foundation for the premise that overcoming pathogen-mediated immune evasion is critical to develop therapies that effectively protect against M. tuberculosis.
Beyond Hip1, the mycobacterial secreted factor CFP-10 and the Ac4SGL prevent the maximal expression of costimulatory molecules on APCs (87, 88). When cells express CFP-10, they are unable to induce CD80 expression following exogenous stimuli. How this inhibition occurs remains unknown (87). Similar to Hip1, Ac4SGL inhibits CD40 surface expression but through a distinct mechanism that suppresses NF-κB activation (88). Thus, M. tuberculosis uses multiple mechanisms to prevent the efficient induction of costimulatory molecules on infected APCs. The consequences of these changes to costimulatory molecules remain to be investigated carefully in vivo. In the future, it will be important to identify the mechanisms M. tuberculosis employs to block costimulatory molecule expression and investigate the regulation of each costimulatory molecule to overcome pathogen-mediated inhibition and improve the overall balance of the host response against M. tuberculosis.
M. tuberculosis induces inhibitory ligands that repress T cell function.
While the primary immune response in most individuals adequately contains M. tuberculosis infection, the lack of sterilizing immunity presents a challenge (1, 4). As effector T cells respond to persistent antigen stimulation, they begin to upregulate inhibitory receptors, such as PD1, Tim3, and CTLA4 (3, 4, 22, 89, 90). These receptors prevent overexuberant inflammation and induce a phenotypic state known as exhaustion (16). While exhaustion is important to ensure that inflammatory damage does not occur, M. tuberculosis likely uses exhaustion to its advantage to prevent effective clearance. As T cells become exhausted, they show dysfunction in their ability to activate upon antigen stimulation, produce cytokines, and directly kill infected cells (16). Several studies have shown that PD1 and Tim3 are upregulated on M. tuberculosis-specific T cells as infection progresses to the chronic phase (22, 89). The induction of these inhibitory molecules reduces the production of proliferative cytokines like IL-2 and effector cytokines such as IFN-γ and TNF. A recent study by Jayaraman and colleagues showed that TB-specific T cells coexpress PD1 and Tim3 and that the removal of Tim3 activity improves T cell function and M. tuberculosis control (89). However, in other studies, genetic deletion of the inhibitory receptor PD1 resulted in the pathological production of IFN-γ that resulted in exacerbated TB disease and early lethality (22, 91). Together, these studies suggest that modulating inhibitory receptors may have pleotropic effects on the host response that must be understood more fully.
To date, most studies examining T cell exhaustion have focused on the expression of the inhibitory molecules on the T cells themselves but not on the ligands present on the M. tuberculosis-infected APCs. It is possible that M. tuberculosis actively exploits the induction of these ligands during infection to promote T cell dysfunction and prevent effective clearance. This prediction is not without support, as other chronic bacterial infections are known to target these pathways. For example, during genital infections with Chlamydia trachomatis, pathogen-specific CD8+ T cells are incapable of contributing to the protective response. The inhibition of CD8+ T cell function was found to be a direct result of C. trachomatis actively inducing the PD1 ligand PDL1 on infected cells (92). Reversing this virulence tactic resulted in more effective bacterial control and T cell function. In addition, Salmonella enterica serovar Typhimurium and Helicobacter pylori also manipulate PD-L1 levels to suppress T cell responses and drive persistent infections (93, 94). In agreement with studies on other pathogens, M. tuberculosis-infected APCs express significantly more inhibitory ligands that may drive T cell dysfunction (95). However, it has yet to be examined whether M. tuberculosis is actively inducing these pathways or if PD-L1 induction is simply a by-product of persistent inflammation. Clearly, the role of T cell exhaustion is important in the balance between control and susceptibility to M. tuberculosis, yet much more study is needed to understand how inhibitory ligands are modulated during infection in the APC and which inhibitory molecules on T cells can be targeted without exacerbating disease.
NEW APPROACHES TO UNRAVEL M. TUBERCULOSIS-MEDIATED T CELL EVASION
As discussed above, M. tuberculosis evades immune clearance by carefully engaging the infected APCs to prevent elimination by T cells. Even though these immune evasion mechanisms are becoming clearer, several key questions remain. These include defining how distinct cell populations in the infected lung drive unique aspects of immune evasion during M. tuberculosis infection, identifying all M. tuberculosis genes that modulate immune detection and developing approaches that bypass M. tuberculosis virulence traits to more effectively activate protective T cell responses. In the second part of this review, we evaluate and discuss new tools and approaches that should be applied to address these outstanding questions in M. tuberculosis-mediated immune evasion.
Expanding ex vivo approaches to reflect cell diversity in the lung environment.
Within the infected lung, M. tuberculosis encounters a variety of cell types, including alveolar macrophages, interstitial macrophages, dendritic cells, and neutrophils (8). To understand how M. tuberculosis impedes T cell responses, it is essential to understand how each distinct host cell population interacts with M. tuberculosis and enables immune evasion. To date, however, most studies examining the mechanisms of M. tuberculosis pathogenesis employ ex vivo models that use primary bone marrow-derived macrophages (BMDMs), peritoneal macrophages, or immortalized cell lines such as RAW or Thp1 cells (7). While these approaches are certainly useful, recent studies suggest that an important aspect of M. tuberculosis disease is the progression of cellular interactions that occur in the complex lung environment (8–10). How M. tuberculosis infection of each cellular population plays into the overall immune evasion remains unknown. As described above, alveolar macrophages are the initial niche for M. tuberculosis following inhalation, and these cells are inherently different than BMDMs. Not only is the innate response distinct between alveolar macrophages and BMDMs, but elegant work by Russell and colleagues also showed that the metabolic differences between macrophage populations in the lungs drive disease progression (10). While interstitial macrophages use glycolysis for energy, alveolar macrophages rely on fatty acid oxidization, which results in a permissive nutrient pool for M. tuberculosis growth. It is likely that the permissive alveolar macrophage niche sequesters M. tuberculosis away from dendritic cells that are more capable of triggering the upregulation of key surface MHC and costimulatory molecules needed for rapid T cell activation (8). Yet, this has remained difficult to test due to a lack of ex vivo models that recapitulate the functions of alveolar macrophages. Effectively examining how M. tuberculosis manipulates distinct APC populations to evade T cell responses requires ex vivo systems that model distinct cell populations from the lung and/or methods to identify and isolate infected cells directly from the infected lung environment.
One reason for the reliance on BMDMs is the ease with which millions of cells can be isolated and manipulated without using large numbers of animals. In contrast, studying alveolar macrophages remains a challenge. These cells must be isolated directly from the bronchoalveolar lavage (BAL) fluid in the lungs, and from each mouse, only 105 cells can be isolated (96, 97). These low numbers and the time involved in isolation limit the scale and reproducibility of primary alveolar macrophage studies. Recently, Fejer et al. discovered an approach to differentiate fetal liver cells, the origin of alveolar macrophages, into alveolar macrophage-like cells called Max Planck Institute (MPI) cells that can be propagated ex vivo for over 100 generations (97). MPI cells have similar morphology and express similar surface markers to alveolar macrophages, differentiating them from bone marrow-derived macrophages. The innate response in these cells was remarkably similar to alveolar macrophages isolated from BAL fluid. Following lipopolysaccharide (LPS) treatment or treatment with heat-killed M. tuberculosis, MPI cells showed a muted IL-10 response and a strong activation of IL-1a that mirrored that of alveolar macrophages but not BMDMs. In a more recent study, Woo et al. showed that MPI cells can phagocytose live M. tuberculosis cells, support M. tuberculosis replication, and activate innate responses, including cytokine production, autophagy, and lipid accumulation (98). Together, these studies suggest that MPI cells may be a useful model for the alveolar macrophages that play a critical role in M. tuberculosis pathogenesis and immune evasion. In the future, it will be important to understand how M. tuberculosis can target MPI cells and alveolar macrophages to alter their antigen presentation and costimulatory capacity during infection.
It is also important to consider how infected cells directly from the lung environment are manipulated by M. tuberculosis. Examining distinct cell types in vivo requires tools that allow the identification and isolation of all M. tuberculosis-infected cell populations. One successful approach is leveraging bacterial reporters that allow the sensitive detection of M. tuberculosis-infected cells while preserving virulence. These reporters range from standard fluorescent strains expressing bright and stable fluorescent proteins to metabolic reporters that discriminate between live and dead bacteria or indicate the activation of particular M. tuberculosis transcriptional responses (8–10, 99). By coupling these sensitive reporters to cell-sorting or whole-tissue-imaging approaches, it is now possible to directly examine how distinct cellular populations are manipulated by M. tuberculosis infection and how this impacts protective T cell responses.
Identifying new mechanisms used by M. tuberculosis to manipulate T cell responses.
M. tuberculosis contains several genes that drive immune evasion mechanisms during infection. However, no global approaches have been employed to systematically define the repertoire of M. tuberculosis immune evasion genes. With proteomic and global genetic approaches now readily available, the time is ripe for those in this field to investigate how individual or groups of M. tuberculosis genes contribute to the evasion of T cell responses.
Global genetic approaches in M. tuberculosis were developed over 15 years ago with the seminal papers using transposon hybridization (TRASH) (100). TRASH uses a pooled library of transposon mutants made with the Mariner transposon. These global transposon experiments determine how the relative representation of each mutant changes between two distinct conditions. Recent advances in deep-sequencing technology (now termed transposon sequencing [Tn-seq]) have accelerated the speed and sensitivity of Tn-seq experiments, yet few studies have focused on immune-related phenotypes (101). Most published studies use M. tuberculosis genetic screens to focus on the pathogen itself by characterizing phenotypes such as physiology, growth, and survival (102–104). However, these bacterial genetic approaches have the power to disentangle exactly how M. tuberculosis inhibits APCs from effectively communicating with T cells. For example, while Hip1 is known to inhibit CD40 induction in APCs, it is likely that other M. tuberculosis genes also contribute to CD40 inhibition (17). By infecting an APC population with a transposon mutant library and isolating host cells that can or cannot induce robust CD40 expression, it is now possible to identify M. tuberculosis mutants that directly impact this important immune cell function. The applications of this approach regarding our understanding of M. tuberculosis factors that drive persistence are endless.
Of course, transposon approaches have their limitations. By definition, transposon libraries cannot assess the role of essential genes, reducing the percentage of the genome probed by a significant margin (102). To address this, recent developments in protein depletion and CRISPR interference (CRISPRi) approaches now allow the investigation into these essential genes (105–107). The recent tour de force study that created a library of barcoded hypomorphic alleles for essential genes throughout the M. tuberculosis genome will be an outstanding tool for immunologists to use in the future to probe even deeper into the M. tuberculosis genome to understand the evasion of the adaptive immune response (107). Additionally, robust yeast two-hybrid screens have proven effective at uncovering unexpected interactions between M. tuberculosis proteins and host networks that influence the ability of APCs to signal to T cells. The interaction between EsxH and the ESCRT system described above was originally found in a yeast two-hybrid screen and suggests that this approach is useful to delineate specific interactions between M. tuberculosis and the host (34). The recent creation of an M. tuberculosis-host protein-protein interaction network to identify M. tuberculosis immune evasion and suppression strategies, such as the LpqN-CBL interaction, is a promising step forward in characterizing the M. tuberculosis-host interface (108).
Altogether, the genetic toolbox to understand how M. tuberculosis directly contributes to immune evasion is now well developed and will inform our understanding of how to overcome M. tuberculosis virulence in the future.
Employing host genetic approaches to bypass M. tuberculosis-mediated immune evasion.
In addition to understanding how M. tuberculosis drives immune evasion, it is important to clarify how host pathways that are targeted by M. tuberculosis become ineffective and devise strategies to overcome M. tuberculosis-mediated evasion. This requires the capability to modify host cells and define what host pathways are necessary and sufficient for immune evasion during M. tuberculosis infection. Much like bacterial genetics, global approaches to modify the host or integrate diversity into host models have exploded over the last decade (109). The application of CRISPR-Cas9 technologies to modulate gene expression in host cells is an exciting future area of research for the field of immune evasion. These approaches can be categorized into loss of function or gain of function. Loss-of-function CRISPR systems generally rely on catalytically active Cas9, or other editing enzymes, that result in double-strand breaks when targeted to the genome by a sequence-specific single guide RNA (sgRNA) (110, 111). By pooling thousands of sgRNAs, knockout libraries targeting all coding genes can be assembled and probed for immunological phenotypes during M. tuberculosis infection that are easily deconvoluted through deep sequencing using similar approaches to Tn-seq (112). The loss-of-function CRISPR approach will be essential to pin down the underlying host pathways that are targeted in APCs by M. tuberculosis to drive distinct immune evasion mechanisms observed during infection. Since many of the changes induced by M. tuberculosis alter the surface expression of MHC molecules and costimulatory molecules, it is straightforward to isolate distinct populations with differential expression of each immune molecule on the cell surface. By comparing the distribution of sgRNAs in a genome-wide library between wild-type M. tuberculosis and an M. tuberculosis deletion or overexpression mutant in immune evasion genes such as EsxH, the host immune pathways that result in reduced MHC-II function can be directly identified.
The loss-of-function CRISPR approaches will be key to understanding how M. tuberculosis evasion mechanisms target the APC to prevent effective T cell detection. However, they are less likely to identify host mechanisms that overcome M. tuberculosis-mediated inhibition and result in improved T cell detection. In contrast, gain-of-function genetic approaches that induce the expression of target host genes have the potential to identify critical host pathways that can overcome M. tuberculosis evasion tactics. Gain-of-function approaches using CRISPR allow the targeted induction of host genes driven by a catalytically dead Cas9 linked to transcriptional activators (113). Two such systems, the synergistic activator mediator (SAM) and the Calabrese system, robustly induce genes in an sgRNA-dependent manner independently of their normal expression patterns (110, 113). The ability to fine-tune gene expression broadly at a genome-wide level opens many experimental opportunities with regard to M. tuberculosis-mediated immune inhibition. As discussed above, CD8+ T cells are unable to detect M. tuberculosis-infected macrophages, resulting in their inability to carry out their effector functions effectively. By infecting a genome-wide gain-of-function library of APCs with M. tuberculosis, it might be possible to identify host pathways that allow the effective detection of M. tuberculosis by CD8+ T cells. These pathways, if effectively targeted, would be capable of overcoming the immune evasion tactics employed by M. tuberculosis and could then be tested for improved disease control in vivo. We strongly believe gain-of-function genetic approaches will be key assets to identify new host targets that are capable of overcoming M. tuberculosis virulence.
Beyond experimental genetic variation induced with CRISPR, another compelling method to investigate M. tuberculosis immune evasion is leveraging the inherent diversity within the mouse population. Resources such as the Collaborative Cross (CC) and Diversity Outbred (DO) collections introduce genetic variability into the mouse populations that can then be examined directly during M. tuberculosis infection (reviewed in reference 114). By modeling the genetic diversity seen in a human population, insights into how M. tuberculosis evades immune detection and clearance in distinct environments can now be understood. These mouse resources are the product of eight founder parental strains that were intercrossed sequentially to introduce genomic loci from all eight founders in each derived mouse strain (115). A subset of these diverse strains was then interbred to homozygosity resulting in the CC panel of recombinant inbred mice. The CC panel models more diversity that is reflective of the human population with the advantage of recombinant lines that can be tested repeatedly and examined for the effectiveness of interventions. The CC panel has already been used to model the variability of BCG vaccination responses in mice, suggesting that the host genetic background plays a significant role in the efficacy of vaccine responses (116). It is not difficult to imagine that a subset of immune evasion strategies employed by M. tuberculosis may only be observable in hosts with a particular genetic background. This would be critical to understand if we are to develop immunizations that are broadly protective in a range of human hosts. The DO collection is similar to the CC panel, except each mouse is an outbred offspring of the intercrossed parental strains (117). While this allows modeling of more genetic variation, it comes with the caveat that each mouse is genetically unique, making follow-up studies challenging. Even so, the DO collection is already being used effectively to understand traits that drive M. tuberculosis disease progression in the mouse model (118, 119). We envision these resources as being central to fully delineating how M. tuberculosis can evade distinct aspects of the host response and in testing new approaches to overcome M. tuberculosis virulence in a diverse host population.
OUTLOOK
Our knowledge of M. tuberculosis-driven immune evasion has expanded rapidly over the last decade. Distinct mechanisms that M. tuberculosis employs to avoid robust activation and detection by the adaptive immune response are now being explored in great detail. These studies have uncovered a range of important immune pathways that are manipulated by M. tuberculosis to avoid robust T cell responses by impeding their activation and effector function. Even with this progress, there are still many unknowns of how M. tuberculosis evades protective host responses. In the next decade, the challenge will be to use these findings to devise immunization approaches that activate protective responses even in the face of M. tuberculosis evasion tactics. There are already promising new vaccine candidates in the pipeline that show encouraging results (3). However, preliminary evidence suggests that taking M. tuberculosis virulence traits into account may improve disease outcomes and should be considered as new vaccine formulations move through clinical trials. Some approaches described here may be useful to develop strategies that bypass immune evasion. Overall, it will be critical to ensure that new vaccines broadly protect the population while overcoming M. tuberculosis immune evasion to result in the most effective protective response.
ACKNOWLEDGMENTS
We thank the Olive lab for helpful discussions. We apologize to all the investigators we were unable to cite and discuss due to space limitations.
This work was supported by the National Institutes of Health (NIH grants AI146504 and AI148961 to A.J.O.).
Biographies

Laurisa Ankley is a Ph.D. student at Michigan State University studying the regulation of MHC-II. In connection with this research, she is particularly interested in discovering mechanisms to increase host-mediated clearance of chronic pathogenic infections. She graduated with a B.S. in biology from Eastern Washington University, where she worked on defining iron regulation mechanisms of manuka honey, a broad-spectrum antimicrobial.

Sean Thomas is a graduate research assistant at Michigan State University. He graduated from Southeast Missouri State University with a B.S. in biochemistry and molecular biology, where he explored the photomagnetic properties of nanoparticles as a treatment for bloodborne illnesses. He currently is working on elucidating the interactions between NADPH phagocyte oxidase and caspase 1 that, when lost, result in loss of tolerance and resistance to infection from Mycobacterium tuberculosis.

Andrew J. Olive is an Assistant Professor at Michigan State University. He graduated with a B.A. from the University of Kansas where he studied the pathogenesis of Shigella flexneri. He continued his studies in graduate school at Harvard Medical in the lab of Michael Starnbach, where he investigated host-pathogen interactions during infections with Chlamydia trachomatis. His interest in the host response motivated him to undertake postdoctoral work with Christopher Sassetti, where he investigated the balance between resistance and tolerance during Mycobacterium tuberculosis infection. He began his own lab at Michigan State University in 2018, where he continues to study host-pathogen interactions during persistent infections, including devising new strategies to overcome pathogen virulence and activate protective immune responses.
REFERENCES
- 1.Olive AJ, Sassetti CM. 2018. Tolerating the unwelcome guest; how the host withstands persistent Mycobacterium tuberculosis. Front Immunol 9:2094. doi: 10.3389/fimmu.2018.02094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Zuniga EI, Macal M, Lewis GM, Harker JA. 2015. Innate and adaptive immune regulation during chronic viral infections. Annu Rev Virol 2:573–597. doi: 10.1146/annurev-virology-100114-055226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ernst JD. 2018. Mechanisms of M. tuberculosis immune evasion as challenges to TB vaccine design. Cell Host Microbe 24:34–42. doi: 10.1016/j.chom.2018.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Nunes-Alves C, Booty MG, Carpenter SM, Jayaraman P, Rothchild AC, Behar SM. 2014. In search of a new paradigm for protective immunity to TB. Nat Rev Microbiol 12:289–299. doi: 10.1038/nrmicro3230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zumla A, Baroness Masham of Ilton, George A, Sharma V, Herbert RH, Baroness Masham Of I, Oxley A, Oliver M. 2015. The WHO 2014 global tuberculosis report–further to go. Lancet Glob Health 3:e10–e12. doi: 10.1016/S2214-109X(14)70361-4. [DOI] [PubMed] [Google Scholar]
- 6.Sia JK, Georgieva M, Rengarajan J. 2015. Innate immune defenses in human tuberculosis: an overview of the interactions between Mycobacterium tuberculosis and innate immune cells. J Immunol Res 2015:747543. doi: 10.1155/2015/747543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Philips JA, Ernst JD. 2012. Tuberculosis pathogenesis and immunity. Annu Rev Pathol 7:353–384. doi: 10.1146/annurev-pathol-011811-132458. [DOI] [PubMed] [Google Scholar]
- 8.Cohen SB, Gern BH, Delahaye JL, Adams KN, Plumlee CR, Winkler JK, Sherman DR, Gerner MY, Urdahl KB. 2018. Alveolar macrophages provide an early Mycobacterium tuberculosis niche and initiate dissemination. Cell Host Microbe 24:439–446.e4. doi: 10.1016/j.chom.2018.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Rothchild AC, Olson GS, Nemeth J, Amon LM, Mai D, Gold ES, Diercks AH, Aderem A. 2019. Alveolar macrophages generate a noncanonical NRF2-driven transcriptional response to Mycobacterium tuberculosis in vivo. Sci Immunol 4:eaaw6693. doi: 10.1126/sciimmunol.aaw6693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Huang L, Nazarova EV, Tan S, Liu Y, Russell DG. 2018. Growth of Mycobacterium tuberculosis in vivo segregates with host macrophage metabolism and ontogeny. J Exp Med 215:1135–1152. doi: 10.1084/jem.20172020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Knudson CJ, Weiss KA, Hartwig SM, Varga SM. 2014. The pulmonary localization of virus-specific T lymphocytes is governed by the tissue tropism of infection. J Virol 88:9010–9016. doi: 10.1128/JVI.00329-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wolf AJ, Linas B, Trevejo-Nunez GJ, Kincaid E, Tamura T, Takatsu K, Ernst JD. 2007. Mycobacterium tuberculosis infects dendritic cells with high frequency and impairs their function in vivo. J Immunol 179:2509–2519. doi: 10.4049/jimmunol.179.4.2509. [DOI] [PubMed] [Google Scholar]
- 13.Wolf AJ, Desvignes L, Linas B, Banaiee N, Tamura T, Takatsu K, Ernst JD. 2008. Initiation of the adaptive immune response to Mycobacterium tuberculosis depends on antigen production in the local lymph node, not the lungs. J Exp Med 205:105–115. doi: 10.1084/jem.20071367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Smith-Garvin JE, Koretzky GA, Jordan MS. 2009. T cell activation. Annu Rev Immunol 27:591–619. doi: 10.1146/annurev.immunol.021908.132706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chen L, Flies DB. 2013. Molecular mechanisms of T cell co-stimulation and co-inhibition. Nat Rev Immunol 13:227–242. doi: 10.1038/nri3405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Attanasio J, Wherry EJ. 2016. Costimulatory and coinhibitory receptor pathways in infectious disease. Immunity 44:1052–1068. doi: 10.1016/j.immuni.2016.04.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sia JK, Bizzell E, Madan-Lala R, Rengarajan J. 2017. Engaging the CD40-CD40L pathway augments T-helper cell responses and improves control of Mycobacterium tuberculosis infection. PLoS Pathog 13:e1006530. doi: 10.1371/journal.ppat.1006530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Srivastava S, Ernst JD. 2013. Cutting edge: direct recognition of infected cells by CD4 T cells is required for control of intracellular Mycobacterium tuberculosis in vivo. J Immunol 191:1016–1020. doi: 10.4049/jimmunol.1301236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ford CB, Shah RR, Maeda MK, Gagneux S, Murray MB, Cohen T, Johnston JC, Gardy J, Lipsitch M, Fortune SM. 2013. Mycobacterium tuberculosis mutation rate estimates from different lineages predict substantial differences in the emergence of drug-resistant tuberculosis. Nat Genet 45:784–790. doi: 10.1038/ng.2656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Comas I, Chakravartti J, Small PM, Galagan J, Niemann S, Kremer K, Ernst JD, Gagneux S. 2010. Human T cell epitopes of Mycobacterium tuberculosis are evolutionarily hyperconserved. Nat Genet 42:498–503. doi: 10.1038/ng.590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Saelens JW, Viswanathan G, Tobin DM. 2019. Mycobacterial evolution intersects with host tolerance. Front Immunol 10:528. doi: 10.3389/fimmu.2019.00528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sakai S, Kauffman KD, Sallin MA, Sharpe AH, Young HA, Ganusov VV, Barber DL. 2016. CD4 T cell-derived IFN-gamma plays a minimal role in control of pulmonary Mycobacterium tuberculosis infection and must be actively repressed by PD-1 to prevent lethal disease. PLoS Pathog 12:e1005667. doi: 10.1371/journal.ppat.1005667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tzelepis F, Blagih J, Khan N, Gillard J, Mendonca L, Roy DG, Ma EH, Joubert P, Jones RG, Divangahi M. 2018. Mitochondrial cyclophilin D regulates T cell metabolic responses and disease tolerance to tuberculosis. Sci Immunol 3:eaar4135. doi: 10.1126/sciimmunol.aar4135. [DOI] [PubMed] [Google Scholar]
- 24.Cadena AM, Hopkins FF, Maiello P, Carey AF, Wong EA, Martin CJ, Gideon HP, DiFazio RM, Andersen P, Lin PL, Fortune SM, Flynn JL. 2018. Concurrent infection with Mycobacterium tuberculosis confers robust protection against secondary infection in macaques. PLoS Pathog 14:e1007305. doi: 10.1371/journal.ppat.1007305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Darrah PA, Zeppa JJ, Maiello P, Hackney JA, Wadsworth MH Jr, Hughes TK, Pokkali S, Swanson PA Jr, Grant NL, Rodgers MA, Kamath M, Causgrove CM, Laddy DJ, Bonavia A, Casimiro D, Lin PL, Klein E, White AG, Scanga CA, Shalek AK, Roederer M, Flynn JL, Seder RA. 2020. Prevention of tuberculosis in macaques after intravenous BCG immunization. Nature 577:95–102. doi: 10.1038/s41586-019-1817-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ernst JD. 2012. The immunological life cycle of tuberculosis. Nat Rev Immunol 12:581–591. doi: 10.1038/nri3259. [DOI] [PubMed] [Google Scholar]
- 27.Patankar YR, Sutiwisesak R, Boyce S, Lai R, Lindestam Arlehamn CS, Sette A, Behar SM. 2019. Limited recognition of Mycobacterium tuberculosis-infected macrophages by polyclonal CD4 and CD8 T cells from the lungs of infected mice. Mucosal Immunol 13:140–148. doi: 10.1038/s41385-019-0217-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yang JD, Mott D, Sutiwisesak R, Lu YJ, Raso F, Stowell B, Babunovic GH, Lee J, Carpenter SM, Way SS, Fortune SM, Behar SM. 2018. Mycobacterium tuberculosis-specific CD4+ and CD8+ T cells differ in their capacity to recognize infected macrophages. PLoS Pathog 14:e1007060. doi: 10.1371/journal.ppat.1007060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Grace PS, Ernst JD. 2016. Suboptimal antigen presentation contributes to virulence of Mycobacterium tuberculosis in vivo. J Immunol 196:357–364. doi: 10.4049/jimmunol.1501494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhang YJ, Reddy MC, Ioerger TR, Rothchild AC, Dartois V, Schuster BM, Trauner A, Wallis D, Galaviz S, Huttenhower C, Sacchettini JC, Behar SM, Rubin EJ. 2013. Tryptophan biosynthesis protects mycobacteria from CD4 T-cell-mediated killing. Cell 155:1296–1308. doi: 10.1016/j.cell.2013.10.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lawrence RE, Zoncu R. 2019. The lysosome as a cellular centre for signalling, metabolism and quality control. Nat Cell Biol 21:133–142. doi: 10.1038/s41556-018-0244-7. [DOI] [PubMed] [Google Scholar]
- 32.Luzio JP, Pryor PR, Bright NA. 2007. Lysosomes: fusion and function. Nat Rev Mol Cell Biol 8:622–632. doi: 10.1038/nrm2217. [DOI] [PubMed] [Google Scholar]
- 33.Armstrong JA, Hart PD. 1975. Phagosome-lysosome interactions in cultured macrophages infected with virulent tubercle bacilli. Reversal of the usual nonfusion pattern and observations on bacterial survival. J Exp Med 142:1–16. doi: 10.1084/jem.142.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mehra A, Zahra A, Thompson V, Sirisaengtaksin N, Wells A, Porto M, Koster S, Penberthy K, Kubota Y, Dricot A, Rogan D, Vidal M, Hill DE, Bean AJ, Philips JA. 2013. Mycobacterium tuberculosis type VII secreted effector EsxH targets host ESCRT to impair trafficking. PLoS Pathog 9:e1003734. doi: 10.1371/journal.ppat.1003734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Portal-Celhay C, Tufariello JM, Srivastava S, Zahra A, Klevorn T, Grace PS, Mehra A, Park HS, Ernst JD, Jacobs WR Jr, Philips JA. 2016. Mycobacterium tuberculosis EsxH inhibits ESCRT-dependent CD4+ T-cell activation. Nat Microbiol 2:16232. doi: 10.1038/nmicrobiol.2016.232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Raiborg C, Stenmark H. 2009. The ESCRT machinery in endosomal sorting of ubiquitylated membrane proteins. Nature 458:445–452. doi: 10.1038/nature07961. [DOI] [PubMed] [Google Scholar]
- 37.Sturgill-Koszycki S, Schlesinger PH, Chakraborty P, Haddix PL, Collins HL, Fok AK, Allen RD, Gluck SL, Heuser J, Russell DG. 1994. Lack of acidification in Mycobacterium phagosomes produced by exclusion of the vesicular proton-ATPase. Science 263:678–681. doi: 10.1126/science.8303277. [DOI] [PubMed] [Google Scholar]
- 38.Clemens DL, Lee BY, Horwitz MA. 2000. Deviant expression of Rab5 on phagosomes containing the intracellular pathogens Mycobacterium tuberculosis and Legionella pneumophila is associated with altered phagosomal fate. Infect Immun 68:2671–2684. doi: 10.1128/iai.68.5.2671-2684.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Vergne I, Chua J, Deretic V. 2003. Tuberculosis toxin blocking phagosome maturation inhibits a novel Ca2+/calmodulin-PI3K hVPS34 cascade. J Exp Med 198:653–659. doi: 10.1084/jem.20030527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zulauf KE, Sullivan JT, Braunstein M. 2018. The SecA2 pathway of Mycobacterium tuberculosis exports effectors that work in concert to arrest phagosome and autophagosome maturation. PLoS Pathog 14:e1007011. doi: 10.1371/journal.ppat.1007011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Chua J, Deretic V. 2004. Mycobacterium tuberculosis reprograms waves of phosphatidylinositol 3-phosphate on phagosomal organelles. J Biol Chem 279:36982–36992. doi: 10.1074/jbc.M405082200. [DOI] [PubMed] [Google Scholar]
- 42.Fratti RA, Chua J, Vergne I, Deretic V. 2003. Mycobacterium tuberculosis glycosylated phosphatidylinositol causes phagosome maturation arrest. Proc Natl Acad Sci U S A 100:5437–5442. doi: 10.1073/pnas.0737613100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Vergne I, Chua J, Lee HH, Lucas M, Belisle J, Deretic V. 2005. Mechanism of phagolysosome biogenesis block by viable Mycobacterium tuberculosis. Proc Natl Acad Sci U S A 102:4033–4038. doi: 10.1073/pnas.0409716102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wong D, Bach H, Sun J, Hmama Z, Av-Gay Y. 2011. Mycobacterium tuberculosis protein tyrosine phosphatase (PtpA) excludes host vacuolar-H+-ATPase to inhibit phagosome acidification. Proc Natl Acad Sci U S A 108:19371–19376. doi: 10.1073/pnas.1109201108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Saini NK, Baena A, Ng TW, Venkataswamy MM, Kennedy SC, Kunnath-Velayudhan S, Carreno LJ, Xu J, Chan J, Larsen MH, Jacobs WR Jr, Porcelli SA. 2016. Suppression of autophagy and antigen presentation by Mycobacterium tuberculosis PE_PGRS47. Nat Microbiol 1:16133. doi: 10.1038/nmicrobiol.2016.133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Deretic V, Levine B. 2018. Autophagy balances inflammation in innate immunity. Autophagy 14:243–251. doi: 10.1080/15548627.2017.1402992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Crotzer VL, Blum JS. 2009. Autophagy and its role in MHC-mediated antigen presentation. J Immunol 182:3335–3341. doi: 10.4049/jimmunol.0803458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Layre E, Lee HJ, Young DC, Martinot AJ, Buter J, Minnaard AJ, Annand JW, Fortune SM, Snider BB, Matsunaga I, Rubin EJ, Alber T, Moody DB. 2014. Molecular profiling of Mycobacterium tuberculosis identifies tuberculosinyl nucleoside products of the virulence-associated enzyme Rv3378c. Proc Natl Acad Sci U S A 111:2978–2983. doi: 10.1073/pnas.1315883111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Buter J, Cheng TY, Ghanem M, Grootemaat AE, Raman S, Feng X, Plantijn AR, Ennis T, Wang J, Cotton RN, Layre E, Ramnarine AK, Mayfield JA, Young DC, Jezek Martinot A, Siddiqi N, Wakabayashi S, Botella H, Calderon R, Murray M, Ehrt S, Snider BB, Reed MB, Oldfield E, Tan S, Rubin EJ, Behr MA, van der Wel NN, Minnaard AJ, Moody DB. 2019. Mycobacterium tuberculosis releases an antacid that remodels phagosomes. Nat Chem Biol 15:889–899. doi: 10.1038/s41589-019-0336-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Celli J, de Chastellier C, Franchini DM, Pizarro-Cerda J, Moreno E, Gorvel JP. 2003. Brucella evades macrophage killing via VirB-dependent sustained interactions with the endoplasmic reticulum. J Exp Med 198:545–556. doi: 10.1084/jem.20030088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Bastidas RJ, Elwell CA, Engel JN, Valdivia RH. 2013. Chlamydial intracellular survival strategies. Cold Spring Harb Perspect Med 3:a010256. doi: 10.1101/cshperspect.a010256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Raines SA, Hodgkinson MR, Dowle AA, Pryor PR. 2017. The Salmonella effector SseJ disrupts microtubule dynamics when ectopically expressed in normal rat kidney cells. PLoS One 12:e0172588. doi: 10.1371/journal.pone.0172588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Reith W, LeibundGut-Landmann S, Waldburger JM. 2005. Regulation of MHC class II gene expression by the class II transactivator. Nat Rev Immunol 5:793–806. doi: 10.1038/nri1708. [DOI] [PubMed] [Google Scholar]
- 54.Kern I, Steimle V, Siegrist CA, Mach B. 1995. The two novel MHC class II transactivators RFX5 and CIITA both control expression of HLA-DM genes. Int Immunol 7:1295–1299. doi: 10.1093/intimm/7.8.1295. [DOI] [PubMed] [Google Scholar]
- 55.Zhong G, Liu L, Fan T, Fan P, Ji H. 2000. Degradation of transcription factor RFX5 during the inhibition of both constitutive and interferon gamma-inducible major histocompatibility complex class I expression in chlamydia-infected cells. J Exp Med 191:1525–1534. doi: 10.1084/jem.191.9.1525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Zhong G, Fan T, Liu L. 1999. Chlamydia inhibits interferon gamma-inducible major histocompatibility complex class II expression by degradation of upstream stimulatory factor 1. J Exp Med 189:1931–1938. doi: 10.1084/jem.189.12.1931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Fortune SM, Solache A, Jaeger A, Hill PJ, Belisle JT, Bloom BR, Rubin EJ, Ernst JD. 2004. Mycobacterium tuberculosis inhibits macrophage responses to IFN-gamma through myeloid differentiation factor 88-dependent and -independent mechanisms. J Immunol 172:6272–6280. doi: 10.4049/jimmunol.172.10.6272. [DOI] [PubMed] [Google Scholar]
- 58.Gehring AJ, Rojas RE, Canaday DH, Lakey DL, Harding CV, Boom WH. 2003. The Mycobacterium tuberculosis 19-kilodalton lipoprotein inhibits gamma interferon-regulated HLA-DR and Fc gamma R1 on human macrophages through Toll-like receptor 2. Infect Immun 71:4487–4497. doi: 10.1128/iai.71.8.4487-4497.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Pai RK, Convery M, Hamilton TA, Boom WH, Harding CV. 2003. Inhibition of IFN-gamma-induced class II transactivator expression by a 19-kDa lipoprotein from Mycobacterium tuberculosis: a potential mechanism for immune evasion. J Immunol 171:175–184. doi: 10.4049/jimmunol.171.1.175. [DOI] [PubMed] [Google Scholar]
- 60.Tobian AA, Potter NS, Ramachandra L, Pai RK, Convery M, Boom WH, Harding CV. 2003. Alternate class I MHC antigen processing is inhibited by Toll-like receptor signaling pathogen-associated molecular patterns: Mycobacterium tuberculosis 19-kDa lipoprotein, CpG DNA, and lipopolysaccharide. J Immunol 171:1413–1422. doi: 10.4049/jimmunol.171.3.1413. [DOI] [PubMed] [Google Scholar]
- 61.Diehl S, Anguita J, Hoffmeyer A, Zapton T, Ihle JN, Fikrig E, Rincon M. 2000. Inhibition of Th1 differentiation by IL-6 is mediated by SOCS1. Immunity 13:805–815. doi: 10.1016/s1074-7613(00)00078-9. [DOI] [PubMed] [Google Scholar]
- 62.Nagabhushanam V, Solache A, Ting LM, Escaron CJ, Zhang JY, Ernst JD. 2003. Innate inhibition of adaptive immunity: Mycobacterium tuberculosis-induced IL-6 inhibits macrophage responses to IFN-gamma. J Immunol 171:4750–4757. doi: 10.4049/jimmunol.171.9.4750. [DOI] [PubMed] [Google Scholar]
- 63.Velásquez LN, Milillo MA, Delpino MV, Trotta A, Fernandez P, Pozner RG, Lang R, Balboa L, Giambartolomei GH, Barrionuevo P. 2017. Brucella abortus down-regulates MHC class II by the IL-6-dependent inhibition of CIITA through the downmodulation of IFN regulatory factor-1 (IRF-1). J Leukoc Biol 101:759–773. doi: 10.1189/jlb.4A0416-196R. [DOI] [PubMed] [Google Scholar]
- 64.Biyikli OO, Baysak A, Ece G, Oz AT, Ozhan MH, Berdeli A. 2016. Role of Toll-like receptors in tuberculosis infection. Jundishapur J Microbiol 9:e20224. doi: 10.5812/jjm.20224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Srivastava S, Ernst JD. 2014. Cell-to-cell transfer of M. tuberculosis antigens optimizes CD4 T cell priming. Cell Host Microbe 15:741–752. doi: 10.1016/j.chom.2014.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Srivastava S, Grace PS, Ernst JD. 2016. Antigen export reduces antigen presentation and limits T cell control of M. tuberculosis. Cell Host Microbe 19:44–54. doi: 10.1016/j.chom.2015.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Stary G, Olive A, Radovic-Moreno AF, Gondek D, Alvarez D, Basto PA, Perro M, Vrbanac VD, Tager AM, Shi J, Yethon JA, Farokhzad OC, Langer R, Starnbach MN, von Andrian UH. 2015. A mucosal vaccine against Chlamydia trachomatis generates two waves of protective memory T cells. Science 348:aaa8205. doi: 10.1126/science.aaa8205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Vlcek KR, Li W, Manam S, Zanotti B, Nicholson BJ, Ramsey KH, Murthy AK. 2016. The contribution of Chlamydia-specific CD8+ T cells to upper genital tract pathology. Immunol Cell Biol 94:208–212. doi: 10.1038/icb.2015.74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Jenkins MK, Schwartz RH. 1987. Antigen presentation by chemically modified splenocytes induces antigen-specific T cell unresponsiveness in vitro and in vivo. J Exp Med 165:302–319. doi: 10.1084/jem.165.2.302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Marckmann S, Wiesemann E, Hilse R, Trebst C, Stangel M, Windhagen A. 2004. Interferon-β up-regulates the expression of co-stimulatory molecules CD80, CD86 and CD40 on monocytes: significance for treatment of multiple sclerosis. Clin Exp Immunol 138:499–506. doi: 10.1111/j.1365-2249.2004.02624.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Vinay DS, Kwon BS. 2009. TNF superfamily: costimulation and clinical applications. Cell Biol Int 33:453–465. doi: 10.1016/j.cellbi.2009.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Bhatt K, Uzelac A, Mathur S, McBride A, Potian J, Salgame P. 2009. B7 costimulation is critical for host control of chronic Mycobacterium tuberculosis infection. J Immunol 182:3793–3800. doi: 10.4049/jimmunol.0802996. [DOI] [PubMed] [Google Scholar]
- 73.Lazarevic V, Myers AJ, Scanga CA, Flynn JL. 2003. CD40, but not CD40L, is required for the optimal priming of T cells and control of aerosol M. tuberculosis infection. Immunity 19:823–835. doi: 10.1016/s1074-7613(03)00324-8. [DOI] [PubMed] [Google Scholar]
- 74.Snelgrove RJ, Cornere MM, Edwards L, Dagg B, Keeble J, Rodgers A, Lyonga DE, Stewart GR, Young DB, Walker B, Hussell T. 2012. OX40 ligand fusion protein delivered simultaneously with the BCG vaccine provides superior protection against murine Mycobacterium tuberculosis infection. J Infect Dis 205:975–983. doi: 10.1093/infdis/jir868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Chen L, Cheng W, Shivshankar P, Lei L, Zhang X, Wu Y, Yeh IT, Zhong G. 2009. Distinct roles of CD28- and CD40 ligand-mediated costimulation in the development of protective immunity and pathology during Chlamydia muridarum urogenital infection in mice. Infect Immun 77:3080–3089. doi: 10.1128/IAI.00611-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Marks E, Verolin M, Stensson A, Lycke N. 2007. Differential CD28 and inducible costimulatory molecule signaling requirements for protective CD4+ T-cell-mediated immunity against genital tract Chlamydia trachomatis infection. Infect Immun 75:4638–4647. doi: 10.1128/IAI.00465-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Schreiber HA, Hulseberg PD, Lee J, Prechl J, Barta P, Szlavik N, Harding JS, Fabry Z, Sandor M. 2010. Dendritic cells in chronic mycobacterial granulomas restrict local anti-bacterial T cell response in a murine model. PLoS One 5:e11453. doi: 10.1371/journal.pone.0011453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Kan-Sutton C, Jagannath C, Hunter RL Jr. 2009. Trehalose 6,6′-dimycolate on the surface of Mycobacterium tuberculosis modulates surface marker expression for antigen presentation and costimulation in murine macrophages. Microbes Infect 11:40–48. doi: 10.1016/j.micinf.2008.10.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Kim JM, Kim JS, Yoo DY, Ko SH, Kim N, Kim H, Kim YJ. 2011. Stimulation of dendritic cells with Helicobacter pylori vacuolating cytotoxin negatively regulates their maturation via the restoration of E2F1. Clin Exp Immunol 166:34–45. doi: 10.1111/j.1365-2249.2011.04447.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Fang F, Wang Y, Li R, Zhao Y, Guo Y, Jiang M, Sun J, Ma Y, Ren Z, Tian Z, Wei F, Yang D, Xiao W. 2010. Transcription factor E2F1 suppresses dendritic cell maturation. J Immunol 184:6084–6091. doi: 10.4049/jimmunol.0902561. [DOI] [PubMed] [Google Scholar]
- 81.Yuan C, Qu ZL, Tang XL, Liu Q, Luo W, Huang C, Pan Q, Zhang XL. 2019. Mycobacterium tuberculosis mannose-capped lipoarabinomannan induces IL-10-producing B cells and hinders CD4+ Th1 immunity. iScience 11:13–30. doi: 10.1016/j.isci.2018.11.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Rengarajan J, Bloom BR, Rubin EJ. 2005. Genome-wide requirements for Mycobacterium tuberculosis adaptation and survival in macrophages. Proc Natl Acad Sci U S A 102:8327–8332. doi: 10.1073/pnas.0503272102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Rengarajan J, Murphy E, Park A, Krone CL, Hett EC, Bloom BR, Glimcher LH, Rubin EJ. 2008. Mycobacterium tuberculosis Rv2224c modulates innate immune responses. Proc Natl Acad Sci U S A 105:264–269. doi: 10.1073/pnas.0710601105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Georgieva M, Sia JK, Bizzell E, Madan-Lala R, Rengarajan J. 2018. Mycobacterium tuberculosis GroEL2 modulates dendritic cell responses. Infect Immun 86:e00387-17. doi: 10.1128/IAI.00387-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Madan-Lala R, Sia JK, King R, Adekambi T, Monin L, Khader SA, Pulendran B, Rengarajan J. 2014. Mycobacterium tuberculosis impairs dendritic cell functions through the serine hydrolase Hip1. J Immunol 192:4263–4272. doi: 10.4049/jimmunol.1303185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Naffin-Olivos JL, Georgieva M, Goldfarb N, Madan-Lala R, Dong L, Bizzell E, Valinetz E, Brandt GS, Yu S, Shabashvili DE, Ringe D, Dunn BM, Petsko GA, Rengarajan J. 2014. Mycobacterium tuberculosis Hip1 modulates macrophage responses through proteolysis of GroEL2. PLoS Pathog 10:e1004132. doi: 10.1371/journal.ppat.1004132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Singh B, Singh G, Trajkovic V, Sharma P. 2003. Intracellular expression of Mycobacterium tuberculosis-specific 10-kDa antigen down-regulates macrophage B7.1 expression and nitric oxide release. Clin Exp Immunol 134:70–77. doi: 10.1046/j.1365-2249.2003.02258.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Blanc L, Gilleron M, Prandi J, Song OR, Jang MS, Gicquel B, Drocourt D, Neyrolles O, Brodin P, Tiraby G, Vercellone A, Nigou J. 2017. Mycobacterium tuberculosis inhibits human innate immune responses via the production of TLR2 antagonist glycolipids. Proc Natl Acad Sci U S A 114:11205–11210. doi: 10.1073/pnas.1707840114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Jayaraman P, Jacques MK, Zhu C, Steblenko KM, Stowell BL, Madi A, Anderson AC, Kuchroo VK, Behar SM. 2016. TIM3 mediates T cell exhaustion during Mycobacterium tuberculosis infection. PLoS Pathog 12:e1005490. doi: 10.1371/journal.ppat.1005490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Barber DL, Mayer-Barber KD, Feng CG, Sharpe AH, Sher A. 2011. CD4 T cells promote rather than control tuberculosis in the absence of PD-1-mediated inhibition. J Immunol 186:1598–1607. doi: 10.4049/jimmunol.1003304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Lázár-Molnár E, Chen B, Sweeney KA, Wang EJ, Liu W, Lin J, Porcelli SA, Almo SC, Nathenson SG, Jacobs WR Jr. 2010. Programmed death-1 (PD-1)-deficient mice are extraordinarily sensitive to tuberculosis. Proc Natl Acad Sci U S A 107:13402–13407. doi: 10.1073/pnas.1007394107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Fankhauser SC, Starnbach MN. 2014. PD-L1 limits the mucosal CD8+ T cell response to Chlamydia trachomatis. J Immunol 192:1079–1090. doi: 10.4049/jimmunol.1301657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Sahler JM, Eade CR, Altier C, March JC. 2018. Salmonella enterica serovar Typhimurium increases functional PD-L1 synergistically with gamma interferon in intestinal epithelial cells via Salmonella pathogenicity island 2. Infect Immun 86:e00674-17. doi: 10.1128/IAI.00674-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Holokai L, Chakrabarti J, Broda T, Chang J, Hawkins JA, Sundaram N, Wroblewski LE, Peek RM Jr, Wang J, Helmrath M, Wells JM, Zavros Y. 2019. Increased programmed death-ligand 1 is an early epithelial cell response to Helicobacter pylori infection. PLoS Pathog 15:e1007468. doi: 10.1371/journal.ppat.1007468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Suarez GV, Melucci Ganzarain CDC, Vecchione MB, Trifone CA, Marin Franco JL, Genoula M, Morana EJ, Balboa L, Quiroga MF. 2019. PD-1/PD-L1 pathway modulates macrophage susceptibility to Mycobacterium tuberculosis specific CD8+ T cell induced death. Sci Rep 9:187. doi: 10.1038/s41598-018-36403-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Nayak DK, Mendez O, Bowen S, Mohanakumar T. 2018. Isolation and in vitro culture of murine and human alveolar macrophages. J Vis Exp 134:57287. doi: 10.3791/57287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Fejer G, Wegner MD, Gyory I, Cohen I, Engelhard P, Voronov E, Manke T, Ruzsics Z, Dolken L, Prazeres da Costa O, Branzk N, Huber M, Prasse A, Schneider R, Apte RN, Galanos C, Freudenberg MA. 2013. Nontransformed, GM-CSF-dependent macrophage lines are a unique model to study tissue macrophage functions. Proc Natl Acad Sci U S A 110:E2191–E2198. doi: 10.1073/pnas.1302877110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Woo M, Wood C, Kwon D, Park KP, Fejer G, Delorme V. 2018. Mycobacterium tuberculosis infection and innate responses in a new model of lung alveolar macrophages. Front Immunol 9:438. doi: 10.3389/fimmu.2018.00438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Olive AJ, Smith CM, Kiritsy MC, Sassetti CM. 2018. The phagocyte oxidase controls tolerance to Mycobacterium tuberculosis infection. J Immunol 201:1705–1716. doi: 10.4049/jimmunol.1800202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Sassetti CM, Boyd DH, Rubin EJ. 2001. Comprehensive identification of conditionally essential genes in mycobacteria. Proc Natl Acad Sci U S A 98:12712–12717. doi: 10.1073/pnas.231275498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Griffin JE, Gawronski JD, Dejesus MA, Ioerger TR, Akerley BJ, Sassetti CM. 2011. High-resolution phenotypic profiling defines genes essential for mycobacterial growth and cholesterol catabolism. PLoS Pathog 7:e1002251. doi: 10.1371/journal.ppat.1002251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.DeJesus MA, Gerrick ER, Xu W, Park SW, Long JE, Boutte CC, Rubin EJ, Schnappinger D, Ehrt S, Fortune SM, Sassetti CM, Ioerger TR. 2017. Comprehensive essentiality analysis of the Mycobacterium tuberculosis genome via saturating transposon mutagenesis. mBio 8:e02133-16. doi: 10.1128/mBio.02133-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Bellerose MM, Baek SH, Huang CC, Moss CE, Koh EI, Proulx MK, Smith CM, Baker RE, Lee JS, Eum S, Shin SJ, Cho SN, Murray M, Sassetti CM. 2019. Common variants in the glycerol kinase gene reduce tuberculosis drug efficacy. mBio 10:e00663-19. doi: 10.1128/mBio.00663-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Carey AF, Rock JM, Krieger IV, Chase MR, Fernandez-Suarez M, Gagneux S, Sacchettini JC, Ioerger TR, Fortune SM. 2018. Tn-seq of Mycobacterium tuberculosis clinical isolates reveals strain-specific antibiotic liabilities. PLoS Pathog 14:e1006939. doi: 10.1371/journal.ppat.1006939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Rock JM, Hopkins FF, Chavez A, Diallo M, Chase MR, Gerrick ER, Pritchard JR, Church GM, Rubin EJ, Sassetti CM, Schnappinger D, Fortune SM. 2017. Programmable transcriptional repression in mycobacteria using an orthogonal CRISPR interference platform. Nat Microbiol 2:16274. doi: 10.1038/nmicrobiol.2016.274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Kim JH, Wei JR, Wallach JB, Robbins RS, Rubin EJ, Schnappinger D. 2011. Protein inactivation in mycobacteria by controlled proteolysis and its application to deplete the beta subunit of RNA polymerase. Nucleic Acids Res 39:2210–2220. doi: 10.1093/nar/gkq1149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Johnson EO, LaVerriere E, Office E, Stanley M, Meyer E, Kawate T, Gomez JE, Audette RE, Bandyopadhyay N, Betancourt N, Delano K, Da Silva I, Davis J, Gallo C, Gardner M, Golas AJ, Guinn KM, Kennedy S, Korn R, McConnell JA, Moss CE, Murphy KC, Nietupski RM, Papavinasasundaram KG, Pinkham JT, Pino PA, Proulx MK, Ruecker N, Song N, Thompson M, Trujillo C, Wakabayashi S, Wallach JB, Watson C, Ioerger TR, Lander ES, Hubbard BK, Serrano-Wu MH, Ehrt S, Fitzgerald M, Rubin EJ, Sassetti CM, Schnappinger D, Hung DT. 2019. Large-scale chemical-genetics yields new M. tuberculosis inhibitor classes. Nature 571:72–78. doi: 10.1038/s41586-019-1315-z. [DOI] [PubMed] [Google Scholar]
- 108.Penn BH, Netter Z, Johnson JR, Von Dollen J, Jang GM, Johnson T, Ohol YM, Maher C, Bell SL, Geiger K, Golovkine G, Du X, Choi A, Parry T, Mohapatra BC, Storck MD, Band H, Chen C, Jager S, Shales M, Portnoy DA, Hernandez R, Coscoy L, Cox JS, Krogan NJ. 2018. An Mtb-human protein-protein interaction map identifies a switch between host antiviral and antibacterial responses. Mol Cell 71:637–648.e5. doi: 10.1016/j.molcel.2018.07.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Pickar-Oliver A, Gersbach CA. 2019. The next generation of CRISPR-Cas technologies and applications. Nat Rev Mol Cell Biol 20:490–507. doi: 10.1038/s41580-019-0131-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Sanson KR, Hanna RE, Hegde M, Donovan KF, Strand C, Sullender ME, Vaimberg EW, Goodale A, Root DE, Piccioni F, Doench JG. 2018. Optimized libraries for CRISPR-Cas9 genetic screens with multiple modalities. Nat Commun 9:5416. doi: 10.1038/s41467-018-07901-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Shalem O, Sanjana NE, Hartenian E, Shi X, Scott DA, Mikkelson T, Heckl D, Ebert BL, Root DE, Doench JG, Zhang F. 2014. Genome-scale CRISPR-Cas9 knockout screening in human cells. Science 343:84–87. doi: 10.1126/science.1247005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Li W, Xu H, Xiao T, Cong L, Love MI, Zhang F, Irizarry RA, Liu JS, Brown M, Liu XS. 2014. MAGeCK enables robust identification of essential genes from genome-scale CRISPR/Cas9 knockout screens. Genome Biol 15:554. doi: 10.1186/s13059-014-0554-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Konermann S, Brigham MD, Trevino AE, Joung J, Abudayyeh OO, Barcena C, Hsu PD, Habib N, Gootenberg JS, Nishimasu H, Nureki O, Zhang F. 2015. Genome-scale transcriptional activation by an engineered CRISPR-Cas9 complex. Nature 517:583–588. doi: 10.1038/nature14136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Smith CM, Sassetti CM. 2018. Modeling diversity: do homogeneous laboratory strains limit discovery? Trends Microbiol 26:892–895. doi: 10.1016/j.tim.2018.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Noll KE, Ferris MT, Heise MT. 2019. The collaborative cross: a systems genetics resource for studying host-pathogen interactions. Cell Host Microbe 25:484–498. doi: 10.1016/j.chom.2019.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Smith CM, Proulx MK, Olive AJ, Laddy D, Mishra BB, Moss C, Gutierrez NM, Bellerose MM, Barreira-Silva P, Phuah JY, Baker RE, Behar SM, Kornfeld H, Evans TG, Beamer G, Sassetti CM. 2016. Tuberculosis susceptibility and vaccine protection are independently controlled by host genotype. mBio 7:e01516-16. doi: 10.1128/mBio.01516-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Churchill GA, Gatti DM, Munger SC, Svenson KL. 2012. The Diversity Outbred mouse population. Mamm Genome 23:713–718. doi: 10.1007/s00335-012-9414-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Harrison DE, Astle CM, Niazi MKK, Major S, Beamer GL. 2014. Genetically diverse mice are novel and valuable models of age-associated susceptibility to Mycobacterium tuberculosis. Immun Ageing 11:24. doi: 10.1186/s12979-014-0024-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Niazi MK, Dhulekar N, Schmidt D, Major S, Cooper R, Abeijon C, Gatti DM, Kramnik I, Yener B, Gurcan M, Beamer G. 2015. Lung necrosis and neutrophils reflect common pathways of susceptibility to Mycobacterium tuberculosis in genetically diverse, immune-competent mice. Dis Model Mech 8:1141–1153. doi: 10.1242/dmm.020867. [DOI] [PMC free article] [PubMed] [Google Scholar]