Peculiar among human RNA viruses, coronaviruses have large genomes containing accessory genes that are not required for replication [1]. Here, we report two large deletions in ORF7a, both of which produce new open reading frames (ORFs) through the fusion of the N-terminus of ORF7a and a downstream ORF.
We recovered genomes from the SARS-CoV-2 isolates from two separate patients as part of an ongoing University of Washington IRB-approved genomic surveillance project [2,3]. Sequencing libraries were generated from double-stranded cDNA [4] using Swift Biosciences’ SARS-CoV-2 Normalase Amplicon Sequencing kit or the Nextera DNA Flex Pre-Enrichment kit (Illumina) followed by enrichment using a SARS-CoV-2 xGen enrichment panel (NC_045512; IDT). Sequence reads were trimmed using Trimommatic v0.38 [5], aligned to the SARS-CoV-2 reference genome (NC_045512.2) using BBMap (https://sourceforge.net/projects/bbmap/), trimmed of synthetic PCR primers using Primerclip (https://github.com/swiftbiosciences/primerclip) if appropriate, and visualized in Geneious v11.1.4 [6]. Sequencing reads and consensus genomes are available under NCBI BioProject PRJNA610428.
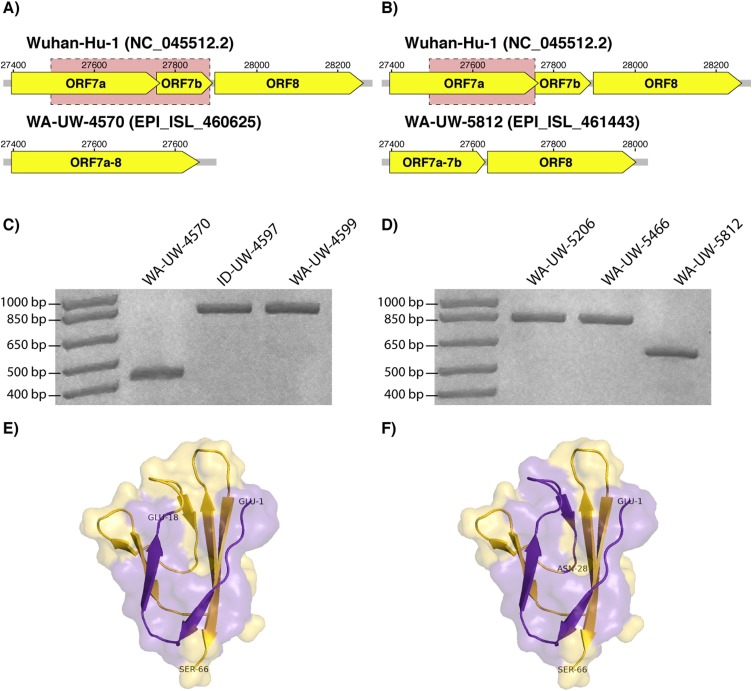
In WA-UW-4570, we identified a 392-nucleotide deletion originating at nt 27,494 in ORF7a. This deletion results in the loss of the entirety of ORF7b and creates a new ORF through the fusion of the N-terminus of ORF7a with ORF8 (Fig. 1 a). Notably, this is the largest deletion recorded to date in SARS-CoV-2. In WA-UW-5812, we identified a 227 nt deletion also resulting a new ORF through the fusion of the N-terminus of ORF7a with ORF7b (Fig. 1 b). Both deletions were confirmed by RT-PCR (Fig. 1 c and d) and Sanger sequencing. In contrast to a previously sequenced SARS-CoV-2 isolate with a deletion in ORF7a [7], our isolates retained the presumptive signal peptide and part of the β sheet of the Ig superfamily fold of ORF7a (Fig. 1 e and f) and lost the protein’s transmembrane and cytoplasmic domains (PDB 6W37) [8].
Fig. 1.
A 392-nucleotide deletion starting at nt 29,424 was identified in a) WA-UW-4570 and resulted in the fusion of ORF7a and ORF8. A 227-nucleotide deletion beginning at nt 27,524 was identified in b) WA-UW-5812 and resulted in the fusion of ORF7a and ORF7b. The deletions in c) WA-UW-4570 and d) WA-UW-5812 were confirmed by RT-PCR. An 826-bp product is expected for strains with an intact ORF7a. 434-bp and 599-bp amplicons were obtained for WA-UW-4570 and WA-UW-5812, respectively. The deletions in e) WA-UW-4570 and f) WA-UW-5812 severely truncate the β sheet and result in the loss of the transmembrane and cytoplasmic domains of ORF7a (PDB 6W37) [8]. Residues retained are highlighted in purple, while those lost are highlighted in gold (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article).
We next assessed the genetic relatedness of the two strains. WA-UW-4570 and WA-UW-5812 differed by 12 single nucleotide variants and a 3-bp deletion in the spike protein of WA-UW-4570 and belonged to the SARS-CoV-2 phylogenetic lineages A.1 and B.1 respectively (https://github.com/hCoV-2019/pangolin) [9]. This suggests that the ORF7a deletions arose independently in the two strains. Both samples were recovered from the only SARS-CoV-2-positive nasopharyngeal swab available from each patient. The ORF1ab qRT-PCR cycle thresholds on the Hologic Panther Fusion for these isolates were 27.6 (WA-UW-4570) and 28.5 (WA-UW-5812). WA-UW-4570 was collected from an individual prophylactically taking hydroxychloroquine and mycophenolate mofetil while the WA-UW-5812 individual was not taking any medications for SARS-CoV-2 infection.
ORF7a of SARS-CoV-2 interacts with the ribosomal transport proteins HEATDR3 and MDN1 [10], and has been demonstrated to inhibit cellular translation in SARS-CoV [11]. Based on the size of the deletions recovered and structure of ORF7a, we hypothesize that most biochemical functions of ORF7a would be inactivated by these deletions. Interestingly, ORF6 of SARS-CoV-2 interacts with the mRNA export proteins NUP98 and RAE1, and may inhibit cellular translation [10]. This redundancy may partially explain the ORF7a deletions observed in our isolates. We predict global sequencing projects may yield additional clinical SARS-CoV-2 isolates with deletions in ORF6 or ORF7a, but not both.
Declaration of Competing Interest
None
References
- 1.Narayanan K., Huang C., Makino S. SARS coronavirus accessory proteins. Virus Res. 2008;133:113–121. doi: 10.1016/j.virusres.2007.10.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Fauver J.R., Petrone M.E., Hodcroft E.B., Shioda K., Ehrlich H.Y., Watts A.G., Vogels C.B.F., Brito A.F., Alpert T., Muyombwe A., Razeq J., Downing R., Cheemarla N.R., Wyllie A.L., Kalinich C.C., Ott I.M., Quick J., Loman N.J., Neugebauer K.M., Greninger A.L., Jerome K.R., Roychoudhury P., Xie H., Shrestha L., Huang M.-L., Pitzer V.E., Iwasaki A., Omer S.B., Khan K., Bogoch I.I., Martinello R.A., Foxman E.F., Landry M.L., Neher R.A., Ko A.I., Grubaugh N.D. Coast-to-Coast spread of SARS-CoV-2 during the early epidemic in the United States. Cell. 2020;181:990–996. doi: 10.1016/j.cell.2020.04.021. e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Peddu V., Shean R.C., Xie H., Shrestha L., Perchetti G.A., Minot S.S., Roychoudhury P., Huang M.-L., Nalla A., Reddy S.B., Phung Q., Reinhardt A., Jerome K.R., Greninger A.L. Metagenomic analysis reveals clinical SARS-CoV-2 infection and bacterial or viral superinfection and colonization. Clin. Chem. 2020 doi: 10.1093/clinchem/hvaa106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Greninger A.L., Zerr D.M., Qin X., Adler A.L., Sampoleo R., Kuypers J.M., Englund J.A., Jerome K.R. Rapid metagenomic next-generation sequencing during an investigation of hospital-acquired human parainfluenza virus 3 infections. J. Clin. Microbiol. 2017;55:177–182. doi: 10.1128/JCM.01881-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bolger A.M., Lohse M., Usadel B. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics. 2014;30:2114–2120. doi: 10.1093/bioinformatics/btu170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kearse M., Moir R., Wilson A., Stones-Havas S., Cheung M., Sturrock S., Buxton S., Cooper A., Markowitz S., Duran C., Thierer T., Ashton B., Meintjes P., Drummond A. Geneious Basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics. 2012;28:1647–1649. doi: 10.1093/bioinformatics/bts199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Holland L.A., Kaelin E.A., Maqsood R., Estifanos B., Wu L.I., Varsani A., Halden R.U., Hogue B.G., Scotch M., Lim E.S. An 81 nucleotide deletion in SARS-CoV-2 ORF7a identified from sentinel surveillance in Arizona (jan-Mar 2020) J. Virol. 2020 doi: 10.1128/JVI.00711-20. JVI.00711-20, jvi;JVI.00711-20v1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Nelson C.A., Pekosz A., Lee C.A., Diamond M.S., Fremont D.H. Structure and intracellular targeting of the SARS-coronavirus Orf7a accessory protein. Structure. 2005;13:75–85. doi: 10.1016/j.str.2004.10.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Rambaut A., Holmes E.C., Hill V., O’Toole Á, McCrone J., Ruis C., du Plessis L., Pybus O.G. A dynamic nomenclature proposal for SARS-CoV-2 to assist genomic epidemiology. Microbiology. 2020 doi: 10.1038/s41564-020-0770-5. Preprint. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gordon D.E., Jang G.M., Bouhaddou M., Xu J., Obernier K., White K.M., O’Meara M.J., Rezelj V.V., Guo J.Z., Swaney D.L., Tummino T.A., Huettenhain R., Kaake R.M., Richards A.L., Tutuncuoglu B., Foussard H., Batra J., Haas K., Modak M., Kim M., Haas P., Polacco B.J., Braberg H., Fabius J.M., Eckhardt M., Soucheray M., Bennett M.J., Cakir M., McGregor M.J., Li Q., Meyer B., Roesch F., Vallet T., Mac Kain A., Miorin L., Moreno E., Naing Z.Z.C., Zhou Y., Peng S., Shi Y., Zhang Z., Shen W., Kirby I.T., Melnyk J.E., Chorba J.S., Lou K., Dai S.A., Barrio-Hernandez I., Memon D., Hernandez-Armenta C., Lyu J., Mathy C.J.P., Perica T., Pilla K.B., Ganesan S.J., Saltzberg D.J., Rakesh R., Liu X., Rosenthal S.B., Calviello L., Venkataramanan S., Liboy-Lugo J., Lin Y., Huang X.-P., Liu Y., Wankowicz S.A., Bohn M., Safari M., Ugur F.S., Koh C., Savar N.S., Tran Q.D., Shengjuler D., Fletcher S.J., O’Neal M.C., Cai Y., Chang J.C.J., Broadhurst D.J., Klippsten S., Sharp P.P., Wenzell N.A., Kuzuoglu D., Wang H.-Y., Trenker R., Young J.M., Cavero D.A., Hiatt J., Roth T.L., Rathore U., Subramanian A., Noack J., Hubert M., Stroud R.M., Frankel A.D., Rosenberg O.S., Verba K.A., Agard D.A., Ott M., Emerman M., Jura N., von Zastrow M., Verdin E., Ashworth A., Schwartz O., d’Enfert C., Mukherjee S., Jacobson M., Malik H.S., Fujimori D.G., Ideker T., Craik C.S., Floor S.N., Fraser J.S., Gross J.D., Sali A., Roth B.L., Ruggero D., Taunton J., Kortemme T., Beltrao P., Vignuzzi M., García-Sastre A., Shokat K.M., Shoichet B.K., Krogan N.J. A SARS-CoV-2 protein interaction map reveals targets for drug repurposing. Nature. 2020 doi: 10.1038/s41586-020-2286-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kopecky-Bromberg S.A., Martinez-Sobrido L., Palese P. 7a protein of severe acute respiratory syndrome coronavirus inhibits cellular protein synthesis and activates p38 mitogen-activated protein kinase. JVI. 2006;80:785–793. doi: 10.1128/JVI.80.2.785-793.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]