Abstract
The crystal structures of 2-(1H-indol-3-yl)-4-phenyl-5H-indeno [( Cheng et al., 2007; Lee et al., 2003) 1,21,2-b]pyridine-3-carbonitrile (Ia) and 2-(1H-indol-3-yl)-4-(4-methoxyphenyl)-5H-indeno [( Cheng et al., 2007; Lee et al., 2003) 1,21,2-b]pyridine-3-carbonitrile (Ib) were determined using single crystal X-ray diffraction. Both the compounds belong to the triclinic system with the P-1 space group. The azafluorene ring system in both the compounds is effectively planar. The intermolecular interactions present in the compounds are discussed using Hirshfeld surface analysis, QTAIM and NCI. Compound Ib formed a strong interaction (−24.174 kJ/mol) with the solvent molecule. Both the compounds were geometry optimized using DFT/B3LYP level of theory. The compound’s drug-like behaviors were studied using HOMO-LUMO analysis. The homology modeling of SARS CoV-2 RdRp was done utilizing the PDB 6NUR_A as a template. The model showed above 99% similarity with its preceder SARS CoV. The molecular docking analysis of the synthesized compounds was carried out along with some suggested drugs for COVID-19 and some phytochemicals. The docking results were then analyzed. The binding free energy of the complexes were calculated using MM-PB(GB)SA and ADMET properties of Ia and Ib were also predicted. Some suggestions are given from this analysis.
Keywords: SARS Coronavirus, Homology modeling, RdRp, Azafluorene, ADMET, MM-GBSA
Graphical abstract
1. Introduction
Coronaviruses (CoV) in humans can cause the common cold to Severe Acute Respiratory Syndrome (SARS). At the beginning of the 21st century, the CoV outbreak was identified in the southern part of China called SARS, spread more than 25 countries with the lethal rate of 10% [1,2]. Next broke out of the corona was happened on 2012 in Saudi Arabia, named Middle East Respiratory Syndrome (MERS) having a fatality rate of 3.5% [3,4]. A new public life threat was identified at the end of 2019, named as novel coronavirus - 2019, and originally emerged from the city Wuhan, China [5]. The World Health Organization (WHO) announced a CoV outbreak on 30th Janurary 2020. On February 11, 2020 the disease was named as COVID-19 (Corona Virus Disease-19) by WHO. According to the situation report-84 released by WHO on April 13, 2020, more than 185 countries and territories were affected by this virus with the number of confirmed cases of 17,73,084 and 1,11,652 deaths. From the situation report-46 released on March 06, 2020, the mortality rate is 3–4%, which is greater than the influenza virus. To date, there are no dedicated drugs or vaccines are approved for treating such CoV’s. Therefore, CoV’s now become a global life threat to humans.
Basically, there are four types of CoV’s namely α, β, γ and δ were identified. Among the four γ and δ affect birds, whereas, α and β affect mammals [6]. This SARS CoV-2 is belonging to β-CoV with four proteins, namely, spike (S) protein (binds with the host cell ACE2 receptor), membrane (M) protein (act as an organizer of CoV assembly), envelope (E) protein (interacts with membrane protein to form an envelope) and nucleocapsid (N) protein (viral RNA genome replication) [7].
Treating such CoV includes, increase the native human immune system and inhibit the replication process of CoV inside the human body. The replication process can be inhibited by targeting one of the proteins, mainly, the nucleocapsid protein. RNA dependent RNA polymerase (RdRp) is one of the enzymes, which catalyze the RNA replication and transcription process. Some nucleotide analogous drugs like Remdesivir, galidesivir target this RdRp and interrupt as a nucleotide in the process of replication. These nucleotide analogous drugs were used as an anti-viral drugs against some diseases caused by RNA viruses like Ebola, HIV and Zika virus [[8], [9], [10], [11]]. Anti-viral drugs such as Lopinavir and Ritonavir showed inhibitory action against SARS CoV-2 [12]. Chloroquine and Hydroxychloroquine are also suggested for the COVID-19 treatment. Chloroquine and Remdesvir showed inhibition activity against SARS CoV-2 in-vitro [13].
Azafluorenes have attracted researchers towards its side by showing some moderate to good biological activities. The alkaloid extracted from the plant Polyalthia debilis contains 4-azafluorene derivatives showed anti-microbial, anti-malarial and cytotoxic activities [14]. Naturally obtained onychine showed anti-fungal activity against candida albicans [15] also anti-microbial activity against Staphylococcus aureus [16]. Further, these derivatives act as an adenosine A2A receptor antagonist [17]. These derivatives are effective for neurodegenerative diseases [18] by having anti-depressant property [19]. Some derivatives are reported to have Phosphodiesterease inhibitory [20], cytotoxic [21], anti-inflammatory [22], anti-oxidant [23] and anti-histamine [24,25] properties. Girgis and co-workers showed the vasodilating and bronchodilating properties of azafluorene derivatives for the treatment of Ashtma [26].
The pyridine skeleton attracts more importance among chemists and biologists because of its abundance in nature. The naturally occurring pyridine containing compounds having vast applications in pharmaceutics. The organic compounds containing pyridine are an important class of HIV drugs, which inhibit RNA dependent DNA polymerase, and hence act as reverse transcriptase inhibitors. Some ruthenium complexes of pyridine show anti-cancer, anti-tumor and anti-viral activities [27]. Pyridine fused with indole compounds were screened for their anti-tumor activities are also shown moderate to good anti-bacterial activity against Staphylococcus aureus and Pseudomonas aeroginosa [28].
Vincristine, Vinblastine, Vinorelbine and Vindesine are vinca alkaloids used as intravenous drugs (anti-mitotic drugs) prescribed for the treatment of various types of cancer, such as, lung cancer, breast cancer, leukaemia, melanoma and lymphoma [29,30]. These drugs bind to the β-tubulin subunit and changes the conformation of tubulin, thus they control the cell mitotic process (cell division) [31].
The repurposing of approved drugs were already done by many research groups [32] and which are still in the trials. Therefore, this work demonstrates the synthesis, structural and packing analysis of two indole containing azafluorene derivatives. The intermolecular hydrogen bonding interactions are qualified and quantified using Hirshfeld surface analysis, QTAIM (Quantum Topological Atoms In Molecules) and NCI (Non-Covalent Interaction) analyses. Also, the homology modeling of the structure of RNA dependent RNA polymerase (RdRp) of SARS CoV-2 is discussed due to the unavailability of three dimensional PDB (Protein Data Bank) structure. Finally, the drugs to treat COVID-19 are suggested based on the results of in-silico molecular docking analysis.
2. Materials and methods
2.1. Synthesis
Compounds C27H17N3 (Ia) and C28H19N3O, C2H6OS (Ib) were synthesized using the following procedure.
2.1.1. 2-(1H-indol-3-yl)-4-phenyl-5H-indeno [1,2-b]pyridine-3-carbonitrile (Ia)
A mixture of 3-(1H-indol-3-yl)-3-oxopropanenitrile (0.1 g, 0.543 mM), benzaldehyde (0.543 mM), ammonium acetate (0.1 g, 1.1 mM) and 2,3-dihydro-1H-inden-1-one (0.543 mM) was dissolved in ethanol (10 ml) and heated to reflux on a heating mantle for 2 h. After completion of the reaction as evident from TLC, the reaction mixture was set aside at ambient temperature for 6–7 h. The precipitate formed was filtered and dried to get a pure product. The crystals were obtained from the slow evaporation technique by dissolving the product in DMSO. (Yield: 81%; m. p: 267–268 °C) (Fig. S1).
2.1.2. 2-(1H-indol-3-yl)-4-(4-methoxyphenyl)-5H-indeno [1,2-b]pyridine-3-carbonitrile (Ib)
A mixture of 3-(1H-indol-3-yl)-3-oxopropanenitrile (0.1 g, 0.543 mM), 4-methoxybenzaldehyde (0.543 mM), ammonium acetate (0.1 g, 1.1 mM) and 2,3-dihydro-1H-inden-1-one (0.543 mM) was dissolved in ethanol (10 ml) and heated to reflux on a heating mantle for 2 h. After completion of the reaction as evident from TLC, the reaction mixture was set aside at ambient temperature for 6–7 h. The precipitate formed was filtered and dried to get a pure product. The crystals were obtained from the slow evaporation technique by dissolving the product in DMSO. (Yield: 77%; m. p: 271–272 °C) (Fig. S1).
2.2. Single crystal X-ray diffraction (SXRD)
A good quality optically clear 0.21 × 0.2 × 0.18 mm3 sized crystal was selected for the intensity data collection using Bruker kappa APEX II diffractometer using the MoKα radiation (λ = 0.71073 Å) source. The intensity data were collected at 20 °C. Absorption correction was carried out using the SADABS program with multi-scan method. Full-matrix least-squares refinement procedure was used for solving structures using SHELXL [33]. All the non-hydrogen atoms were refined anisotropically and hydrogen atoms were positioned from the difference fourier maps and refined isotropically. Hydrogen atoms were placed in calculated positions, with C–H = 0.93–0.98 Å and N–H = 0.86 Å, and allowed to ride on their respective carrier atoms, Uiso(H) = 1.2Ueq(C) for CH2, CH and NH groups.
Initial structural solution of Ib showed co-crystallized completely disordered solvent molecule (DMSO) which was modeled and refined using PART command along with a free variable. The solvent molecule is disordered over two sets of sites in a 0.515(2):0.485(3) ratio. The final refined structure was validated using PLATON [34] and CheckCIF routine from IuCr. Thermal ellipsoidal image and molecular packing diagrams were generated using ORTEP [35] and Mercury [36]. CCDC 1997850 (Ia) and CCDC 1997851 (Ib) contain the supplementary crystallographic data. The data can be obtained free of charge from The Cambridge Crystallographic Data Center via www.ccdc.cam.ac.uk/structures. The crystallographic data and refinement parameters were listed in Table 1 .
Table 1.
Crystallographic table of Compounds Ia and Ib.
| Ia | Ib | |
|---|---|---|
| Empirical formula | C27 H17 N3 | C28 H19 N3 O, C2 H6O S |
| Molecular weight | 383.43 | 491.59 |
| Temperature (K) | 293 (2) | 293 (2) |
| Wavelength (Å) | 0.71073 | 0.71073 |
| Crystal system | Triclinic | Triclinic |
| Space group | P −1 | P −1 |
| a (Å) | 9.5662 (5) | 9.9760 (6) |
| b (Å) | 10.4295 (5) | 11.0573 (5) |
| c (Å) | 11.4163 (6) | 12.9317 (7) |
| α (°) | 90.300 (3)° | 76.737 (3)° |
| β (°) | 105.005 (3)° | 68.739 (3)° |
| γ (°) | 115.950 (3)° | 78.469 (3)° |
| Volume (Å3) | 980.00 (9) | 1283.17 (12) |
| Z | 2 | 2 |
| Density (calculated) (mg/m3) | 1.299 | 1.272 |
| Absorption coefficient (mm−1) | 0.078 | 0.158 |
| F (000) | 400 | 516 |
| Crystal size (mm3) | 0.3 × 0.25 x 0.2 | 0.15 × 0.15 x 0.1 |
| Theta range for data collection (°) | 2.192 to 29.514 | 2.209 to 27.267 |
| Index ranges | −13 ≤ h<=13, −14 ≤ k<=14, −15 ≤ l<=15 |
−12 ≤ h<=12, −14 ≤ k<=14, −16 ≤ l<=16 |
| Reflections collected | 26957 | 26682 |
| Independent reflections | 5451 [R (int) = 0.0537] | 5689 [R (int) = 0.0574] |
| Completeness to theta = 25.242° | 100.0% | 100.0% |
| Refinement method | Full-matrix least-squares on F2 | Full-matrix least-squares on F2 |
| Data/restraints/parameters | 5451/0/271 | 5689/0/367 |
| Goodness-of-fit on F2 | 1.000 | 1.006 |
| Final R indices [I > 2sigma(I)] | R1 = 0.0510, wR2 = 0.1081 | R1 = 0.0499, wR2 = 0.1105 |
| R indices (all data) | R1 = 0.1146, wR2 = 0.1366 | R1 = 0.1264, wR2 = 0.1513 |
| Extinction coefficient | n/a | 0.0091 (18) |
| Largest diff. peak and hole (e.Å−3) | 0.174 and −0.246 | 0.193 and −0.196 |
2.3. DFT and intermolecular interaction analysis
Theoretical DFT calculations were done using the computational package ORCA 3.0.3 [37]. To obtain optimized geometry, the input file for ORCA was generated from experimental crystal data as initial coordinates with the DFT B3LYP level of theory with 6-311G (d,p) [38] as a basis set. The frontier molecular orbital analysis was carried out using this optimized geometry. Hirshfeld surface (HS) along with the fingerprint plot analysis has been done using the software CrystalExplorer 3.1 [39]. For the analysis of QTAIM (Quantum Theory of Atoms In Molecules) and NCI (Non-Covalent Interaction), the Single Point energy calculation was done using the single crystal X-ray geometry as the input. The output wavefunction file was used for the analysis. For QTAIM analysis Multiwfn [40] software was used and for NCI analysis, NCIPLOT [41] program was used. The outputs were combined and visualized through VMD [42] software.
The principles behind the Hirshfeld surface analysis, QTAIM and NCI can be found somewhere else [43].
2.4. Homology modeling SARS CoV-2 RdRp and molecular docking
2.4.1. Template protein identification
The nucleotide sequence of RdRp of the novel SARS CoV (QIQ08767.1) was first made available on march 24, 2020 at the National Center for Biotechnology Information (NCBI) database (https://www.ncbi.nlm.nih.gov/protein/QIQ08767.1). The template protein structure containing this similar sequence was searched using Basic Local Alignment Search Tool program (BLAST) [44,45]. The best Protein Data Bank (PDB) template was chosen on the basis of the percentage of sequence identity, maximum score query coverage and E-value. The BLAST program displayed a protein (PDB id: 6NUR) with a 100% sequence query coverage, 99.11% identity match, a minimum E-value of 6 × 10−74 and a maximum score of 236. This protein structure is retrieved from the Research Collaboratory for Structural Bioinformatics (RCSB) database (https://www.rcsb.org/structure/6NUR). This protein structure was solved using single particle cryo-electron microscopy with a resolution of 3.1 Å. This protein is a tetramer (four chains), RNA polymerase of SARS CoV nsp12 (A-chain) along with the co-factors nsp7 (C-chain) and nsp8 (B & D-chain) [46]. Since SARS CoV-2 RdRp is homologous with nsp12 of the previous SARS CoV RNA polymerase. Therefore nsp12 (A chain) was chosen as the template for homology modeling.
2.4.2. Homology modeling of RdRp of SARS CoV-2
The three dimensional models of the RdRp of SARS CoV-2 were produced using the homology modeling software MODELLER 9.23 [47]. As stated earlier, the target sequence has 99.11% identical with the template; the coordinates from the template to the Structurally Variable Regions (SVR), Structurally Conserved Regions (SCR), C-termini and N-termini were allotted on the basis of the fulfillment of the spatial restraints. Ten models were produced, out of which the model having lowest energy was chosen for the next step of energy minimization. This step is essential to eliminate the geometrical errors occurred during the modeling stage. This process utilizes the software GROMACS-5.1.2 [48]. The minimization process uses Steepest descent algorithm using GROMOS96-43a1 as a force field. The modeled structure was put inside a cubic box of 8.061 nm × 8.168 nm x 8.383 nm size, containing 57,689 SPC216 water molecules. Eight sodium ions were added to neutralize the system. Physicochemical parameters that is, atomic and amino acid composition, isoelectric point, instability index, Grand Average of hydropathicity (GRAVY) were analyzed using ProtParam tool of ExPASy [49].
2.4.3. Validation and active site prediction of SARS CoV-2
The RdRp of SARS CoV-2 model is validated using PROCHECK [50], PROVE [51], ERRAT [52], Verify 3D [53] and along with Ramachandran plot [54]. Since it is a new virus, its active sites can be predicted using some web servers. Here 3DLigandSite [55] web server (http://www.sbg.bio.imperial.ac.uk/3dligandsite/) was used to predict the binding sites.
2.4.4. Preparation of ligands and its docking
A database containing more than 500 molecules including suggested drugs for COVID-19 treatment (www.drugbank.ca/), some phytochemicals (https://phytochem.nal.usda.gov/; https://cb.imsc.res.in/imppat/) and some modeled derivatives of azafluorene. All the ligand molecules were energy minimized using Steepest descent algorithm employed in Avogadro. The water molecules were removed, if any, from the protein molecule. A grid box with the size of 22.3508 × 27.3405 x 27.5145 was used. The box was fixed on the active site having the coordinates (x,y,z) is 148.032, 140.6072, 157.7467. The docking Lamarckian Genetic Algorithm (LGA) was allowed to produce 10 docked positions for each ligand. Molecular docking studies were performed using AutoDock vina [56] in PyRx [57] software. The final results were analyzed and visualized on the basis of docking scores using Chimera [58] and PyMol [59] software’s.
2.4.5. Pharmacophore modeling and ADMET analysis
PharmaGIST [60] webserver was used to generate pharmacophore model with the training set includes the suggested inhibitors of COVID-19 [61]. The ADMET (Absorption, Distribution, Metabolism, Excretion and Toxicity) properties of the compounds Ia and Ib were then predicted using SwissADME server [62], admetSAR [63] and OSIRIS property explorer [64].
2.4.6. Binding free energy calculation using MM-PB(GB)SA
The binding free energy was calculated using farPPI server [65]. The input files were generated using Chimera [58]. The lowest binding energy, protein-ligand complex from the docking analysis was selected. The partial charges of the ligands were calculated using the AM1-BCC method [66] using the DockPrep tool in Chimera. The force fields, GAFF2 [67] and ff14SB [68] were used for ligand and protein respectively.
3. Results and discussion
3.1. Single crystal X-ray diffraction studies
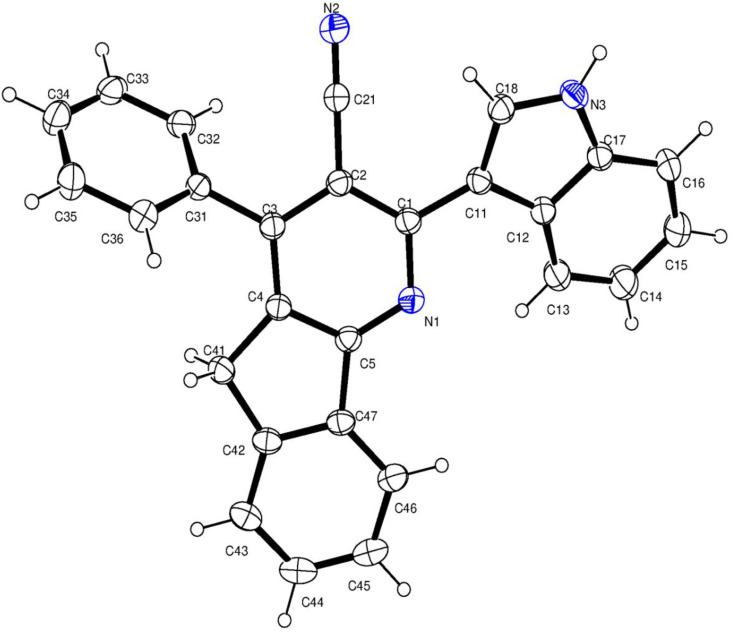
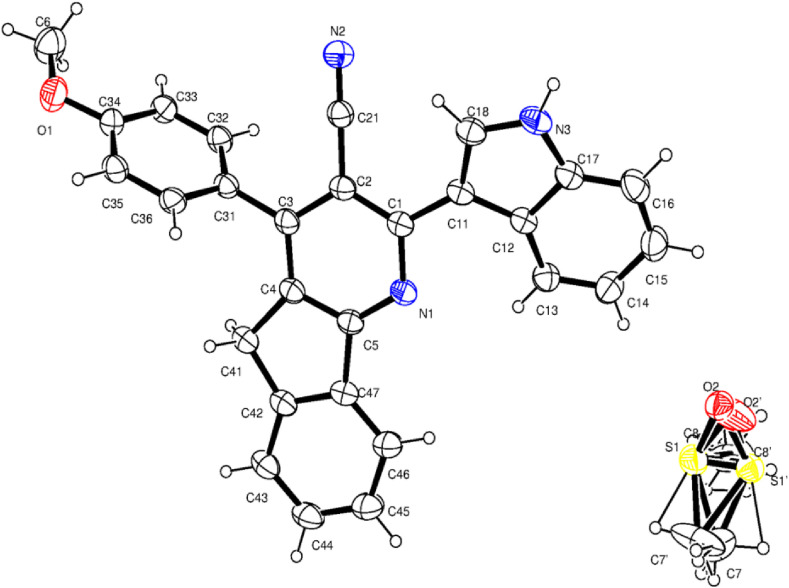
The two compounds differ by the substituent at fourth position of azafluorene ring. This substitution does not affect the crystal system. The ORTEP diagrams with thermal ellipsoids at 30% probability with an atom numbering scheme for Ia and Ib are shown in Fig. 1, Fig. 2 .
Fig. 1.
ORTEP diagram of Ia showing 30% probability and atom-numbering scheme.
Fig. 2.
ORTEP diagram of Ib showing 30% probability and atom-numbering scheme.
In both the compounds, the azafluorene ring is essentially planar with the deviation of 0.0198 (2) Å in Ia and 0.019 (2) Å in Ib with the fitted atoms. The bond lengths and angles of the azafluorene ring system are very much similar to the reported compounds ([26,[69], [70], [71], [72]] and CSD ref. code: CCDC 1481137, 1448345, 1470253, 1459303) and gives the strongest proof for the aromatic type delocalization within the pyridine moiety and the fused aryl ring. The bond lengths of C4 – C41 [1.507 (2) in Ia & 1.504 (3) in Ib], C41 – C42 [1.508 (2) in Ia & 1.508 (2) in Ib] and C47 – C5 [1.46 (2) in Ia & 1.469 (3) in Ib] are relatively longer indicates the lack of π delocalization within the cyclopentane ring [71,73,74]. The shortening of the bond lengths C4 – C5 [1.401 (2) Å in (Ia) and 1.396 (3) Å in (Ib)], C42 – C47 [1.393 (2) Å in (Ia) and 1.391 (3) Å in (Ib)] and reduction in the angle around C41 [102.63 (13)° and 102.03 (19)°] are the key indicators of the fusion of phenyl and pyridine rings with the cyclopentane.
The C2 – C21 bond length in Ia and Ib are 1.434 (2) Å and 1.438 (3) Å, which signifies the aromatic type bond length, the bond distance N2 – C21 is 1.146 (2) Å in (Ia) and 1.148 (3) Å in (Ib) indicates the triple bond nature [26]. The angle around C21 [179.6 (2)° in (Ia) and 179.3 (3)° in (Ib)] defines the linearity of the nitrile group. The deviation of C21 and N2 atoms from the mean plane of pyridine [0.007 (2) Å & 0.02 (3) Å in (Ia) and 0.092 (3) Å & 0.166 (4) Å in (Ib)] exemplifies the coplanarity. These deviations are due to the nearby substitutions in the pyridine ring.
The indole ring (N3/C11–C18) in both the compounds is planar with the r.m.s deviation of 0.0078 (1) Å in (Ia) and 0.0068 (2) Å in (Ib) with the fitted atoms. The sum of the angles around N3 is 360 (1)° in both the compounds, which indicates the sp2 hydridization of nitrogen. The indole ring in both the compounds are in (+) syn-periplanar conformation with the pyridine ring evidenced from the torsion N1–C1–C11–C12 [20.8 (2)° in (Ia) and 20.2 (3)° in (Ib)]. The indole in all the compounds are nearly coplanar with the pyridine which may be seen from the dihedral angle [23.68 (2)° and 23.48 (3)° in Ia and Ib respectively]. The endocyclic angles at C12 and C16 are contracted, while at C15 and C17 are expanded in Ia and Ib. This may due to the fusion of the pyrrole ring with the phenyl ring, with the strain taken up by angular distortion rather than by bond length distortions [75].
The bond lengths and angles of phenyl [69] and anisole [70] rings of Ia and Ib are consistent with the similar structures. From the torsion C2–C3–C31–C32 the aryl substituent at C3 in compound Ia [53.4 (2)°] and in Ib [66.5 (2)°] are (+) syn-clinal.
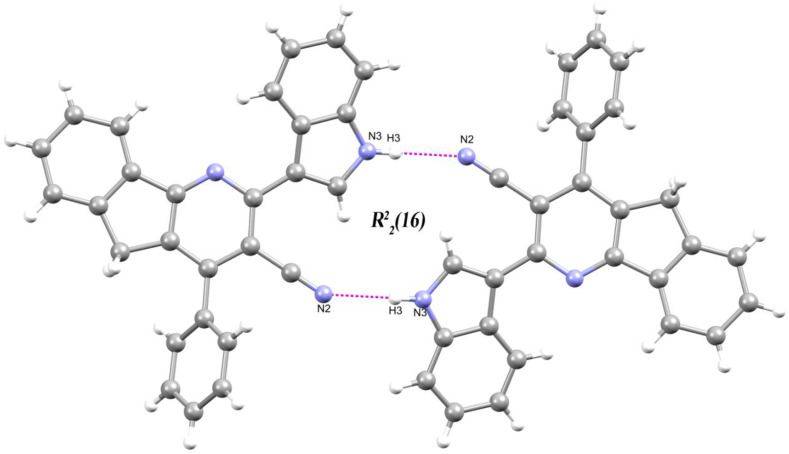
The molecular structure of compound Ia is stabilized through an intermolecular interaction N3–H3⋯N2. (Table 2 and Fig. 3 ). The interaction from indole (N3–H3) to cyano nitrogen (N2i [symmetry code: (i)-2–x, -y, 1-z]) connects the inversely related molecules thus forming an motif lying along the (110) plane. This motif is a characteristic motif of indole and nitrile containing compounds [[76], [77], [78]].
Table 2.
Hydrogen bonds for Ia [Å and °].
| D-H … A | d (D-H) | d (H … A) | d (D … A) | <(DHA) |
|---|---|---|---|---|
| N (3)–H (3) … N (2)#1 | 0.86 | 2.16 | 3.014 (3) | 174 |
Symmetry transformations used to generate equivalent atoms: #1 2–x, -y, 1-z.
Fig. 3.
Partial packing diagram of Ia shows ring motifs.
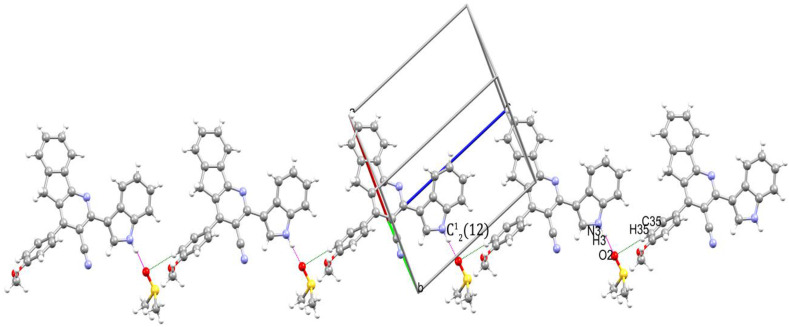
In Ib, the host and guest molecules are forming an intermolecular interaction thus stabilizing the crystal. Here the characteristic ring motif was not found because of the strong interaction with the solvent (DMSO) molecule. The atoms N3 and C35 act as donors and O2 in DMSO act as an acceptor (Table 3 ). The two interactions N3–H3⋯O2i [symmetry code: (i)-x, 1-y, 1-z] and C35–H35⋯O2ii [symmetry code: (ii)-x+1,-y+1,-z] forming a chain motif of (Fig. 4 ).
Table 3.
Hydrogen bonds for Ib [Å and °].
| D-H … A | d (D-H) | d (H … A) | d (D … A) | <(DHA) |
|---|---|---|---|---|
| N (3)–H (3) … O (2)#1 | 0.86 | 1.96 | 2.777 (12) | 158 |
| C (35)–H (35) … O (2)#2 | 0.93 | 2.66 | 3.549 (4) | 158 |
Symmetry transformations used to generate equivalent atoms: #1 -x, 1-y, 1-z #2 -x+1,-y+1,-z.
Fig. 4.
Partial packing diagram of Ib shows chain motifs.
3.2. Hirshfeld surface, QTAIM and NCI analysis
Hirshfeld surface of compounds Ia and Ib are mapped over dnorm surface shown in Figs. S2 and S3 respectively. Table 4 shows the individual contributions. The colour coded NCI plot along with QTAIM critical points of Ia and Ib are shown in Figs. S4 and S5 and the values of QTAIM descriptors are shown in Table 5, Table 6 .
Table 4.
Contribution of individual interactions to Hirshfeld surface.
| Compound | H – H (%) | C–H/H–C (%) | N–H/H–N (%) | O–H/H–O (%) | C–C (%) | C–N/N–C (%) |
|---|---|---|---|---|---|---|
| Ia | 44.4 | 35.8 | 11.2 | – | 5.6 | 3.0 |
| Ib | 46.2 | 29.4 | 10.0 | 9.1 | 2.7 | – |
Table 5.
QTAIM descriptors for Ia.
| Interaction | ρ a.u |
a.u |
G(r) a.u | V(r) a.u | H(r) a.u | Ebond kJ.mol−1 | |
|---|---|---|---|---|---|---|---|
| N3–H3⋯N2 #1 | 0.0177 | 0.0569 | 0.0126 | −0.011 | 0.0016 | 0.873 | −14.856 |
Symmetry transformations used to generate equivalent atoms: #1 2–x, -y, 1-z.
Table 6.
QTAIM descriptors for Ib.
| Interaction | ρ a.u |
a.u |
G(r) a.u | V(r) a.u | H(r) a.u | Ebond kJ.mol−1 | |
|---|---|---|---|---|---|---|---|
| N3–H3⋯O2 #1 | 0.0231 | 0.0928 | 0.0206 | −0.0179 | 0.0026 | 0.869 | −24.174 |
| C35–H35⋯O2 #2 | 0.0069 | 0.0233 | 0.0053 | −0.005 | 0.0006 | 0.943 | −6.783 |
Symmetry transformations used to generate equivalent atoms: #1 -x, 1-y, 1-z #2 -x+1,-y+1,-z.
In both the structures, apparently short contacts seem to influence H–H interaction (Table 4). Other significant contributions are from C–H, N–H and O–H interactions. These arise due to the intermolecular interactions present in the compounds. Further contributions are may be due to van der Waals interactions. Hirshfeld surface shows two and four red spots for Ia and Ib respectively (Figs. S2 and S3).
In compound Ia, for N–H⋯N interaction, di + de ≈ 2.0 Å which is less than the sum of the van der Waals radii 2.74 Å [79,80] indicated as bright red spots (Fig. S2). This interaction is colour coded as blue in NCI index indicating strong interactions (Fig. S4). The energy of this interaction from QTAIM descriptors is found to be −14.856 kJ/mol. The ‘sign ()ρ’ value is −0.0177 a. u.
In compound Ib, for N–H⋯O interaction, di + de ≈ 1.7 Å which is less than the sum of the van der Waals radii 2.61 Å. The energy of this interaction is −24.174 kJ/mol with the ‘sign ()ρ’ value of −0.023 a. u. and there is an additional C – H ⋯ O interaction, which is indicated as a light red spot in Hirshfeld surface and in NCI index it is green (Figs. S3 and S5). For the C – H ⋯ O interactions, di + de ≈ 1.8 Å with the energy of −6.783 kJ/mol.
From this analysis, it can be concluded that the compound Ib forms a stronger interaction (N3–H3⋯O2) with the solvent molecule.
3.3. HOMO-LUMO analysis
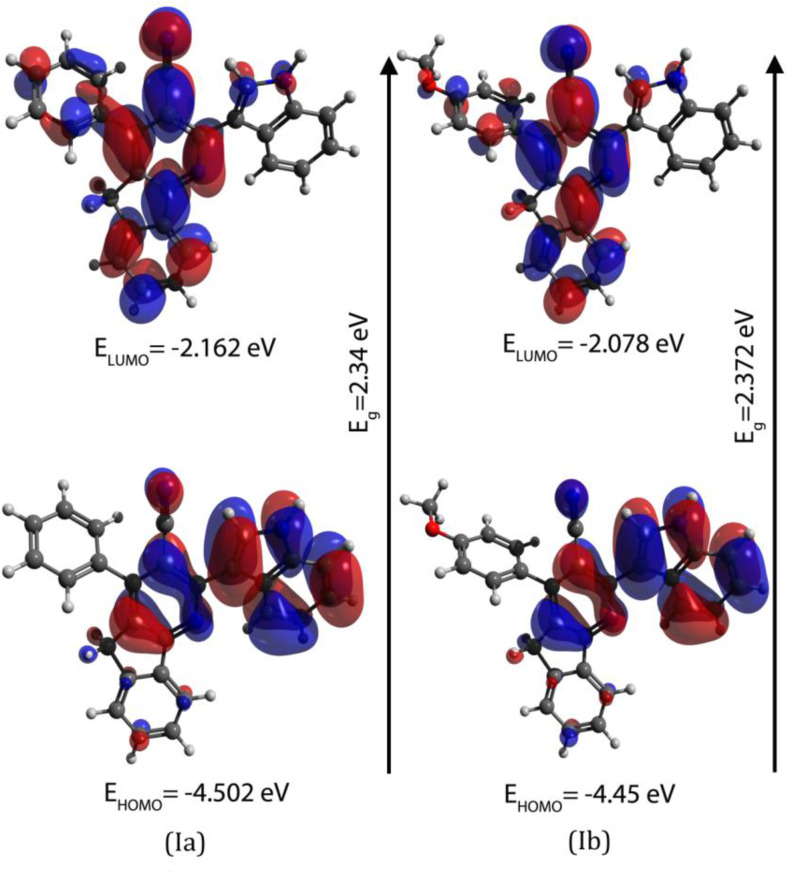
The studies on frontier molecular orbital reveal the chemical reactivity, chemical hardness or softness of the molecule and kinetic stability [81]. All the calculated energy parameters along with a partition coefficient (log P) and dipole moment are given in Table 7 . It can be seen from Fig. 5 that the HOMO orbitals are located on both cyanopyridine and indole moieties while LUMO orbitals are located on azafluorene ring, nitrile group and substituted aryl rings. From the molecular orbital analysis, the substitution has an influence on the electron accepting ability of the compounds. The intra-molecular charge transfer interactions of both the compounds are nearly the same as evidenced from their energy gaps.
Table 7.
Calculated energy values by B3LYP/6-311G (d,p) level.
| Parameters | Ia | Ib |
|---|---|---|
| EHOMO (eV) | −4.502 | −4.45 |
| ELUMO (eV) | −2.162 | −2.078 |
| Ionization potential (I) (I = -EHOMO) (eV) | 4.502 | 4.45 |
| Electron affinity (A) (A = -ELUMO) (eV) | 2.162 | 2.078 |
| Energy gap (ΔE) (eV) | 2.34 | 2.372 |
| Electronegativity (χ) () (eV) | 3.332 | 3.264 |
| Chemical potential (μ) () (eV) | −3.332 | −3.264 |
| Chemical hardness (η) () (eV) | 1.17 | 1.186 |
| Chemical softness (S) () (eV) | 0.427 | 0.422 |
| Electrophilicity index (ω) () (eV) | 4.745 | 4.491 |
| log P | 3.68 | 3.29 |
| Dipole moment (debye) | 1.863 | 1.982 |
Fig. 5.
Frontier molecular orbitals and energies of the HOMO, LUMO.
The compound Ia is more reactive than Ib. This is evident from low chemical hardness (η) and high softness (S) for Ia than in Ib. Based on the values of electronegativity (χ) and chemical potential (μ) Ia has more electron attracting ability than Ib and this is supplemented by a high electrophilicity index (ω) of Ia. The cell membrane permeability of Ib is higher than Ia indicated by ‘log P’ values. The value of dipole moment which tells about the ligand-protein interaction on the basis of electrostatic interaction is higher in Ib than Ia. Both the compounds are reducers based on the negative HOMO and LUMO values and may undergo oxidative reactions with cytochrome P450 enzyme [82].
3.4. Pharmacophore mapping and drug-likeness properties
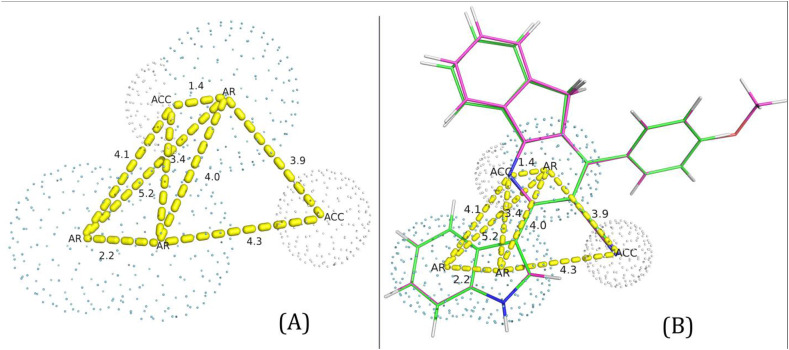
The best model given by the PharmaGIST revealed four pharmacophric features includes two hydrogen bonding acceptors and two aromatic rings (Fig. 6 ). Further, the Drug-likeliness properties like physicochemical properties, lipophilicity, pharmacokinetics and toxicity were predicted for compounds Ia and Ib and given in Table 8 . Both the compounds do not violate the Lipinski’s rule of 5 [83] inferred from the physico-chemical and lipophilicity properties. Both the compounds do not show mutagenicity, tumorigenicity, irritating and reproductive effects. Also, these compounds have no Blood-Brain Barrier permeation ability. However, these compounds have CYP450 inhibition effects except CYP3A4 and 2D6.
Fig. 6.
Pharmacophore map: (A) 3D spatial arrangement and the distance constraints between the chemical features. Acc – Hydrogen bond acceptor; AR – Aromatic ring. (B) Superimposition of compounds Ia and Ib with the pharmacophore model. Green represents Ia and Pink represents Ib respectively.
Table 8.
Drug-likeness properties of Azafluorene derivatives.
| Drug-likeness Properties | Compound – I | Compound - II |
|---|---|---|
| Physicochemical properties | ||
| Molecular weight (g/mol) | 383.44 | 413.47 |
| Number of Hydrogen bond donors | 1 | 1 |
| Number of Hydrogen bond acceptors | 2 | 3 |
| Molar refractivity | 120.13 | 126.62 |
| TPSA (Topological Polar Surface Area) | 52.47 Å2 | 61.7 Å2 |
| Lipophilicity | ||
| Log PO/W (iLogP) | 3.2 | 3.43 |
| Log PO/W (XLogP3) | 5.65 | 5.62 |
| Log PO/W (WLogP) | 6.34 | 6.35 |
| Log PO/W (MLogP) | 3.68 | 3.29 |
| Log PO/W (SILICOS-IT) | 7.03 | 7.07 |
| Consensus Log PO/W | 5.18 | 5.15 |
| Solubility | ||
| Log S (SILICOS-IT) | −10.76 | −10.86 |
| SILICOS-IT solubility (mg/ml) | 6.74E-09 | 5.73E-09 |
| SILICOS-IT solubility (mol/l) | 1.76E-11 | 1.39E-11 |
| Solubility class | Insoluble | Insoluble |
| Pharmacokinetics | ||
| Druglikeness | −3.19 | −2.81 |
| Drug-score | 0.15 | 0.15 |
| Blood-Brain-Barrier permeant | No | No |
| Gastrointestinal absorption | Yes | Yes |
| Caco-2 permeant | No | No |
| P-glycoprotein substrate | No | No |
| CYP450 1A2 inhibitor | Yes | Yes |
| CYP450 2C9 inhibitor | Yes | Yes |
| CYP450 2D6 inhibitor | No | Yes |
| CYP450 2C19 inhibitor | Yes | Yes |
| CYP450 3A4 inhibitor | No | No |
| CYP Inhibitory Promiscuity | Yes | Yes |
| Toxicity | ||
| Carcinogens | No | No |
| Biodegradation | No | No |
| Acute Oral Toxicity (kg/mol) | III, 2.317 | III, 2.394 |
| Mutagenicity | No | No |
| Tumorigenicity | No | No |
| Irritating effects | No | No |
| Reproductive effects | No | No |
3.5. Molecular docking analysis
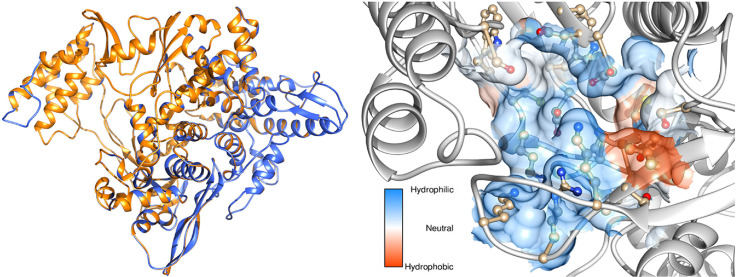
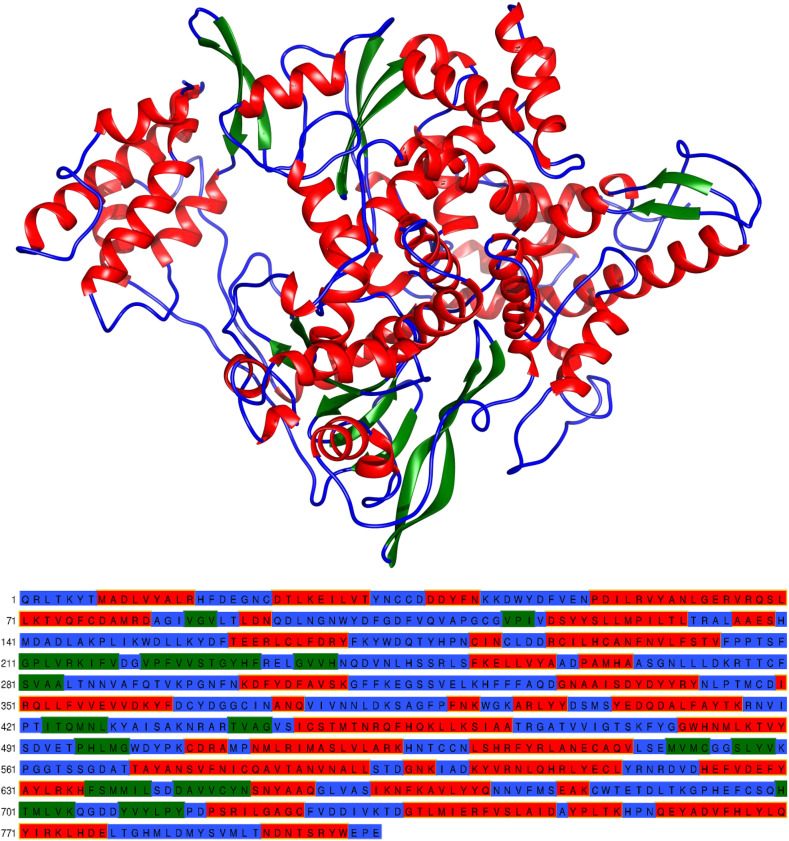
The final minimized potential energy of RdRp model is −418539 kJ/mol. The root mean square deviation between the template (6NUR_A) and model is 0.0016 Å. The superimposed figure of template and model is shown in Fig. S6. The final protein model contains thirty six α-helices, fifteen β-strands and fifty one coils (Fig. 7 ).
Fig. 7.
Top: The secondary structural elements such as α-helices (Red), β-strands (Green) and coils (Blue). Bottom: The amino acid sequence colour coded as corresponding secondary structural elements.
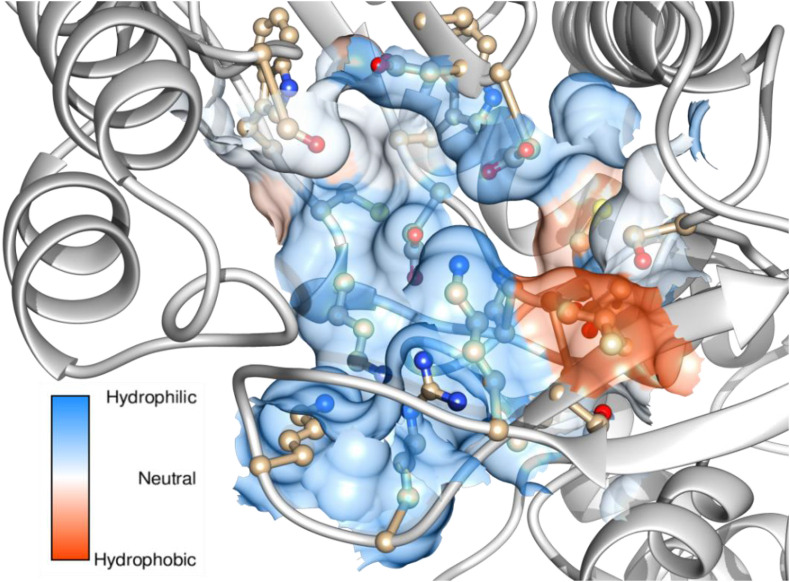
Table S1 shows the values obtained from ProtParam analysis. This revealed that this protein contains 803 amino acids with a molecular weight of 91796.61 Da. Also the instability index was computed as 29.14 and a GRAVY index of −0.18 indicates that this protein is highly stable, hydrophilic molecule with hydrogen bonding capability. From the ERRAT calculation, the quality factor of the model was found as 94.9871. This is a very good indication of an acceptable 3D profile. The model passed the Verify 3D and PROCHECK evaluations. The Ramachandran plot was shown in Fig. S7. From this plot it is inferred that, 91.7% residues are in allowed regions, 8.0% are in additionally allowed regions and 0.3% are in generally allowed regions. There is no residues are situated outside of the allowed regions. This clearly indicates that the model is highly reliable for further analysis. The amino acids within the predicted binding site are LYS-429, LYS-435, ARG-437, THR-440, VAL-441, ASP-502, LYS-505, CYS-506, ASP-507, SER-566, ASP-644, ASP-645 and LYS-682 (Fig. 8 ). This active site is used for the docking analysis with ligands.
Fig. 8.
Colour coded active site residues of the RdRp model.
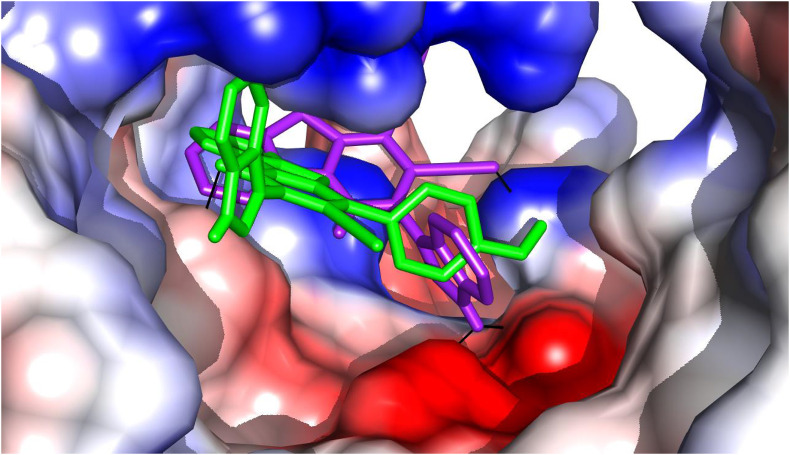
The novel corona virus is a beta-corona virus having single positive strand RNA [84]. The multiplication/replication and transcription from an RNA template of this virus is facilitated by multi-subunit non-structural proteins (nsp7, 8 & 12). The co-factors nsp7 and 8 accelerates nsp12, which is a core catalytic unit of the RdRp, increases the activity of template binding and processing [85]. The study around the structural features of RdRp is a key to design a drug that can interact with this protein and hence to suppress its activity of replication. This nsp12 resembles a cupped “right hand” structure having fingers domain (amino acids (a.a) 180–465 & 512–564), a palm domain (a.a 466–511 & 565–699) and a thumb domain (a.a 700–803) (Fig. S8) [46]. The enzymatic activity of this polymerase is highly dependent on the conserved active site SER643-ASP644-ASP645 residues located at the palm domain [85]. The residues in the finger domain LYS429 and ARG439 arrange the incoming NTP (Nucleotide Tri Phosphate) in a perfect manner and provide a gateway to the catalytic center and LYS384 and SER385 arrange themselves to hold the template RNA strand [86]. At the catalytic center, the base of the NTP binds with the template though the 2′ and 3′ hydroxyl groups form hydrogen bonds with the residues THR564, ASN575 and ASP507. Also, VAL441 stacks the +1 template RNA base to support the base pairing of NTP’s [46]. Therefore, targeting the above mentioned residues may greatly affect the catalytic activity and processivity of the RdRp. The docked poses of compounds Ia and Ib are shown in Fig. 9 .
Fig. 9.
The docked pose of Ia (purple) and Ib (green) within the active pocket. The black solid line indicates the interaction.
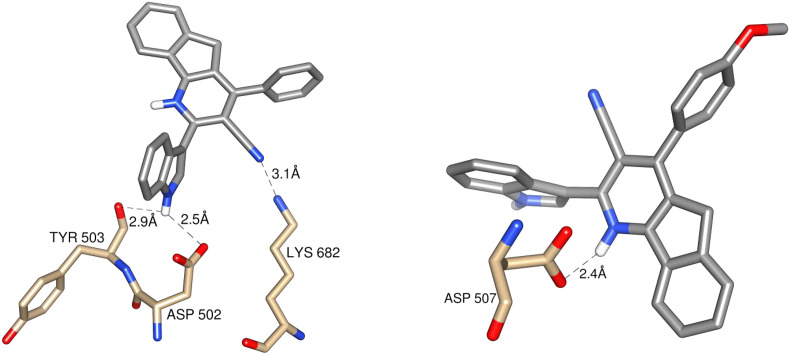
Among the approved drugs, Lopinavir has the lowest binding energy (B.E) of −9.4 kcal/mol but no interactions with the mentioned important residues. Favipiravir, Galidesivir, Hydroxychloroquine and Ritonavir showed at least one interaction with the mentioned residues (Table S2). Moreover, Ritonvir (−8.3 kcal/mol) can bind tightly with the polymerase and interact with one of the catalytic residues ASP644 (Table S2). Therefore, these drugs can directly interfere with the enzymatic activity, thus preventing the replication. The similar interaction is observed in some vinca alkaloids like, vindesine (−8.7 kcal/mol), vincathine (−7.9 kcal/mol), vincristine (−7.8 kcal/mol), vinblastine (−7.2 kcal/mol) and vindoline (−6.8 kcal/mol) (Table S3). The modeled compounds M − 1 and M − 3 have interactions with ASP644 and ASP645 (Table S4). Thus, these compounds may control the catalytic activity.
Galidesivir (−7.6 kcal/mol), Hydroxychloroquine (−6.3 kcal/mol), and Favipiravir (−5.8 kcal/mol), make one or more hydrogen bonds with 2′ and 3′ hydroxyl group of NTP interacting residues, thus obstructing the incoming NTP (Table S2). The similar behavior is observed in compound Ib having the binding energy of −7.8 kcal/mol (Table 9 ; Fig. 10 ). The binding free energy of this complex using MM-PB(GB)SA is −39.42 kcal/mol. Also, this compound Ib does not violate the Lipinski’s rule of 5 [83], suggests the drug-likeness property. Further, log P and dipole moment values of Ib confirmed the drug-like activity. The phytochemicals possess the same interaction type are vindesine, vincristine, quercetin and vincamine with B.E of −8.7, −7.8, −7.8 and-6.6 respectively.
Table 9.
Binding energy of Ia and Ib.
| Compound | Hydrogen bond (docking) | Distance (Å) | B.E (kcal/mol) |
|---|---|---|---|
| Ia | N–H⋯O (ASP-502) N–H⋯O (TYR-503) N–H⋯N (LYS-682) |
2.5 2.9 3.1 |
−8.3 |
| Ib | N–H⋯O (ASP-507) | 2.4 | −7.8 |
Fig. 10.
The interactions between the synthesized compounds Ia and Ib with the amino acids of SARS CoV-2 RdRp.
The proper orientation of NTP is maintained by the residues LYS429 and ARG439. Compound Ia interacts with the residues that are nearby the above mentioned residues (Table 9; Fig. 10). Therefore, this compound may block the space for the NTP molecules to stay. The binding energy and binding free energy of this complex is −8.3 kcal/mol and −11.04 kcal/mol respectively. The decrease in binding free energy of Ia is may be due to the comparatively weak interactions with the residues as that of Ib. Vincathine (−7.9 kcal/mol), quercetin (−7.8 kcal/mol) and vincaleukoblastine (−7.3 kcal/mol) also exhibits the same interactions.
4. Conclusion
In this work, two azafluorene derivatives having different substituents at fourth position were synthesized and structural parameters were analyzed using SXRD. The change in substituent brought changes in physicochemical properties. The intermolecular interactions were analyzed qualitatively and quantitatively using Hirshfeld surface analysis, QTAIM and NCI index. The results suggest that, compound Ib forms strong interaction (−24.174 kJ/mol) with the solvent. Both the compounds were geometry optimized using DFT/B3LYP methods and its frontier molecular orbital analysis was done. The energy gap and other properties related to molecular interacting abilities were predicted. From docking analysis, the drugs, Lopinavir, Favipiravir, Galidesivir, Hydroxychloroquine and Ritonavir may be used against COVID-19. The mentioned phytochemicals showed good binding affinities with the target indicating its potential efficacy. The compounds Ia, Ib and modeled derivatives (M − 1 and M − 3) interact with the RdRp indicating its potential activity. However, further in-vitro and in-vivo analyses around these mentioned compounds are required to ascertain these suggestions.
CRediT authorship contribution statement
M. Venkateshan: Conceptualization, Methodology, Visualization, Formal analysis, Resources, Writing - original draft. M. Muthu: Resources, Investigation. J. Suresh: Supervision, Funding acquisition. R. Ranjith Kumar: Supervision.
Declaration of competing interest
The authors declare no conflict of interest.
Acknowledgements
JS and MV thank the management of The Madura College for their encouragement and support.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.molstruc.2020.128741.
Appendix A. Supplementary data
The following is the Supplementary data to this article:
References
- 1.Cheng V.C., Lau S.K., Woo P.C., Yuen K.Y. Severe acute respiratory syndrome coronavirus as an agent of emerging and reemerging infection. Clin. Microbiol. Rev. 2007;20:660–694. doi: 10.1128/CMR.00023-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lee N., Hui D., Wu A., Chan P., Cameron P., Joynt G.M., Ahuja A., Yung M.Y., Leung C.B., To K.F., Lui S.F., Szeto C.C., Chung S., Sung J.J. A major outbreak of severe acute respiratory syndrome in Hong Kong. N. Engl. J. Med. 2003;348:1986–1994. doi: 10.1056/NEJMoa030685. [DOI] [PubMed] [Google Scholar]
- 3.Zaki A.M., van Boheemen S., Bestebroer T.M., Osterhaus A.D., Fouchier R.A. Isolation of a novel coronavirus from a man with pneumonia in Saudi Arabia. N. Engl. J. Med. 2012;367:1814–1820. doi: 10.1056/NEJMoa1211721. [DOI] [PubMed] [Google Scholar]
- 4.de Groot R.J., Baker S.C., Baric R.S., Brown C.S., Drosten C., Enjuanes L., Fouchier R.A.M., Galiano M., Gorbalenya A.E., Memish Z.A., Perlman S., Poon L.L.M., Snijder E.J., Stephens G.M., Woo P.C.Y., Zaki A.M., Zambon M., Ziebuhr J. Middle East respiratory syndrome coronavirus (MERS-CoV): announcement of the Coronavirus Study Group. J. Virol. 2013;87:7790–7792. doi: 10.1128/JVI.01244-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lee P.R., Hsueh P.I. Emerging threats from zoonotic coronaviruses-from SARS and MERS to 2019-nCoV. J. Microbiol. Immunol. Infect. 2020:1–3. doi: 10.1016/j.jmii.2020.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Guo Y., Cao Q., Hong Z., Tan Y., Chen S., Jin H., Tan K., Wang D., Yan Y. The origin, transmission and clinical therapies on coronavirus disease 2019 (COVID-19) outbreak – an update on the status. Military Med. Res. 2020;7:11. doi: 10.1186/s40779-020-00240-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Seah I., Su X., Lingam G. 2020. Revisiting the Dangers of the Coronavirus in the Ophthalmology Practice Eye. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tchesnokov E.P., Feng J.Y., Porter D.P., Gotte M. Mechanism of inhibition of Ebola virus RNA-dependent RNA polymerase by Remdesivir. Viruses. 2019;4:11. doi: 10.3390/v11040326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.De Clercq E. The design of drugs for HIV and HCV. Nat. Rev. Drug Discov. 2007;6:1001–1018. doi: 10.1038/nrd2424. [DOI] [PubMed] [Google Scholar]
- 10.Xu H.-T., Hassounah S.A., Colby-Germinario S.P., Oliveira M., Fogarty C., Quan Y., Han Y., Golubkov O., Ibanescu I., Brenner B., Stranix B.R., Wainberg M.A. Purification of Zika virus RNA-dependent RNA polymerase and its use to identify small-molecule Zika inhibitors. J. Antimicrob. Chemother. 2017;72:727–734. doi: 10.1093/jac/dkw514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ganeshpurkar A., Gutti G., Singh S.K. RNA-dependent RNA polymerases and their emerging roles in antiviral therapy. Viral Polym. 2019;1–42 doi: 10.1016/b978-0-12-815422-9.00001-2. [DOI] [Google Scholar]
- 12.Lu H. Drug treatment options for the 2019-new coronavirus (2019-nCoV), Biosci. Trends. 2020;14:69–71. doi: 10.5582/bst.2020.01020. [DOI] [PubMed] [Google Scholar]
- 13.Wang M., Cao R., Zhang L., Yang X., Liu J., Xu M., Shi Z., Hu Z., Zhong W., Xiao G. Remdesivir and chloroquine effectively inhibit the recently emerged novel coronavirus (2019-nCoV) in vitro. Cell Res. 2020;30:269–271. doi: 10.1038/s41422-020-0282-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Prachayasittikul S., Manam P., Chinworrungsee M., Isarankura-Na-Ayudhya C., Ruchirawat S., Prachayasittikul V. Bioactive azafluorenone alkaloids from Polyalthia debilis (pierre) finet & gagnep. Molecules. 2009;14:4414–4424. doi: 10.3390/molecules14114414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hufford C.D., Liu S., Clark A.M., Oguntimein B.O. Anticandidal activity of eupolauridine and onychine, alkaloids from Cleistopholis patens. J. Nat. Prod. 1987;50:961. doi: 10.1021/np50053a037. [DOI] [PubMed] [Google Scholar]
- 16.Kraus G.A., Kempema A. Synthesis of azafluorenone antimicrobial agents. J. Nat. Prod. 2010;73:1967. doi: 10.1021/np100536a. [DOI] [PubMed] [Google Scholar]
- 17.Heintzelman G.R., Averill K.M., Dodd J.H., Demarest K.T., Tang Y., Jackson P.E., Pat. Appl. Publ. (2004) US 20040082578 A1 200400429.
- 18.Heintzelman G.R., Averill K.M., Dodd J.H., Demarest K.T., Tang Y., Jackson P.E., PCT Int. Appl. (2003) WO 2003088963 A1 20031030.
- 19.Kunstmann R., Fischer R. Molecular analysis of hexahydro-1H-indeno[1,2-b]pyridines: potential antidepressants. J. Med. Chem. 1984;27:1312–1316. doi: 10.1021/jm00376a015. [DOI] [PubMed] [Google Scholar]
- 20.Heintzelman G.R., Averill K.M., Dodd J.H., PCT Int. Appl. (2002) WO 2002085894 A1 20021031.
- 21.Miri R., Javidnia K., Hemmateenejad B., Azarpira A., Amirghofran Z. Synthesis, cytotoxicity, QSAR, and intercalation study of new diindenopyridine derivatives. Bioorg. Med. Chem. 2004;12:2529. doi: 10.1016/j.bmc.2004.03.032. [DOI] [PubMed] [Google Scholar]
- 22.Cooper K., Fray M.J., Cross P.E., Richardson K., Eur. Pat. Appl. (1989) EP 299727 A1 19890118.
- 23.Almeida M.E.L., de F.R., Braz, von Bulow M.V., Gottlieb O.R., Maia J.G.S. Onychine, an alkaloid from Onychopetalum amazonicum. Phytochemistry. 1976;15:1186–1187. [Google Scholar]
- 24.Earl R.A., Zaczek R., Teleha C.A., Fisher B.N., Maciag C.M., Marynowski M.E., Logue A.R., Tam S.W., Tinker W.J., Huang S.M., Chorvat R.J. 2-Fluoro-4-pyridinylmethyl analogues of linopirdine as orally active acetylcholine release-enhancing agents with good efficacy and duration of action. J. Med. Chem. 1998;41:4615–4622. doi: 10.1021/jm9803424. [DOI] [PubMed] [Google Scholar]
- 25.Padwa A., Heidelbaugh T.M., Kuethe J.T. Using the Pummerer cyclization deprotonation-cycloaddition cascade of imidosulfoxides for alkaloid synthesis. J. Org. Chem. 2000;65:2368–2378. doi: 10.1021/jo9915729. [DOI] [PubMed] [Google Scholar]
- 26.a Girgis A.S., Aziz M.N., Shalaby E.M., Saleh D.O., Mishriky N., El-Eraky W.I., Ahmed Farag I.S. Molecular structure studies of novel bronchodilatory-active 4-azafluorenes. Z. Kristallogr. 2016;231:179–187. [Google Scholar]; b Girgis A.S., Saleh D.O., George R.F., Srour A.M., Pillai G.G., Panda C.S., Katritzky A.R. Synthesis, bioassay, and QSAR study of bronchodilatory active 4H-pyrano[3,2-c]pyridine-3-carbonitriles. Eur. J. Med. Chem. 2015;89:835. doi: 10.1016/j.ejmech.2013.12.032. [DOI] [PubMed] [Google Scholar]; c Girgis A.S., Aziz M.N., Shalaby E.M., Asaada F.M., Ahmed Farag I.S. Synthesis and X-ray studies of novel azaphenanthrenes. J. Chem. Res. 2018;42:90–95. [Google Scholar]
- 27.Vrábel M., Hocek M., Havran L., Fojta M., Votruba I., Klepetářová B., Pohl R., Rulíšek L., Zendlová L., Hobza P., Shih I.H., Mabery E., Mackman R. Purines bearing phenanthroline or bipyridine ligands and their RuII complexes in position 8 as model compounds for electrochemical DNA labeling – synthesis, crystal structure, electrochemistry, quantum chemical calculations, cytostatic and antiviral activity. Eur. J. Inorg. Chem. 2007;2007:1752–1769. [Google Scholar]
- 28.Rajput C.S., Sharma S. Synthesis of new pyridine derivatives as potent antifungal agents. Int. J. Pharma Bio Sci. 2011;2:200–209. [Google Scholar]
- 29.Duflos A., Kruczynski A., Barrat J.M. Novel aspects of natural and modified vinca alkaloids. Curr. Med. Chem. Anticancer Agent. 2002;2:55–70. doi: 10.2174/1568011023354452. [DOI] [PubMed] [Google Scholar]
- 30.Beckers T., Mahboobi S. Natural, semisynthetic and synthetic microtubule inhibitors for cancer therapy. Drugs Future. 2003;28:767–785. [Google Scholar]
- 31.Jordan M.A., Wilson L. Microtubules as a target for anticancer drugs. Nat. Rev. Canc. 2004;4:253–265. doi: 10.1038/nrc1317. [DOI] [PubMed] [Google Scholar]
- 32.a Pawar A.Y. Combating devastating COVID -19 by drug repurposing. Int. J. Antimicrob. Agents. 2020 doi: 10.1016/j.ijantimicag.2020.105984. [DOI] [PMC free article] [PubMed] [Google Scholar]; b Shah B., Modi P., Sagar S.R. In silico studies on therapeutic agents for COVID-19: drug repurposing approach. Life Sci. 2020;252:117652. doi: 10.1016/j.lfs.2020.117652. [DOI] [PMC free article] [PubMed] [Google Scholar]; c Ciliberto G., Mancini R., Paggi M.G. Drug repurposing against COVID-19: focus on anticancer agents. J. Exp. Clin. Canc. Res. 2020;39:86. doi: 10.1186/s13046-020-01590-2. [DOI] [PMC free article] [PubMed] [Google Scholar]; d Guy R.K., DiPaola R.S., Romanelli F., Dutch R.E. Rapid repurposing of drugs for COVID-19. Science. 2020;368:829–830. doi: 10.1126/science.abb9332. [DOI] [PubMed] [Google Scholar]
- 33.Sheldrick G.M. Crystal structure refinement with SHELXL. Acta Crystallogr. C. 2015;71:3–8. doi: 10.1107/S2053229614024218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Spek A.L. Structure validation in chemical crystallography. Acta Crystallogr. D. 2009;65:148–155. doi: 10.1107/S090744490804362X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Farrugia L.J. ORTEP-3 for windows - a version of ORTEP-III with a graphical user interface (GUI) J. Appl. Crystallogr. 1997;30:565. [Google Scholar]
- 36.Macrae C.F., Bruno I.J., Chisholm J.A., Edgington P.R., McCabe P., Pidcock E., Rodriguez-Monge L., Taylor R., van de Streek J., Wood P.A. Mercury CSD 2.0 - new features for the visualization and investigation of crystal structures. J. Appl. Crystallogr. 2008;41:466–470. [Google Scholar]
- 37.Neese F. The ORCA program system Wiley Interdiscip. Rev.: Comput. Mol. Sci.. 2012;2:73–78. [Google Scholar]
- 38.Frisch M.J., Pople J.A., Binkley J.S. Self-consistent molecular orbital methods 25. Supplementary functions for Gaussian basis sets. J. Chem. Phys. 1984;80:3265. [Google Scholar]
- 39.Wolff S.K., Grimwood D.J., McKinnon J.J., Turner M.J., Jayatilaka D., Spackman M.A. University of Western Australia.; 2012. CrystalExplorer. [Google Scholar]
- 40.Lu T., Chen F. MULTIWFN: a Multifunctional wavefunction analyzer. J. Comput. Chem. 2012;33:580–592. doi: 10.1002/jcc.22885. [DOI] [PubMed] [Google Scholar]
- 41.Contreras-García J., Johnson E.R., Keinan S., Chaudret R., Piquemal J.-P., Beratan D.N., Yang W. NCIPLOT: a program for plotting non covalent interaction regions. J. Chem. Theor. Comput. 2011;7:625–632. doi: 10.1021/ct100641a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Humpherey W., Dalke A., Schulten K. VMD: visual molecular dynamics. J. Mol. Graph. 1996;14:33–38. doi: 10.1016/0263-7855(96)00018-5. [DOI] [PubMed] [Google Scholar]
- 43.Venkateshan M., Suresh J. Synthesis, physicochemical and quantum chemical studies on a new organic NLO crystal: Cinnamoylproline. J. Mol. Struct. 2019;1180:826–838. [Google Scholar]
- 44.Altschul S.F., Gish W., Miller W., Myers E.W., Lipman V. Basic local alignment search tool. J. Mol. Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 45.Altschul S.F., Madden T.L., Schaffer A.A., Zhang J., Zhang Z., Miller W., Lipman D.J. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;50:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kirchdoerfer R.N., Ward A.B. Structure of the SARS-CoV nsp12 polymerase bound to nsp7 and nsp8 co-factors. Nat. Commun. 2019;10:2342. doi: 10.1038/s41467-019-10280-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Šali A., Blundell T.L. Comparative protein modelling by satisfaction of spatial restraints. J. Mol. Biol. 1993;234:779–815. doi: 10.1006/jmbi.1993.1626. [DOI] [PubMed] [Google Scholar]
- 48.Berendsen H.J.C., van der Spoel D., van Drunen R. GROMACS: a message-passing parallel molecular dynamics implementation. Comput. Phys. Commun. 1995;91:43–56. [Google Scholar]
- 49.Gasteiger E., Gattiker A., Hoogland C., Ivanyi I., Appel R.D., Bairoch A. ExPASy: the proteomics server for in-depth protein knowledge and analysis. Nucleic Acids Res. 2003;31:3784–3788. doi: 10.1093/nar/gkg563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Laskowski R.A., Rullmann J.A.C., MacArthur M.W., Kaptein R., Thornton J.M. AQUA and PROCHECK-NMR: programs for checking the quality of protein structures solved by NMR. J. Biomol. NMR. 1996;8:477–486. doi: 10.1007/BF00228148. [DOI] [PubMed] [Google Scholar]
- 51.Ra J., Joan Pontius S.J.W. Deviations from standard atomic volumes as a quality measure for protein crystal structures. J. Mol. Biol. 1996;264:121–136. doi: 10.1006/jmbi.1996.0628. [DOI] [PubMed] [Google Scholar]
- 52.Hooft R.W., Vriend G., Sander C., Abola E.E. Errors in protein structures. Nature. 1996;381:272. doi: 10.1038/381272a0. [DOI] [PubMed] [Google Scholar]
- 53.Eisenberg D., Lüthy R., Bowie J.U. VERIFY3D: assessment of protein models with three-dimensional profiles. Methods Enzymol. 1997;277:396–404. doi: 10.1016/s0076-6879(97)77022-8. [DOI] [PubMed] [Google Scholar]
- 54.Ramachandran G.N., Ramakrishnan C., Sasisekharan V. Stereochemistry of polypeptide chain configurations. J. Mol. Biol. 1963;7:95–99. doi: 10.1016/s0022-2836(63)80023-6. [DOI] [PubMed] [Google Scholar]
- 55.Wass M.N., Kalley L.A., Sternberg M.J.E. 3DLigandSite: predicting ligand-binding sites using similar structures. Nucleic Acids Res. 2010;38:W469–W473. doi: 10.1093/nar/gkq406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Trott O., Olson A.J., Vina AutoDock. Improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. J. Comput. Chem. 2010;31:455–461. doi: 10.1002/jcc.21334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Dallakyan S., Olson A.J. Small-molecule library screening by docking with PyRx. Methods Mol. Biol. 2015;1263:243–250. doi: 10.1007/978-1-4939-2269-7_19. [DOI] [PubMed] [Google Scholar]
- 58.Pettersen E.F., Goddard T.D., Huang C.C., Couch G.S., Greenblatt D.M., Meng E.C., Ferrin T.E. UCSF Chimera - a visualization system for exploratory research and analysis. J. Comput. Chem. 2004;25:1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- 59.DeLano W.L. 2002. The PyMOL Molecular Graphics System.http://www.pymol.org [Google Scholar]
- 60.Schneidman-Duhovny D., Dror O., Inbar Y., Nussinov R., Wolfson H.J. PharmaGist: a webserver for ligand-based pharmacophore detection. Nucleic Acids Res. 2008;36:W223–W228. doi: 10.1093/nar/gkn187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Dong L., Hu S., Gao J. Discovering drugs to treat coronavirus disease 2019 (COVID-19) Drug Discov. Ther. 2020;14:58–60. doi: 10.5582/ddt.2020.01012. [DOI] [PubMed] [Google Scholar]
- 62.Daina A., Michielin O., Zoete V. SwissADME: a free web tool to evaluate pharmacokinetics, drug-likeness and medicinal chemistry friendliness of small molecules. Sci. Rep. 2017;7:42717. doi: 10.1038/srep42717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Yang H., Lou C., Sun L., Li J., Cai Y., Wang Z., Li W., Liu G., Tang Y. admetSAR 2.0: web-service for prediction and optimization of chemical ADMET properties. Bioinformatics. 2019;35:1067–1069. doi: 10.1093/bioinformatics/bty707. [DOI] [PubMed] [Google Scholar]
- 64.Organic Chemistry Portal. 2012. http://www.organic-chemistry.org/prog/peo/ [Google Scholar]
- 65.Wang Z., Wang X., Li Y., Lei T., Wang E., Li D., Kang Y., Zhu F., Hou T. farPPI: a webserver for accurate prediction of protein-ligand binding structures for small-molecule PPI inhibitors by MM/PB(GB)SA methods. Bioinformatics. 2019;35:1777–1779. doi: 10.1093/bioinformatics/bty879. [DOI] [PubMed] [Google Scholar]
- 66.Jakalian A., Bush B.L., Jack D.B., Bayly C.I. Fast, efficient generation of high-quality atomic Charges. AM1-BCC model: I. Method. J. Comput. Chem. 2000;21:132–146. doi: 10.1002/jcc.10128. [DOI] [PubMed] [Google Scholar]
- 67.Maier J.A., Martinez C., Kasavajhala K., Wickstrom L., Hauser K.E., Simmerling C. ff14SB: improving the accuracy of protein side chain and backbone parameters from ff99SB. J. Chem. Theor. Comput. 2015;11:3696–3713. doi: 10.1021/acs.jctc.5b00255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Wang J.M., Wolf R.M., Caldwell J.W., Kollman P.A., Case D.A. Development and testing of a general amber force field. J. Comput. Chem. 2004;25:1157–1174. doi: 10.1002/jcc.20035. [DOI] [PubMed] [Google Scholar]
- 69.Gonzalez H.F., Blanco-Lomas M., Rivado-Casas L., Rodriguez M.A., Campos P.J., Sampedro D. Addition of oxime derivatives to alkynyl fischer carbine complexes. Organometallics. 2012;31:6572–6581. [Google Scholar]
- 70.Portilla J., Lizarazo C., Cobo J., Glidewel C. A [pi]-stacked chain of hydrogen-bonded dimers in 3-tert-butyl-1-(4-chloro-phen-yl)-4-phenyl-indeno-[1,2-b]pyrazolo-[4,3-e]pyridin-5(1H)-one and a [pi]-stacked sheet of hydrogen-bonded chains in 3-tert-butyl-1-(4-chloro-phen-yl)-4-(4-meth-oxy-phen-yl)indeno-[1,2-b]pyrazolo-[4,3-e]pyridin-5(1H)-one. Acta Crystallogr. C. 2011;67:o479–o483. doi: 10.1107/S0108270111045082. [DOI] [PubMed] [Google Scholar]
- 71.Cobo D., Quiroga J., Cobo J., Low J.N., Glidewel C. 4-(4-Fluorophenyl)-3-methyl-1-phenylindeno[1,2- b ]pyrazolo[4,3- e ]pyridin-5(1 H )-one: sheets built from C—H...N, C—H...O and C—H...π(arene) hydrogen bonds. Acta Crystallogr. E. 2006;62:5176–5178. [Google Scholar]
- 72.a Tu S.J., Zhang Y., Jia R.H. 7-(4-Fluoro-phen-yl)-8H-benzo[h]indeno[1,2-b]quinolin-8-one. Acta Crystallogr. E. 2006;62:o3928–o3929. [Google Scholar]; b Tu S.J., Zhang Y., Jia R.H. 13-(4-Fluoro-phen-yl)-12H-benzo[f]indeno[1,2-b]quinolin-12-one. Acta Crystallogr. E. 2006;62:o3930–o3931. [Google Scholar]
- 73.Lemmerer A., Fernandes M.A. Adventures in co-crystal land: high Z′, stoichiometric variations, polymorphism and phase transitions in the co-crystals of four liquid and solid cyclic carboxylic acids with the supramolecular reagent isonicotinamide. New J. Chem. 2012;36:2242–2252. [Google Scholar]
- 74.Uhl W., Emden C.H., Geiseler G., Harms K. Z. Anorg. Allg. Chem. 2003;629:2157–2167. [Google Scholar]
- 75.Suresh J., Suresh Kumar R., Perumal S., Natarajan S. 1′-Methylcyclo-octane-1-spiro-3′-pyrrolidine-2′-spiro-3′′-indoline-2,2′′-dione and 1,1′-di-methylpiper-idine-3-spiro-3′-pyrrolidine-2′-spiro-3′′-indoline-2′′,4-dione. Acta Crystallogr. C. 2007;63:o538–o542. doi: 10.1107/S0108270107035779. [DOI] [PubMed] [Google Scholar]
- 76.a Ramesh P., Subbiahpandi A., Thirumurugan P., Perumal P.T., Ponnuswamy M.N. 4-(4-Bromo-phen-yl)-6-(1H-indol-3-yl)-2,2′-bi-pyridine-5-carbo-nitrile. Acta Crystallogr. E. 2009;65:o450. doi: 10.1107/S1600536809001354. [DOI] [PMC free article] [PubMed] [Google Scholar]; b Ramesh P., Subbiahpandi A., Thirumurugan P., Perumal P.T., Ponnuswamy M.N. 4-(2,4-Di-chloro-phenyl)-2-(1H-indol-3-yl)-6-(2-pyridyl)-1,4-di-hydro-pyridine-4-carbo-nitrile. Acta Crystallogr. E. 2008;64:o1891. doi: 10.1107/S1600536808027669. [DOI] [PMC free article] [PubMed] [Google Scholar]; c Ramesh P., Subbiahpandi A., Thirumurugan P., P, Perumal T., Ponnuswamy M.N. 4-(2,4-Di-chloro-phen-yl)-6-(1H-indol-3-yl)-2,2′-bi-pyridine-5-carbo-nitrile. Acta Crystallogr. E. 2009;65:o996–o997. doi: 10.1107/S1600536809012392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Zhu W., Xiang Y., Zhu S. 6-(1H-Indol-3-yl)-4-phenyl-2,2′-bi-pyridine-5-carbo-nitrile. Acta Crystallogr. E. 2009;65:o1187. doi: 10.1107/S1600536809016195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.a Vimala G., Poomathi N., AaminaNaaz Y., Perumal P.T., SubbiahPandi A. Crystal structure of 5-(5-chloro-2-hy-droxy-benzo-yl)-2-(2-methyl-1H-indol-3-yl)nicotino-nitrile. Acta Crystallogr. E. 2015;71:o822–o823. doi: 10.1107/S2056989015018058. [DOI] [PMC free article] [PubMed] [Google Scholar]; b Vishnupriya R., Suresh J., Bharkavi S., Perumal S., Nilantha Lakshman P.L. Crystal structure of 2-(2-bromo-phen-yl)-4-(1H-indol-3-yl)-6-(thio-phen-2-yl)pyridine-3-carbonitrile. Acta Crystallogr. E. 2014;70:o968–o969. doi: 10.1107/S1600536814017188. [DOI] [PMC free article] [PubMed] [Google Scholar]; c Vishnupriya R., Suresh J., Gunasekaran P., Perumal S., Nilantha Lakshman P.L.N. Crystal structure of 2-(4-chloro-phen-yl)-4-(1H-indol-3-yl)-6-phenyl-pyridine-3-carbo-nitrile. Acta Crystallogr. E. 2014;70:o978. doi: 10.1107/S1600536814017693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Bondi A. van der Waals volumes and radii. J. Phys. Chem. 1964;68:441–451. [Google Scholar]
- 80.Rowland R.S., Taylor R. Intermolecular nonbonded contact distances in organic crystal structures: comparison with distances expected from van der Waals radii. J. Phys. Chem. 1996;100:7384–7391. [Google Scholar]
- 81.Babu B., Chandrasekaran J., Mohanbabu B., Matsushita Y., Saravanakumar M. Growth, physicochemical and quantum chemical investigations on 2-amino 5-chloropyridinium 4-carboxybutanoate – an organic crystal for biological and optoelectronic device applications. RSC Adv. 2016;6:110884–110897. [Google Scholar]
- 82.Salihovic M., Huseinovic S., Spirtovic-Halilovic S., Osmanovic A., Dedic A., Asimovic Z., Zavrsnik D. DFT study and biological activity of some methylxanthines. Bulletin of the Chemists and Technologists of Bosnia and Herzegovina. 2014;42:31–36. [Google Scholar]
- 83.Lipinski C.A., Lombardo F., Dominy B.W., Feeney P.J. Experimental and computational approaches to estimate solubility and permeability in drug discovery and development settings. Adv. Drug Deliv. Rev. 2001;46:3–26. doi: 10.1016/s0169-409x(00)00129-0. [DOI] [PubMed] [Google Scholar]
- 84.Elfiky A.A. Ribavirin, Remdesivir, sofosbuvir, galidesivir, and tenofovir against SARS-CoV-2 RNA dependent RNA polymerase (RdRp): a molecular docking study. Life Sci. 2020;253:117592. doi: 10.1016/j.lfs.2020.117592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Ahn D.G., Choi J.K., Taylor D.R., Oh J.W. Biochemical characterization of a recombinant SARS coronavirus nsp12 RNA-dependent RNA polymerase capable of copying viral RNA templates. Arch. Virol. 2012;157:2095–2104. doi: 10.1007/s00705-012-1404-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Yin W., Mao C., Luan X., Shen D.D., Shen Q., Su H., Wang X., Zhou F., Zhao W., Gao M., Chang S., Xie Y.C., Tian G., Jiang H.W., Tao S.C., Shen J., Jinag Y., Jiang H., Xu Y., Zhang S., Zhang Y., Xu H.E. 2020. Structural Basis for the Inhibition of the RNA-dependent RNA Polymerase from SARS-CoV-2 by Remdesivir. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.