Abstract
Base editing is a genome editing strategy that induces specific single nucleotide changes within genomic DNA. Two major DNA base editors, Cytosine base editors (CBEs) and Adenine base editors (ABEs), have been developed that consist of a Cas9 protein linked to a deaminase enzyme that catalyzes targeted base conversion directed by a sgRNA. This strategy has been used widely for precise genome editing because, unlike CRISPR-Cas nuclease-based genome editing systems, this strategy does not create double strand DNA breaks (DSBs) that often result in high levels of undesirable indels. However, recent papers have reported that DNA base editors can cause substantial off-target editing in both genomic DNA and RNA. The off-target editing described in these studies is primarily guide RNA-independent arising from promiscuous reactivity of the deaminase enzymes used in DNA base editors. In this perspective, we discuss the development of DNA base editors, the guide RNA independent off-target activity reported in recent studies, and strategies that improve the selectivity of DNA base editors.
Keywords: genome editing, CRISPR-Cas, APOBEC, TadA, cytidine deaminase, adenosine deaminase
Graphical Abstract

Introduction
The majority of human genetic diseases arise from single nucleotide changes1–2. Thus, therapeutic strategies that could allow one to correct single point mutations hold promise as powerful tools to treat genetic disorders. CRISPR (clustered regularly interspaced short palindromic repeat)-Cas(CRISPR-associated protein nuclease)-based systems have revolutionized the field of genome engineering3–5. Several CRISPR-Cas systems have been developed to induce double strand DNA breaks (DSBs) at specific sequences directed by single guide RNAs (sgRNAs)5–11. Such induced DSBs are then repaired by cellular DNA repair pathways including the non-homologous end-joining (NHEJ) pathway and the homology-directed repair (HDR) pathway12. Indeed, CRISPR-Cas-mediated selective cleavage of duplex DNA coupled with HDR using appropriately designed donor DNA fragments has become a popular approach for introducing specific sequence changes in genomes4, 13–14. However, HDR competes with NHEJ which often results in unwanted indels (insertions or deletions)15–16.
Unwanted off-target changes in the genome resulting from DSBs and subsequent NHEJ is a significant concern for the therapeutic application of typical CRISPR-Cas genome editing strategies17–18. Therefore, manipulating the genome sequence without DSBs could be more powerful. Base editing is a genome editing strategy originally developed in the laboratories of David Liu at Harvard University that changes a specific single nucleotide within genomic DNA directly without introducing DSBs19–20. The base editors are composed of a catalytically defective Cas protein fused to a deaminase enzyme capable of direct modification of the target nucleotide guided by the sequence of a sgRNA. Since base editing does not introduce DSBs, creation of undesired indels is minimized.19–21 In addition, DNA base editing can be applied more broadly since the application of CRISPR-Cas genome editing is largely limited to actively dividing cells where the HDR pathway is most efficient12, 22. However, off-target editing by DNA base editors has been observed19–21, 23–27. Earlier work described approaches to measure the extent of guide RNA-dependent off-target editing and to reduce this type of off target activity23–25, 28–29. However, more recent reports suggest that promiscuous reactivity of deaminase domains present in the DNA base editors can lead to guide RNA-independent off target editing in both DNA and RNA30–34. In this Perspective, we review the development of the base editor genome editing enzymes, recent reports of a guide RNA-independent off target activity and discuss strategies for improving target selectivity.
DNA Base editors
Two main classes of base editors have been developed; Cytosine Base Editors (CBEs) that convert C-to-T19, 21 and Adenine Base Editors (ABEs) that convert A-to-G20.
Cytosine Base Editors (CBEs)
Cytosine Base Editors (CBEs) are composed of catalytically inactive Cas nuclease and an APOBEC/AID cytidine deaminase19, 21. The Cas protein is directed by a sgRNA to bring the deaminase to a specific base editing target site4–5. APOBECs (Apolipoprotein B mRNA Editing Catalytic Polypeptide-like) and AID (activation-induced cytidine deaminase) are members of a family of enzymes that catalyze hydrolytic deamination of cytosine (C) to generate uracil (U) in DNA and RNA35. Since uracil is replicated like thymine, the deamination of cytidine leads to a C-to-T change in DNA. Fusion of rat APOBEC1 and catalytically dead Cas9 nuclease (dCas9) from Streptococcus pyogenes created the first generation of cytosine base editor (BE1)19. Since the development of BE1, additional studies led to incorporation of uracil DNA glycosylase inhibitor (UGI) to prevent removal of U by Uracil DNA glycosylase (UDG) activity (BE2)19 and replacing dCas9 with Cas9 D10A nickase (nCas9) (BE3)19 which creates a nick on the non-edited DNA strand leading to preferential replication of the edited strand (Figure 1). When the BE3-sgRNA complex binds to target genomic DNA, this results in formation of a DNA-RNA hybrid structure near the target site and a ssDNA loop that is not bound to sgRNA19. This ssDNA is then available for rAPOBEC1-catalyzed cytosine deamination within a 5-nucleotide window19. Although the BE3-sgRNA system still induces a small number of indels, this level is substantially lower than CRISPR-Cas systems that induce DSBs19, 24. Therefore, it has been used widely for various applications27, 36.
Figure 1.

Cytosine base editor and Adenine base editor. (Top) Cytosine base editor can be directed to target DNA by nCas9 complex19. A cytidine deaminase (rAPOBEC1) deaminates a target cytidine (C) to uridine (U). This modification results in conversion of the original C•G base pair to T•A base pair by DNA repair or replication process19. (Bottom) When Adenine base editor is directed to target DNA by the nCas9 complex, an adenosine deaminase (ecTadA) deaminates a target adenosine (A) to inosine (I)20. Inosine is recognized as guanosine (G) by cellular machinery37. Therefore, base modification by adenine base editor leads to conversion of the original A•T base pair to G•C base pair by DNA repair or replication process20.
Adenine Base Editors (ABEs)
To increase the scope of genomic DNA base editing, Adenine Base Editors (ABEs) were developed for Adenosine (A) to Guanosine (G) conversion20. Adenosine deaminases carryout hydrolytic deamination of Adenosine(A) to generate Inosine (I)37. Although A is not directly converted to G by adenosine deaminase activity, inosine is recognized as G during replication37. Thus, A-to-I editing is interpreted as an A-to-G conversion. The major challenge in the development of ABEs was the fact that there are no known adenosine deaminases that act on ssDNA. E. coli TadA (ecTadA) is a tRNA adenosine deaminase that catalyzes the conversion of A43 in tRNAArg2 to inosine38. In an impressive feat of in vitro evolution, the Liu lab converted ecTadA into an enzyme that can readily deaminate 2’-deoxyadenosine in ssDNA20. This created the first generation of ABE, TadA*-dCas9 (TadA* = evolved TadA)20. ABEs were further optimized to increase the efficiency of editing by incorporating wild-type TadA20, accounting the fact that wild-type ecTadA catalytic activity requires homodimer formation39. This resulted in ABE 7.10 (ecTadA-ecTadA*-nCas9)20 (Figure 1). Like BE3, when treated with ABE, the base editor complex is directed to target a specific site in genomic DNA. After formation of the sgRNA-DNA complex, ssDNA is available for base modification by evolved ecTadA* within a 4–7 nucleotide window20.
Genome and transcriptome wide off-target editing by DNA base editors
There have been several studies looking at the off-target effects of CRISPR-Cas-based genome engineering methods40–43. However, less was known about the genome wide off-target activity of DNA base editors. Recently, two papers in Science reported analyses of genome-wide off-target mutation induced by DNA base editors31–32. Each study took a different approach to accurately analyze the off-target mutations. Zuo et al.32 developed a method called Genome-wide Off-target analysis by Two-cell embryo Injection (GOTI) to determine genome wide off-target mutations in mice (Figure 2A). Their GOTI method uses two-cell embryos derived from Ai9 (CAG-LoxP-Stop-LoxP-tdTomato) mice44 which display a strong tdTomato fluorescence when Cre recombinase is expressed. They injected a total of 11 different combinations of CRISPR-Cas9, BE3, or ABE with or without sgRNAs along with Cre mRNA into one blastomere of two-cell embryos, which they referred to as the edited blastomere32. Then, the edited blastomere and non-edited blastomere were sorted by FACS based on the tdTomato expression level at embryonic day 14.5 (E14.5)32. This process enabled them to distinguish spontaneous mutations from off-target mutations generated by CRISPR-Cas9 or DNA base editors32. Cells with CRISPR-Cas9 or base editors (tdTomato+) and without CRISPR-Cas9 or base editors (tdTomato-) were sequenced separately by Whole genome sequencing (WGS) followed by variant calling processes for the tdTomato+ samples to detect single nucleotide changes and indels using tdTomato- samples as reference32.
Figure 2.

Experimental designs to study off-target editing of DNA base editors. (A) Zuo et al. developed a method called Genome-wide Off-target analysis by Two-cell embryo Injection (GOTI) to determine genome wide off-target mutations in mouse32. (B) Workflow used in Jin et al. to identify identified the unintended genome wide off-target conversions in rice plants31.
The method of Jin et al.31 used a clonally derived rice plant system to rule out spontaneous mutations caused by cellular processes from off-target mutations by DNA base editors (Figure 2B). In addition, they included control plants in their analysis to remove background mutations possibly caused by tissue culture and transformation processes31. A total of 14 combinations of different base editors with or without sgRNAs were transformed into rice via Agrobacterium transformation31. Off-target mutations from each regenerated rice plant with or without sgRNAs along with control plants were investigated by WGS followed by variant calling to identify single nucleotide changes and indels31.
Although the two studies used different strategies to investigate genome wide off-target mutations caused by DNA base editors, both studies found that there were no significant changes in the number of single nucleotide changes upon treatment with ABE compared to that of controls31–32. However, the amount of unintended off-target single nucleotide changes was significantly increased by BE3, predominately having C>T mutations, compared to that of ABE or controls31–32. Interestingly, both studies indicated that the observed off-target mutations were sgRNA independent31–32. This suggests that off-target mutations caused by BE3 originate from promiscuous reaction of the cytidine deaminase fused to BE3. In fact, both groups reported that off-target mutations caused by BE3 were high in transcribed regions where ssDNA is available due to R-loop formation during transcription31–32. This ssDNA could be accessible to the cytidine deaminase domain of the base editor, resulting in off-target mutations by BE3 throughout the genome. Therefore, both studies suggest that the rAPOBEC1 present in cytosine base editors should be optimized to increase the specificity of target base editing and reduce unintended off-target mutations31–32.
The deaminase enzymes rAPOBEC1 and mutant ecTadA* were fused to nCas9 to yield site-specific Cytosine base editors and Adenine base editors, respectively19–20. However, the extent to which the APOBEC1 or TadA domains in these proteins might edit different sites in the transcriptome was not fully defined when the DNA base editors were originally developed19–20. Indeed, rAPOBEC1 is capable of converting C-to-U in both ssDNA and RNA35, 45. Also, ecTadA was selected for efficient A-to-I modification on ssDNA resulting in an evolved enzyme bearing several mutations20. Although these mutations convert wild type ecTadA to mutant ecTadA* suitable for DNA base editing, these mutations do not guarantee the loss of RNA editing activity of ecTadA*. Furthermore, RNA editing activity could originate from wild type ecTadA that is also a part of the ABE design33–34.
Three recent papers investigated the transcriptome wide RNA off-target mutations caused by DNA base editors30, 33–34. For instance, in a paper recently published in Nature by Grünewald30, the authors first transfected cultured mammalian cells with BE3 and ABE plasmids fused to eGFP along with sgRNA for specific genome editing30. Controls for each DNA base editor lacking deaminase domains (BE3 without rAPOBEC1 and ABE without ecTadA dimer) were also tested30. Transfected cells were sorted based on eGFP level and DNA and total RNA samples from fluorescent cells were analyzed for on-target DNA editing as well as off-target RNA editing30 (Figure 3A). They identified BE3-induced C-to-U modifications transcriptome wide, having a preference for the sequence motif ACW (W=A or U) which is identical to the preferred editing context for wild-type rAPOBEC130. They further analyzed RNA off-target sites by Whole-exome sequencing (WES) to rule out the possibility of mutations caused by DNA editing30. These experiments confirmed that the RNA off-target modifications were not caused by corresponding genomic DNA editing30. They also showed that these RNA off-target modifications are sgRNA-independent by comparing off-target modifications caused by BE3 with sgRNAs targeting a specific human genome sequence and sgRNA having a sequence that cannot be found in human genome30. To reduce RNA off-target modifications, this group introduced specific mutations in the rAPOBEC1 domain previously reported to reduce C-to-U editing on RNA (R33A or R33A/K34A)30, 46–47. They refer to these as SECURE (SElective Curbing of Unwanted RNA Editing) variants of the Cytidine base editor30. These new variants showed a substantial reduction in RNA off-targets, while maintaining similar DNA on-target editing with a narrowed editing window30. In addition to BE3, ABE RNA off-target editing was also addressed in this paper30. These authors showed that ABE induces a significant amount of RNA off-target modifications throughout the transcriptome, showing a UA motif editing preference matching that of wild-type ecTadA30. These results were corroborated by two additional recent papers on this topic, Rees et al.33 and Zhou et al.34, both reporting off target modifications in RNA induced by ABEs. Furthermore, both of these studies indicated that RNA off-target activity of ABEs is transient and can be controlled by mutagenesis in the TadA domains. Rees et al.33 showed that ABE RNA off-target editing originates from both the wild-type ecTadA domain and the evolved ecTadA* domain. Thus, wild-type ecTadA, evolved ecTadA*, or both were inactivated by point mutation (E59A). Only when wild-type ecTadA was inactivated with this mutation could the RNA off-target activity be reduced and the DNA on-target editing be maintained33. The DNA editing specificity of the evolved ecTadA* was further optimized by mutating three residues identified from the crystal structure of S. aureus TadA39 bound to RNA. These authors hypothesized that introducing a steric clash between one of these three residues and RNA by mutating each to bulky or hydrophobic amino acids could reduce unintended RNA editing activity (Figure 3B). As a result, a new ABE variant, ABEmaxAW with ecTadA E59A and ecTadA* V106W mutations, showed a substantial reduction in RNA off-target editing while maintaining efficient DNA on-target base editing33. Similarly, Zhou et al.34 incorporated a single point mutation (D53E or F148A) identified from previous studies38, 48–50 within both wild-type ecTadA and evolved ecTadA* to prevent high level RNA off-target activity while retaining a similar level of DNA on-target activity. Taken together, these studies indicate that one must consider not only genome-wide but also transcriptome-wide off-target effects from the DNA base editors. In addition, this work clearly indicates that one effective strategy to reduce transcriptome off-targets is by strategic mutation of the deaminase domains present in the base editors30, 33–34.
Figure 3.
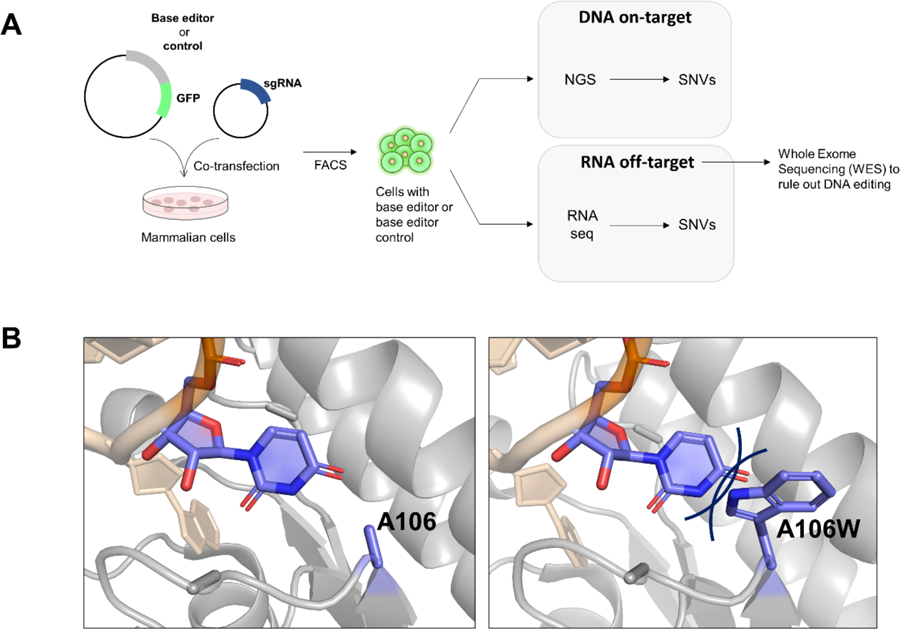
(A) General experimental workflow from Grünewald et al.30 to investigate the transcriptome wide RNA off-target mutations caused by DNA base editors. (B) Crystal structure of S. aureus TadA bound to a minimized version of its native substrate (tRNAArg2) (PDB id: 2B3J)39 showing Ala 106, a residue that corresponds to V106 in ecTadA*. The V106W mutation in ecTadA* (shown here as an A106W mutation in S. aureus TadA) is expected to result in a steric clash with RNA reducing undesired RNA off-target editing33.
Each of the studies discussed above indicated that substantial guide RNA-independent off-target base editing arises from promiscuous reactivity of the deaminase enzymes present in each base editor (rAPOBEC1 in BE3 and ecTadA in ABE). Thus, additional control of the enzymatic activity (i.e. catalysis and substrate binding) of these domains is required to achieve precise targeted base editing without unwanted genome or transcriptome modification (Figure 4). Fortunately, there are several strategies available to modulate the activity of the base editor deaminase domains. Since the development of the first DNA base editors, variants have been reported with different deaminases, linker lengths, and Cas nucleases27. Each system has a slightly different editing window in the ssDNA in the Cas-sgRNA-target complex. For instance, BE3 has a 5-nucleotide editing window and ABE has a 4–7 nucleotide window19–20, 27. Within each editing window, any C or A base can be modified by the corresponding deaminase domain, resulting in bystander off-target modifications. Therefore, a narrow editing window is preferred, ideally targeting a single, specific nucleotide for modification. Recent reports suggest that this type of off-target editing can be controlled by tuning the activity of the deaminase in the editor enzyme. For example, different cytosine base editors (YE1-BE3, EE-BE3, YE2-BE3, and YEE-BE3) were developed by incorporating two or three mutations in rAPOBEC1 that reduce catalytic activity and substrate binding of rAPOBEC1 enzyme resulting in a narrow editing window.23 Mutations used in these base editors were identified from the APOBEC3G enzyme that has 42% sequence similarity with rAPOBEC145. With those mutations, off-target editing was reduced while still maintaining similar base modification efficiency for on-target DNA base editing. In addition, the SECURE variants of cytosine base editors reported by Grünewald et al.30 described above reduced transcriptome off-target editing with mutations that were previously shown to decrease RNA editing activity of wild-type APOBEC1. They used the R33A and K34A mutations for this purpose. Interestingly, R33 and K34 are part of the N-terminal basic amino acid cluster of APOBEC147, 51. It has been reported that this part of protein binds the ACF cofactor, which is an RNA binding protein52–54. Mutating these residues likely inhibits interaction with ACF, reducing off target RNA binding affinity and hyper editing activity. Also, it is likely to continue to be useful to test other deaminase enzymes in the context of new base editors, particularly deaminases for which structural information is available. For example, hA3A-BE3 was developed using engineered human APOBEC3A (hA3A) guided by available high resolution structures for hA3A55. hA3A-BE3 shows higher editing specificity while reducing bystander off-target editing compared to cytosine base editor variants with rAPOBEC1 mentioned above. Furthermore, it has been reported that BE3 fused with an hA3A mutant (R128A or Y130F) further reduces unwanted RNA off-target editing, while retaining similar DNA on-target editing34.
Figure 4.

Components of base editors and what can be optimized to minimize off-target editing, while maximizing the efficiency of base editing by DNA base editors.
Other factors should also be considered to further increase on-target specificity for DNA base editors (Figure 4). As mentioned before, UGI was incorporated into cytosine base editors to prevent the removal of U in DNA, thus increasing the overall efficiency of cytosine base editor activity. However, the presence of UGI can also increase the level of unwanted C-to-T mutations by blocking Uracil N-glycosylase (UNG) activity56–57. Therefore, the development of highly efficient cytosine base editors without UGI should be considered. Also, the level of overexpression and the cellular half life of DNA base editors provide additional means of optimization since high cellular concentrations of long-lived deaminase enzymes are likely to lead to undesirable off-target editing58–59. Indeed, RNP delivery of DNA base editors improves on-target specificity compared to plasmid delivery24. Thus, different delivery strategies should be considered carefully to control exposure time as a means of further minimizing off-target editing24, 33. Finally, increasing the targetable sequences by using different Cas nuclease variants with different PAM requirements will continue to expand the application of the base editors23, 60.
Additional insight from site-directed RNA editing approaches
Since substantial off-target editing by DNA base editors originates from their deaminase activity on RNA, experience with existing site-directed RNA editing strategies (RNA base editors) could provide some insight for reducing RNA off-target editing by DNA base editors. There have been several site-directed RNA editors developed using the Adenosine Deaminases Acting on RNA (ADARs) that convert A-to-I within duplex RNA. Site-directed RNA editing strategies can be classified into two approaches. One approach uses an antisense oligonucleotide to redirect endogenous ADARs for site-specific editing. For example, both RESTORE61 and LEAPER62 systems use antisense oligonucleotides to recruit endogenous ADARs to the target editing site of interest. Because these methods harness endogenous ADARs rather than overexpressing ADARs, RNA off-target editing from high expression levels of the deaminase is reduced61–63. The other approach uses an antisense guide RNA (gRNA) to direct engineered ADAR fusion proteins for site-specific editing. Several systems have been developed that link ADAR fusions to a gRNA including SNAP tagging (SNAP-ADAR)64–68, λN peptide (λN-ADAR)69–71, or a Cas protein (dCas13b-ADAR or REPAIR)72. These ADAR fusions are then directed by the gRNA that forms a duplex at the target site for site-specific RNA editing. It is noteworthy that both approaches take advantage of ADARs’ preference for duplex RNA and editing A in the context of an A•C mismatch61–62, 66, 70, 72. Site directed RNA editing strategies that use fusion proteins containing ADAR catalytic domains must also contend with guide RNA-independent off targeting. For instance, Rosenthal addressed this issue for λN-ADAR by targeting the fusion proteins to the nucleus where off-target editing is less efficient63. To reduce off-target RNA editing in the REPAIR system, Zhang and colleagues made use of high-resolution structures of human ADAR2 bound to RNA73 to introduce mutations at sites known to contact RNA, thus reducing substrate binding affinity of the deaminase domain itself72. Finally, our lab described a bump-hole approach for increasing directed RNA editing specificity74. This approach uses a mutant deaminase domain of human ADAR2 with a bulky residue that causes a steric clash with the nucleobase opposite the targeted A74. Only when this clash is relieved with a gRNA containing an abasic site opposite the A can efficient reaction occur, thus increasing the specificity of directed RNA editing74. While high resolution structures that include human APOBECs and S. aureus TadA have been helpful in guiding mutations in DNA base editors, no structures are currently available for the DNA base editors themselves bound to DNA. Such structures would be highly valuable for guiding further optimization of editor efficiency and specificity.
Conclusion and future outlook
DNA editing systems can be powerful tools with various applications from basic research to therapeutics. The DNA base editors described here are promising systems that introduce specific C-to-T or A-to-G changes in genomes. However, optimization of DNA base editors has been necessary to reduce guide RNA-independent base modifications in cellular DNA and RNA30–34. The promiscuous editing activity of the deaminase enzymes used in base editor systems appears to be a primary cause of off-target base editing30–34. rAPOBEC1 in BE3 retains ssDNA and RNA editing activity which results in off-target modifications in both the genome and transcriptome, whereas ecTadA in ABE retains RNA editing activity, leading to transcriptome wide off-target base modifications30–34. Initial efforts have shown that strategic use of mutation within the deaminase domains can decrease guide RNA-independent off-target editing for DNA base editors30, 33–34. However, it is often true that mutations that reduce off-target base editing can also lead to a decrease in on-target base editing. Therefore, it is important to fully understand features of these enzymes that control catalysis and substrate binding such that maximum editing activity can be delivered precisely to the desired site of reaction. Additional biochemical and structural studies with nucleic acid modifying deaminases, alone or in the context of base editors, will inform these efforts.
Acknowledgments
The authors acknowledge funding from the US National Institutes of Health (NIH) grant R01GM061115 (P.A.B). The contents of this publication are solely the responsibility of the authors and do not necessarily represent the official views of NIH.
Footnotes
P.A.B. is a consultant for Beam Therapeutics, a company developing therapeutic CRISPR-guided DNA base editors.
References
- 1.Landrum MJ; Lee JM; Riley GR; Jang W; Rubinstein WS; Church DM; Maglott DR, ClinVar: public archive of relationships among sequence variation and human phenotype. Nucleic Acids Res 2013, 42 (D1), D980–D985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Landrum MJ; Lee JM; Benson M; Brown G; Chao C; Chitipiralla S; Gu B; Hart J; Hoffman D; Hoover J; Jang W; Katz K; Ovetsky M; Riley G; Sethi A; Tully R; Villamarin-Salomon R; Rubinstein W; Maglott DR, ClinVar: public archive of interpretations of clinically relevant variants. Nucleic Acids Res 2015, 44 (D1), D862–D868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Doudna JA; Charpentier E, The new frontier of genome engineering with CRISPR-Cas9. Science 2014, 346 (6213), 1258096. [DOI] [PubMed] [Google Scholar]
- 4.Hsu Patrick D.; Lander Eric S.; Zhang F, Development and Applications of CRISPR-Cas9 for Genome Engineering. Cell 2014, 157 (6), 1262–1278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sternberg Samuel H.; Doudna Jennifer A., Expanding the Biologist’s Toolkit with CRISPR-Cas9. Mol Cell 2015, 58 (4), 568–574. [DOI] [PubMed] [Google Scholar]
- 6.Wright AV; Nunez JK; Doudna JA, Biology and Applications of CRISPR Systems: Harnessing Nature’s Toolbox for Genome Engineering. Cell 2016, 164 (1–2), 29–44. [DOI] [PubMed] [Google Scholar]
- 7.Cho SW; Kim S; Kim JM; Kim JS, Targeted genome engineering in human cells with the Cas9 RNA-guided endonuclease. Nat Biotechnol 2013, 31 (3), 230–2. [DOI] [PubMed] [Google Scholar]
- 8.Cong L; Ran FA; Cox D; Lin S; Barretto R; Habib N; Hsu PD; Wu X; Jiang W; Marraffini LA; Zhang F, Multiplex genome engineering using CRISPR/Cas systems. Science 2013, 339 (6121), 819–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Garneau JE; Dupuis ME; Villion M; Romero DA; Barrangou R; Boyaval P; Fremaux C; Horvath P; Magadan AH; Moineau S, The CRISPR/Cas bacterial immune system cleaves bacteriophage and plasmid DNA. Nature 2010, 468 (7320), 67–71. [DOI] [PubMed] [Google Scholar]
- 10.Jinek M; East A; Cheng A; Lin S; Ma E; Doudna J, RNA-programmed genome editing in human cells. Elife 2013, 2, e00471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mali P; Yang L; Esvelt KM; Aach J; Guell M; DiCarlo JE; Norville JE; Church GM, RNA-guided human genome engineering via Cas9. Science 2013, 339 (6121), 823–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chapman JR; Taylor MR; Boulton SJ, Playing the end game: DNA double-strand break repair pathway choice. Mol Cell 2012, 47 (4), 497–510. [DOI] [PubMed] [Google Scholar]
- 13.Nunez JK; Harrington LB; Doudna JA, Chemical and Biophysical Modulation of Cas9 for Tunable Genome Engineering. ACS Chem Biol 2016, 11 (3), 681–8. [DOI] [PubMed] [Google Scholar]
- 14.Ran FA; Hsu PD; Wright J; Agarwala V; Scott DA; Zhang F, Genome engineering using the CRISPR-Cas9 system. Nat Protoc 2013, 8 (11), 2281–2308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lin S; Staahl BT; Alla RK; Doudna JA, Enhanced homology-directed human genome engineering by controlled timing of CRISPR/Cas9 delivery. Elife 2014, 3, e04766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Paquet D; Kwart D; Chen A; Sproul A; Jacob S; Teo S; Olsen KM; Gregg A; Noggle S; Tessier-Lavigne M, Efficient introduction of specific homozygous and heterozygous mutations using CRISPR/Cas9. Nature 2016, 533 (7601), 125–9. [DOI] [PubMed] [Google Scholar]
- 17.Kosicki M; Tomberg K; Bradley A, Repair of double-strand breaks induced by CRISPR-Cas9 leads to large deletions and complex rearrangements. Nat Biotechnol 2018, 36 (8), 765–771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhang L; Jia R; Palange NJ; Satheka AC; Togo J; An Y; Humphrey M; Ban L; Ji Y; Jin H; Feng X; Zheng Y, Large genomic fragment deletions and insertions in mouse using CRISPR/Cas9. PLoS One 2015, 10 (3), e0120396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Komor AC; Kim YB; Packer MS; Zuris JA; Liu DR, Programmable editing of a target base in genomic DNA without double-stranded DNA cleavage. Nature 2016, 533 (7603), 420–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gaudelli NM; Komor AC; Rees HA; Packer MS; Badran AH; Bryson DI; Liu DR, Programmable base editing of A*T to G*C in genomic DNA without DNA cleavage. Nature 2017, 551 (7681), 464–471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Nishida K; Arazoe T; Yachie N; Banno S; Kakimoto M; Tabata M; Mochizuki M; Miyabe A; Araki M; Hara KY; Shimatani Z; Kondo A, Targeted nucleotide editing using hybrid prokaryotic and vertebrate adaptive immune systems. Science 2016, 353 (6305). [DOI] [PubMed] [Google Scholar]
- 22.Cox DB; Platt RJ; Zhang F, Therapeutic genome editing: prospects and challenges. Nat Med 2015, 21 (2), 121–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kim YB; Komor AC; Levy JM; Packer MS; Zhao KT; Liu DR, Increasing the genome-targeting scope and precision of base editing with engineered Cas9-cytidine deaminase fusions. Nat Biotechnol 2017, 35 (4), 371–376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Rees HA; Komor AC; Yeh WH; Caetano-Lopes J; Warman M; Edge ASB; Liu DR, Improving the DNA specificity and applicability of base editing through protein engineering and protein delivery. Nat Commun 2017, 8, 15790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kim D; Lim K; Kim ST; Yoon SH; Kim K; Ryu SM; Kim JS, Genome-wide target specificities of CRISPR RNA-guided programmable deaminases. Nat Biotechnol 2017, 35 (5), 475–480. [DOI] [PubMed] [Google Scholar]
- 26.Liang P; Xie X; Zhi S; Sun H; Zhang X; Chen Y; Chen Y; Xiong Y; Ma W; Liu D; Huang J; Songyang Z, Genome-wide profiling of adenine base editor specificity by EndoV-seq. Nat Commun 2019, 10 (1), 67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rees HA; Liu DR, Base editing: precision chemistry on the genome and transcriptome of living cells. Nat Rev Genet 2018, 19 (12), 770–788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yeh WH; Chiang H; Rees HA; Edge ASB; Liu DR, In vivo base editing of post-mitotic sensory cells. Nat Commun 2018, 9 (1), 2184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kleinstiver BP; Pattanayak V; Prew MS; Tsai SQ; Nguyen NT; Zheng Z; Joung JK, High-fidelity CRISPR–Cas9 nucleases with no detectable genome-wide off-target effects. Nature 2016, 529, 490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Grunewald J; Zhou R; Garcia SP; Iyer S; Lareau CA; Aryee MJ; Joung JK, Transcriptome-wide off-target RNA editing induced by CRISPR-guided DNA base editors. Nature 2019, 569 (7756), 433–437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Jin S; Zong Y; Gao Q; Zhu Z; Wang Y; Qin P; Liang C; Wang D; Qiu J-L; Zhang F; Gao C, Cytosine, but not adenine, base editors induce genome-wide off-target mutations in rice. Science 2019, 364 (6437), 292–295. [DOI] [PubMed] [Google Scholar]
- 32.Zuo E; Sun Y; Wei W; Yuan T; Ying W; Sun H; Yuan L; Steinmetz LM; Li Y; Yang H, Cytosine base editor generates substantial off-target single-nucleotide variants in mouse embryos. Science 2019, 364 (6437), 289–292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rees HA; Wilson C; Doman JL; Liu DR, Analysis and minimization of cellular RNA editing by DNA adenine base editors. Sci Adv 2019, 5 (5), eaax5717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zhou C; Sun Y; Yan R; Liu Y; Zuo E; Gu C; Han L; Wei Y; Hu X; Zeng R; Li Y; Zhou H; Guo F; Yang H, Off-target RNA mutation induced by DNA base editing and its elimination by mutagenesis. Nature 2019, 571 (7764), 275–278. [DOI] [PubMed] [Google Scholar]
- 35.Salter JD; Bennett RP; Smith HC, The APOBEC Protein Family: United by Structure, Divergent in Function. Trends Biochem Sci 2016, 41 (7), 578–594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hess GT; Tycko J; Yao D; Bassik MC, Methods and Applications of CRISPR-Mediated Base Editing in Eukaryotic Genomes. Mol Cell 2017, 68 (1), 26–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yasui M; Suenaga E; Koyama N; Masutani C; Hanaoka F; Gruz P; Shibutani S; Nohmi T; Hayashi M; Honma M, Miscoding properties of 2’-deoxyinosine, a nitric oxide-derived DNA Adduct, during translesion synthesis catalyzed by human DNA polymerases. J Mol Biol 2008, 377 (4), 1015–23. [DOI] [PubMed] [Google Scholar]
- 38.Wolf J; Gerber AP; Keller W, tadA, an essential tRNA-specific adenosine deaminase from Escherichia coli. EMBO J 2002, 21 (14), 3841–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Losey HC; Ruthenburg AJ; Verdine GL, Crystal structure of Staphylococcus aureus tRNA adenosine deaminase TadA in complex with RNA. Nat Struct Mol Biol 2006, 13 (2), 153–9. [DOI] [PubMed] [Google Scholar]
- 40.Akcakaya P; Bobbin ML; Guo JA; Malagon-Lopez J; Clement K; Garcia SP; Fellows MD; Porritt MJ; Firth MA; Carreras A; Baccega T; Seeliger F; Bjursell M; Tsai SQ; Nguyen NT; Nitsch R; Mayr LM; Pinello L; Bohlooly YM; Aryee MJ; Maresca M; Joung JK, In vivo CRISPR editing with no detectable genome-wide off-target mutations. Nature 2018, 561 (7723), 416–419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Fu Y; Foden JA; Khayter C; Maeder ML; Reyon D; Joung JK; Sander JD, High-frequency off-target mutagenesis induced by CRISPR-Cas nucleases in human cells. Nat Biotechnol 2013, 31 (9), 822–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kim D; Bae S; Park J; Kim E; Kim S; Yu HR; Hwang J; Kim JI; Kim JS, Digenome-seq: genome-wide profiling of CRISPR-Cas9 off-target effects in human cells. Nat Methods 2015, 12 (3), 237–43, 1 p following 243. [DOI] [PubMed] [Google Scholar]
- 43.Liang P; Xu Y; Zhang X; Ding C; Huang R; Zhang Z; Lv J; Xie X; Chen Y; Li Y; Sun Y; Bai Y; Songyang Z; Ma W; Zhou C; Huang J, CRISPR/Cas9-mediated gene editing in human tripronuclear zygotes. Protein Cell 2015, 6 (5), 363–372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Madisen L; Zwingman TA; Sunkin SM; Oh SW; Zariwala HA; Gu H; Ng LL; Palmiter RD; Hawrylycz MJ; Jones AR; Lein ES; Zeng H, A robust and high-throughput Cre reporting and characterization system for the whole mouse brain. Nat Neurosci 2010, 13 (1), 133–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Harris RS; Petersen-Mahrt SK; Neuberger MS, RNA editing enzyme APOBEC1 and some of its homologs can act as DNA mutators. Mol Cell 2002, 10 (5), 1247–53. [DOI] [PubMed] [Google Scholar]
- 46.Teng B-B; Ochsner S; Zhang Q; Soman KV; Lau PP; Chan L, Mutational analysis of apolipoprotein B mRNA editing enzyme (APOBEC1): structure–function relationships of RNA editing and dimerization. J Lipid Res 1999, 40 (4), 623–635. [PubMed] [Google Scholar]
- 47.Chen Z; Eggerman TL; Bocharov AV; Baranova IN; Vishnyakova TG; Csako G; Patterson AP, Hypermutation induced by APOBEC-1 overexpression can be eliminated. RNA 2010, 16 (5), 1040–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kim J; Malashkevich V; Roday S; Lisbin M; Schramm VL; Almo SC, Structural and kinetic characterization of Escherichia coli TadA, the wobble-specific tRNA deaminase. Biochemistry 2006, 45 (20), 6407–16. [DOI] [PubMed] [Google Scholar]
- 49.Poulsen LK; Larsen NW; Molin S; Andersson P, Analysis of an Escherichia coli mutant strain resistant to the cell-killing function encoded by the gef gene family. Mol Microbiol 1992, 6 (7), 895–905. [DOI] [PubMed] [Google Scholar]
- 50.Xiang S; Short SA; Wolfenden R; Carter CW Jr., The structure of the cytidine deaminase-product complex provides evidence for efficient proton transfer and ground-state destabilization. Biochemistry 1997, 36 (16), 4768–74. [DOI] [PubMed] [Google Scholar]
- 51.Chester A; Somasekaram A; Tzimina M; Jarmuz A; Gisbourne J; O’Keefe R; Scott J; Navaratnam N, The apolipoprotein B mRNA editing complex performs a multifunctional cycle and suppresses nonsense-mediated decay. EMBO J 2003, 22 (15), 3971–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Lellek H; Kirsten R; Diehl I; Apostel F; Buck F; Greeve J, Purification and molecular cloning of a novel essential component of the apolipoprotein B mRNA editing enzyme-complex. J Biol Chem 2000, 275 (26), 19848–56. [DOI] [PubMed] [Google Scholar]
- 53.Mehta A; Kinter MT; Sherman NE; Driscoll DM, Molecular cloning of apobec-1 complementation factor, a novel RNA-binding protein involved in the editing of apolipoprotein B mRNA. Mol Cell Biol 2000, 20 (5), 1846–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Lerner T; Papavasiliou FN; Pecori R, RNA Editors, Cofactors, and mRNA Targets: An Overview of the C-to-U RNA Editing Machinery and Its Implication in Human Disease. Genes (Basel) 2018, 10 (1). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Gehrke JM; Cervantes O; Clement MK; Wu Y; Zeng J; Bauer DE; Pinello L; Joung JK, An APOBEC3A-Cas9 base editor with minimized bystander and off-target activities. Nat Biotechnol 2018, 36 (10), 977–982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Radany EH; Dornfeld KJ; Sanderson RJ; Savage MK; Majumdar A; Seidman MM; Mosbaugh DW, Increased spontaneous mutation frequency in human cells expressing the phage PBS2-encoded inhibitor of uracil-DNA glycosylase. Mutat Res 2000, 461 (1), 41–58. [DOI] [PubMed] [Google Scholar]
- 57.Di Noia J; Neuberger MS, Altering the pathway of immunoglobulin hypermutation by inhibiting uracil-DNA glycosylase. Nature 2002, 419 (6902), 43–8. [DOI] [PubMed] [Google Scholar]
- 58.Burns MB; Lackey L; Carpenter MA; Rathore A; Land AM; Leonard B; Refsland EW; Kotandeniya D; Tretyakova N; Nikas JB; Yee D; Temiz NA; Donohue DE; McDougle RM; Brown WL; Law EK; Harris RS, APOBEC3B is an enzymatic source of mutation in breast cancer. Nature 2013, 494, 366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Okazaki IM; Hiai H; Kakazu N; Yamada S; Muramatsu M; Kinoshita K; Honjo T, Constitutive expression of AID leads to tumorigenesis. J Exp Med 2003, 197 (9), 1173–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Li X; Wang Y; Liu Y; Yang B; Wang X; Wei J; Lu Z; Zhang Y; Wu J; Huang X; Yang L; Chen J, Base editing with a Cpf1–cytidine deaminase fusion. Nature Biotechnology 2018, 36, 324. [DOI] [PubMed] [Google Scholar]
- 61.Merkle T; Merz S; Reautschnig P; Blaha A; Li Q; Vogel P; Wettengel J; Li JB; Stafforst T, Precise RNA editing by recruiting endogenous ADARs with antisense oligonucleotides. Nat Biotechnol 2019, 37 (2), 133–138. [DOI] [PubMed] [Google Scholar]
- 62.Qu L; Yi Z; Zhu S; Wang C; Cao Z; Zhou Z; Yuan P; Yu Y; Tian F; Liu Z; Bao Y; Zhao Y; Wei W, Programmable RNA editing by recruiting endogenous ADAR using engineered RNAs. Nat Biotechnol 2019. [DOI] [PubMed]
- 63.Vallecillo-Viejo IC; Liscovitch-Brauer N; Montiel-Gonzalez MF; Eisenberg E; Rosenthal JJC, Abundant off-target edits from site-directed RNA editing can be reduced by nuclear localization of the editing enzyme. RNA Biol 2018, 15 (1), 104–114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Stafforst T; Schneider MF, An RNA-deaminase conjugate selectively repairs point mutations. Angew Chem Int Ed Engl 2012, 51 (44), 11166–9. [DOI] [PubMed] [Google Scholar]
- 65.Hanswillemenke A; Kuzdere T; Vogel P; Jekely G; Stafforst T, Site-Directed RNA Editing in Vivo Can Be Triggered by the Light-Driven Assembly of an Artificial Riboprotein. J Am Chem Soc 2015, 137 (50), 15875–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Schneider MF; Wettengel J; Hoffmann PC; Stafforst T, Optimal guideRNAs for re-directing deaminase activity of hADAR1 and hADAR2 in trans. Nucleic Acids Res 2014, 42 (10), e87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Vogel P; Hanswillemenke A; Stafforst T, Switching Protein Localization by Site-Directed RNA Editing under Control of Light. ACS Synth Biol 2017, 6 (9), 1642–1649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Vogel P; Moschref M; Li Q; Merkle T; Selvasaravanan KD; Li JB; Stafforst T, Efficient and precise editing of endogenous transcripts with SNAP-tagged ADARs. Nat Methods 2018, 15 (7), 535–538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Montiel-Gonzalez MF; Vallecillo-Viejo I; Yudowski GA; Rosenthal JJ, Correction of mutations within the cystic fibrosis transmembrane conductance regulator by site-directed RNA editing. Proc Natl Acad Sci U S A 2013, 110 (45), 18285–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Montiel-Gonzalez MF; Vallecillo-Viejo IC; Rosenthal JJ, An efficient system for selectively altering genetic information within mRNAs. Nucleic Acids Res 2016, 44 (21), e157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Sinnamon JR; Kim SY; Corson GM; Song Z; Nakai H; Adelman JP; Mandel G, Site-directed RNA repair of endogenous Mecp2 RNA in neurons. Proc Natl Acad Sci U S A 2017, 114 (44), E9395–E9402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Cox DBT; Gootenberg JS; Abudayyeh OO; Franklin B; Kellner MJ; Joung J; Zhang F, RNA editing with CRISPR-Cas13. Science 2017, 358 (6366), 1019–1027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Matthews MM; Thomas JM; Zheng Y; Tran K; Phelps KJ; Scott AI; Havel J; Fisher AJ; Beal PA, Structures of human ADAR2 bound to dsRNA reveal base-flipping mechanism and basis for site selectivity. Nat Struct Mol Biol 2016, 23 (5), 426–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Monteleone LR; Matthews MM; Palumbo CM; Thomas JM; Zheng Y; Chiang Y; Fisher AJ; Beal PA, A Bump-Hole Approach for Directed RNA Editing. Cell Chem Biol 2019, 26 (2), 269–277 e5. [DOI] [PMC free article] [PubMed] [Google Scholar]


