Highlights
-
•
ORF6, ORF8 and nucleocapsid proteins of SARS-CoV-2 strongly inhibited type I interferon (IFN-β) activation and NF-κB pathway.
-
•
ORF6 and ORF8, but not nucleocapsid proteins, were capable of inhibiting ISRE-driven transcription activated by IFN-β.
-
•
ORF6, ORF8 and nucleocapsid proteins of SARS-CoV-2 inhibited interferon-stimulated genes (ISGs) such as ISG54 and ISG56.
-
•
ORF6, ORF8 and nucleocapsid proteins of SARS-CoV-2 played critical roles in innate immune suppression during viral infection.
Keywords: COVID-19, SARS-CoV-2, Structural proteins, Accessory proteins, Interferon
Abstract
The severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) is a novel human coronavirus causing the pandemic of severe pneumonia (Coronavirus Disease 2019, COVID-19). SARS-CoV-2 is highly pathogenic in human, having posed immeasurable public health challenges to the world. Innate immune response is critical for the host defense against viral infection and the dysregulation of the host innate immune responses probably aggravates SARS-CoV-2 infection, contributing to the high morbidity and lethality of COVID-19. It has been reported that some coronavirus proteins play an important role in modulating innate immunity of the host, but few studies have been conducted on SARS-CoV-2. In this study, we screened the viral proteins of SARS-CoV-2 and found that the viral ORF6, ORF8 and nucleocapsid proteins were potential inhibitors of type I interferon signaling pathway, a key component for antiviral response of host innate immune. All the three proteins showed strong inhibition on type I interferon (IFN-β) and NF-κB-responsive promoter, further examination revealed that these proteins were able to inhibit the interferon-stimulated response element (ISRE) after infection with Sendai virus, while only ORF6 and ORF8 proteins were able to inhibit the ISRE after treatment with interferon beta. These findings would be helpful for the further study of the detailed signaling pathway and unveil the key molecular player that may be targeted.
1. Short communication
The pandemic of coronavirus disease 2019 (COVID-19) caused by the 2019 novel coronavirus (2019-nCoV or SARS-CoV-2) infection has become a Public Health Emergency of International Concern (PHEIC) with more than 6 million cases and 376,320 deaths as of June 2, 2020 (WHO, 2020, https://covid19.who.int). SARS-CoV-2 infection causes disorder of natural and adaptive immunity, leading to tissue damage and systemic inflammation, which is the main reason for death of COVID-19 patients (Huang et al., 2020). Up to now, the mechanism underlying the modulation of immune signaling pathways by SARS-CoV-2 is still unclear. Viral proteins usually play critical roles in interfering with host immune response. In this study, we aimed to screen potential SARS-CoV-2 proteins modulating host immune response, especially the type I interferon (IFN) pathways.
Upon virus infection, several transcription factors, such as IRF-3 and NF-κB, bind to the interferon promoter to stimulate type I IFN (IFN-α/β) expression (García-Sastre and Biron, 2006). Then the interferon is secreted and binds to the interferon receptors, initiating the JAK/STAT pathway and inducing the nucleus translocation of IFN-responsive transcriptional factors. These transcriptional factors activate genes containing interferon-stimulated response elements (ISREs) in their promoters, resulting in the expression of a set of IFN-stimulated genes (ISGs) which establish an antiviral state (Catanzaro et al., 2020). In response to this powerful selective environment, many viruses from diverse families, including filoviruses, poxviruses, influenza viruses, flaviviruses, and coronaviruses (CoVs), have evolved multiple passive and active mechanisms to avoid induction of the antiviral type I interferon, and they could optimize the intracellular resource for efficient virus replication (Volk et al., 2020). For the case of highly pathogenic coronaviruses, the structural and nonstructural proteins (nsp16−2-O MTase, nsp14-ExoN, nsp1, nsp7, envelope (E) protein, nucleocapsid (N) protein, membrane (M) protein, SARS-CoV-ORF6, MERS-CoV-ORF3−5, MERS-CoV-4a, and MERS-CoV-4b) have been shown to antagonize the innate immune response (Volk et al., 2020). In addition, inactivating viral interferon antagonists, such as MERS-CoV ΔORF3−5 mutant virus (Menachery et al., 2018), nsp14 and nsp16 of SARS-CoV (Menachery et al., 2017) and nsp15 in PEDV (Deng et al., 2019), would prompt earlier and more robust type I interferon responses to suppress viruses replication. Thus, systematic elimination of IFN-modulating functions from the virus is supposed to become a promoting approach for vaccine development. However, functions of the proteins encoded by SARS-CoV-2 have not been revealed clearly yet, some of them may be immunoregulator against host innate immune system.
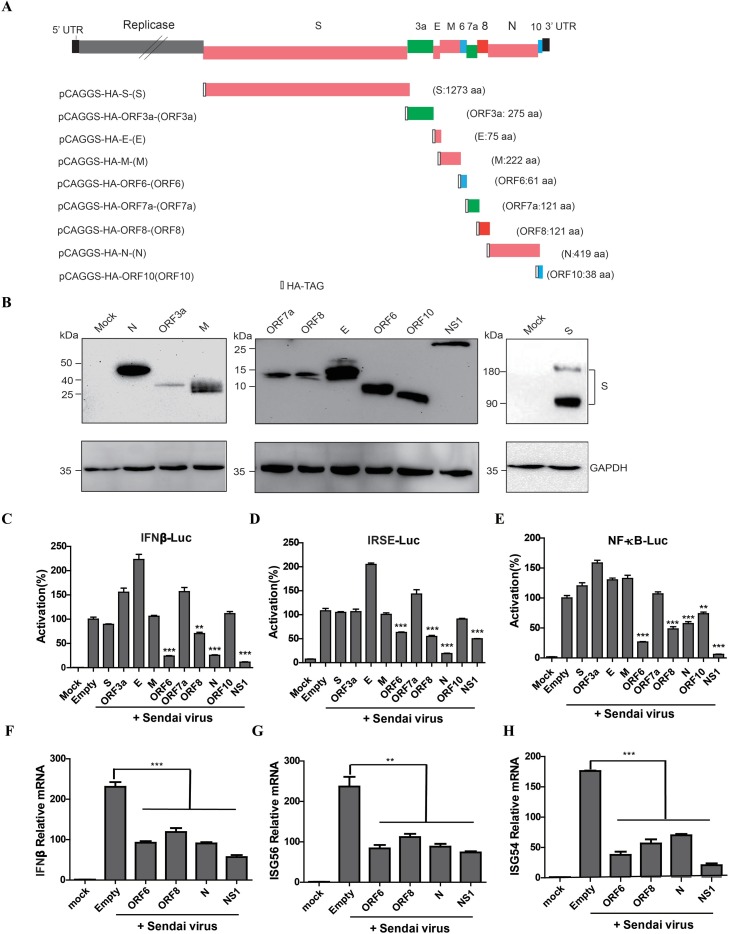
To study the gene function of new coronavirus, SARS-CoV-2 ORFs of the structural genes (S, E, M, and N) and the accessory genes (3a, 6, 7a, 8 and 10) (GenBank ID MN908947.3) were synthesized by Sangon Biotechnology Co., Ltd. (Shanghai, China), and then cloned into pCAGGS vector with a tag encoding hemagglutinin (HA) at the N-terminus of each protein (Fig. 1 A). The plasmids were transfected into 293 T cells individually, and the proteins were collected at 48 h post transfection. The expression of each gene is measured by Western blotting using the anti-HA tag antibody, confirming the correct expression of all proteins (Fig. 1B).
Fig. 1.
SARS-CoV-2 ORF6, ORF8, and N proteins inhibit the expression of IFN-β and the activation of ISGs. (A) Schematic diagram of the genome organization of SARS-CoV-2 and expression constructs used in this study. The synthesized coding genes of individual structural and accessory proteins were inserted into the expression vector pCAGGS with an HA-tag at the N-terminus of each protein. (B) Expression verification of the individual structural and accessory proteins of SARS-CoV-2 in HEK-293 T cells. Samples were collected from cells transiently transfected with individual ORF expression plasmids or pCAGGS (indicated as Mock) at 48 h post transfection. Proteins were separated by SDS-PAGE and the SARS-CoV-2 proteins were detected by Western blotting using mouse anti-HA and anti-GAPDH monoclonal antibodies. (C, D, E) HEK-293 T cells were co-transfected with IFN-β-Luc (C), ISRE-Luc (D) or NF-κB-Luc (E) together with the pRL-TK plasmid, and then transfected with the plasmid expressing the indicated SARS-CoV-2 protein. 24 h after the initial transfection, the cells were infected with Sendai virus. Luciferase assays were performed 18 h after infection. The results were presented as the means and standard deviations of data from three independent experiments. The relative firefly luciferase activity was normalized to the Renilla reniformis luciferase activity, and the treated empty vector control value was set to 100. (F, G, H) HEK-293 T cells were transfected with empty vector- or expressing plasmids for 24 h and then mock infected or infected with SeV for 18 h. mRNA expression levels of IFN-β (F), ISG56 (G), and ISG54 (H) in the collected cells were detected by qPCR. *, P < 0.05; **, P < 0.01; *** P < 0.001 versus empty (Student’s t-test).
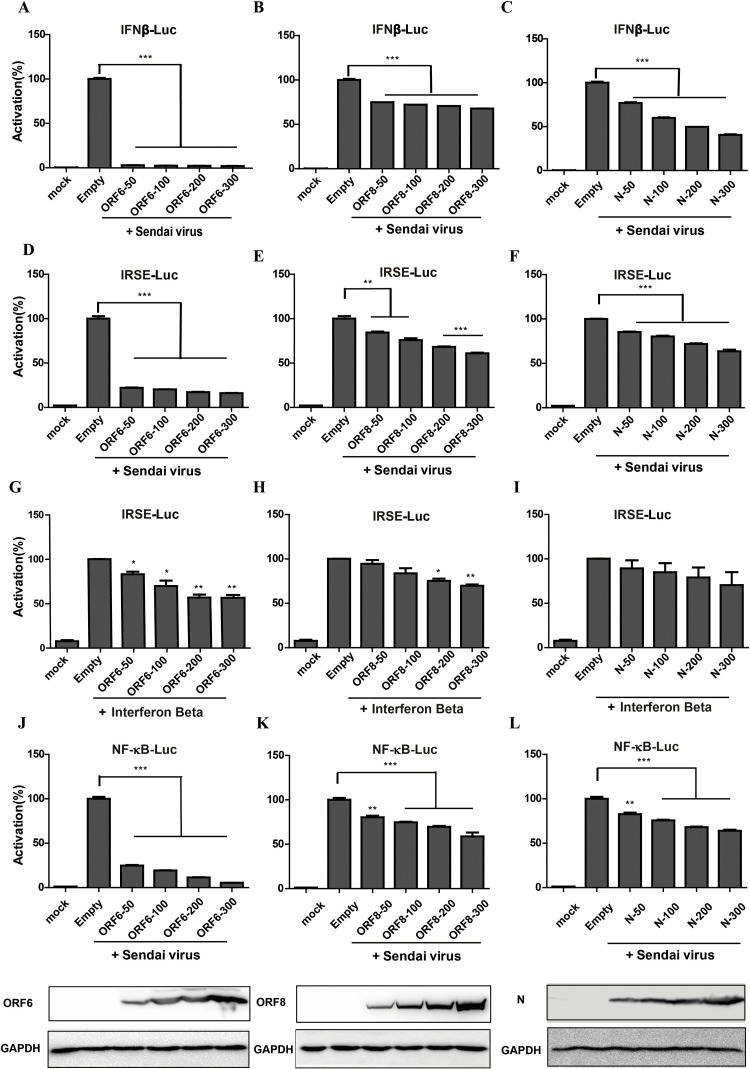
To examine the potential role of SARS-CoV-2 proteins in modulating IFN signaling pathway, each viral protein was individually tested using the luciferase reporter gene assay system. HEK 293 T cells were transfected with the luciferase reporter plasmids IFN-β, ISRE or NF-κB and the plasmid pRL-TK as an internal control. The cells were co-transfected with plasmids expressing individual SARS-CoV-2 structural or accessory proteins, with empty pCAGGS plasmid as negative control and an NS1 expression plasmid of influenza (A/Puerto Rico/8/34, H1N1) virus as positive control. At 24 h post transfection, cells were infected with Sendai virus (SeV) to induce interferon synthesis, and at 18 h postinfection, cells were lysed and firefly luciferase and Renilla luciferase activities were determined with the Dual-Luciferase reporter assay system. As expected, the positive control (NS1) demonstrated the strongest inhibition of IFN-β. The SARS-CoV-2 ORF6, ORF8, and N proteins could inhibit of IFN-β promoter (Fig. 1C), ISRE promoter (Fig. 1D) and NF-κB element (Fig. 1E). A dose dependent assay was performed to confirm the antagonistic IFN-β promoter activity (Fig. 2 A-C), ISRE promoter (Fig. 2D-F) or NF-κB responsive promoter (Fig. 2J-L) of ORF6, ORF8, and N protein with SeV infection. To clarify whether these viral proteins impaired IFN synthesis or the downstream signals, we substituted SeV with the recombinant IFN-β protein and found that ORF6 and ORF8, but not N proteins inhibited expression from the ISRE promoter (Fig. 2G-I), indicating that they play a role in different nodes and may have different mechanisms to regulate host interferon pathway.
Fig. 2.
Dose-dependent inhibition the activation of IFN-β promoter, ISRE and NF-κB promoter by SARS-CoV-2 ORF6, ORF8, and N proteins. HEK-293 T cells were co-transfected with IFN-β-Luc, ISRE-Luc or NF-κB together with the pRL-TK plasmid (a plasmid constitutively expressing Renilla luciferase), and then were transfected with the plasmid pCAGGS-HA-ORF6 (A, D, G, J) or pCAGGS-HA-ORF8 (B, E, H, K) or pCAGGS-HA-N (C, F, I, L) of 0, 0.05, 0.1, 0.2 and 0.3 μg. The IFN-β promoter activity was measured upon SeV infection (A-C). The ISRE promoter activity was measured upon SeV infection (D-F) or the recombinant IFN-β protein treatment (G-I). The NF-κB promoter activity was measured upon SeV infection and equal amounts of lysates were also used for Western blotting analysis to ensure equal expression for each transfection (J-L).
It is well known that infection by various RNA viruses activates the RIG-I Like Receptor pathway and initiates the expression of IFN-β and a set of ISGs. To further confirm the activation of the IFN-β pathway, 293 T cells were transfected with the plasmids expressing SARS-CoV-2 proteins and then infected with SeV. The cells were collected for the quantitative PCR (qPCR) detection of the expression levels of IFN-β and some ISGs. The results showed that ORF6, ORF8, and N protein significantly suppressed SeV-induced mRNA expression of IFN-β (Fig. 1F), ISG56 (Fig. 1G) and ISG54 (Fig. 1H).
We identified ORF6, ORF8, and N of SARS-CoV-2 as type I IFN antagonists. Compared with ORF6 and N, the antagonistic IFN-β promoter activity of ORF8 was the weakest (Fig. 1C and 2A-C). Recent literature reported that the ORF8 deletion mutant (Δ382) displayed a greater level of transcripts per million (TPM) in the ORF6 and N genes compared to WT, having no effect on viral RNA replication (Yvonne et al., 2020).We therefore hypothesized that the ability of evading the host innate immune of ORF8 deletion mutant may be stronger than WT. In fact, deletions in ORF8 were also observed during the SARS-CoV outbreak in 2003 (Muth et al., 2018). Similarly, the SARS-CoV-2 strains with the ORF8 deletion discovered in different regions of the world (Gong et al., 2020; Yvonne et al., 2020), such variations are believed to be still evolving and facilitate the successful adaption of the virus to various hosts. Furthermore, SARS-CoV-2 ORF3b was also identified as a potent interferon antagonist whose activity is further increased by a naturally occurring elongation variant (Yoriyuki et al., 2020). ORF3b is not performed in this study, since the first sequence published in the database did not annotate the ORF3b gene.
SARS-CoV-2 ORF6, ORF8, N and ORF3b are potent interferon antagonist, in the early stages of SARS-CoV-2 infection, delayed release of IFNs would hinder the host's antiviral response and then benefit virus replication. Afterward, the rapidly increased cytokine and chemokine attract inflammatory cells, such as neutrophils and monocytes, resulting in excessive immune infiltration causing tissue damage. Inactivating viral interferon antagonists is an approach of acquiring live-attenuated vaccines, which is supported by recent reports about the development of influenza A virus and yellow fever virus by impairing the interferon antagonists (Laurent-Rolle et al., 2014; Du et al., 2018). For SARS-CoV-2, it is not yet clear if disabling a single interferon antagonist, such as the accessory protein (ORF6, ORF8), will be sufficient to attenuate viruses that infect different cell types in different species. Nevertheless, this study could lay the theoretical foundation for exploring detailed signal pathways and generating safe and protective live-attenuated coronavirus vaccines.
CRediT authorship contribution statement
Jin-Yan Li: Conceptualization, Methodology, Formal analysis, Investigation, Writing - original draft, Writing - review & editing. Ce-Heng Liao: Methodology, Formal analysis, Investigation. Qiong Wang: Formal analysis, Investigation. Yong-Jun Tan: Resources. Rui Luo: Resources. Ye Qiu: Writing - review & editing, Supervision, Funding acquisition. Xing-Yi Ge: Conceptualization, Writing - review & editing, Supervision, Funding acquisition.
Declaration of Competing Interest
The authors declare that they have no conflict of interest.
Acknowledgements
This work was jointly funded by National Natural Science Foundation of China (grant number 32041001 and 81902070), the Provincial Natural Science Foundation of Hunan Province (grant number 2019JJ20004, 2019JJ50035, and 2020SK3001).
Contributor Information
Jin-Yan Li, Email: lijinyan@hnu.edu.cn.
Ce-Heng Liao, Email: liaoceheng@hnu.edu.cn.
Qiong Wang, Email: qw@hnu.edu.cn.
Yong-Jun Tan, Email: yjtan@hnu.edu.cn.
Rui Luo, Email: luorui@mail.hzau.edu.cn.
Ye Qiu, Email: qiuye@hnu.edu.cn.
Xing-Yi Ge, Email: xyge@hnu.edu.cn.
References
- Catanzaro M., Fagiani F., Racchi M., Corsini E., Govoni S., Lanni C. Immune response in COVID-19: addressing a pharmacological challenge by targeting pathways triggered by SARS-CoV-2. Signal Transduct. Target. Ther. 2020;5(1):84. doi: 10.1038/s41392-020-0191-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng X., van Geelen A., Buckley A.C., O’Brien A., Pillatzki A., Lager K.M., Faaberg K.S., Baker S.C. Coronavirus endoribonuclease activity in porcine epidemic diarrhea virus suppresses type I and type III interferon responses. J. Virol. 2019;93(8) doi: 10.1128/JVI.02000-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du Y., Xin L., Shi Y., Zhang T.H., Wu N.C., Dai L., Gong D., Brar G., Shu S., Luo J., Reiley W., Tseng Y.W., Bai H., Wu T.T., Wang J., Shu Y., Sun R. Genome-wide identification of interferon-sensitive mutations enables influenza vaccine design. Science. 2018;359(6373):290–296. doi: 10.1126/science.aan8806. [DOI] [PubMed] [Google Scholar]
- García-Sastre A., Biron C.A. Type 1 interferons and the virus-host relationship: a lesson in détente. Science. 2006;312(5775):879–882. doi: 10.1126/science.1125676. [DOI] [PubMed] [Google Scholar]
- Gong Y.N., Tsao K.C., Hsiao M.J., Huang C.G., Huang P.N., Huang P.W., Lee K.M., Liu Y.C., Yang S.L., Kuo R.L., Chen K.F., Liu Y.C., Huang S.Y., Huang H.I., Liu M.T., Yang J.R., Chiu C.H., Yang C.T., Chen G.W., Shih S.R. SARS-CoV-2 genomic surveillance in Taiwan revealed novel ORF8-deletion mutant and clade possibly associated with infections in Middle East. Emerg. Microbes & Infec. 2020:1–37. doi: 10.1080/22221751.2020.1782271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang C., Wang Y., Li X., Ren L., Zhao J., Hu Y., Zhang L., Fan G., Xu J., Gu X., Cheng Z., Yu T., Xia J., Wei Y., Wu W., Xie X., Yin W., Li H., Liu M., Xiao Y., Gao H., Guo L., Xie J., Wang G., Jiang R., Gao Z., Jin Q., Wang J., Cao B. Clinical features of patients infected with 2019 novel coronavirus in Wuhan. China. Lancet. 2020;395(10223):497–506. doi: 10.1016/S0140-6736(20)30183-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laurent-Rolle M., Morrison J., Rajsbaum R., Macleod J.M.L., Pisanelli G., Pham A., Ayllon J., Miorin L., Martinez C., tenOever B.R., García-Sastre A. The interferon signaling antagonist function of yellow fever virus NS5 protein is activated by type I interferon. Cell Host Microbe. 2014;16(3):314–327. doi: 10.1016/j.chom.2014.07.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Menachery V.D., Mitchell H.D., Cockrell A.S., Gralinski L.E., Yount B.L., Jr., Graham R.L., McAnarney E.T., Douglas M.G., Scobey T., Beall A., Dinnon K., 3rd, Kocher J.F., Hale A.E., Stratton K.G., Waters K.M., Baric R.S. MERS-CoV accessory ORFs play key role for infection and pathogenesis. mBio. 2017;8(4) doi: 10.1128/mBio.00665-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Menachery V.D., Gralinski L.E., Mitchell H.D., Dinnon K.H., 3rd, Leist S.R., Yount B.L., Jr., McAnarney E.T., Graham R.L., Waters K.M., Baric R.S. Combination attenuation offers strategy for live attenuated coronavirus vaccines. J. Virol. 2018;92(17) doi: 10.1128/JVI.00710-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muth D., Corman V.M., Roth H., Binger T., Dijkman R., Gottula L.T., Gloza-Rausch F., Balboni A., Battilani M., Rihtarič D., Toplak I., Ameneiros R.S., Pfeifer A., Thiel V., Drexler J.F., Müller M.A., Drosten C. Attenuation of replication by a 29 nucleotide deletion in SARS-coronavirus acquired during the early stages of human-to-human transmission. Sci. Rep. 2018;8(1):15177. doi: 10.1038/s41598-018-33487-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Volk A., Hackbart M., Deng X., Cruz-Pulido Y., O’Brien A., Baker S.C. Coronavirus endoribonuclease and deubiquitinating interferon antagonists differentially modulate the host response during replication in macrophages. J. Virol. 2020;94(11) doi: 10.1128/JVI.00178-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoriyuki K., Izumi K., Keiya U., Masaya F., Takashi I., Yoshio K., So N., Kei S. SARS-CoV-2 ORF3b is a potent interferon antagonist whose activity is further increased by a naturally occurring elongation variant. bioRxivorg. 2020 doi: 10.1101/2020.05.11.088179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yvonne C.S., Danielle E.A., Barnaby E.Y., Feng Z., Martin L., Shirin K., Jenny G.H.L., Zhuang Y., Jayanthi J., Louisa S., Gabriel Z.Y., Ian H.M., Yee-Sin L., David C.L., Wang L.F., Gavin J.D.S. Discovery of a 382-nt deletion during the early evolution of SARS-CoV-2. bioRxivorg. 2020 doi: 10.1101/2020.03.11.987222. [DOI] [Google Scholar]