Abstract
During an epidemic with a new virus, we depend on modelling to plan the response: but how good are the data? The aim of our work was to better understand the impact of misclassification errors in identification of true cases of COVID-19 on epidemic curves. Data originated from Alberta, Canada (available on 28 May 2020). There is presently no information of sensitivity (Sn) and specificity (Sp) of laboratory tests used in Canada for the causal agent for COVID-19. Therefore, we examined best attainable performance in other jurisdictions and similar viruses. This suggested perfect Sp and Sn 60–95%. We used these values to re-calculate epidemic curves to visualize the potential bias due to imperfect testing. If the sensitivity improved, the observed and adjusted epidemic curves likely fall within 95% confidence intervals of the observed counts. However, bias in shape and peak of the epidemic curves can be pronounced, if sensitivity either degrades or remains poor in the 60–70% range. These issues are minor early in the epidemic, but hundreds of undiagnosed cases are likely later on. It is therefore hazardous to judge progress of the epidemic based on observed epidemic curves unless quality of testing is better understood.
Electronic supplementary material
The online version of this article (10.17269/s41997-020-00367-6) contains supplementary material, which is available to authorized users.
Keywords: Epidemic, Misclassification, Probabilistic bias analysis, SARS-CoV-2
Résumé
Au cours d’une épidémie causée par un nouveau virus nous dépendons de la modélisation pour établir un plan de gestion de la situation : mais les données sont-elles de bonne qualité? Le but de ce travail était de mieux comprendre l’impact que les erreurs de classement du virus COVID-19 ont sur les courbes qui décrivent l’épidémie. Les données proviennent de l’Alberta, au Canada (datées du 28 mai, 2020). Il n’y a actuellement pas d’information sur la sensibilité (Sn) et sur la spécificité (Sp) des tests de dépistage de la COVID-19 utilisés en laboratoire au Canada. De ce fait, nous avons examiné la performance optimale observée dans d’autres juridictions et pour d’autres virus semblables. Ceci a mené à une spécificité parfaite et une sensibilité de 60-95%. Nous avons donc utilisé ces valeurs pour refaire le calcul des courbes épidémiques afin de visualiser les risques potentiels de biais dus au testage moins que parfait. Si la sensibilité s’améliore, les courbes épidémiques ajustées et celles observées seront probablement incluses dans les intervalles de confiance (95%) entourant le nombre observé de cas. Néanmoins, la forme et le sommet des courbes épidémiques peuvent être biaisées de façon considérable si la sensibilité se dégrade ou si elle se tient aux alentours de 60-70%. Ce problème n’est pas un problème majeur au tout début d’une épidémie mais peut le devenir après un certain temps. Il est donc risqué de porter jugement sur le progrès de la propagation du virus en se fiant sur les courbes épidémiques à moins que la qualité et la fiabilité des méthodes de dépistage soient bien connues.
Mots-clés: Épidémie, classification erronée, analyse des biais probabilistique, SARS-CoV-2
Introduction
It is well known that outcome misclassification can bias epidemiologic results, yet is infrequently quantified and adjusted. In the context of infectious disease outbreaks, such as during the COVID-19 pandemic of 2019–2020, false positive diagnoses may lead to a waste of limited resources, such as testing kits, hospital beds, and loss of healthcare workforce. On the other hand, false negative diagnoses contribute to uncontrolled spread of contagion. In an ongoing epidemic, where test sensitivity (Sn) and specificity (Sp) of case ascertainment are fixed, prevalence of the outcome (infection) determines whether false positives or negatives dominate. Poor sensitivity is more tolerable when prevalence is low early in the epidemic but less so as the number of infected grows (Goldstein and Burstyn 2020). At present, the accuracy of testing for SARS-CoV-2 viral infection, the causal agent for COVID-19, is unknown in Canada. All available research suggest that near-perfect Sp is justified but Sn can range from 60 to 95% (Binsaeed et al. 2011; COVID-19 Science Report: Diagnostics 2020; Fang et al. 2020; Ai et al. 2020; Konrad et al. 2020; US FDA 2020; He et al. 2020). Full details of our efforts to learn more about Sn and Sp are detailed in Burstyn et al. (2020). We conducted a Monte Carlo (probabilistic) sensitivity analysis of the impact of the plausible extent of this misclassification on bias in epidemic curves in Alberta, Canada.
Methods
Data on observed counts of “confirmed” positive diagnoses versus presumed incident dates (“date reported to Alberta Health”) were obtained on May 28, 2020 from their “Figure 3: COVID-19 cases in Alberta by day status” posted on https://www.alberta.ca/stats/covid-19-alberta-statistics htm under “Case counts” tab. Samples (e.g., nasopharyngeal swab; bronchial wash) undergo nucleic acid testing that use primers/probes targeting the E (envelope protein; Corman et al. 2020) and RdRp (RNA-dependent RNA polymerase; qualitative detection method developed at the Alberta Public Health Laboratory (Alberta Precision Laboratories (APL)) genes of the COVID-19 virus. The relevant data notes are reproduced in full here: “Data sources: The Provincial Surveillance Information system (PSI) is a laboratory surveillance system which receives positive results for all Notifiable Diseases and diseases under laboratory surveillance from Alberta Precision Labs (APL). The system also receives negative results for a subset of organisms such as COVID-19. … Disclaimer: The content and format of this report are subject to change. Cases are under investigation and numbers may fluctuate as cases are resolved. Data included in the interactive data application are up-to-date as of midday of the date of posting.” The data were digitized as shown in Table 1. Counts from May 28, 2020 were excluded because data notes state that “current day is partial.” Dates are the “date reported to Alberta Health.”
Table 1.
Timeline of counts of COVID-19 cases by onset date in Alberta, Canada, on May 28, 2020
| Lab report date/date of onset for incident case (t) | Observed incident cases of COVID-19 | Lab report date/date of onset for incident case (t) | Observed incident cases of COVID-19 | Lab report date/date of onset for incident case (t) | Observed incident cases of COVID-19 |
|---|---|---|---|---|---|
| 3/6/2020 | 1 | 4/1/2020 | 80 | 5/1/2020 | 120 |
| 3/7/2020 | 0 | 4/2/2020 | 85 | 5/2/2020 | 112 |
| 3/8/2020 | 0 | 4/3/2020 | 37 | 5/3/2020 | 67 |
| 3/9/2020 | 6 | 4/4/2020 | 38 | 5/4/2020 | 74 |
| 3/10/2020 | 9 | 4/5/2020 | 36 | 5/5/2020 | 60 |
| 3/11/2020 | 7 | 4/6/2020 | 20 | 5/6/2020 | 62 |
| 3/12/2020 | 2 | 4/7/2020 | 39 | 5/7/2020 | 81 |
| 3/13/2020 | 8 | 4/8/2020 | 24 | 5/8/2020 | 76 |
| 3/14/2020 | 18 | 4/9/2020 | 32 | 5/9/2020 | 84 |
| 3/15/2020 | 9 | 4/10/2020 | 37 | 5/10/2020 | 70 |
| 3/16/2020 | 21 | 4/11/2020 | 48 | 5/11/2020 | 48 |
| 3/17/2020 | 8 | 4/12/2020 | 54 | 5/12/2020 | 69 |
| 3/18/2020 | 27 | 4/13/2020 | 58 | 5/13/2020 | 62 |
| 3/19/2020 | 27 | 4/14/2020 | 119 | 5/14/2020 | 60 |
| 3/20/2020 | 34 | 4/15/2020 | 118 | 5/15/2020 | 71 |
| 3/21/2020 | 28 | 4/16/2020 | 134 | 5/16/2020 | 54 |
| 3/22/2020 | 30 | 4/17/2020 | 201 | 5/17/2020 | 46 |
| 3/23/2020 | 38 | 4/18/2020 | 175 | 5/18/2020 | 38 |
| 3/24/2020 | 51 | 4/19/2020 | 185 | 5/19/2020 | 43 |
| 3/25/2020 | 49 | 4/20/2020 | 179 | 5/20/2020 | 27 |
| 3/26/2020 | 26 | 4/21/2020 | 251 | 5/21/2020 | 30 |
| 3/27/2020 | 71 | 4/22/2020 | 285 | 5/22/2020 | 27 |
| 3/28/2020 | 37 | 4/23/2020 | 336 | 5/23/2020 | 32 |
| 3/29/2020 | 18 | 4/24/2020 | 225 | 5/24/2020 | 27 |
| 3/30/2020 | 26 | 4/25/2020 | 223 | 5/25/2020 | 12 |
| 3/31/2020 | 110 | 4/26/2020 | 190 | 5/26/2020 | 19 |
| 4/27/2020 | 172 | 5/27/2020 | 27 | ||
| 4/28/2020 | 244 | ||||
| 4/29/2020 | 224 | ||||
| 4/30/2020 | 215 |
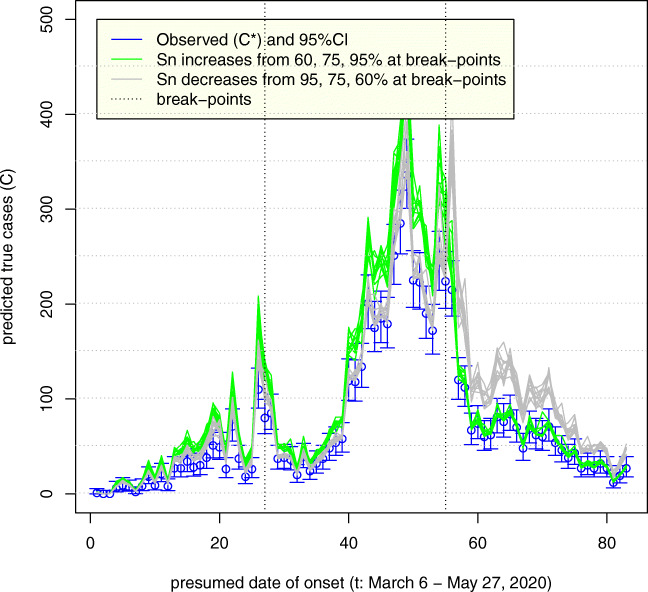
For each observed count of incident cases, we estimated true counts as (observed count)/Sn under the assumption that specificity is indistinguishable from perfect. Because values of Sn are not estimated or known, we simulated various values of Sn as arising from Beta distribution, ranging in means from 0.60 to 0.95 and fixed standard deviation of 0.05. The results of 10 simulation realizations for each scenario are plotted in Fig. 1. We assumed that Sn either improved (realistic best case: green) or degraded (pessimistic worst case: brown). Our analysis only reflects systematic errors for illustrative purposes and under the common assumptions (and experience) that they dwarf random errors. Calculations were performed in R (R Core Team 2019) and code is available via Burstyn et al. (2020).
Fig. 1.
Uncertainty in the epidemic curve of COVID-19 on May 28, 2020 in Alberta, Canada, due to imperfect sensitivity (Sn) that varies in time; standard deviation of Sn = 5%; assumes specificity 100% (10 simulation realizations plotted)
Figure 1 indicates how, as expected, if sensitivity improves over time with the natural refinement of laboratory procedures (green lines), then the true epidemic curve is expected to be flatter than the observed. It also appears that observed and true curves may well fall within the range of 95% confidence intervals around the observed counts (blue lines). If sensitivity decreases over time (grey lines) (e.g., due to increased pressures of work, scaling up of the analyses with less experienced staff), then the true epidemic curve is expected to be steeper than the observed. In either scenario, there can be an under-counting of cases by nearly a factor of two, most apparent as the incidence grows, such that on day April 23, 2020, there may have been almost 500 cases vs. 336 observed. Even at the tail end of the epidemic, on May 27, 2020, our simulation predicts that there may have been as many as 45 to 50 cases vs. 27 observed. This is alarming, because misdiagnosed patients who have not self-isolated can spread infection unimpeded and it is impossible to know who they are among symptomatic persons tested negative around that time per day.
Alberta initially restricted COVID-19 testing to “any person exhibiting any symptom of COVID-19, all close contacts of confirmed COVID-19 cases, all workers and/or residents at specific outbreak sites, all workers and residents at long-term care and level 4 supportive living facilities … , all patients admitted to continuing care or transferred between continuing care and hospital settings” (Government of Alberta 2020). Currently, testing is available to any person who wishes to be tested (Government of Alberta 2020). Consequently, the sensitivity of surveillance has likely improved over time, with numbers of test-positive persons more closely resembling occurrence of the disease in the population. However, as the capture for test of cases of truly infected improves, the sensitivity of laboratory tests, which is the focus of our work, may or may not have improved, leaving bias of unknown magnitude in the surveillance data.
Conclusion
We conclude that it is of paramount importance to validate laboratory tests and to share this knowledge, especially as the epidemic matures into its full force. In absence of such knowledge, the observed epidemic curves can be dangerously misleading for surveillance purposes.
Electronic supplementary material
(DOCX 14 kb)
Acknowledgements
The authors thank Isaac R. Burstyn and Marguerite R. Burstyn for their diligence in perfect extraction of Alberta data from the online charts. The authors thank Jean-Michel Galarneau for translating the abstract into French.
Funding information
Research reported in this publication was partially supported by the National Institute of Allergy and Infectious Diseases of the National Institutes of Health under Award Number K01AI143356 (to NDG).
Compliance with ethical standards
Conflict of interest
The authors declare that they have no conflict of interest.
Footnotes
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- Ai, T., Yang, Z., Hou, H., et al. (2020). Correlation of chest CT and RT-PCR testing in coronavirus disease 2019 (COVID-19) in China: a report of 1014 cases. Radiology. 10.1148/radiol.2020200642. [DOI] [PMC free article] [PubMed]
- Binsaeed AA, Al-Khedhairy AA, Mandil, et al. A validation study comparing the sensitivity and specificity of the new Dr. KSU H1N1 RT-PCR kit with real-time RT-PCR for diagnosing influenza A (H1N1) Annals of Saudi Medicine. 2011;31(4):351–355. doi: 10.4103/0256-4947.83212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burstyn, I. Goldstein, N., Gustafson, P. (2020). Towards reduction in bias in epidemic curves due to outcome misclassification through Bayesian analysis of time-series of laboratory test results: case study of COVID-19 in Alberta, Canada and Philadelphia, USA. BMC Medical Research Methdology (in press); OSF Preprints, doi:10.31219/osf.io/qpgbk. [DOI] [PMC free article] [PubMed]
- Core Team R. R: A language and environment for statistical computing. Vienna: R Foundation for Statistical Computing; 2019. [Google Scholar]
- Corman VM, Olfert L, Marco K, et al. Detection of 2019 novel coronavirus (2019-nCoV) by real-time RT-PCR. Euro Surveill. 2020;25(3):2000045. doi: 10.2807/1560-7917.ES.2020.25.3.2000045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- COVID-19 Science Report: Diagnostics. (2020). NUS Saw Swee Hock School of Public Health, Singapore. 10.25540/e3y2-aqye.
- Fang, Y., Zhang, H., Xie, J., et al. (2020). Sensitivity of chest CT for COVID-19: comparison to RT-PCR. Radiology. 10.1148/radiol.2020200432. [DOI] [PMC free article] [PubMed]
- Goldstein, N. D., & Burstyn, I. (2020). On the importance of early testing even when imperfect in a pandemic such as COVID-19. 10.31219/osf.io/9pz4d. [DOI] [PMC free article] [PubMed]
- Government of Alberta (2020). Symptoms and testing. https://www.alberta.ca/covid-19-testing-in-alberta.aspx (accessed 6/2/2020)).
- He JL, Luo L, Luo ZD, et al. Diagnostic performance between CT and initial real-time RT-PCR for clinically suspected 2019 coronavirus disease (COVID-19) patients outside Wuhan, China. Respir Med. 2020;168:105980. doi: 10.1016/j.rmed.2020.105980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Konrad, R., Eberle, U., Dangel, A., et al. (2020). Rapid establishment of laboratory diagnostics for the novel coronavirus SARS-CoV-2 in Bavaria, Germany, February 2020. Euro Surveill, 9. 10.2807/1560-7917.ES.2020.25.9.2000173. [DOI] [PMC free article] [PubMed]
- US FDA (2020). Accelerated emergency use authorization (EUA) summary COVID-19 RT-PCR test (Laboratory Corporation of America). https://www.fda.gov/media/136151/download. Accessed 20 March 2020).
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(DOCX 14 kb)