Abstract
Purpose:
Head and Neck Squamous Cell Carcinomas (HNSCC) cause more than 300,000 deaths worldwide each year. Locoregional and distant recurrences represent worse prognostic events and accepted surrogate markers of patients’ overall survival. No valid biomarker and salvage therapy exist to identify and treat patients at high-risk of recurrence. We aimed to verify if selected microRNAs (miRs) could be used as biomarkers of recurrence in HNSCC.
Experimental Design:
A Nanostring array was used to identify miRs associated with locoregional recurrence in 44 HNSCC patients. Bioinformatic approaches validated the signature and identified potential miR targets. Validation experiments were performed using an independent cohort of primary HNSCC samples and a panel of HNSCC cell lines. In vivo experiments validated the in vitro results.
Results:
Our data identified a four-miR signature that classified HNSCC patients at high- or low-risk of recurrence. These miRs collectively impinge on the epithelial-mesenchymal transition process. In silico and wet lab approaches showed that miR-9, expressed at high levels in recurrent HNSCC, targets SASH1 and KRT13, while miR-1, miR-133 and miR-150, expressed at low levels in recurrent HNSCC, collectively target SP1 and TGFβ pathways. A six-genes signature comprising these targets identified patients at high risk of recurrences, as well.
Combined pharmacological inhibition of SP1 and TGFβ pathways induced HNSCC cell death and, when timely administered, prevented recurrence formation in a preclinical model of HNSCC recurrence.
Conclusions:
By integrating different experimental approaches and competences, we identified critical mediators of recurrence formation in HNSCC that may merit to be considered for future clinical development.
Translational Relevance.
Most of HNSCC patients are diagnosed with a locally advanced disease and are treated with the combination of surgery, radiotherapy, and chemotherapy. This highly toxic approach is curative in about half of the cases but recurrent patients do not have effective salvage therapies. Therefore there is the urgency to identify and validate solid biomarkers able to classify patients at high risk that may benefit for specific targeted approaches.
Our work tackled these two unmet clinical needs and identified a microRNA signature of locoregional recurrence in HNSCC patients. Starting from this signature, we identified two druggable pathways (i.e. SP1 and TGFβ) that when timely and concomitantly targeted efficiently prevented recurrence formation in a preclinical model.
Both SP1 and TGFβ inhibitors have been already used to treat human patients, thus our work is of potential immediate translational relevance.
Keywords: Head and neck squamous carcinomas, epithelial to mesenchymal transition, recurrence, SP1, TGFβ, microRNA
Introduction
Head and Neck Squamous Cell Carcinomas (HNSCC) comprehend a relatively common group of neoplasms, with about 550,000 new cases/year worldwide (1). Most patients are diagnosed with a locally advanced potentially curable cancer, but 40–60% of these patients eventually recur (2,3). A recent meta-analysis demonstrated that the combination of chemo- and radio-therapy is a valid, although highly toxic, therapeutic option (4). Despite this aggressive schedule, the 5-year survival of HNSCC patients ranges from 35 to 55% (4).
Local and distant recurrences represent valid surrogate endpoints to estimate the efficacy of radiotherapy and chemotherapy on patients’ survival (5). This observation implies that identifying patients that will recur could be extremely beneficial for the management of HNSCC patients, to avoid unnecessary toxicity and improve patients’ survival. To date no validated biomarkers exist to identify HNSCC patients with higher probability to develop recurrences and that, therefore, may merit a closer follow-up or different therapeutic approaches (6).
HNSCC progression is a stepwise process, resulting from the accumulation of molecular alterations in the squamous epithelium, which eventually drive the progression from premalignant lesions to invasive disease (3). While inactivation of p53 and RB pathways are considered an early, nearly universal event in HNSCC progression, either through somatic mutation/inactivation of critical tumor suppressor genes (e.g. TP53 and CDKN1A) or through HPV infection (3,6–8), less is known about the subsequent molecular events necessary for the progression of HNSCC to invasive, metastatic carcinomas. Among others, it has been hypothesized that the regulation of cancer cell plasticity through reversible reprogramming of Epithelial to Mesenchymal Transition (EMT) and Mesenchymal to Epithelial Transition (MET) could play a primary role (3,9–12).
Accumulating evidences suggest that EMT favors the distant dissemination of single carcinoma cells from the site of the primary tumor (10,13) and, more recently, EMT has been proposed as an escape mechanism mediating drug resistance (13). It is therefore conceivable that EMT could play a pivotal role in recurrence formation in HNSCC, either by stimulating local and distant cancer cell spreading or by inducing chemoresistance.
Converging evidences suggest that cancer cell plasticity is regulated epigenetically (11). In this context, microRNAs (miRs) could play a primary role also in HNSCC (12,14). After the seminal demonstration that a reciprocal feedback loop exists between the miR-200 family and the ZEB transcription factors to tightly control EMT (15,16), the number of miRs that has been directly or indirectly associated with EMT is becoming an extensive list.
The role of miR expression in HNSCC has been widely investigated and several differentially expressed miRs in normal/peritumoral mucosa versus primary tumors have been identified (12,17,18). However, no study has specifically compared the expression profile of miRs in primary tumors from patients who have recurred versus patients that have not.
Here, we report the identification of a 4-miRs signature able to identify HNSCC patients at high risk of recurrence and describe the mechanism whereby they orchestrate the expression of genes regulating cancer cell plasticity via EMT modulation.
Materials and Methods
Patient samples
Specimens from primary HNSCC were collected from patients who underwent surgery at our institution and at Santa Maria degli Angeli Hospital, Pordenone, Italy. HNSCC specimens were immediately frozen and stored at −80°C. The study was approved by the Internal Review Board of the Centro di Riferimento Oncologico (CRO) of Aviano (#IRB-08/2013) and all patients provided written informed consent.
Bioinformatic analyses
Computational analysis.
Uni-variant Significance Test by The Permutation Test (19) was used to calculate the statistical significance of each of the four miRs individually. Testing of Sample Classification includes building and testing a computational prediction model to predict recurrence based on the miRs’ expressions using the Weka software (20). We adopt the under-sampling technique described in (21), to counter the effects of class imbalance and the potential of over-fitting due to a limited and small minority class data (only 11 recurrent samples). Both methods are further described in Supplementary Material and Methods.
Network analysis.
After downloading predicted miR–gene interactions for the genes in our network from the mirDIP portal ver. 1 (http://ophid.utoronto.ca/mirDIP) (22), which integrates 12 microRNA prediction datasets, we kept only those interactions that were identified in at least three independent datasets. We then integrated the analysis using genes up- or down-regulated in head neck recurrences from CDIP, the Cancer Data Integration Portal ver. 1 (http://ophid.utoronto.ca/cdip), a collection of gene expression data from published studies. We also used a list of genes associated with recurrence formation in HNSCC from a published cohort (23).
Next, we uploaded this list of gene IDs into NAViGaTOR 3 as our network visualization tool (http://ophid.utoronto.ca/navigator (24) and retrieved known, publicly available human physical protein interaction using the I2D 2.2 portal (http://ophid.utoronto.ca/i2d (25). Our goal was to explore the relationships between our 4 miRs and genes known to regulate recurrence formation in HNSCC. Network nodes represent miRs and proteins respectively, while edges represent physical protein-protein interactions (PPIs) and microRNA:gene regulation. We downloaded the list of the 4 miRs targets from the mirDIP database ver. 1 (http://ophid.utoronto.ca/mirDIP/) and then we looked at the intersection among genes that control recurrence formation (23), genes associated to a pro-survival signature (23) and the 4 miRs targets.
PathDIP analyses.
We analyzed 56 genes, involved in recurrence formation (23), as potential miR-9 targets. These pathway analyses were conducted using pathDIP Ver. 2.4.3.12 (http://ophid.utoronto.ca/pathDIP; (26).
Analysis of the TCGA dataset.
Correlation analysis using HNSCC TCGA RNAseq and microRNA-seq data for SASH1, KRT13 and hsa-miR-9 were performed using Spearman correlation (27). All statistical analyses were performed using [R] (https://www.r-project.org/). KRT13/SASH1 correlation analysis using HNSCC TCGA RNAseq data were performed using cBioPortal for Cancer Genomics (http://www.cbioportal.org/) (28).
Clustering of RNAseq values was performed using Ward linkage. This approach identified cluster 1 with high expression of KRT13/SASH1 and low expression of TGFβR1/2, SMAD3 and SP1, while cluster 2 is characterized by high expression of TGFβR1/2, SMAD3, SP1 and low expression of SASH1/KRT13. The associations of the 2 clusters with survival was evaluated with the log-rank test using the survival package in R. HNSCC TCGA patients were stratified either by high mRNA expression of KRT13/SASH1 and low SMAD3 (above median of mRNA expression, below median of mRNA expression, respectively) or by high mRNA expression of SMAD3 and low mRNA expression of KRT13/SASH1. Cox proportional hazards regression analysis based on the KMsurv package in R was used to assess the hazard ratios. All HNSCC TCGA data were downloaded using the TCGA data portal (https://tcga-data.nci.nih.gov/tcga/) and the cBioPortal for Cancer Genomics (http://www.cbioportal.org/)
To validate prognostic properties of the four-miRNA signature, we used SurvMicro v.0.9 (http://bioinformatica.mty.itesm.mx:8080/Biomatec/Survmicro.jsp) (29). Signature was validated on TCGA LUAD Ilumina HiSeq dataset, comprising 311 patient samples. All settings were used as default.
Wet lab analyses
All wet lab analyses were performed according to procedures commonly used in our lab (30,31) and are described in detail in the Supplementary Material and Methods.
Molecular Biology Experiments.
microRNA expression profile was performed using the Nanostring™ technology (NanoString nCounter Human miRNA assay (v1.1) that allowed to evaluate the expression of 746 miRs (664 Human 82 Viral) along with the one of housekeeping genes (GeneBank GSE89000). Nanostring technology, DNA and RNA extraction, quantification and analysis, evaluation of TP53 and HPV status, protein extraction and western blotting are described in the Supplementary Material and Methods.
Cell culture, transfection and transduction.
All cell lines were authenticated by BMR Genomics srl Padova, Italia according to Cell ID ™ System (Promega) protocol and using Genemapper ID Ver 3.2.1, to identify DNA STR profiles. UMSCC74b and UMSCC1 cells were kindly provided by Dr. Thomas Carey (University of Michigan, Ann Arbor, MI). All other head and neck squamous cell lines were obtained from ATCC (LGC Standards).
Xenograft growth in mouse flanks and treatment.
Animal experimentation was approved by local ethic committee for animal welfare (OPBA) and experiments performed according to committee’s guidelines. Athymic nude mice (Harlan, Foxn1nu, females, 6 weeks old) were injected with 2×106 FaDu cells bilaterally in the flanks. The evaluation of local relapse and the administration on Mythramicin A and SB-525334 are provided in the Supplementary Material and Methods.
Statistical analyses
All graphs and statistical analyses were performed using PRISM (version 6, GraphPad, Inc.) and R, SAS Software 9.2 and R for statistical analyses. In all experiments, differences were considered significant when p was <0.05. Statistical analyses included paired and un-paired t-tests, Mann-Whitney un-paired t-test and Spearman correlation test, used as appropriate and as specified in each figure. Differences in miR expression between patients’ groups were evaluated by non-parametric Wilcoxon test (two groups) or Kruskal-Wallis test (three groups). Correlation between Array and PCR quantification of miRs was evaluated through Spearman correlation coefficient.
Results
Identification of miRs differentially expressed in recurrent (R) and non-recurrent (NR) HNSCC.
Primary HNSCC fresh frozen surgical samples collected from 44 patients (Table 1) who experienced (n=11) or not (n=33) local recurrence within 2 years from the first surgery were analyzed by Nanostring™ technology for the expression of 746 human and 82 viral miRs. Statistical analyses of normalized miR expression demonstrated that 7 miRs were significantly different between the two groups (data not shown). Validation of these data by qRT-PCR analyses confirmed significant differences for miR-1, miR-133a, miR-150 and miR-9 between recurrent and non-recurrent HNSCC (data not shown). In particular, univariant significance testing confirmed that miR-9 was upregulated, while miR-1, miR-133a and miR-150 were downregulated in tumors from recurrent patients (Supplementary Fig. S1 and data not shown). No significant associations were found between expression of these miRs and other clinical and biological variables of the tumors, including the presence of TP53 mutation and the positivity for HPV infection (Table 1 and Supplementary Table S1). Accordingly, miR-9 upregulation and miR-1, miR-133a and miR-150 downregulation in recurrent tumors was also confirmed when only TP53 mutant or HPV-negative cases were considered (Supplementary Fig. S1). Bioinformatic validation by data reiteration confirmed the significance of this interaction (Fig. 1A and B and Supplementary Fig. S2). Using the classifier testing, we calculated the Area Under the Curve (AUC) to estimate the ability of the four miRs, each one alone or in combination, to predict recurrence. Models 1–4, built using a Naïve Bayes or a Logistic Regression models, in which the classifier combines miR-133a with miR-150, with or without miR-9, achieved a high AUC (80–81%), high sensitivity (82–88%) and low false positive rates (29–35%), which translated into 65–71% specificity (Fig. 1A and B and Supplementary Fig. S2). Our analyses also suggested that the addition of miR-1 does little to improve the classification accuracy (Supplementary Fig. S2C).
Table 1.
Distribution of relapsed/not relapsed patients and mean level of miR expression according to selected covariates.
| Relapsed | Not relapsed | miR-9 | miR-1 | miR-133a | miR-150 | |||
|---|---|---|---|---|---|---|---|---|
| n | (%) | n | (%) | |||||
| Sex | ||||||||
| Men | 11 | (100.0) | 24 | (72.7) | 319.0 | 2193.8 | 396.7 | 537.7 |
| Women | 0 | (0.0) | 9 | (27.3) | 180.9 | 186.5 | 132.0 | 1752.8 |
| P=0.0849 | P=0.6812 | P=0.4459 | P=0.9266 | P=0.0358 | ||||
| Age | ||||||||
| <60 years | 8 | (72.7) | 15 | (46.9) | 310.8 | 512.3 | 86.2 | 542.4 |
| ≥60 years | 3 | (27.3) | 17 | (53.1) | 288.2 | 3777.7 | 721.0 | 896.1 |
| P=0.1753 | P=0.5217 | P=0.6463 | P=0.6575 | P=0.4025 | ||||
| Cancer site | ||||||||
| Tongue Oral cavity |
5 | (45.5) | 23 | (69.7) | 149.9 | 724.8 | 110.0 | 684.6 |
| Oropharynx Hypopharynx |
5 | (45.5) | 6 | (18.2) | 329.9 | 3425.2 | 662.3 | 499.5 |
| Larynx | 1 | (9.0) | 4 | (12.1) | 648.3 | 78.7 | 18.4 | 1452.2 |
| P=0.2104 | P=0.3178 | P=0.3296 | P=0.5829 | P=0.2347 | ||||
| cT | ||||||||
| T1-T2 | 3 | (27.3) | 21 | (65.6) | 209.0 | 941.5 | 169.3 | 584.8 |
| T3-T4 | 8 | (72.7) | 11 | (34.7) | 341.2 | 2365.4 | 446.6 | 744.9 |
| P=0.0383 | P=0.9737 | P=0.0409 | P=0.1021 | P=0.9211 | ||||
| cN | ||||||||
| 0 | 3 | (27.3) | 13 | (40.6) | 321.0 | 5360.7 | 1024.5 | 626.5 |
| 1–2 | 8 | (72.7) | 19 | (59.4) | 292.2 | 432.0 | 72.5 | 726.6 |
| P=0.4942 | P=0.7667 | P=0.4503 | P=0.4409 | P=0.7168 | ||||
| Adjuvant radio/chemotherapy | ||||||||
| No | 6 | (54.6) | 26 | (78.8) | 160.0 | 638.9 | 126.2 | 489.1 |
| Yes | 5 | (45.4) | 7 | (21.2) | 391.6 | 2763.3 | 513.9 | 829.3 |
| P=0.1387 | P=0.3709 | P=0.9752 | P=0.4672 | P=0.4043 | ||||
cT = Clinical Evaluation of Tumor size
cN = Clinical Evaluation of Node status
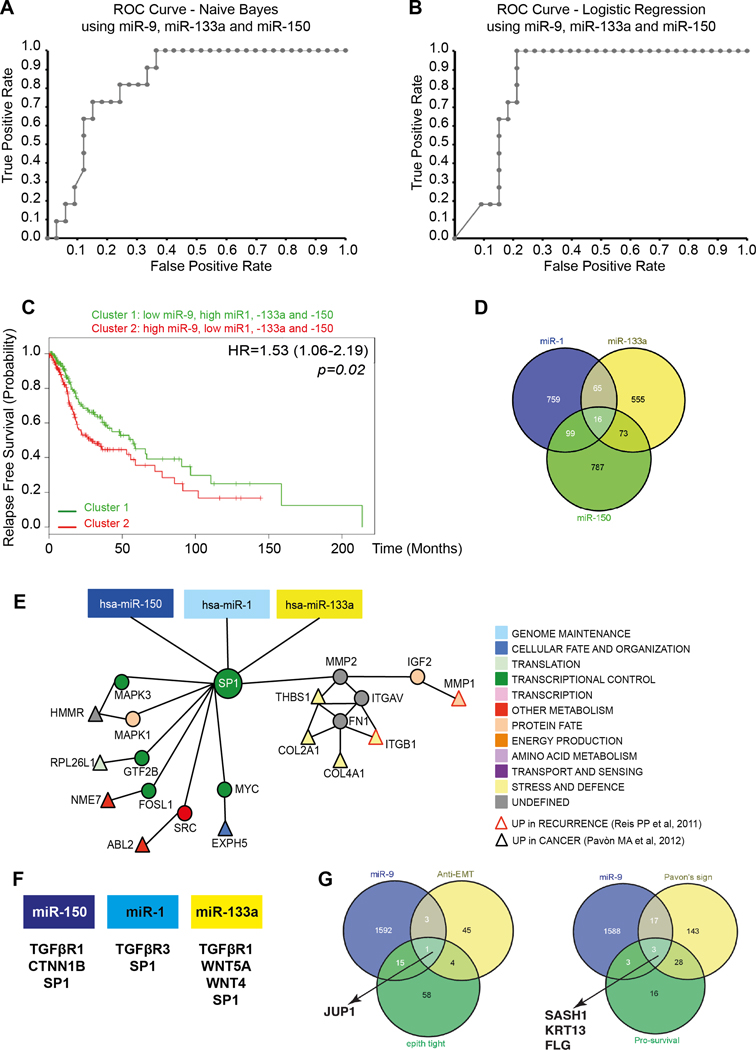
FIGURE 1. Identification of miR-1, −133a, −150 and −9 potential targets in HNSCC.
A/B. ROC curve predicting recurrence formation, using miR-9, 133a and −150 data iteration, applying the Naïve Bayes (A) or the Logistic Regression (B) models. The AUC is 81.3% (Sensitivity 87% and Specificity 75%) in A and 80.3% (Sensitivity 82% and Specificity 71%) in B.
C. Kaplan-Mayer curve evaluating progression free survival of HNSCC patients clustered based on the expression of miR-1, −9, −133a and 150.
D. Venn Diagram showing the number of miR-1, −133a and −150 common potential targets, among the EMT genes, altered in HNSCC.
E. Visualization of miR-1, 133a and −150 and SP1 network in HNSCC. Direct interactions are shown by edges. Red-border triangles identify genes upregulated in recurrent HNSCC and black-border triangles identify genes upregulated in HNSCC primary tumors
F. List of miR-1, 133a and −150 targets belonging to the TGFβ and WNT pathways.
G. Venn Diagram showing the number of miR-9 potential targets, among the anti-EMT genes and among the genes altered in HNSCC.
These computational analyses are in accord with the notion that miR-1 and miR-133a belongs to the same cluster (32) and, consequently, their expression highly correlates, as we observed in our samples set (Spearman correlation value r=0.9621 and p<0.0001). Correlation analyses also indicated that miR-133a expression directly correlates with miR-150 (Spearman correlation value r=0.2984 and p=0.049) and that miR-9 expression inversely correlates with both miR-133a (Spearman correlation value r=−0.1715, p=n.s.) and miR-150 (Spearman correlation value r=−0.4757 and p=0.0011).
Accordingly, using the HNSCC TCGA dataset (27) we confirmed that i) miR-1 and 133a expression strongly correlate (R= 0.79; p<0.0001); ii) miR-9 inversely correlates with both miR-1 (R= −0.27; p<0.0001) and miR-133a (R= −0.34; p<0.0001) and iii) the expression of miR-1, −133a, −150 and −9 classify patients at high risk of relapse (Fig. 1C).
Bioinformatic analyses identified miR targets involved in the regulation of cell plasticity.
Confirmed targets of the identified 4-miRs impinge on EMT process. Specifically, miR-9, promotes EMT by targeting E-Cadherin (33) and miR-1, −133 and −150, act as EMT suppressors by targeting SLUG (34), SNAIL (35) and ZEB1 (36), respectively.
Since EMT plays a pivotal role in HNSCC progression and recurrence formation (3,12), we applied the mirDIP (22) and NAVIGaTOR (24) bioinformatic tools to integrate miR-target predictions with experimentally determined proteins-protein interactions (PPIs) from I2D as described (25,37), focusing on genes regulating EMT.
We first identified 16 possible common targets of miR-1, −133a and 150 (Fig. 1D). Then, we built an interaction network by integrating these analyses with the genes upregulated in HNSCC, as determined by the TCGA consortium. These analyses led us to the identification of SP1 as a common target of these 3 miRs (data not shown). Refinement of these results, using two other available HNSCC datasets of coding gene signatures that could predict recurrences in HNSCC (23,38), also identified SP1 (Fig. 1E) together with other genes belonging to the TGFβ and β-Catenin pathways, as potential targets of miR-1,−133a and −150 (Fig. 1F).
A similar approach was used to screen the 1611 potential miR-9 targets. By intersecting these targets with genes associated with EMT negative regulators and HNSCC recurrence formation and/or patients’ survival, we defined a complex interaction network that highlighted potential targets altered in HNSCC (data not shown). Further refinement of these analyses led to the identification of Plakoglobin (JUP1), SASH1, Keratin 13 (KRT13) and Filaggrin (FLG), as the genes with the highest probability to represent miR-9 targets in recurrent HNSCC (Fig. 1G).
Pathway enrichment analyses using pathDIP (26), to integrate miR-1, −133a, and −150 with miR-9 networks, identified significantly enriched pathways (Supplementary Table S2). The most frequently occurring genes belonged to Signal Transduction (30 genes; p<0.05), EGFR1 (29 genes; p=0.01), Immune System (29 genes; p<0.02), Integrin α6β4 (26 genes; p=0.001), TNFα (23 genes; p=0.05), Developmental Biology (22 genes; p=0.02), and TGFβ Signaling (22 genes; p=0.04) pathways. Interestingly, four prognostic genes from the work of Reiss et al., (38) are predicted to be regulated by miR-9–5p and miR-150–5p (using an updated mirDIP 3.0.1; http://ophid.utoronto.ca/mirDIP) (22)
Experimental validation of identified miRs targets involved in the regulation of EMT.
Overall these analyses suggested that the relative expression of miR-1, −133, −150 and miR-9 could play a functional role in HNSCC progression possibly by regulating EMT.
To experimentally validate miRs targets identified in silico, we screened a panel of HNSCC-derived cell lines for endogenous miRs levels to choose the most appropriate in vitro model. Generally, all cell lines expressed low levels of miR-1, −133a and −150 and higher levels of miR-9 (Supplementary Fig. S3A).
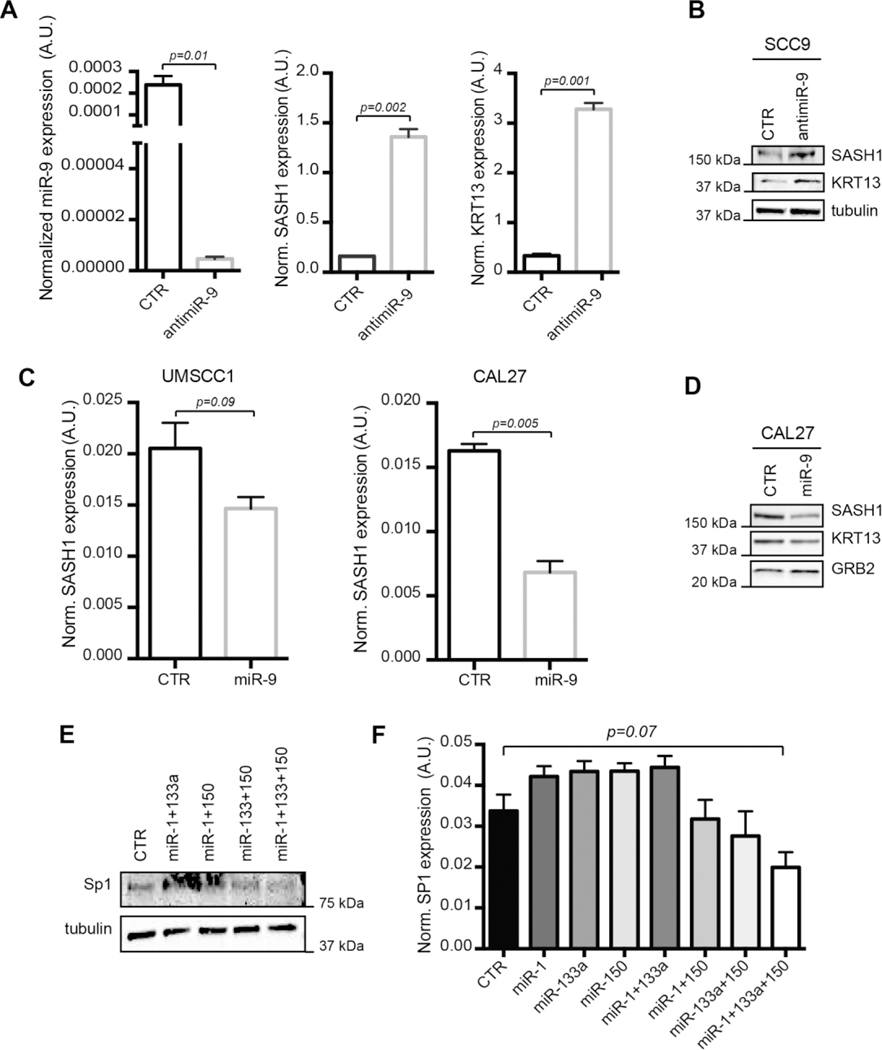
First we focused on the four most probable miR-9 targets, namely JUP1, SASH1, KRT13 and FLG (Fig. 1G). FaDu and SCC9 cells expressed highest level of miR-9 and low levels of SASH1, KRT13 and of the epithelial marker E-Cadherin (Supplementary Fig. S3A and B). In both cell lines, miR-9 knock-down resulted in the upregulation of SASH1 and KRT13, but not of JUP1, and FLG mRNAs (Fig. 2A and Supplementary Fig. S3C and D). The increased SASH1 and KRT13 expression was also confirmed at protein level (Fig. 2B). In CAL27 and UMSCC1 cells, expressing low endogenous miR-9, its overexpression decreased mRNA and protein expression of SASH1 and KRT13 (Fig. 2C and D). As a proof of principle, we also tested whether miR-9 could directly regulate SASH1 expression acting on its 3’-UTR, that contains three seed sites for miR-9 (Supplementary Fig. S4A). Luciferase assay in FaDu cells demonstrated that miR-9 knockdown significantly increased the luciferase activity when the first seed site (position 217–223) was tested (Supplementary Fig. S4B). Overall, these data suggest that SASH1 (and likely KRT13) could represent reliable markers of miR-9 activity in HNSCC cells.
FIGURE 2. Validation of SASH1 and KRT13 as miR-9 targets and of SP1 as common target of miR-1, −133a and −150, in HNSCC cells.
A. Graphs showing (from left to right) the normalized expression of miR-9, SASH1 and KRT13 RNA in control and miR-9 knock-down SCC9 cells. Data report the median value (±S.D.) of 3 independent experiments performed in duplicate.
B. Western blot analysis of SASH1 and KRT13 protein expressions in control and miR-9 knock-down SCC9 cells. Tubulin was used as loading control.
C. Graphs showing the normalized expression of SASH1 mRNA expressions in control and miR-9 overexpressing UMSCC1 (left) and CAL27 (right) cells. Graphs report the median value (±S.D.) of 3 independent experiments performed in duplicate.
D. Western blot analysis of SASH1 and KRT13 protein expressions in control and miR-9 overexpressing CAL27 cells. GRB2 was used as loading control.
E Western blot analysis of SP1 protein expression in UMSCC74b cells expressing miR-1, −133a and −150 alone or in combination, as indicated. Tubulin was used as loading control.
F. Graph showing the normalized expression of SP1 mRNA in UMSCC74b cells transfected as in (E). Data report the median value (±S.D.) of 3 independent experiments performed in duplicate. Statistical significance was calculated using unpaired two-tailed Student’s t-test.
We next experimentally validated SP1 as common target of miR-1, miR-133a and miR-150. Overexpression of miR-1, miR-133a or miR-150 alone did not affect SP1 mRNA or protein expression in different HNSCC cell lines. However, the combined overexpression of two and, even better, of the three miRs together significantly reduced SP1, at both protein and mRNA levels (Fig. 2E and F and Supplementary Fig. S5A and B). Expression data paralleled luciferase assay on SP1 3’-UTR, that contains one seed site for miR-1 and miR-133a and two seed sites for miR-150 (Supplementary Fig. S4C). Each miR alone slightly reduced the luciferase activity of each construct (Supplementary Fig. S4D), further supporting the possibility that the three miRs need to work together to reduce SP1 expression.
Members of TGFβ and β-Catenin pathways were also predicted targets of miR-1, miR-133a and miR-150. Among the predicted targets of miR-1, −133a and −150 (i.e. TGFβ-R1, -R2 and -R3, WNT4, WNT5A and CTNN1B), the expression of WNT5A strongly decreased in cells overexpressing miR-133a (Supplementary Fig. S5C) while the expression of TGFβ-R3 decreased following the combined overexpression of the three miRs (Supplementary Fig. S5D).
From a functional point of view, we observed that combined inhibition of miR-9 and overexpression of miR-1, −133a and −150 (Supplementary Fig. S5E) up-regulated the epithelial marker E-Cadherin in FaDu cells (Supplementary Fig. S5F), supporting the hypothesis that balanced expression of miR-9 and miR-1, miR-133a and miR-150 plays a functional role in HNSCC recurrence formation by modulating the EMT process.
In vivo validation of identified miRs targets involved in the regulation of EMT.
To verify if the predicted targets of the 4 miRs were effectively differentially regulated in HNSCC samples, we tested by qRT-PCR the expression SP1, TGFβ-R1, R2 and R3, WNT4 and WNT5A, CTNN1B, SASH1 and KRT13 in the 44 samples described in Table S1 and analyzed for the expression of miRs.
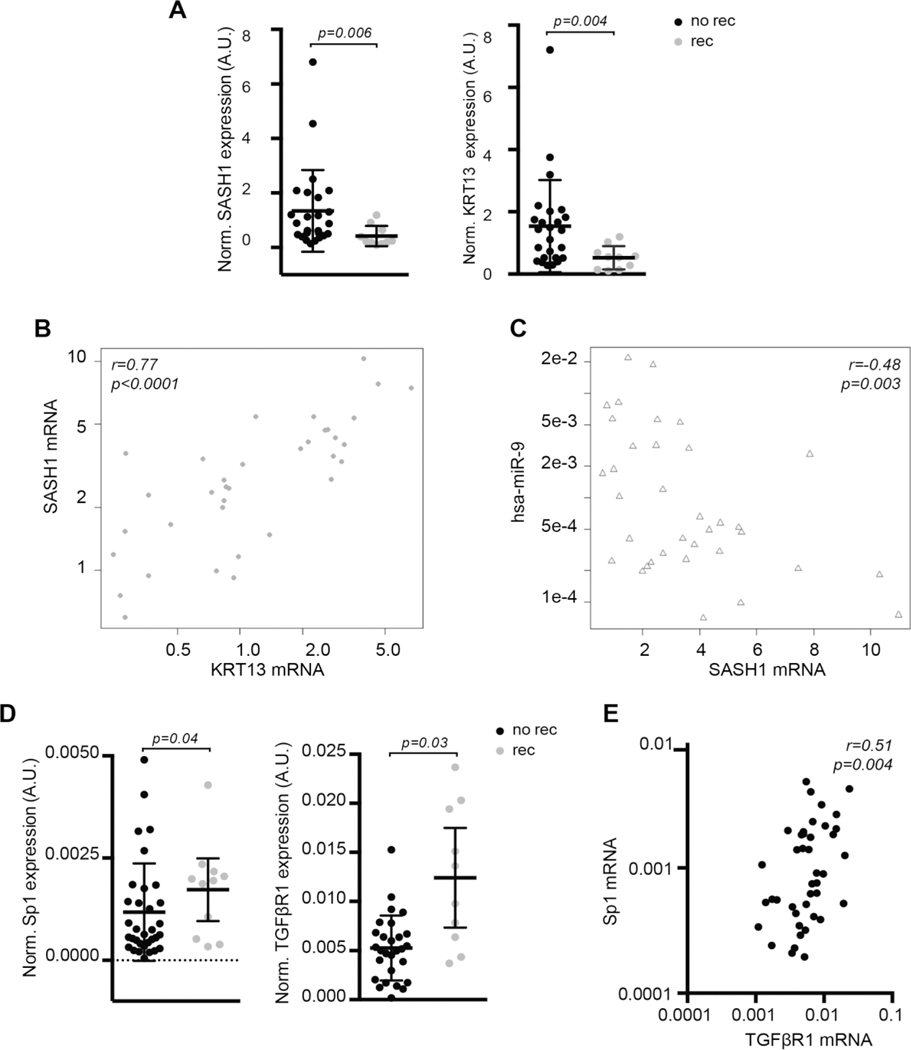
In line with in vitro results, we observed that miR-9 targets, SASH1 and KRT13, were both significantly downregulated in patients who experienced recurrence (Fig. 3A) and the expression of SASH1 correlated directly with KRT13 and inversely with miR-9 in primary HNSCC (Fig. 3B–C). Among miR-1, −133a and −150 targets, only SP1 and TGFβ-R1 were significantly upregulated in HNSCC samples from recurrent patients (Fig. 3D) and their expression directly correlated (Fig. 3E). No differential expression was observed for TGFβ-R2, and R3, WNT4, WNT5A and CTNN1B between patients with or without recurrence (data not shown).
FIGURE 3. Expression of identified miRs targets in primary HNSCC from recurrent and non-recurrent patients.
A. Dot plots showing the normalized expression of SASH1 (left) and KRT13 (right) mRNA, in samples from primary HNSCC from recurrent and non-recurrent patients. Bars indicate the mean and the 95% confidence interval. Statistical significance was calculated using the Mann-Whitney test.
B/C. Dot plot showing the correlation of SASH1 and KRT13 (B) or SASH1 and miR-9 (C) mRNA expression in HNSCC samples described in (A). Correlation value (r) and statistical significance (p) were calculated using the Spearman correlation test.
D. Dot plots showing the normalized expression of SP1 (left) and TGFβ-R1 (right) mRNA expressions in samples from primary and recurrent HNSCC samples. Bars indicate the mean and the 95% confidence interval. Statistical significance reported in the graph was calculated using the Mann-Whitney test.
E Dot plot showing the correlation of SP1 and TGFβ-R1 mRNA expression in HNSCC samples as in A. Correlation value (r) and statistical significance (p) were calculated using the Spearman correlation test.
To validate our findings in an independent cohort, we collected 78 HNSCC samples in our Institutes (Supplementary Table S3) and evaluated the expression of the 4 miRs, SASH1, KRT13, SP1 and TGFβR1. In accord with previous results, miRs expression did not correlate with any biological variable of HNSCC patients, including sex, age, cancer site, T and N stages (Supplementary Table S3).
The expression of SASH1 inversely correlated with miR-9 and directly correlated with KRT13 (Supplementary Fig. S6A and B). An inverse, although not significant, correlation was noticeable between miR-1 or miR-133a and SP1 expression, as expected from the in vitro results (Supplementary Fig. S6C and D).
MiRs targets predict prognosis in the TCGA HNSCC dataset.
We hypothesized that the expression of SASH1, KRT13, SP1 and members of TGFβ pathway could be used in HNSCC as readout of miR activity and tested the expression and the correlation of these genes with miR-9 and miR-1, −133a and −150, in the TCGA dataset.
In accord with the data obtained using our discovery- and validation-cohorts, correlation analyses confirmed that SP1 positively correlated with miR-9 and inversely with miR-133a (Supplementary Fig. S7A and B). Similar results were observed for SMAD2 and SMAD3 expression, used as readouts of SP1 and TGFβ pathway activity (Supplementary Fig. S7C-H). Moreover, SASH1 expression directly correlated with KRT13 (Supplementary Fig. S7I) and both were higher in tumor-free cohort (Supplementary Fig. S7L).
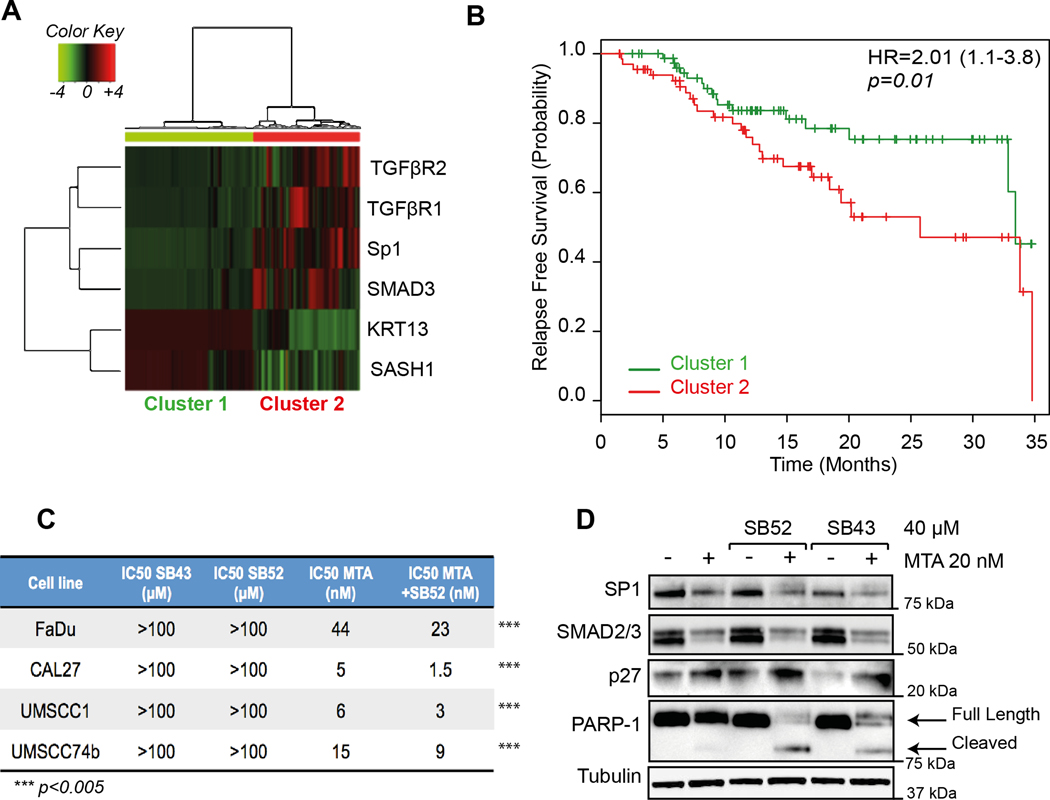
Cluster analyses, using the expression of SASH1, KRT13, SP1, TGFβR1, TGFβR2, SMAD2 and SMAD3, divided the HNSCC patients included in the TCGA dataset in two groups: one with low expression of SP1 and members of TGFβ pathway and high expression of KRT13 and SASH1 (Cluster 1) and the other with the opposite gene expression profile (Cluster 2) (Fig. 4A). This clustering had prediction power, since patients included in Cluster 2 displayed worse relapse free survival than patients included in Cluster 1 (HR=2.01; 95% CI 1.1–3.8; p=0.01; Fig. 4B).
FIGURE 4. Expression of miRs targets predict prognosis of HNSCC patients in the TCGA dataset.
A. Cluster analyses of TGFβR1, TGFβR2, SP1, SMAD3, KRT13 and SASH1 in HNSCC samples included in the TCGA dataset.
B. Kaplan-Mayer curve evaluating progression free survival of HNSCC patients divided based on the cluster analysis shown in (A). Hazard ratio (HR), Confidence Interval (between brackets) and significance (p) were evaluated with the log-rank test using the survival package in R.
C. Table reports the IC50 values of SB431542 (SB43), SB525334 (SB52) and Mitramycin A (MTA) alone or in combination with SB52 (40 µM) in the indicated HNSCC cell lines. Significant difference (*** = p<0.0001) between the IC50 of MTA, in the presence or not of SB52, was calculated with unpaired two-tailed Student’s t-test.
D. Western blot analysis evaluating the expression of SP1, SMAD2/3 (readouts of MTA activity), p27 (readout of SB52/SB43 activity) and PARP-1 (cleaved form used as marker of apoptosis) in FaDu cells, treated for 24 hours with 20 nM MTA, 40μM SB52 or SB43 or their combination, as indicated. Tubulin was used as loading control.
Peri-surgical treatment with SP1 and TGFβR1 inhibitors prevents local relapse in a xenograft model of HNSCC.
The above data suggested that SP1 and TGFβ pathway could act together in the establishment of recurrence in HNSCC patients. To evaluate this possibility, we first tested the in vitro efficacy of Mithramycin A (MTA), a validated SP1 inhibitor, and of two different TGFβR1 inhibitors (SB525334, SB52 and SB431542, SB43) on HNSCC cell survival. While MTA was highly active in decreasing HNSCC cell survival in the nanomolar range, both SB52 and SB43 did not significantly affect cell survival, when used up to 100 μM (Fig. 4C). Yet, in all tested cell lines SB52, used at the ineffective dose of 40 μM, reduced by the half the IC50 of MTA (Fig. 4C), suggesting that they could have synergistic effects. Accordingly, MTA used at the concentration of 20nM effectively induced PARP1 cleavage (marker of apoptosis) only in combination with SB52 or SB43 (40µM) in FaDu cells (Fig. 4D).
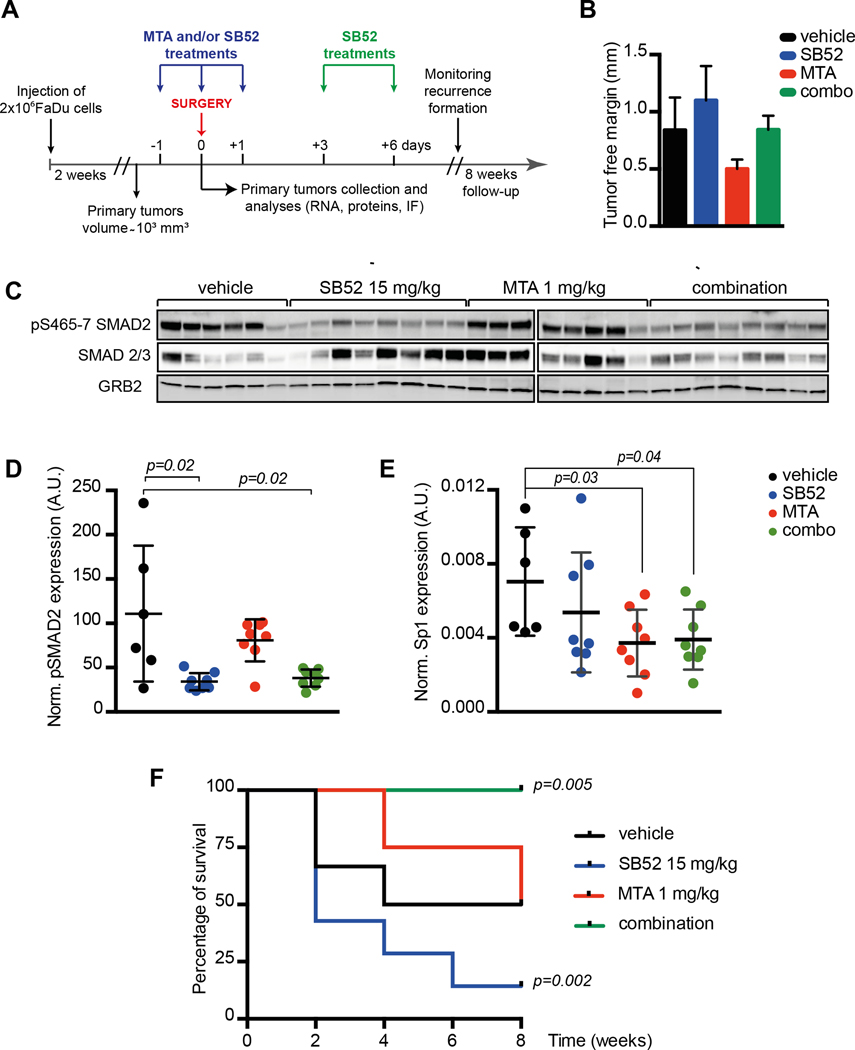
Based on these results we tested if combined pharmacological inhibition of SP1 and TGFβ pathways could restrain the formation of local recurrences in vivo, using a model of HNSCC recurrences and a peri-surgical treatment schedule (Fig. 5A). Mice were subcutaneously injected with FaDu cells and tumor mass allowed to grow up to ~1cm3. Mice were then randomly divided in four groups: 1) sham-treated; 2) treated with MTA 1 mg/kg; 3) SB52 15 mg/kg; 4) the combination of MTA and SB52. All groups were treated for 3 consecutive days (day − 1, day 0 and day +1, with respect to surgery) and two more doses (day +3 and +6) only for SB52 treatment. Tumors were surgically removed at day 0 and appearance of local recurrence was monitored over the subsequent 8 weeks of follow-up (Fig. 5A). Pathological analyses of explanted tumors, evaluating the presence and the width of resection margins, demonstrated that no significant differences existed among the four groups in the extent of radical surgery (Fig. 5B)
FIGURE 5. In vivo inhibition of SP1 and TGFβ pathway effectively reduces recurrence formation in mice.
A. Schematic representation of the experimental workflow used for the evaluation of recurrence formation in mice xenografted with HNSCC cells. Tumor bearing mice received the treatments the day before surgery (day −1), the day of surgery (day 0) and the days +1, +3 and +6 after surgery, as indicated. Mice have been then followed up to 8 weeks to monitor recurrence formation. Control mice were treated with the vehicle alone.
B The graph reports the evaluation of surgical resection margins in explanted tumors, measured with an optical microscope equipped with an ocular micrometer. Tumor free margins varied from 0.3 to 1.6 mm. No significant differences were observed among the four groups of treatment.
C Western blot analysis of tumors explanted from mice treated as described in (A) showing the expression and inhibition of pSMAD2 (S465/7) and SMAD2/3. GRB2 was used as loading control.
D/E Dot plot reporting the normalized expression of pSMAD2 (S465/7) (D) or SP1 mRNA (E) in tumors explanted at day 0 from mice treated as described in (A). Each dots in the graph represents a primary tumor that received the one dose of vehicle (black) MTA 1mg/kg (red), SB52 15mg/kg (blue) or their combination of MTA + SB52 (green).
F Disease-free survival of mice subjected to surgery to remove primary tumors (1000–1200 mm3) and then treated with vehicle (black line), MTA at 1mg/kg (red line), SB52 15mg/kg (blue line) or combination of MTA + SB52 (green line). Kaplan–Meier test has been used to calculate the significance of combo-treatment respect to control and to SB52-treated mice.
Explanted tumors (6 from controls and 8/group of treatment, respectively) were analyzed for the expression of phosphorylated SMAD2 (pSMAD2) (Fig. 5C and D) and SP1 levels (Fig. 5E), as readouts of SB52 and MTA activity, respectively. A single administration of SB52 and/or MTA (administered the day prior to surgery) significantly inhibited SMAD2 phosphorylation and SP1 expression in the tumors (Fig. 5C–E), confirming the in vivo efficacy of these drugs.
Control mice developed local recurrence in 50% of injected sites and MTA used alone did not significantly alter the rate of recurrence formation (4/8, 50%) (Fig. 5F). Interestingly, treatment with TGFβR1 inhibitor, SB52, as single agent considerably increased local recurrence formation, with recurrence occurring in almost all cases (7/8, 90%) (Fig. 5F). However, the combination of SB52 + MTA completely prevented recurrence formation (0/8, 0%) (p=0.005 in log-rank test; HR = 10.01; by comparing control- and MTA+SB treated-mice) (Fig. 5F), suggesting that peri-surgical treatment with MTA and SB52 was sufficient to efficiently suppress HNSCC recurrence in vivo.
Discussion
Here, we show that four miRs can identify in HNSCC patients those at high risk of developing recurrence. Using multiple approaches, we have linked the activity of these miRs to the regulation of EMT, a process considered an critical step in the progression of HNSCC from a pre-invasive to a frankly invasive stage (3). We identified two principal pathways regulated by these miRs that likely play a role in the regulation of HNSCC recurrence via the modulation of EMT, the SP1 and the TGFβ pathways. On one side, miR-1, miR-133a and miR-150 acted together to regulate SP1 and, possibly, TGFβ pathway members, two pathways largely involved in cancer progression and EMT (39,40). On the other, miR-9 acted by lowering the expression of SASH1 and KRT13, two known tumor suppressors with potential anti-EMT roles in HNSCC progression (41–43). Using two independent HNSCC samples cohorts and the TCGA datasets, we confirmed the inverse correlation between the identified miRs and their targets’ expression. This finding supports the possibility that the activity of miR-1, miR-9, miR-133a and miR-150 could be estimated in primary HNSCC by evaluating the expression of SP1, members of the TGFβ pathway, SASH1 and KRT13 (our six-gene signature) and that this balanced activity, more than the sole miRs expression, could be used as a marker for identifying patients at high risk of developing recurrence.
From a biological point of view, it is interesting to note that we describe here two new targets of miR-9, SASH1 and KRT13, both already linked to the regulation of cell motility and invasion, (41,44). Although the aim of this study was limited to the assessment of their role as potential readouts of high miR-9 activity in vivo, it will be worth testing if SASH1 and KRT13 may partially mediate the miR-9-induced local relapse in HNSCC, also in light of the recently proposed role for miR-9 in promoting EMT and a cancer stem cell phenotype in squamous cell carcinomas (45).
Importantly, our data point to the simultaneous inhibition of SP1 and TGFβ pathway by the concerted action of three miRs, miR-1, miR-133a and miR-150, as a possible mechanism to prevent disease relapse. At a first glance, our data do not fit in the current literature showing that, in mouse models, the inhibition of TGFβ pathway combined with K-Ras activation is linked to the onset of SCC in the skin and in the oral mucosa (45,46). However, increasing evidences clearly show that, depending on the stages of tumor progression, TGFβ pathway can exert either pro- or anti-tumorigenic effects (40). For instance, increased TGFβ signaling, in benign tumors or during the course of cancer induction, selects for more aggressive cells and contributes to metastasis formation, in different models of HNSCC (47–49). Our data in the mouse model, showing that TGFβR1 inhibition could result in different outcomes, depending on the simultaneous inhibition of SP1 by MTA or not (Fig. 5F), confirms the hypothesis that TGFβ pathway could act as tumor suppressor or tumor promoter in a context-dependent manner (40,47). In strict accord with our data, SP1 is required for TGFβ–induced EMT in pancreatic cancer (50) and SP1 and SMAD2 proteins have been reported to directly interact in different models (39), supporting the possibility that SP1 and TGFβ pathway may act together to drive EMT, local invasion and, eventually, recurrence formation in HNSCC. The experimental and in silico analyses performed on our and on TCGA samples support this possibility and point to the expression of miR-1, miR-133a and -miR150 as possible switches of the TGFβ activity.
We are aware that our study has limitations that should be taken into account. The cohort of patients used as discovery set contains a relatively small number of heterogeneous patients that received different post-surgery treatments. These variables could impact on the prognostic value of the miR-signature and also prevented the possible evaluation of its independent prognostic role in multivariate analyses. Although we experimentally validated the correlation between miRs expression and their target, we could not test the potential prognostic value of these miRs in a second independent cohort of samples for the absence of a precise follow-up in this group of patients.
Furthermore, it is important to point out that we focused our bioinformatic analyses on the relation between miRs expression and EMT regulators, and this represents a possible limitation of our study, since other biological pathways are also significantly altered by the same miRs in HNSCC.
In perspective, it will be important to verify if these 4 miRs and/or 6 genes signatures could be prospectively validated to identify patients at high risk of recurrence who may merit to be treated specific targeted therapies. Since both SP1 and TGFβ inhibitors have been already tested in cancer patients, our data are of potential immediate translational relevance.
Supplementary Material
Acknowledgements
We are grateful to the patients who participated to this study. We thank all members of the SCICC Lab of The Molecular Oncology Unit for supportive scientific discussions.
Grant Support
This work was partially supported by grants from Associazione Italiana Ricerca sul Cancro (AIRC) IG 12854 to GB and IG 16862 to AV and by CRO 5‰ grant to GB.
Footnotes
Footnotes
Supplementary data accompanies this article.
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
REFERENCES
- 1.Ferlay J, Shin H-R, Bray F, Forman D, Mathers C, Parkin DM. Estimates of worldwide burden of cancer in 2008: GLOBOCAN 2008. Int J Cancer J Int Cancer. 2010;127:2893–917. [DOI] [PubMed] [Google Scholar]
- 2.Adelstein DJ, Li Y, Adams GL, Wagner H Jr, Kish JA, Ensley JF, et al. An intergroup phase III comparison of standard radiation therapy and two schedules of concurrent chemoradiotherapy in patients with unresectable squamous cell head and neck cancer. J Clin Oncol. 2003;21:92–8. [DOI] [PubMed] [Google Scholar]
- 3.Haddad RI, Shin DM. Recent advances in head and neck cancer. N Engl J Med. 2008;359:1143–54. [DOI] [PubMed] [Google Scholar]
- 4.Blanchard P, Baujat B, Holostenco V, Bourredjem A, Baey C, Bourhis J, et al. Meta-analysis of chemotherapy in head and neck cancer (MACH-NC): a comprehensive analysis by tumour site. Radiother Oncol J Eur Soc Ther Radiol Oncol. 2011;100:33–40. [DOI] [PubMed] [Google Scholar]
- 5.Michiels S, Le Maître A, Buyse M, Burzykowski T, Maillard E, Bogaerts J, et al. Surrogate endpoints for overall survival in locally advanced head and neck cancer: meta-analyses of individual patient data. Lancet Oncol. 2009;10:341–50. [DOI] [PubMed] [Google Scholar]
- 6.Hammerman PS, Hayes DN, Grandis JR. Therapeutic Insights from Genomic Studies of Head and Neck Squamous Cell Carcinomas. Cancer Discov. 2015;5:239–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Stransky N, Egloff AM, Tward AD, Kostic AD, Cibulskis K, Sivachenko A, et al. The mutational landscape of head and neck squamous cell carcinoma. Science. 2011;333:1157–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Agrawal N, Frederick MJ, Pickering CR, Bettegowda C, Chang K, Li RJ, et al. Exome sequencing of head and neck squamous cell carcinoma reveals inactivating mutations in NOTCH1. Science. 2011;333:1154–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Rothenberg SM, Ellisen LW. The molecular pathogenesis of head and neck squamous cell carcinoma. J Clin Invest. 2012;122:1951–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tsai JH, Yang J. Epithelial-mesenchymal plasticity in carcinoma metastasis. Genes Dev. 2013;27:2192–206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tam WL, Weinberg RA. The epigenetics of epithelial-mesenchymal plasticity in cancer. Nat Med. 2013;19:1438–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Babu JM, Prathibha R, Jijith VS, Hariharan R, Pillai MR. A miR-centric view of head and neck cancers. Biochim Biophys Acta. 2011;1816:67–72. [DOI] [PubMed] [Google Scholar]
- 13.Nieto MA, Huang RY-J, Jackson RA, Thiery JP. EMT: 2016. Cell. 2016;166:21–45. [DOI] [PubMed] [Google Scholar]
- 14.Ceppi P, Peter ME. MicroRNAs regulate both epithelial-to-mesenchymal transition and cancer stem cells. Oncogene. 2014;33:269–78. [DOI] [PubMed] [Google Scholar]
- 15.Gregory PA, Bert AG, Paterson EL, Barry SC, Tsykin A, Farshid G, et al. The miR-200 family and miR-205 regulate epithelial to mesenchymal transition by targeting ZEB1 and SIP1. Nat Cell Biol. 2008;10:593–601. [DOI] [PubMed] [Google Scholar]
- 16.Park S-M, Gaur AB, Lengyel E, Peter ME. The miR-200 family determines the epithelial phenotype of cancer cells by targeting the E-cadherin repressors ZEB1 and ZEB2. Genes Dev. 2008;22:894–907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sethi N, Wright A, Wood H, Rabbitts P. MicroRNAs and head and neck cancer: Reviewing the first decade of research. Eur J Cancer. 2014 [DOI] [PubMed] [Google Scholar]
- 18.Jamali Z, Asl Aminabadi N, Attaran R, Pournagiazar F, Ghertasi Oskouei S, Ahmadpour F. MicroRNAs as prognostic molecular signatures in human head and neck squamous cell carcinoma: A systematic review and meta-analysis. Oral Oncol. 2015;51:321–31. [DOI] [PubMed] [Google Scholar]
- 19.Corcoran CD, Senchaudhuri P, Mehta CR, Patel NR. Exact Inference for Categorical Data Encycl Biostat. John Wiley & Sons, Ltd; 2005. doi/ 10.1002/0470011815.b2a10019/ [DOI] [Google Scholar]
- 20.Hall M, Frank E, Holmes G, Pfahringer B, Reutemann P, Witten IH. The WEKA Data Mining Software: An Update. [Google Scholar]
- 21.Klement W, Wilk S, Michalowski W, Matwin S. Classifying Severely Imbalanced Data In: Butz C, Lingras P, editors. Adv Artif Intell. Springer Berlin Heidelberg; 2011. page 258–64. [Google Scholar]
- 22.Shirdel EA, Xie W, Mak TW, Jurisica I. NAViGaTing the micronome--using multiple microRNA prediction databases to identify signalling pathway-associated microRNAs. PloS One. 2011;6:e17429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Pavón MA, Parreño M, Téllez-Gabriel M, Sancho FJ, López M, Céspedes MV, et al. Gene expression signatures and molecular markers associated with clinical outcome in locally advanced head and neck carcinoma. Carcinogenesis. 2012;33:1707–16. [DOI] [PubMed] [Google Scholar]
- 24.Brown KR, Otasek D, Ali M, McGuffin MJ, Xie W, Devani B, et al. NAViGaTOR: Network Analysis, Visualization and Graphing Toronto. Bioinforma Oxf Engl. 2009;25:3327–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Brown KR, Jurisica I. Unequal evolutionary conservation of human protein interactions in interologous networks. Genome Biol. 2007;8:R95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rahmati S, Abovsky M, Pastrello C, Jurisica I. pathDIP: an annotated resource for known and predicted human gene-pathway associations and pathway enrichment analysis. Nucleic Acids Res. 2017;45:D419–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.The Cancer Genome Atlas Network. Comprehensive genomic characterization of head and neck squamous cell carcinomas. Nature. 2015;517:576–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gao J, Aksoy BA, Dogrusoz U, Dresdner G, Gross B, Sumer SO, et al. Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Sci Signal. 2013;6:pl1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Aguirre-Gamboa R, Trevino V. SurvMicro: assessment of miRNA-based prognostic signatures for cancer clinical outcomes by multivariate survival analysis. Bioinforma Oxf Engl. 2014;30:1630–2. [DOI] [PubMed] [Google Scholar]
- 30.Fabris L, Berton S, Pellizzari I, Segatto I, D’Andrea S, Armenia J, et al. p27kip1 controls H-Ras/MAPK activation and cell cycle entry via modulation of MT stability. Proc Natl Acad Sci U S A. 2015;112:13916–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Fabris L, Berton S, Citron F, D’Andrea S, Segatto I, Nicoloso MS, et al. Radiotherapy-induced miR-223 prevents relapse of breast cancer by targeting the EGF pathway. Oncogene. 2016;35:4914–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chan W-C, Ho M-R, Li S-C, Tsai K-W, Lai C-H, Hsu C-N, et al. MetaMirClust: Discovery of miRNA cluster patterns using a data-mining approach. Genomics. 2012; 100:141–8. [DOI] [PubMed] [Google Scholar]
- 33.Ma L, Young J, Prabhala H, Pan E, Mestdagh P, Muth D, et al. miR-9, a MYC/MYCN-activated microRNA, regulates E-cadherin and cancer metastasis. Nat Cell Biol. 2010;12:247–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Liu Y-N, Yin JJ, Abou-Kheir W, Hynes PG, Casey OM, Fang L, et al. MiR-1 and miR-200 inhibit EMT via Slug-dependent and tumorigenesis via Slug-independent mechanisms. Oncogene. 2013;32:296–306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Muraoka N, Yamakawa H, Miyamoto K, Sadahiro T, Umei T, Isomi M, et al. MiR-133 promotes cardiac reprogramming by directly repressing Snai1 and silencing fibroblast signatures. EMBO J. 2014;33:1565–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yokobori T, Suzuki S, Tanaka N, Inose T, Sohda M, Sano A, et al. MiR-150 is associated with poor prognosis in esophageal squamous cell carcinoma via targeting the EMT inducer ZEB1. Cancer Sci. 2013;104:48–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Pastrello C, Otasek D, Fortney K, Agapito G, Cannataro M, Shirdel E, et al. Visual Data Mining of Biological Networks: One Size Does Not Fit All. PLoS Comput Biol. 2013; 9:e1002833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Reis PP, Waldron L, Perez-Ordonez B, Pintilie M, Galloni NN, Xuan Y, et al. A gene signature in histologically normal surgical margins is predictive of oral carcinoma recurrence. BMC Cancer. 2011;11:437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Beishline K, Azizkhan-Clifford J. Sp1 and the “hallmarks of cancer.” FEBS J. 2015; 282:224–58. [DOI] [PubMed] [Google Scholar]
- 40.Inman GJ. Switching TGFβ from a tumor suppressor to a tumor promoter. Curr Opin Genet Dev. 2011;21:93–9. [DOI] [PubMed] [Google Scholar]
- 41.Zeller C, Hinzmann B, Seitz S, Prokoph H, Burkhard-Goettges E, Fischer J, et al. SASH1: a candidate tumor suppressor gene on chromosome 6q24.3 is downregulated in breast cancer. Oncogene. 2003;22:2972–83. [DOI] [PubMed] [Google Scholar]
- 42.Martini M, Gnann A, Scheikl D, Holzmann B, Janssen K-P. The candidate tumor suppressor SASH1 interacts with the actin cytoskeleton and stimulates cell-matrix adhesion. Int J Biochem Cell Biol. 2011;43:1630–40. [DOI] [PubMed] [Google Scholar]
- 43.Yanagawa T, Yoshida H, Yamagata K, Onizawa K, Tabuchi K, Koyama Y, et al. Loss of cytokeratin 13 expression in squamous cell carcinoma of the tongue is a possible sign for local recurrence. J Exp Clin Cancer Res CR. 2007;26:215–20. [PubMed] [Google Scholar]
- 44.Naganuma K, Hatta M, Ikebe T, Yamazaki J. Epigenetic alterations of the keratin 13 gene in oral squamous cell carcinoma. BMC Cancer. 2014;14:988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.White RA, Neiman JM, Reddi A, Han G, Birlea S, Mitra D, et al. Epithelial stem cell mutations that promote squamous cell carcinoma metastasis. J Clin Invest. 2013;123:4390–404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Leemans CR, Braakhuis BJM, Brakenhoff RH. The molecular biology of head and neck cancer. Nat Rev Cancer. 2011;11:9–22. [DOI] [PubMed] [Google Scholar]
- 47.Glick AB, Glick AB. The Role of TGF Signaling in Squamous Cell Cancer: Lessons from Mouse Models, The Role of TGF Signaling in Squamous Cell Cancer: Lessons from Mouse Models. J Skin Cancer J Skin Cancer. 2012;, 2012:e249063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Du L, Chen X, Cao Y, Lu L, Zhang F, Bornstein S, et al. Overexpression of PIK3CA in murine head and neck epithelium drives tumor invasion and metastasis through PDK1 and enhanced TGFβ signaling. Oncogene. 2016; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Dasgupta S, Bhattacharya-Chatterjee M, O’Malley BW, Chatterjee SK. Tumor metastasis in an orthotopic murine model of head and neck cancer: Possible role of TGF-beta 1 secreted by the tumor cells. J Cell Biochem. 2006;97:1036–51. [DOI] [PubMed] [Google Scholar]
- 50.Jungert K, Buck A, von Wichert G, Adler G, König A, Buchholz M, et al. Sp1 is required for transforming growth factor-beta-induced mesenchymal transition and migration in pancreatic cancer cells. Cancer Res. 2007;67:1563–70. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.