Abstract
This first International Alliance for Biological Standardization Covid-19 webinar brought together a broad range of international stakeholders, including academia, regulators, funders and industry, with a considerable delegation from low- and middle-income countries, to discuss the virology, epidemiology and immunology of, and the vaccine development for SARS-CoV-2.
Keywords: SARS-CoV-2, Virology, Epidemiology, Immunology, Vaccine development
Highlights
-
•
In SARS-CoV-2, genetic diversity exists but has limited impact on antigenicity.
-
•
Second and possibly third SARS-CoV-2 waves are expected, only to be controlled with safe and effective vaccines.
-
•
Immunopathology plays a role in severe Covid-19, with a role for macrophages, T lymphocytes and potentially antibodies.
-
•
Normally taking many years, under the outbreak paradigm, the goal is to achieve vaccine development within 12–18 months.
1. Introduction
The International Alliance for Biological Standardization (IABS,1 https://www.iabs.org) is devoted to the scientific and medical advancement of biologicals, by facilitating communication among those who develop, produce and regulate biological products for human and animal health. Towards this end, IABS organized a webinar on Covid-19 to provide open access information to a broad range of stakeholders from all continents, including nearly 200 participants from 26 low- and middle-income countries. The webinar consisted of four presentations, followed by ample time for discussion between speakers and participants.
1.1. Virology
Bruno Lina, professor of Virology at the University Claude Bernard Lyon and Director of the research laboratory VirPath, France, provided an overview of the virologic aspects of the new pandemic coronavirus (CoV) strain SARS-CoV-2. The origins of the virus lie in China. Despite rumors, there is no scientific evidence that it is a laboratory-produced virus. The major risk factor is contact of humans with live wild animals at markets, which is not uncommon in East Asia.
At a very early stage, compared to previous pandemics, electron microscopy images and the full genome sequence were available, providing targets for vaccine development. The GISAID Initiative [https://www.gisaid.org], which promotes the rapid sharing of data from SARS-CoV-2 (as well as data from influenza viruses) has collected more than 34,000 SARS-CoV-2 genomes, from labs around the world. Data show that the virus has a degree of similarity to viruses obtained from bats and pangolins. Based on genetic diversity, three genogroups are defined, groups G, V and S, of which the G group is currently most prominent and has been additionally subdivided in 3 sub-groups (G, GR and GH). However, the genetic diversity seems to have limited impact on antigenicity.
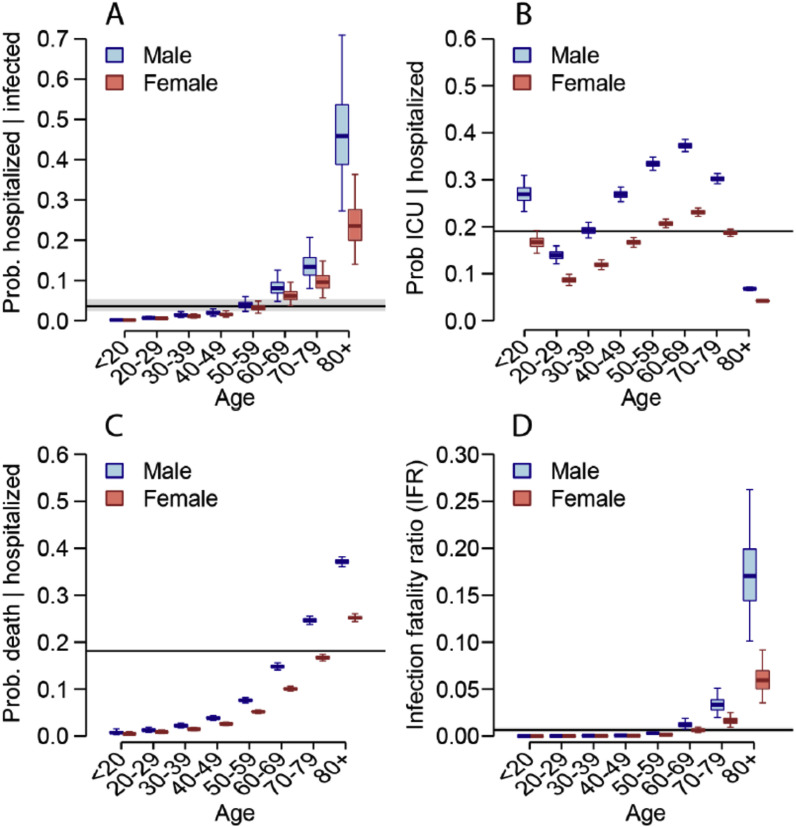
Based on French data [1], the mean age of hospitalized patients was 68 years of age, while the mean age at time of death was 79 years of age. Half of the hospitalizations and more than 80% of deaths occurred in individuals older than 70 years. Furthermore, 56.2% of hospitalizations and 60.3% of deaths were male (Fig. 1 ) [1].
Fig. 1.
Sex differences in hospitalization, ICU admissions and death due to Covid-19
Source: Salje et al. Science, 2020.
The availability of the sequence data has also enabled development of diagnostic tools. Diagnosis is usually based on nasopharyngeal swabs (a sample from the upper respiratory tract). Using real-time (RT-)PCR, a high accuracy can be obtained (combining high sensitivity with high specificity) but variation exists between different tests [2]. This molecular assay can be used to identify infected patients but also to monitor individuals to monitor/prevent spread of the infection. While the virus can also be found in stool using molecular assays, the value of this finding is as yet unclear, but detection does not seem to be related to disease severity.
Using a semi-quantitative RT-PCR on different samples (nasopharyngeal swab, blood, urine, and stool samples), three different clinical and biological types of evolution in five patients infected with SARS-CoV-2 could be observed [3]: (1) paucisymptomatic individuals, with high nasopharyngeal titres within the first 24 h of the illness onset but early recovery; (2) a two-step disease progression, with a secondary worsening around 10 days after disease onset with a decreasing viral load in nasopharyngeal samples and a shift to detection of virus in the lower respiratory tract; and (3) a rapid evolution towards multiple organ failure and a persistent high viral load in lower and upper respiratory tract with systemic virus dissemination and virus detection in plasma [3].
Antibody detection methods have been developed to investigate seroepidemiology, including ELISAs, lateral flow assays and virus neutralization assays, although the latter need to be performed in biosafety level 3 facilities. These assays provide insight into the number of previously infected individuals. However, different assays and different antigens within an assay will provide different pictures, therefore, it is essential to know which test was used, to be able to interpret the data.
Regarding the immunological response to infection, in three patients, viral loads were low with no obvious quantitative difference related to severity [4]. Interleukin(IL)-6, C-reactive protein and interferon(IFN) γ-induced protein 10 were elevated in the two symptomatic patients. Strikingly, no IFN-α2 was detectable in these two patients. In contrast, IL-6, C-reactive protein and IFN γ-induced protein 10 remained low during the hospital isolation stay for the asymptomatic individual and a significant elevation of plasmatic IFN-α2 was observed [4].
Tracking immunological markers from illness onset, it was shown that the baseline lymphocyte count was significantly higher in survivors than in non-survivors [5]. In survivors, lymphocyte count was lowest on day 7 after illness onset and improved during hospitalization, whereas severe lymphopenia was observed in non-survivors. Levels of IL-6 were clearly elevated in non-survivors compared with survivors throughout the clinical course and increased with illness deterioration.
Towards a diagnostic strategy, the data seem to show all infected individuals will develop an immune response (seroconversion), although earlier with higher severity. After 14–21 days, neutralizing antibodies can be detected but it is as yet unclear whether these neutralizing antibodies persist.
In all infected individuals, the virus can be detected in the upper respiratory tract at high loads from day 0. In mild cases, the virus will no longer be detectable after 20 days, whereas severe cases will shed virus beyond this time point. Furthermore, in severe cases there will be a shift from upper to lower respiratory tract.
1.2. Epidemiology
Arnaud Fontanet, Director of the Department of Global Health, Head of the Emerging Diseases Epidemiology Unit, Institut Pasteur, France, discussed the epidemiology of SARS-CoV-2, using France as an example. As other countries, France was hit hard by the epidemic, particularly in the Eastern parts and in the capital, Paris. Overall, 4.4% of the population was infected and the reproduction number, R0, was estimated at 2.9, indicating 66% of the population (100 – [1/R0]) should become infected to achieve herd immunity. Now that the first wave has passed, what can be expected in the coming months? Looking at human CoV types OC43, 229E, HKU1, and NL63, in a cohort from Ann Harbor, Michigan, U.S., distinct seasonality in the identifications was observed. Only 9 (2.5%) of the total 364 CoV-associated acute respiratory infections occurred between June and September. The number of identifications for each virus increased in December, peaked in January or February, and began to decrease in March. Furthermore, the seasonal similarity between the four types is striking, with only the peak aggregate month differing between January and February [6]. Seasonality was also observed for SARS-CoV-1 [7], whereas no seasonality could be observed for MERS, showing several peaks without a clear pattern [8].
The virus is highly stable at 4 °C, but sensitive to heat. At 4 °C, there was only around a 0·7 log-unit reduction of infectious titre on day 14, compared to a 3 log-unit reduction at 22 °C after 14 days, and a 3 log-unit reduction at 37 °C after 1 day [9]. This suggests that the summer season in the northern hemisphere may have an impact on spread, also because people spend time outdoors during summertime, while respiratory viruses are known to spread more effectively indoors. Nevertheless, it cannot be excluded that a second wave already occurs during summer.
In Copenhagen, the first wave of the 1918–1919 Spanish flu pandemic was marked by high transmissibility, substantial morbidity, and low mortality [10]. Despite the few deaths attributed to influenza in the summer wave, its general patterns were otherwise characteristic of the 1918 influenza pandemic overall. First, its peak morbidity rate was approximately 300-fold higher than that of any other summer during 1910–1917 and was approximately 50% that of the fall wave. Second, the mortality rate was highest in young adults and lower in elderly. In contrast, the second wave during autumn had a lower R0 but a higher case fatality rate. The third wave had an intermediate case fatality rate, and deaths were more equally spread over age groups.
In 2009–2010, despite the low level of susceptibility to H1N1 expected to be remaining in the population after the second wave, the H1N1 pandemic flu also showed third waves in New York, USA [11] and the UK [12]. This was explained by substantially increased transmissibility of the H1N1 in combination with cold weather at the time of transmission [12].
Based on these findings, SARS-CoV-2 is expected to be milder in summer, due to seasonality combined with a lower R0 due to spending more time outside. However, second and possibly third waves are expected and cannot be controlled until one or more safe and effective vaccines have been developed. Until that time, the only option is to maintain social distancing and focus on the development of therapeutics and vaccines.
1.3. Immunology
Arnaud Marchant, Director of the Institute for Medical Immunology, Université libre de Bruxelles, Belgium, discussed the disease from an immunologist's perspective. While it starts with a viral infection, Covid-19 shows clear signs of an inflammatory disease, with massive infiltration of immune cells in the tissue, most prominently the lungs [13]. But markers of inflammation are also found in the blood: cytokines, chemokines, C-reactive protein and fibrin degradation products, and correlate with disease severity.
When cells are infected by SARS-CoV-2 and the virus starts replicating, in a healthy immune response, the cells are rapidly destroyed by innate and adaptive immune effectors, and the released viral particles inactivated by neutralizing antibodies [14]. Although the immunological determinants of severe disease are not yet established, it is associated with tissue infiltration of inflammatory cells, including macrophages and T lymphocytes. These cells generate pro-inflammatory cytokines and chemokines, attracting further inflammatory cells to the site, creating a pro-inflammatory feedback loop, potentially leading to damage to the tissues. Moreover, the cytokine storm may lead to multi-organ damage [14]. Macrophages are thought to play an important role in this cascade. Monocytes are recruited to the site of infection by chemokines produced by epithelial and endothelial cells. Reaching the tissue, they differentiate in macrophages and are potentially activated by multiple signals, including viral RNA, inflammatory cytokines produced by neighboring inflammatory cells as well as complexes formed by viral antigens and antibodies [15]. They then start contributing themselves to the excessive inflammation and to the activation of intravascular coagulation.
Similarly, T lymphocytes could play an important role in control of SARS-CoV-2 and in the development of immunopathology, as severe Covid-19 is associated with lymphopenia, which might reflect recruitment of lymphocytes to the organs, or depletion of lymphocytes. Severe disease is associated with T cell exhaustion, IL-6 producing T-cells, and a reduction in the production of regulatory cytokines. Together, these alterations may impair viral control and promote inflammatory responses [16].
While a prompt and efficient type 1 IFN leads to viral control, a delayed response allows viral replication, and may pave the way for an exacerbated response resulting in tissue damage. As SARS-CoV-1 and MERS interfere with type 1 IFN responses, SARS-CoV-2 is likely to impact this key anti-viral pathway as well [16].
Our current understanding of the role of the immune system in the pathogenesis of COVID-19 suggests a potential for immune-based therapeutic interventions, including a strive for immune modulation by the use of anti-cytokine antibodies; or antibody-based therapies. Additional immune targets will be identified as our understanding of the disease progresses.
Some key questions remain open. Are people who were previously infected protected against reinfection? The potential for reinfection has been discussed [17]. Studies should determine the persistence of antibodies as well as memory B-cells and T-cells following symptomatic as well as asymptomatic infection. Nevertheless, the aim of vaccines is to induce neutralizing antibodies and there could be an advantage of inducing cytotoxic T-lymphocytes. Currently, around 200 vaccine candidates are in development based on one of the following strategies: live-attenuated or inactivated virus; recombinant subunits in combination with adjuvants; recombinant viral vectors or bare nucleic acids. If all these strategies have the potential to induce neutralizing antibodies, they differ in their capacity to induce cytotoxic T cells. Another approach involves the induction of pathogen-agnostic immunity using live-attenuated vaccines e.g. BCG. Studies are ongoing to determine the potential of this approach [18,19]. When developing vaccines, it will be essential to avoid disease enhancement, as observed with dengue or inactivated respiratory syncytial virus vaccines. Finally, an impact of vaccination on virus replication in the upper respiratory tract would increase its efficacy at the population level.
In conclusion, there is strong evidence for a role of immunopathology in severe Covid-19, with a dual role for macrophages, T lymphocytes and potentially antibodies. Although the determinants of severe disease are as yet unknown in relation to risk factors, there is a high potential for therapeutic interventions targeting the immune system. Defining how long natural immunity protects against reinfection in diverse patient populations will be key until vaccines become available.
1.4. Vaccine development
Melanie Saville, Head of Vaccine R&D, CEPI Vaccines (www.cepi.net), described the role of CEPI in vaccine development. CEPI is an innovative global partnership between public, private, philanthropic, and civil society organizations, launched in Davos in 2017, with the mission to accelerate the development of vaccines against emerging infectious diseases and enable equitable access to these vaccines for people during outbreaks. In the pre-Covid era, research was focused on five pathogens: Lassa, MERS, Nipah, Rift Valley Fever and Chikungunya. Of these, MERS is also a CoV, which has helped in accelerating vaccine development for SARS-CoV-2. For the advancement of vaccine candidates for each of the pathogens, a science portfolio is needed, including an antibody standard, standardization of assays, the antigen, an animal model for preclinical testing, diagnostics and epidemiological studies.
CEPI aims to get a vaccine through to clinical testing in 16 weeks, which is an extremely ambitious timeline, unprecedented in the field of infectious diseases.
Partnerships and programmes of work have been announced which will leverage rapid response platforms already supported by CEPI. The aim is to advance Covid-19 vaccine candidates into clinical testing as quickly as possible. This includes:
-
1)
A DNA platform with electroporation for delivery (Inovio). Already having developed a MERS vaccine candidate currently in clinical trials, the SARS CoV2 builds on that knowledge to deliver optimized synthetic antigenic genes into cells. These will then be translated into antigens that activate an individual's immune system to generate robust immune response. The SARS CoV2 vaccine candidate is in Phase I clinical trials.
-
2)
A recombinant protein platform (University of Queensland). This “molecular clamp” technology works by synthesizing viral surface proteins and “clamping” the protein in a pre-fusion form to optimize the immune response.
-
3)
An mRNA platform (Moderna). An mRNA vaccine has been manufactured against the novel CoV spike protein to conduct investigational drug studies to decide whether it is safe to progress to the next stage of clinical trials. This vaccine moved into phase I clinical trial testing in just 10 weeks after sequence identification and is currently in phase II
-
4)
A second mRNA platform (CureVac). This platform aims to optimize the properties of mRNA therapeutics and vaccines. The technology can be tailored to induce varying degrees of immune responses against antigens of choice, potentially providing potent prophylactic vaccines for the prevention of infectious diseases.
Other platforms include nanoparticles (Novavax) in phase I clinical testing, viral vectors including a measles vector (Institut Pasteur/Themis) in preclinical testing and ChadOx1 adenovirus vector (university of Oxford/AstraZeneca) in phase I/II and a live attenuated influenza vector (University of Hong Kong) in preclinical testing, as well as subunit platforms (Clover Biopharma) in preclinical testing. Most vaccines in development are based on the viral spike protein.
On the 31st of December 2019, the WHO notified a pneumonia-like case cluster in Wuhan, China. On 7 January 2020, CEPI activated a response and within 4 months more than 200 vaccine candidates are in development, of which at least ten are in phase 1+ clinical testing, including nine supported by CEPI, using different platforms.
Normally, vaccine development takes many years, if not decades, with all steps in sequence. Under the outbreak paradigm, the goal is to achieve this within 12–18 months, running preclinical development parallel to early clinical development and starting manufacturing scale up and scale out early in development and even starting manufacturing of doses before licensure.
This can only be achieved if the whole process, including regulation is streamlined. CEPI will prepare a portfolio for regulatory support and guidance for vaccine candidates. Furthermore, CEPI will reach out to National Regulatory Agencies, for the evaluation of regulatory aspects for rapid access to licensed/marketed vaccine and framing of product agnostic questions for official review and comment.
First-in-human studies will be performed on healthy adults for dose selection, safety and reactogenicity, excluding the at-risk population of older adults). During Phase Ib/IIa trials, the population will be expanded to include at risk populations. For vaccine efficacy, cases of COVID19 will be captured from phase I, to support development of a case definition and potential integrated analyses across early stage trials. Furthermore, an adaptive design will be used, with a number-of-events approach, subject to Covid-19 incidence. Finally, regarding safety, a safety database will be set up prior to market access; the number of subjects followed for safety will depend on discussions with regulators. Data on Vaccine-Mediated Enhanced Disease will be derived from animal models, immune response characterisation as well as monitoring throughout clinical development (and post-licensure). CEPI, together with the Safety Platform for Emergency vACcines (SPEAC) will develop case definitions for adverse events of special interest and set up a meta Data Safety Monitoring Board to evaluate safety across projects. Simultaneously, manufacturing is also scaled-up at risk (before clinical trial data is available) and scaled-out (the best candidates will be produced in multiple countries, potentially by multiple manufacturers) to maximize doses available upon licensure.
To support vaccine development projects and prepare for vaccine use in clinical trials and in outbreak response settings, biological standards and assays will be developed and access will be provided to laboratory analyses of preclinical and clinical samples. Secondly, animal models will be developed, and access to laboratories for vaccine testing will be secured. Thirdly, epidemiological modelling of the public health impact of a vaccine on the disease burden and transmission, including the possibility of herd immunity will be done using the following parameters: target populations, duration of protection, onset of immune response, dosing schedules, dosing regimen, adherence, vaccine coverage, vaccine effectiveness, and others. Global seroprevalence, incidence and mortality data of Covid-19 will be gathered.
Finally, CEPI is fully committed to fair allocation of the vaccine upon licensure. Equitable access to epidemic vaccines — in the context of an outbreak — means that appropriate vaccines are first available to populations when and where they are needed to end an outbreak or curtail an epidemic or pandemic, regardless of ability to pay. Global health authorities and manufacturers will have to work closely to increase the supply and ensure access to the vaccine for the most vulnerable populations. It is anticipated that vaccine will initially be in short supply and distribution will be prioritized to those who are identified as being at the highest risk.
2. Discussion
Regarding seasonality, it was discussed whether seasonal vaccines would be needed, as for influenza. But it is too early to say, first of all, the genetic drift is currently unknown, and secondly the duration of post-vaccine protection is unclear. In analogy, MERS antibodies do wane, but it is unclear how quickly. As previously discussed, some reinfections with SARS-CoV-2 have been described but this could be due to waning or inadequate antibody levels after infection. An annual booster may therefore be necessary, perhaps with a (slightly) adapted vaccine. It is as yet unclear whether overcoming a natural infection is protective and whether we can rely on herd immunity. However, T lymphocytes are frequent, so we may benefit from previous exposure.
The need for a booster may also depend on the platform used. Live attenuated vaccines generally provide long-term protection but take a long time to develop, so is unlikely to be readily available soon. The platform also has consequences for production: billions of doses will be needed. This is theoretically possible for the nucleic acid-based vaccines, although production is not at that scale yet.
Towards speeding up vaccine development, the number of people needed to be included in clinical trials was discussed. The classic number of people in safety studies is at least 3000 people but this may depend on the vaccine platform, and how well characterized the platform is for safety.
Regarding potential risk populations, it was discussed whether a vaccine would be suitable for pregnant women. Some platforms may have better safety profile, but this will have to be tested in real life.
Will Africa become the next epicenter? It is often the last continent to be hit, as it is less connected, with fewer travelers. On the other hand, Africans may have a more responsive immune response, the virus may be impaired by the higher temperature, and due to more outdoor living and lower population density, the virus may have a lower infectivity. However, Africa also has some big cities. It will be interesting to see what happens there.
The infectious dose is currently unknown, as there are no good animal models yet. Using high doses, 108 plaque forming units, animals can become infected, but transmission is less investigated. The early stage of disease, subjects are highly contagious, much more than in the case of flu or RSV.
The virus databank, currently holding over 20,000 genomes provides a good global picture with lots of data from Asia, Europe and the US. Data from Africa are slower to come in.
The difference between males and females regarding disease severity was also discussed. Although there is no clear explanation, in infectious diseases, males are often more severely ill than women. Women have a qualitatively different immune response than men, possibly it is more active against SARS-CoV-2.
Children are at a lower risk of severe disease. Most children are asymptomatic and (consequently) seroconversion is also less frequent. However, no epidemics have been retraced to schools, suggesting children do not seem to spread the infection.
Concerning the approach taken by different countries, South Korea had more experience with epidemics, and hence, was better prepared. However, the virus is still circulating and may re-appear. Taiwan, being an island, could more easily shut down travel and avoid importation of new cases. In Europe, Germany followed the South Korean approach in isolating cases at the earliest possibility. Nevertheless, the biggest difference between Germany on one hand and Italy and Spain on the other hand seems to be contact with the elderly population: a difference in frequency of contact with elderly directly impacts mortality in this elderly population, which is most vulnerable.
Vaccine Mediated Enhanced Disease (VMED) was the focus of discussion, could it be antigen associated? The spike protein is the antigen of choice for the vaccine, associated with a strong (neutralizing) antibody response. This antigen was also shown to be the best antigen in MERS. Using the nucleoprotein may be less risky in eliciting VMED but this antigen was not successful in SARS. To be able to look at VMED, good animal models are needed. Nevertheless, even with those, there will be some remaining risk going into clinical trials, which is impossible to avoid as animal models may not fully predict effects in humans. Antibody Dependent Enhancement (ADE) as a second mechanism for enhanced disease has been hypothesized as having a potential link to enhanced disease for coronaviruses and is mostly an in vitro phenomenon, but people may be at risk after waning of the antibody response, with dengue as an example. ADE will remain a difficult topic, for which careful monitoring will remain necessary.
The role of human challenge trials in SARS-CoV-2 development was discussed. A World Health Organization Working Group is currently discussing this topic, which is an ethical challenge [20]. A recommendation is expected soon. However, in the absence of clear treatment of disease, human challenge trials are not recommended [21].
Finally, the most effective treatment was discussed. It was strongly advocated that hydroxychloroquine should not be used based on rumors but only within the setting of a clinical trial. This will lead to a comparison of benefits and risks. So far, the picture is that there is no clear benefit (no reduction in disease severity), whereas there are risks associated with the use (increased number of cardiological events in treatment group). Meta-analysis of ongoing clinical trials should provide a definitive answer. On the other hand, monoclonal antibodies do seem to be able to play a key role and can be manufactured in suitable quantities. These monoclonal antibodies are almost in phase 1 clinical trials which will hopefully show their value.
In conclusion, vaccine development is continuing at unprecedented speed, but vaccines are not to be expected in large volumes until 2021. Therefore, until that time, we will have to stick to social distancing and perhaps monoclonal antibodies as treatment. New waves of SARS-CoV-2 infections are likely to occur, with unpredictable height and breadth of the waves, although the experience from previous pandemics is not reassuring.
Declaration of competing interest
The authors have no competing interests to declare.
Acknowledgements
The authors are grateful for the support by a grant of Wellcome Trust to organize the webinar; and would like to thank Madinina Cox and Abigail Charlet (International Alliance for Biological Standardization) for logistic management.
Footnotes
ADE – antibody-dependent enhancement; CoV – coronavirus; IABS – International Alliance for Biological Standardization; ICU – intensive care unit; IFN – interferon; IL – interleukin; RT-PCR – real-time PCR; SPEAC –Safety Platform for Emergency vACcines; VMED - Vaccine Mediated Enhanced Disease.
Contributor Information
Marc Baay, Email: marc.baay@p-95.com.
Bruno Lina, Email: bruno.lina@univ-lyon1.fr.
Arnaud Fontanet, Email: arnaud.fontanet@pasteur.fr.
Arnaud Marchant, Email: arnaud.marchant@ulb.be.
Melanie Saville, Email: melanie.saville@cepi.net.
Philippe Sabot, Email: philippe.sabot@iabs.org.
Philippe Duclos, Email: Philippe.Duclos@unige.ch.
Joris Vandeputte, Email: joris.vandeputte@iabs.org.
Pieter Neels, Email: Pieter.neels@vaccine-advice.be.
References
- 1.Salje H., Tran Kiem C., Lefrancq N., Courtejoie N., Bosetti P., Paireau J. Science; New York, NY): 2020. Estimating the burden of SARS-CoV-2 in France. eabc3517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Etievvant S., al e. Performance assessment of SARS-CoV-2 PCR assays developed by WHO referral laboratories. J Clin Med. 2020;9(6):E1871. doi: 10.3390/jcm9061871. In Press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lescure F.X., Bouadma L., Nguyen D., Parisey M., Wicky P.H., Behillil S. Clinical and virological data of the first cases of COVID-19 in Europe: a case series. Lancet Infect Dis. 2020;20(6):697–706. doi: 10.1016/S1473-3099(20)30200-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Trouillet-Assant S., Viel S., Gaymard A., Pons S., Richard J.C., Perret M. Type I IFN immunoprofiling in COVID-19 patients. J Allergy Clin Immunol. 2020 doi: 10.1016/j.jaci.2020.04.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zhou F., Yu T., Du R., Fan G., Liu Y., Liu Z. Clinical course and risk factors for mortality of adult inpatients with COVID-19 in Wuhan, China: a retrospective cohort study. Lancet. 2020;395:1054–1062. doi: 10.1016/S0140-6736(20)30566-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Monto A.S., DeJonge P., Callear A.P., Bazzi L.A., Capriola S., Malosh R.E. Coronavirus occurrence and transmission over 8 years in the HIVE cohort of households in Michigan. J Infect Dis. 2020;222(1):9–16. doi: 10.1093/infdis/jiaa161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.World Health Organization Epidemic curves - severe acute respiratory syndrome (SARS) https://www.who.int/csr/sars/epicurve/epiindex/en/index1.html Available from: Last accessed: (last accessed 28 May 2020.
- 8.World Health Organization Middle East respiratory syndrome coronavirus (MERS-CoV) https://www.who.int/emergencies/mers-cov/en/Last Available from: accessed: (last accessed 28 May 2020.
- 9.Chin A., Chu J., Perera M., Hui K., Yen H., Chan M. Stability of SARS-CoV-2 in different environmental conditions. Lancet Microb. 2020;1:e10. doi: 10.1016/S2666-5247(20)30003-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Andreasen V., Viboud C., Simonsen L. Epidemiologic characterization of the 1918 influenza pandemic summer wave in Copenhagen: implications for pandemic control strategies. J Infect Dis. 2008;197:270–278. doi: 10.1086/524065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Viboud C., Charu V., Olson D., Ballesteros S., Gog J., Khan F. Demonstrating the use of high-volume electronic medical claims data to monitor local and regional influenza activity in the US. PloS One. 2014;9 doi: 10.1371/journal.pone.0102429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dorigatti I., Cauchemez S., Ferguson N.M. Increased transmissibility explains the third wave of infection by the 2009 H1N1 pandemic virus in England. Proc Natl Acad Sci U S A. 2013;110:13422–13427. doi: 10.1073/pnas.1303117110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ackermann M., Verleden S.E., Kuehnel M., Haverich A., Welte T., Laenger F. Pulmonary vascular endothelialitis, thrombosis, and angiogenesis in Covid-19. N Engl J Med. 2020 doi: 10.1056/NEJMoa2015432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tay M.Z., Poh C.M., Rénia L., MacAry P.A., Ng L.F.P. The trinity of COVID-19: immunity, inflammation and intervention. Nat Rev Immunol. 2020:1–12. doi: 10.1038/s41577-020-0311-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Merad M., Martin J.C. Pathological inflammation in patients with COVID-19: a key role for monocytes and macrophages. Nat Rev Immunol. 2020:1–8. doi: 10.1038/s41577-020-0331-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Vabret N., Britton G.J., Gruber C., Hegde S., Kim J., Kuksin M. Immunity; 2020. Immunology of COVID-19: current state of the science. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kellam P., Barclay W. The dynamics of humoral immune responses following SARS-CoV-2 infection and the potential for reinfection. J Gen Virol. 2020 doi: 10.1099/jgv.0.001439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Miller A., Reandelar M.J., Fasciglione K., Roumenova V., Li Y., Otazu G.H. Correlation between universal BCG vaccination policy and reduced morbidity and mortality for COVID-19: an epidemiological study. medRxiv. 2020:2020. 03.24.20042937. [Google Scholar]
- 19.Hamiel U., Kozer E., Youngster I. SARS-CoV-2 rates in BCG-vaccinated and unvaccinated young adults. Jama. 2020;323(22):2340–2341. doi: 10.1001/jama.2020.8189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.WHO Working Group for Guidance on Human Challenge Studies in COVID-19 Key criteria for the ethical acceptability of COVID-19 human challenge studies. https://www.who.int/ethics/publications/key-criteria-ethical-acceptability-of-covid-19-human-challenge/en/ Available from: [DOI] [PMC free article] [PubMed]
- 21.Baay M.F.D., Richie T.L., Neels P. vol. 61. Biologicals; Rockville, MD, USA: 2019. pp. 85–94. (Human challenge trials in vaccine development). September 28-30, 2017. [DOI] [PubMed] [Google Scholar]