Highlights
-
•
Coronaviruses are wide-ranging diseases in humans and animals.
-
•
SARS-CoV-2 is a bat-origin zoonotic coronavirus causing the COViD-19 pandemic.
-
•
SARS-CoV-2 can also infect cats and ferrets and has been detected in dogs.
-
•
The SARS-CoV-2 receptor ACE2 is highly similar between cats and humans.
Keywords: Coronavirus, Feline coronavirus, SARS-CoV-2, ACE-2
Abstract
Coronaviruses (CoVs) cause disease in a range of agricultural and companion animal species, and can be important causes of zoonotic infections. In humans, several coronaviruses circulate seasonally. Recently, a novel zoonotic CoV named SARS-CoV-2 emerged from a bat reservoir, resulting in the COVID-19 pandemic. With a focus on felines, we review here the evidence for SARS-CoV-2 infection in cats, ferrets and dogs, describe the relationship between SARS-CoV-2 and the natural coronaviruses known to infect these species, and provide a rationale for the relative susceptibility of these species to SARS-CoV-2 through comparative analysis of the ACE-2 receptor.
1. Introduction
Coronaviruses (CoVs) are part of a complex and diverse group of viruses grouped into four different genera Alpha-, Beta-, Gamma- and Delta-, based on their evolutionary and genetic characteristics (Masters and Perlman, 2013). In animals, CoVs cause major disease in a wide range of agricultural and companion animal species (MacLaughlan and Dubovi, 2016) (Table 1 ). In humans several coronaviruses circulate seasonally, including HCoV-NL63, HCoV-229E, HCoV-OC43, and HCoV-HKU1, and can result in the common cold. The zoonotic severe acute respiratory syndrome-CoV (SARS-CoV) and the Middle East respiratory syndrome-CoV (MERS-CoV), however, are broadly recognized as they have caused significant outbreaks in humans during 2002−3 and 2012−13 respectively, with the MERS-CoV outbreak still ongoing (Bennett et al., 2019). SARS-CoV and MERS-CoV are both bat-origin viruses with an intermediate host of masked palm civets/raccoon dogs and dromedary camels respectively (Song et al., 2019). Recently, a novel CoV named SARS-CoV-2 emerged and has now caused the global COVID-19 pandemic. The disease outbreak started in Wuhan, China, with the virus origin attributed to populations of SARS-like viruses that circulate in wild bats (Zhou et al., 2020). While pangolins and other animal species from live animal markets have been implicated, there is currently no strong evidence regarding the possible intermediate host(s) that could have facilitated the transmission of SARS-CoV-2 between its bat reservoir and humans (Jaimes et al., 2020a; Zhang and Holmes, 2020).
Table 1.
Select CoVs and the species they infect.
| Genus | Virus | Infected species |
|---|---|---|
| Alphacoronavirus | Feline coronaviruses (FCoV) types I and II | Cats (domestic and wild) |
| Canine coronaviruses (CCoV) types I and II | Dogs | |
| Ferret coronavirus (FRECV and FRSCV) | Ferrets | |
| Transmissible gastroenteritis virus (TEGV) | Pigs | |
| Human coronavirus NL63 (HCoV-NL63) | Humans | |
| Human coronavirus 229E (HCoV-229E) | Humans | |
| Porcine epidemic diarrhea (PEDV) | Pigs | |
| Mink coronavirus (MCoV) | Mink | |
| Betacoronavirus | Bovine coronavirus (BCoV) | Cattle |
| Human coronavirus OC43 (HCoV-OC43) | Humans | |
| Canine respiratory coronavirus (CRCoV) | Dogs | |
| Severe acute respiratory syndrome coronavirus (SARS-CoV) | Humans | |
| Middle East respiratory syndrome coronavirus (MERS-CoV) | Humans | |
| Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) | Humans | |
| Equine coronavirus (ECoV) | Horses | |
| Human coronavirus HKU1 (HCoV-HKU1) | Humans | |
| Gammacoronavirus | Infectious bronchitis virus (IBV) | Poultry |
| Turkey coronavirus | Turkeys | |
| Deltacoronavirus | Porcine deltacoronavirus (PDCoV) | Pigs |
In recent weeks, cats, ferrets and dogs have all drawn attention in the public health battle against COVID-19, based on their proposed infection by owners and handlers and possibility to act as an intermediate host. With a focus on felines, we review here the evidence for SARS-CoV-2 infection in cats, ferrets and dogs, describe the relationship between SARS-CoV-2 and the natural coronaviruses known to infect these species, and provide a rationale for the relative susceptibility of these species to SARS-CoV-2.
2. Feline coronaviruses and SARS-CoV-2 in felines
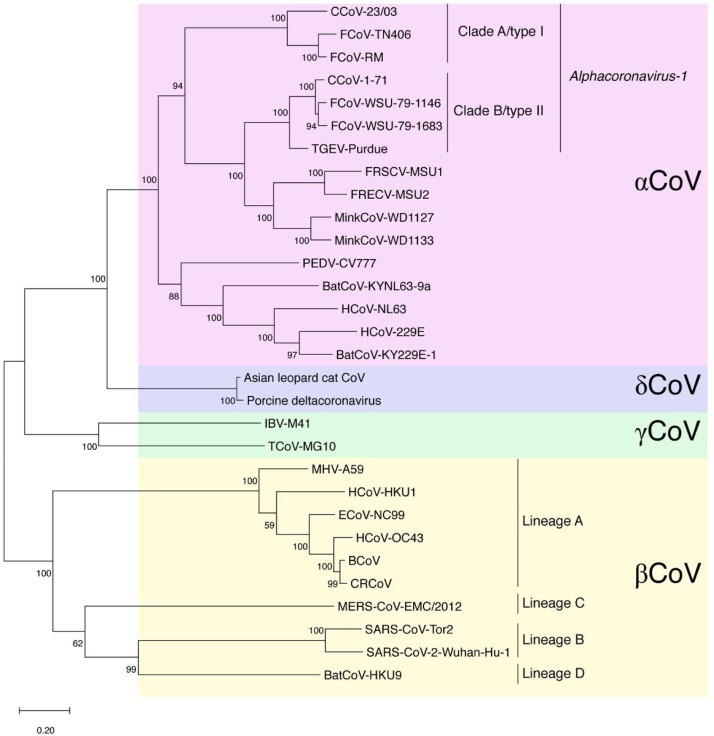
Feline coronaviruses (FCoVs) are widely known as agents capable of infection in both domestic and wild felids (Pedersen, 2014). Initial infection causes only a mild localized disease; however, the virus can convert also exist in a form that is responsible for the severe and mostly lethal feline infectious peritonitis (FIP). This is most likely through a process of “internal mutation”; however circulating virus variants with higher propensity to cause FIP may also exist (Brown et al., 2009; Vennema et al., 1998). The virus is suggested to infect all types of felids and infections have been reported in numerous non-domestic felids; among the big cats, cheetahs are highly susceptible to FIP (Evermann et al., 1989). Two different types of FCoV have been described with distinct epidemiological distribution and specific biological and molecular features that prove the diversity of these agents (Jaimes et al., 2020b). The type I FCoV is considered to be the ancestor virus of the feline lineage. It is the most common of the two types and is currently circulating in domestic cat reservoirs globally. In contrast, the type II FCoV has been suggested to have emerged from recombination events between feline and canine CoVs, which is less epidemiologically distributed and displays its own biological and molecular mechanisms of infection, diverting from the FCoV type I viruses (Herrewegh et al., 1998; Jaimes et al., 2020b). We recently reported the complex evolutionary pathway of the Alphacoronavirus-1 species, which includes both FCoV types, as well as canine coronaviruses and transmissible gastroenteritis virus of swine (TGEV) (Whittaker et al., 2018). A summary of the relationship of the spike proteins of the Alphacoronavirus-1 species in comparison to animal and human coronaviruses, including four human endemic viruses (human CoVs 229E, OC43, NL63 and HKU1) is shown in Fig. 1 . A distinct feline coronavirus found in a wild Asian leopard cat is also included in Fig. 1 (Dong et al., 2007).
Fig. 1.
Phylogenetic tree of selected coronaviruses, based on spike protein sequences. Selected coronavirus spike protein sequences were aligned using MUSCLE and a maximum-likelihood (ML) phylogenetic tree was generated using MEGAX. Bootstrap values shown at nodes were calculated from 1000 replicates. The tree is drawn to scale, with branch lengths measured in the number of substitutions per site. Abbreviations used: FCoV, feline coronavirus; CCoV, canine coronavirus; TGEV, transmissible gastroenteritis virus; FRECV, ferret enteric coronavirus; FRSCV, ferret systemic coronavirus; PEDV, porcine epidemic coronavirus; BatCoV, bat coronavirus; HCoV, human coronavirus; IBV, infectious bronchitis virus; TCoV: turkey coronavirus; MHV, murine hepatitis virus; ECoV, equine coronavirus; BCoV, bovine coronavirus; CRCoV, canine respiratory coronavirus; MERS-CoV, Middle East respiratory syndrome coronavirus; SARS-CoV, severe acute respiratory syndrome coronavirus.
While typically considered to be an enteric infection, FCoVs may in fact have more systemic distribution in their host (including in the respiratory tract) before the critical mutation(s) in the viral genome that results in the acquisition of macrophage tropism and progression to FIP, with its effusive (“wet”) and/or non-effusive (“dry”) clinical outcome often accompanied by extensive granulomatous lesions (Kipar and Meli, 2014; Pedersen et al., 2009; Sykes, 2014). FIP is a complex disease syndrome that is further complicated by the process of antibody-dependent enhancement (ADE) of infection, whereby sub-neutralizing antibodies can recognize the virus spike protein and lead to enhanced macrophage infection, leading to more rapid disease progression (Olsen et al., 1992; Takano et al., 2008).
As described in more detail below, both natural and experimental SARS-CoV and SARS-CoV-2 infection of cats have been reported, generally resulting in mild respiratory signs (van den Brand et al., 2008; Martina et al., 2003; Shi et al., 2020). Additionally, an antibody response has been demonstrated in several cats in Wuhan, China (Zhang et al., 2020). It is important to note that that SARS-like viruses (betacoronaviruses in lineage B) and feline coronaviruses (alphacoronaviruses) are quite distinct (Fig. 1). As such, there is currently no definitive evidence that prior exposure to feline coronaviruses (which are widespread) will protect against SARS-like viruses; however serological testing will need to carefully evaluate any potential cross-reaction. Whether antibodies to FCoV can induce ADE upon subsequent infection with a SARS-like virus remains an open question. Preliminary studies, however, indicate the potential for transmission between individual cats (Halfmann et al., 2020).
3. Ferret coronaviruses and SARS-CoV-2 in ferrets
Two distinct alphacoronaviruses have been described in ferrets: ferret enteric coronavirus (FRECV) and ferret systemic coronavirus (FRSCV) (Garner et al., 2008; Williams et al., 2000) (Fig. 1). Infection with FRECV was originally termed epizootic catarrhal enteritis and is characterized by profuse, green mucoid diarrhea, in addition to non-specific signs (Williams et al., 2000). In contrast, FRSCV infection is associated with systemic disease with granulomatous lesions, often described as similar to the dry form of feline infectious peritonitis (FIP) and has been described in laboratory, farm-raised, and pet ferrets (Autieri et al., 2015; Williams et al., 2000). Currently, there is no obvious mechanistic link between FRECV and FRSV that is equivalent to the “internal mutation” of FCoV. It is unknown whether antibodies against FRECV or FRSCV would be able to neutralize SARS-CoV-2 or contribute to further disease in ferrets via ADE. Generally speaking, infection with ferret coronavirus is not considered to be respiratory.
4. Canine coronaviruses and SARS-CoV-2 in canines
The alphacoronavirus canine coronavirus (CCoV) is widely known to cause enteric infection of dogs and as with FCoV these infections are caused by two distinct types CCoV type I and CCoV type II (Fig. 1). Infection is generally mild in the absence of co-infecting pathogens such as parvovirus, although severe gastroenteritis can occur. CCoVs with systemic signs have been also reported, although FIP-like disease does not occur in dogs. Dogs also have a distinct respiratory coronavirus (CRCoV), which often causes mild clinical signs, though severe clinical signs have been reported (Erles et al., 2003). CRCoV is closely related to bovine coronavirus but as a betacoronavirus in lineage A, relatively distant from SARS-like viruses, including SARS-CoV, which are in lineage B. It is unlikely that prior exposure to CRCoV or CCoVs (which are widespread) will protect against SARS-like viruses; ADE is not known to occur with canine coronaviruses.
5. Community and experimental SARS-CoV and SARS-CoV-2 infection of cats and other companion animals
Before addressing recent concerns about SARS-CoV-2 in companion animals, we first briefly review data from the original SARS outbreak in 2002−03. During the initial SARS outbreak in Hong Kong, it was first recognized that cats in the Amoy Garden apartment complex (often considered an epicenter of the outbreak) could be infected by SARS-CoV (Martina et al., 2003). Subsequently inoculation of experimental cats and ferrets was performed, which did not result in clinical signs in cats, but did lead to lethargy in a subset of ferrets (3/6) and death in a single ferret. Across both species, the virus was observed to be shed from the pharynx. While fecal shedding was not observed, RT-PCR of the GI tract was positive. Seroconversion and the presence of a viral titer were observed in naïve animals group housed with experimentally inoculated animals (Martina et al., 2003). Experimentally, SARS-CoV has been shown to cause respiratory damage in both cats and ferrets, though only ferrets demonstrated clinical disease (van den Brand et al., 2008). The identification of virus in ferrets was commonly found in areas of high receptor (ACE2) expression, as would be expected, though outside of the respiratory tract, ACE2 expression has also been demonstrated by RT-PCR in the lung, heart, kidney and small intestine of the ferret (van den Brand et al., 2008; Zamoto et al., 2006). In vitro, HeLa cells expressing ferret ACE2 are additionally permissive to SARS-CoV (Zamoto et al., 2006). In comparison, Middle East respiratory syndrome coronavirus (MERS-CoV)—a betacoronavirus in lineage C—has not been shown to cause active infection in ferrets, although the viral receptor DPP4 shares a high level of homology across humans and ferrets (Peck et al., 2017; Raj et al., 2014). There are no reports of SARS- or MERS-like viruses transmitting to dogs.
Recent reports in regard to COVID-19 and the identification of SARS-CoV-2 viral RNA in two dogs from Hong Kong, a dog in New York, a cat in Belgium, and a tiger at the Bronx Zoo in New York City have raised many new questions in regard to SARS-CoV-2 (Mallapaty, 2020; USDA APHIS, 2020). These reports also been augmented by two PCR-positive cases in cats in different locations in New York State (SARS-CoV-2/COVID-19, United States of America, 2020). In all cases, infection appears to be limited and restricted to the upper respiratory tract, although viral shedding in the feces is also apparent. It has been suggested that these species became infected by their owners or handlers. No evidence of transmission to other animals or to humans has been reported so far, although other tigers and lions in the vicinity of the SARS-CoV-2-positve tiger also had respiratory signs. The finding of animals infected with the SARS-CoV-2 is not particularly surprising, given the widespread nature of the disease in humans and the precedent set by SARS-CoV. While being distinct viruses, SARS-CoV and SARS-CoV-2 share a common receptor (ACE2) (Hoffmann et al., 2020). The feline ACE2 protein is among the most closely related to human ACE2, in particular within the receptor binding interface region, which may explain why cross-species transmission may occur with this particular species (see below).
Recent experimental investigations of SARS-CoV-2 infections of domestic species, isolation of the virus from upper respiratory samples from a small sample of ferrets was consistent with infection, in addition to the development of clinical signs (Kim et al., 2020; Shi et al., 2020). In cats that were experimentally challenged with SARS-CoV-2, viral RNA was detectable in both respiratory tissues and the small intestines of cats sacrificed at day 3 and day 6, however, only at day 3 was viral RNA detectable in the lungs (Shi et al., 2020). Across both cats and ferrets, viral RNA has detected in feces, similar to observations in human disease and additionally, animals in close proximity to infected conspecifics have detectable levels of viral RNA (Kim et al., 2020; Shi et al., 2020). Dogs have also been experimentally inoculated with SARS-CoV-2 and while seroconversion was observed, no virus was isolated and other dogs in close proximity did not show infection (Shi et al., 2020).
A seroprevalence study has shown that nearly 15 % of cats (15/102) sampled after the start of the COVID-19 outbreak in Wuhan, China were positive for antibody against SARS-CoV-2 (Zhang et al., 2020). Among these cats, information regarding owner health status was not provided, nor how this sample compares to the population as a whole as far as owned pets versus strays. The spread of SARS-CoV-2 was additionally investigated in France in a small cohort of veterinary students (n = 18) and their 21 pets (9 cats, 12 dogs), of which 11 students had symptoms consistent with COVID-19, though only two were confirmed positive (Temmam et al., 2020). While three of these cats showed clinical signs of respiratory or gastrointestinal disease, no single animal was considered positive via RT-PCR of nasal or rectal swabs, or by the presence of SARS-CoV-2 specific antibodies (Temmam et al., 2020). However, a case of an asymptomatic cat testing positive for viral antigen after cohabitating with a COVID-19 patient has been reported (Ruiz-Arrondo et al., 2020).
6. Receptor utilization by SARS-CoV-2 in cats and other mammalian species
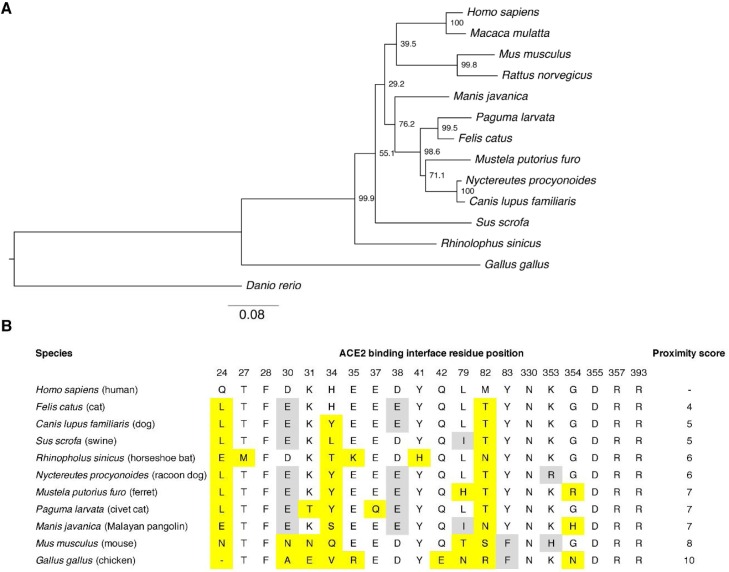
As described above, the receptor for both SARS-CoV and SARS-CoV-2 is ACE2. Protein alignment and phylogenetic analysis of full-length ACE2 protein in representative mammalian species reveal that as expected simian ACE2 from macaque (Macaca mulatta) and chimpanzee (Pan troglodytes) are the closest to human ACE2 with 94.9–99 % sequence identity (Table 2 ). Interestingly, amongst the other mammalian ACE2 sequences analyzed, the next most closely-related sequence is from the domestic cat (Felis catus) with 85.2 % overall identity compared to human ACE2. Other closely related ACE2 sequences found were from Malayan pangolin (Manis javanica, 84.8 % identity) and European rabbit (Oryctolagus cuniculus, 84.8 %). The ACE2 proteins of the intermediate hosts of SARS-CoV raccoon dog (Nyctereutes procyonoides) and masked palm civet (Paguma larvata) also featured a high degree of conservation in sequence with 83.9 % and 83.5 % sequence identity, respectively. Domestic dog (Canis lupus familiaris) and domestic ferret (Mustela putorius furo) sequences displayed ACE2 sequence identities of 83.4 % and 82.6 % respectively. The bat ACE2 from Rhinolophus sinicus and Hipposideros armiger species known to harbor bat-SARS-related coronaviruses, had lower sequence identities at 80.7 % and 80.5 % respectively. A phylogenetic tree of the relationship between these and other species is shown in Fig. 2 A.
Table 2.
Percent identity of ACE2 sequence, compared to human (Homo sapiens).
| % identity | |
|---|---|
| Pan troglodytes (chimpanzee) | 99 |
| Macaca mulatta (macaque) | 94.9 |
| Felis catus (cat) | 85.2 |
| Manis javanica (Malayan pangolin) | 84.8 |
| Oryctolagus cuniculus (rabbit) | 84.8 |
| Nyctereutes procyonoides (racoon dog) | 83.9 |
| Paguma larvata (masked palm civet) | 83.5 |
| Canis lupus familiaris (dog) | 83.4 |
| Camelus dromedarius (dromedary camel) | 83.2 |
| Mustela putorius furo (ferret) | 82.6 |
| Rattus norvegicus (rat) | 82.5 |
| Mus musculus (mouse) | 82.1 |
| Sus scrofa (swine) | 81.4 |
| Bos taurus (cattle) | 80.9 |
| Rhinolophus sinicus (horseshoe bat) | 80.7 |
| Hipposideros armiger (great roundleaf bat) | 80.5 |
| Gallus gallus (chicken) | 65.6 |
Fig. 2.
Comparative analysis of ACE2 in different mammalian species. A. Phylogenetic tree of ACE2 sequences from different species. The ACE2 sequences from various animal species were aligned using MUSCLE and a maximum-likelihood (ML) phylogenetic tree was generated using MEGAX. Bootstrap values shown at nodes were calculated from 1000 replicates. The tree is drawn to scale, with branch lengths measured in the number of substitutions per site. B. Comparative analysis of ACE2 amino acid composition at residue positions involved in the ACE2-virus binding interface. The ACE2 sequences of ACE2 from selected species were aligned and the amino acid found at each of the residue positions involved in the binding interface are shown. Yellow highlight indicates a substituted residue compared to the human ACE2 sequence. A grey highlight indicates a conservative change in residue composition compared to human ACE2 sequence. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article).
The structure of SARS-CoV-2 spike receptor binding motif (RBM) bound to the human ACE2 receptor has recently been resolved (Lan et al., 2020; Shang et al., 2020). Notably, the ACE2 residues that directly contact the SARS-CoV-2 spike receptor binding domain (RBD) have been identified (Lan et al., 2020). To gauge potential use by SARS-CoV-2 of the ACE2 receptor from various mammalian species, a comparative analysis of the amino acid composition of the ACE2 contacting residues in different species was performed (Fig. 2B). Such analysis allows the estimation of a score of proximity to human ACE2 contacting residues for a given mammalian ACE2 sequence. Notably feline ACE2 has a score of 4, indicating that only 4 out of a total of 20 contacting residues were different between feline and human ACE2, with two of those differences conservative substitutions (D30E) and (D38E). This suggests that SARS-CoV-2 spike is highly likely to bind to feline ACE2. The contacting residues found in canine ACE2 and the ACE2 of the bat Rhinolophus sinicus were also shown to have a relatively high degree of conservation with scores in the 5–6 range. In contrast to the relative overall sequence identity, mouse ACE2 contained a relatively high number of different residues at the ACE2 binding interface, with a score of 8. This is in line with a report showing that mouse ACE2 cannot be functionally used by SARS-CoV-2 (Zhou et al., 2020). Based on these analyses the susceptibility of cats to infection with SARS-CoV-2 can likely be explained by the high similarities in human and feline ACE2. The reason for the susceptibility of ferrets to SARS-CoV-2 remains uncertain, but is unlikely due to the similarities in ACE2, with ferret ACE2 being more divergent that canine ACE2, an apparently less susceptible species for SARS-CoV-2.
7. Summary and perspectives
It is now becoming clear that cats are susceptible hosts for the human virus SARS-CoV-2. However, much remains to be determined about the extent of infection in the cat population and level of infection and clinical signs in individual cats, as well at the potential for transmission between cats or to other animals, including humans. A likely explanation for this susceptibility lies in the high degree of similarity between the human and feline forms of the SARS-CoV-2 receptor, ACE2. However, there are still many unanswered questions related to interactions between the naturally occurring feline coronaviruses (types I and II) and SARS-CoV-2, and what the outcome of co-infection with both human and feline coronaviruses might be. Ferrets are also susceptible hosts for SARS-CoV-2, but the reasons behind this are unclear and do not appear to the linked to receptor similarity. It may be that ferrets and human share common properties in relation to the architecture of their respective respiratory tracts, which makes them susceptible hosts (Lakdawala et al., 2015). Dogs are currently not considered to be susceptible hosts for SARS-CoV-2, despite some positive test results in dogs. Recent reports that SARS-CoV-2 may have originated in stray dogs based on similar genetic signatures between systemic forms of CCoV and SARS-CoV-2 are unlikely to be substantiated (Xia, 2020).
In the public health battle against coronaviruses, it is important to note that both cats and ferrets are known hosts of human (and avian) influenza viruses yet are not considered to be a significant risk for human infections. In contrast, dogs maintain their own pool of influenza viruses. Overall, both cats and ferrets may be part of a common pool of human respiratory infections. Whether this is due to molecular similarities at the pathogen-host interface, social connection between humans and their companion animals (or with zoo animals) or based on more physiological or immunological parameters within the respiratory tract (or elsewhere) remains to be determined. There is also the issue of viral pathogenesis, by which it is known both cats and ferrets (but not other species) can allow the conversion of what is normally to be a benign yet transmissible coronavirus (FCoV and FRECV) into a lethal, systemic (but typically not transmissible) form. Clearly, many important investigations lie ahead in understanding feline (and ferret) coronaviruses in their natural host and at the human-animal interface, both within the context of the current COVID-19 pandemic and for its likely continued circulation in the future.
Funding
AS is supported by NIH Comparative Medicine Training Program T32OD011000. Work in the author’s lab is funded in part by the Cornell Feline Health Center and the Winn Feline Foundation.
Footnotes
Supplementary material related to this article can be found, in the online version, at doi:https://doi.org/10.1016/j.vetmic.2020.108777.
Appendix A. Supplementary data
The following is Supplementary data to this article:
References
- Autieri C.R., Miller C.L., Scott K.E., Kilgore A., Papscoe V.A., Garner M.M., Haupt J.L., Bakthavatchalu V., Muthupalani S., Fox J.G. Systemic Coronaviral Disease in 5 Ferrets. Comp. Med. 2015;65(6):508–516. [PMC free article] [PubMed] [Google Scholar]
- Bennett J.E., Dolin R., Blaser M.J. 9th ed. Elsevier; 2019. Mandell, Douglas, and Bennett’s Principles and Practice of Infectious Diseases. [Google Scholar]
- Brown M.A., Troyer J.L., Pecon-Slattery J., Roelke M.E., O’Brien S.J. Genetics and pathogenesis of feline infectious peritonitis virus. Emerging Infect. Dis. 2009;15(9):1445–1452. doi: 10.3201/eid1509.081573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong B.Q., Liu W., Fan X.H., Vijaykrishna D., Tang X.C., Gao F., Li L.F., Li G.J., Zhang J.X., Yang L.Q., Poon L.L.M., Zhang S.Y., Peiris J.S.M., Smith G.J.D., Chen H., Guan Y. Detection of a novel and highly divergent coronavirus from Asian Leopard Cats and Chinese Ferret Badgers in Southern China. J. Virol. 2007;81(13):6920–6926. doi: 10.1128/JVI.00299-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erles K., Toomey C., Brooks H.W., Brownlie J. Detection of a group 2 coronavirus in dogs with canine infectious respiratory disease. Virology. 2003;310(2):216–223. doi: 10.1016/S0042-6822(03)00160-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evermann J.F., Heeney J.L., McKeirnan A.J., O’Brien S.J. Comparative features of a coronavirus isolated from a cheetah with feline infectious peritonitis. Virus Res. 1989;13(1):15–27. doi: 10.1016/0168-1702(89)90084-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garner M.M., Ramsell K., Morera N., Juan-Sallés C., Jiménez J., Ardiaca M., Montesinos A., Teifke J.P., Löhr C.V., Evermann J.F., Baszler T.V., Nordhausen R.W., Wise A.G., Maes R.K., Kiupel M. Clinicopathologic features of a systemic coronavirus-associated disease resembling feline infectious peritonitis in the domestic ferret (Mustela putorius) Vet. Pathol. 2008;45(2):236–246. doi: 10.1354/vp.45-2-236. [DOI] [PubMed] [Google Scholar]
- Halfmann P.J., Hatta M., Chiba S., Maemura T., Fan S., Takeda M., Kinoshita N., Hattori S., Sakai-Tagawa Y., Iwatsuki-Horimoto K., Imai M., Kawaoka Y. Transmission of SARS-CoV-2 in Domestic Cats. N. Engl. J. Med. 2020 doi: 10.1056/NEJMc2013400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herrewegh A.A.P.M., Smeenk I., Horzinek M.C., Rottier P.J.M., Groot J. de R. Feline coronavirus type II strains 79-1683 and 79-1146 originate from a double recombination between feline coronavirus type I and canine coronavirus. J. Virol. 1998;72(5):4508–4514. doi: 10.1128/JVI.72.5.4508-4514.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffmann M., Kleine-Weber H., Schroeder S., Krüger N., Herrler T., Erichsen S., Schiergens T.S., Herrler G., Wu N.-H., Nitsche A., Müller M.A., Drosten C., Pöhlmann S. SARS-CoV-2 cell entry depends on ACE2 and TMPRSS2 and is blocked by a clinically proven protease inhibitor. Cell. 2020;181(2):271–280.e8. doi: 10.1016/j.cell.2020.02.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaimes J.A., André N.M., Chappie J.S., Millet J.K., Whittaker G.R. Phylogenetic analysis and structural modeling of SARS-CoV-2 spike protein reveals an evolutionary distinct and proteolytically sensitive activation loop. J. Mol. Biol. 2020;432(10):3309–3325. doi: 10.1016/j.jmb.2020.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaimes J.A., Millet J.K., Stout A.E., André N.M., Whittaker G.R. A tale of two viruses: the distinct spike glycoproteins of feline coronaviruses. Viruses. 2020;12(1) doi: 10.3390/v12010083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim Y.-I., Kim S.G., Kim S.M., Kim E.H., Park S.J., Yu K.M., Chang J.H., Lee S. Infection and rapid transmission of SARS-CoV-2 in ferrets. Cell. 2020;27(5) doi: 10.1016/j.chom.2020.03.023. https://www.cell.com/pb-assets/journals/research/cell-host-microbe/PDFs/chom_2285_preproof.pdf [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kipar A., Meli M.L. Feline infectious peritonitis: Still an enigma? Vet. Pathol. 2014;51(2):505–526. doi: 10.1177/0300985814522077. [DOI] [PubMed] [Google Scholar]
- Lakdawala S.S., Jayaraman A., Halpin R.A., Lamirande E.W., Shih A.R., Stockwell T.B., Lin X., Simenauer A., Hanson C.T., Vogel L., Paskel M., Minai M., Moore I., Orandle M., Das S.R., Wentworth D.E., Sasisekharan R., Subbarao K. The soft palate is an important site of adaptation for transmissible influenza viruses. Nature. 2015;526(7571):122–125. doi: 10.1038/nature15379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lan J., Ge J., Yu J., Shan S., Zhou H., Fan S., Zhang Q., Shi X., Wang Q., Zhang L., Wang X. Structure of the SARS-CoV-2 spike receptor-binding domain bound to the ACE2 receptor. Nature. 2020;581(7807):215–220. doi: 10.1038/s41586-020-2180-5. [DOI] [PubMed] [Google Scholar]
- MacLaughlan N.J., Dubovi E.J. 5th ed. Acdaemic Press; 2016. Fenner’s Veterinary Virology. [Google Scholar]
- Mallapaty S. Coronavirus can infect cats—dogs, not so much. Nature. 2020 doi: 10.1038/d41586-020-00984-8. [DOI] [PubMed] [Google Scholar]
- Martina B.E.E., Haagmans B.L., Kuiken T., Fouchier R.A.M., Rimmelzwaan G.F., Van Amerongen G., Peiris J.S.M., Lim W., Osterhaus A.D.M.E. Virology: SARS virus infection of cats and ferrets. Nature. 2003;425(6961):915. doi: 10.1038/425915a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masters P.S., Perlman S. Fields Virology. 6th ed. Lippincott, Williams and Wilkins; 2013. Coronaviridae; pp. 825–858. [Google Scholar]
- Olsen C.W., Corapi W.V., Ngichabe C.K., Baines J.D., Scott F.W. Monoclonal antibodies to the spike protein of feline infectious peritonitis virus mediate antibody-dependent enhancement of infection of feline macrophages. J. Virol. 1992;66(2):956–965. doi: 10.1128/jvi.66.2.956-965.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peck K.M., Scobey T., Swanstrom J., Jensen K.L., Burch C.L., Baric R.S., Heise M.T. Permissivity of dipeptidyl peptidase 4 orthologs to middle east respiratory syndrome coronavirus is governed by glycosylation and other complex determinants. J. Virol. 2017;91(19) doi: 10.1128/JVI.00534-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pedersen N.C. An update on feline infectious peritonitis: diagnostics and therapeutics. Vet. J. (Lond. Engl.: 1997) 2014;201(2):133–141. doi: 10.1016/j.tvjl.2014.04.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pedersen N.C., Liu H., Dodd K.A., Pesavento P.A. Significance of coronavirus mutants in feces and diseased tissues of cats suffering from feline infectious peritonitis. Viruses. 2009;1(2):166–184. doi: 10.3390/v1020166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raj V.S., Osterhaus A.D.M.E., Fouchier R.A.M., Haagmans B.L. MERS: emergence of a novel human coronavirus. Curr. Opin. Virol. 2014;5:58–62. doi: 10.1016/j.coviro.2014.01.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruiz-Arrondo I., Portillo A., Palomar A.M., Santibanez S., Santibanez P., Cervera C., Oteo J.A. Detection of SARS-CoV-2 in pets living with COVID-19 owners diagnosed during the COVID-19 lockdown in Spain: a case of an asymptomatic cat with SARS-CoV-2 in Europe [Preprint] medRXiV. 2020 doi: 10.1101/2020.05.14.20101444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- SARS-CoV-2/COVID-19, United States of America. (2020). https://www.oie.int/wahis_2/public/wahid.php/Reviewreport/Review?page_refer=MapFullEventReport&reportid=34086.
- Shang J., Ye G., Shi K., Wan Y., Luo C., Aihara H., Geng Q., Auerbach A., Li F. Structural basis of receptor recognition by SARS-CoV-2. Nature. 2020;581(7807):221–224. doi: 10.1038/s41586-020-2179-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi J., Wen Z., Zhong G., Yang H., Wang C., Huang B., Liu R., He X., Shuai L., Sun Z., Zhao Y., Liu P., Liang L., Cui P., Wang J., Zhang X., Guan Y., Tan W., Wu G. Susceptibility of ferrets, cats, dogs, and other domesticated animals to SARS–coronavirus 2. Science. 2020;368(6494):1016–1020. doi: 10.1126/science.abb7015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song Z., Xu Y., Bao L., Zhang L., Yu P., Qu Y., Zhu H., Zhao W., Han Y., Qin C. From SARS to MERS, thrusting coronaviruses into the spotlight. Viruses. 2019;11(1) doi: 10.3390/v11010059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sykes J.E. Canine and Feline Infectious Diseases. Elsevier; 2014. Feline coronavirus infection; pp. 195–208. [DOI] [Google Scholar]
- Takano T., Kawakami C., Yamada S., Satoh R., Hohdatsu T. Antibody-dependent enhancement occurs upon re-infection with the identical serotype virus in feline infectious peritonitis virus infection. J. Vet. Med. Sci. 2008;70(12):1315–1321. doi: 10.1292/jvms.70.1315. [DOI] [PubMed] [Google Scholar]
- Temmam S., Barbarino A., Maso D., Behillil S., Enouf V., Huon C., Jaraud A., Chevallier L., Backovic M. Absence of SARS-CoV-2 infection in cats and dogs in close contact with a cluster of COVID-19 patients in a veterinary campus. BioRXiV. 2020 doi: 10.1016/j.onehlt.2020.100164. https://www.biorxiv.org/content/10.1101/2020.04.07.029090v1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- USDA APHIS . USDA Animal and Plant Health Inspection Service; 2020. Confirmation of COVID-19 in Pet Dog in New York.https://content.govdelivery.com/accounts/USDAAPHIS/bulletins/28eae2e [Google Scholar]
- van den Brand J.M.A., Haagmans B.L., Leijten L., van Riel D., Martina B.E.E., Osterhaus A.D.M.E., Kuiken T. Pathology of experimental SARS coronavirus infection in cats and ferrets. Vet. Pathol. 2008;45:551–562. doi: 10.1354/vp.45-4-551. [DOI] [PubMed] [Google Scholar]
- Vennema H., Poland A., Foley J., Pedersen N.C. Feline infectious peritonitis viruses arise by mutation from endemic feline enteric coronaviruses. Virology. 1998;243(1):150–157. doi: 10.1006/viro.1998.9045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whittaker G.R., André N.M., Millet J.K. Improving virus taxonomy by recontextualizing sequence-based classification with biologically relevant data: the case of the alphacoronavirus 1 species. MSphere. 2018;3(1):e00463-17. doi: 10.1128/mSphereDirect.00463-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams B.H., Kiupel M., West K.H., Raymond J.T., Grant C.K., Glickman L.T. Coronavirus-associated epizootic catarrhal enteritis in ferrets. J. Am. Vet. Med. Assoc. 2000;217(4):526–530. doi: 10.2460/javma.2000.217.526. [DOI] [PubMed] [Google Scholar]
- Xia X. Extreme genomic CpG deficiency in SARS-CoV-2 and evasion of host antiviral defense. Mol. Biol. Evol. 2020 doi: 10.1093/molbev/msaa094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zamoto A., Taguchi F., Fukushi S., Morikawa S., Yamada Y.K. Identification of ferret ACE2 and its receptor function for sars-coronavirus. In: Perlman S., Holmes K.V., editors. Vol. 581. Springer; US: 2006. pp. 519–522. (The Nidoviruses). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y.-Z., Holmes E.C. A genomic perspective on the origin and emergence of SARS-CoV-2. Cell. 2020;181(2):223–227. doi: 10.1016/j.cell.2020.03.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Q., Zhang H., Huang K., Yang Y., Hui X., Gao J., He X., Li C., Gong W., Zhang Y., Peng C., Gao X., Chen H., Zou Z., Shi Z., Jin M. SARS-CoV-2 neutralizing serum antibodies in cats: A serological investigation [Preprint] bioRXiv. 2020 doi: 10.1101/2020.04.01.021196. [DOI] [Google Scholar]
- Zhou P., Yang X.-L., Wang X.-G., Hu B., Zhang L., Zhang W., Si H.-R., Zhu Y., Li B., Huang C.-L., Chen H.-D., Chen J., Luo Y., Guo H., Jiang R.-D., Liu M.-Q., Chen Y., Shen X.-R., Wang X. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature. 2020;579:270–273. doi: 10.1038/s41586-020-2012-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.