Abstract
Objectives
The present review systematically summarized existing publications regarding the genetic associations between voltage-gated calcium channels (VGCCs) and autism spectrum disorder (ASD).
Methods
A comprehensive literature search was conducted to gather pertinent studies in three online databases. Two authors independently screened the included records based on the selection criteria. Discrepancies in each step were settled through discussions.
Results
From 1163 resulting searched articles, 28 were identified for inclusion. The most prominent among the VGCCs variants found in ASD were those falling within loci encoding the α subunits, CACNA1A, CACNA1B, CACNA1C, CACNA1D, CACNA1E, CACNA1F, CACNA1G, CACNA1H, and CACNA1I as well as those of their accessory subunits CACNB2, CACNA2D3, and CACNA2D4. Two signaling pathways, the IP3-Ca2+ pathway and the MAPK pathway, were identified as scaffolds that united genetic lesions into a consensus etiology of ASD.
Conclusions
Evidence generated from this review supports the role of VGCC genetic variants in the pathogenesis of ASD, making it a promising therapeutic target. Future research should focus on the specific mechanism that connects VGCC genetic variants to the complex ASD phenotype.
Keywords: Autism spectrum disorder, Voltage-gated calcium channels, Calcium signaling, Calcium pathway
Background
Autism spectrum disorder
Autism spectrum disorder (ASD) is a neurodevelopmental disorder characterized by deficits in social-communication skills along with repetitive stereotypic behaviors that manifest in early postnatal life [24]. The incidence of ASD has markedly increased 10 to 17% per year during the last two decades [4], affecting approximately 1 out of 68 children in the United States [3]. The rapid rise in the prevalence of ASD has stimulated numerous research interests into the potential etiology of this disorder, but the underlying molecular and cellular mechanisms have yet to be determined. Advances in human genetics and sequencing technologies have identified a stunning number of newly arising mutations associated with ASD [15], some of which are located in genes encoding voltage-gated calcium channels (VGCCs).
Voltage-gated calcium channels
Voltage-gated calcium channels (VGCCs) are transmembrane proteins that activate in response to depolarization of the cell membrane and mediate the flux of calcium (Ca2+) into excitable cells [1, 20]. The VGCCs can be divided into high voltage-activated channels (HVA) and low voltage-activated channels (LVA) based on their electrophysiological properties [6]. HVA channels include L-, N-, P−/Q- and R-type channels, and LVA channels are also known as T-type calcium channels [13, 44, 55].
The L-type calcium channel is activated by a strong depolarization voltage and inactivated with a slow time course and is expressed in neurons, endocrine, cardiac, and smooth muscle [12]. The N-, P−/Q-, and R-type calcium channels also require strong depolarization for activation and are expressed mainly in neurons [28]. The T-type calcium channel is opened with weak depolarization voltages and remains active for a short time and is widely expressed in various cell types [49].
VGCCs are hetero-oligomeric proteins composed of a pore-forming α1 subunit and a group of auxiliary subunits known as α2δ and β [14, 16]. The α1 subunit is encoded by the CACNA1x genes (CACNA1A to CACNA1I and CACNA1S), and there are 10 isoforms in the human genome [7, 20]. The L-type channels, including Cav1.1, Cav1.2, Cav1.3, and Cav1.4, are encoded by CACNA1S, C, D, and F, respectively [8]. The P/Q-, N- and R-type channels, corresponding to Cav2.1, Cav2.2, and Cav2.3, are encoded by CACNA1A, B and E, respectively [8]. The T-type channels, including Cav3.1, Cav3.2, and Cav3.3, are encoded by CACNA1G, H and I, respectively [35]. Four genes exist for the Cavα2δ subunits (CACNA2D1–4) and encode Cavα2δ-1-4 proteins. Four genes exist for the Cavβ subunits (CACNB1–4) and encode Cavβ1–4 proteins.
Both the Cav1 and Cav2 channels form multiprotein complexes comprising the Cavα1 pore-forming co-assembly with one of four α2δ subunits and one of four β subunits. The Cav3 channels form functional channels using the α1 subunits alone but may also associate with other proteins. The kinetics, voltage-dependence, and pharmacological properties of the calcium channels are principally determined by the α1 subunits [20]. Although α1 subunits determine the main properties of the calcium channels, their functions are modified by the two auxiliary subunits. These auxiliary subunits have profound effects on the biophysical properties and membrane targeting of the α1 subunit [14, 16]. The different channel isoforms and possible combinations make for considerable potential diversity in the properties and function of the calcium channels.
Objectives
Although the association between VGCCs and ASD has been reported in both correlation studies and informative reviews, no systematic review has been conducted to comprehensively synthesize the literature to determine whether this is fully supported by the totality of all available evidence. Therefore, the present review aimed to systematically summarize existing evidence regarding the role of VGCC genetic variants in the etiology of ASD.
Methods
Literature search
Electronic research was conducted to identify pertinent English articles examining the genetic associations between VGCCs and ASD. Three online databases, PubMed, Embase, and Web of Science, were searched using the following search terms: (ASD OR autism OR autistic disorder OR autism spectrum disorder OR Asperger syndrome OR pervasive developmental disorder) AND (Ca2+ channel OR Ca2+ signal OR Ca2+ pathway OR Ca2+ homeostasis OR calcium channel OR calcium signal OR calcium pathway OR calcium homeostasis OR CACNA1* OR CACNB* OR CACNA2D*).
A brief review of the articles was used to identify preliminary keywords. No restrictions were placed on the study location or the publication year. All authors achieved a consensus on the search strategies. References cited by all eligible citations and previous reviews were manually searched for additional relevant studies that might have been omitted.
Study selection
The following criteria were required for articles to be eligible: focused on associations between VGCC gene variants and ASD; presented variants in the VGCC gene or reported deregulation of calcium signal; performed on human beings or human cells. Reviews, editorials and commentaries were excluded.
Data extraction
Two authors independently extracted key information from each eligible study using a predefined abstraction form. Extracted data included first author, publication year, country setting, sample size, studied gene, identified mutation, adopted methods, and main findings.
Data synthesis
Study characteristics were descriptively reported and study results were qualitatively reviewed due to the considerable heterogeneities across the included studies.
Results
Literature search
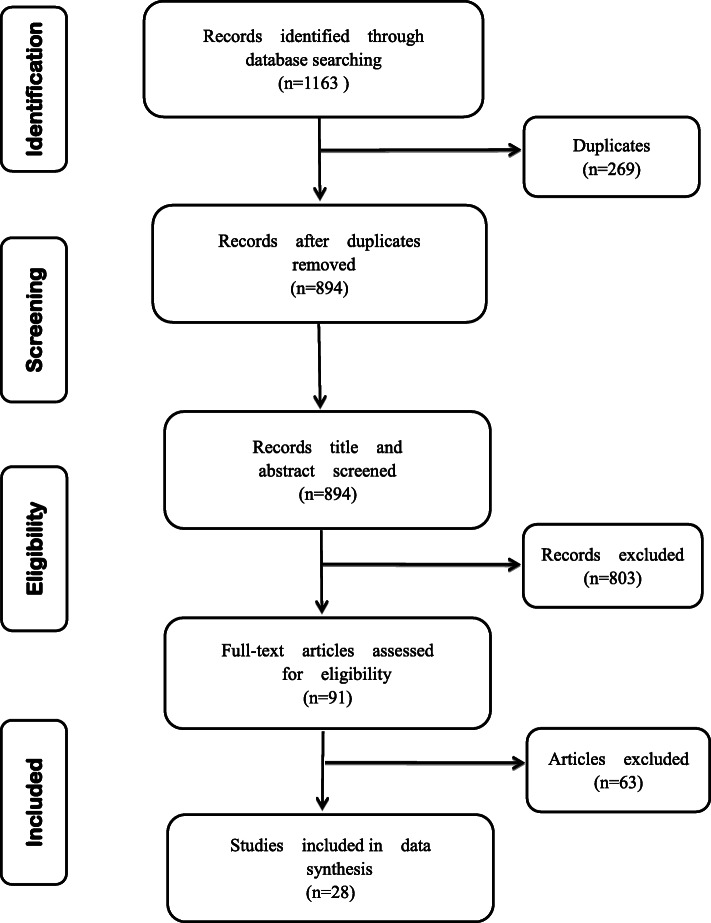
Figure 1 presented the process of literature screening. A total of 1163 records were generated from the literature search. After eliminating 269 duplicates, 894 were considered as potentially relevant for subsequent analysis. The titles and abstracts of the remaining 894 were preliminary checked by two authors, and 803 were excluded as lacking of relevance. The full-text of the leaving 91 citations were further appraised independently by two authors, and 63 were excluded as failing to meet the selection criteria. A final total of 28 articles were selected as source documents for this review.
Fig. 1.
Study selection flowchart
Study characteristics
Table 1 summarized the key information of included studies. Animal models of psychiatric disorders are often unable to reflect typical features of human psychiatric disorders, although there is consensus on some animal correlates of clinical manifestations [2]. While there is an increasing number of ASD mouse models, studies regarding the role of VGCCs in the etiology of ASD are sparse [30]. Thus, the present review focused exclusively on human studies regarding the genetic association between VGCCs and ASD.
Table 1.
Key information of included studies
| Reference | Country | Sample | Gene | Mutation | Gain/loss of function | Method | Main Findings |
|---|---|---|---|---|---|---|---|
| Damaj et al. [10] | Canada | 16 individuals with ASD-like behavioral deficits | CACNA1A |
(1)deletion (2) point mutation |
loss-of-function | whole genome sequencing | Results from sequencing revealed one CACNA1A gene deletion as well as two deleterious CACNA1A point mutations including one known stop-gain and one new frameshift variant and a new splice-site variant, which pointed to an association between CACNA1A variants and ASD symptoms. |
| Pinggera et al .[33] | Austria | tsA-201 cells | CACNA1D | de novo variant | gain-of-function | whole-cell patch-clamp |
The study identified two de novo mutation, p.A749G and p.G407R, in ASD subjects, both demonstrated a pronounced gain-of-function. |
| Splawski et al .[43] | USA |
461ASD 480controls |
CACNA1H | rare missense variant | lose of function | (1) Genotypic Analyses (2) DNA Sequence Analyses | The study identified missense mutations in CACNA1H in 6 of 461 individuals with ASD. |
| Splawski et al .[42] | USA |
13 TS 13 controls |
CACNA1C | de novo missense variant | gain-of-function | (1) Genotypic Analyses (2) DNA Sequencing | The result showed that the CACNA1C gain-of-function mutation causes the diverse physiological and developmental defects in TS. |
| Li et al .[23] | China | 553 trios | CACNA1A | SNPs |
(1) SNP selection (2) SNP genotyping |
The study identified association of rs7249246 and rs12609735 with ASD. | |
| Strom et al .[45] | USA | 284 trios | CACNA1G | SNPs |
(1) SNP selection (2) SNP genotyping |
Markers within an interval containing the gene, CACNA1G, were found to be associated with ASD at a locally significant level. | |
| Smith et al .[41] | USA |
(1)69 ASD (2)35 parents (3)89 CEU HapMap controls |
CACNA2D4 | CNVs | Microarray analyses | The study identified a rare homozygous deletion in a male proband that removed one copy of the CACNA2D4 calcium channel genes(12p13.33) | |
| Pinggera et al .[34] | Germany |
(1)1ASD (2)tsA-201 cells |
CACNA1D | de novo missense variant | gain-of-function |
(1) DNA Sequencing (2)whole-cell patch-clamp |
The study identified a de novo missense mutation in CACNA1D (V401L) in a patient with ASD. |
| Breitenkamp et al .[5] | Germany |
(1)155 ASD (2)259 controls |
CACNB2 | rare missense variant | gain-of-function |
(1) Genotypic Analyses (2) DNA Sequencing |
The study identified three missense mutations in CACNB2 gene in ASD subjects that result in a decelerated inactivation of the Cav1.2 subunit |
| Li et al .[23] | China | 553 trios | CACNA1C | SNPs |
(1) SNP selection (2) SNP genotyping |
The result found a nominal significant association between two SNPs (rs1006737 and rs4765905) in CACNA1C and ASD. | |
| Lu et al .[25] | USA | 2781 trios |
(1)CACNA1C (2)CACNA1G (3)CACNA1I |
SNPs |
(1) SNP selection (2) SNP genotyping |
Four SNPs in three CCGs were associated with ASD. One, rs10848653, is located in CACNA1C.Two others, rs198538 and rs198545, located in CACN1G, and a fourth, rs5750860, located in CACNA1I. | |
| Iossifov et al .[21] | USA | 343 families with ASD | CACNA2D3 | de novo variant | DNA Sequencing | The result identified a variation in CACNA2D3 that disrupted a splice junction associated with ASD (A/G). | |
| Wang et al .[47] | USA |
(1)780 families with ASD (2)1204 affected subjects (3)6491 control subjects |
CACNA1C | SNPs | genome-wide SNP genotyping | The result found an relationship between SNPs in CACNA1C and ASD | |
| Cross-Disorder Group [9] | USA |
(1)33,332ASD (2)27,888 controls |
(1)CACNA1C (2)CACNB2 | SNPs | genome-wide SNP genotyping | The result showed that SNPs within CACNA1C and CACNB2 were associated with ASD. | |
| Hemara-Wahanui et al .[18] | New Zealand | 27 families with ASD | CACNA1F | nucleotide substitution | gain-of-function |
(1) DNA Sequencing (2) Functional Analyses |
The molecular genetic analyses revealed an I745T CACNA1F allele in a New Zealand family, some male probands also affected with ASD. |
| Myers et al .[27] | USA |
(1)142 ASD (2)143 SCZ (3) 240controls |
CACNA1F | rare missense variant | DNA Sequencing | The study found a rare causal CACNA1F variants associated with ASD. | |
| Palmieri et al .[32] | USA |
(1)6ASD (2)6controls |
Ca2+ concentration | Fluorimetric measure of Ca2+ | The study found a significantly higher level of Ca2+ in ASD patients as compared to healthy controls. | ||
| Schmunk et al .[39] | USA | skin fibroblast cultures | IP3-mediated Ca 2+ release | High-throughput Ca2+ imaging | The study found a significantly reduction in IP3-mediated Ca2+ release from the ER. | ||
| Wen et al .[48] | USA | ASD genes |
(1) Gene set enrichment analysis (2)Pathway-pathway interactions |
The results showed that the process “calcium-PRC (protein kinase C)-Ras-Raf-MAPK/ERK” was a major contributor to ASD pathophysiology. | |||
| Skafidas et al .[40] | Australia |
(1) Index sample:2609 ASD (2) Vlidation sample:737 ASD |
CACNA1A | SNPs | Gene set enrichment analysis | The result showed that SNPs in CACNA1A to be among the top 15 SNPs contributing to the ASD diagnosis | |
| Schmunk et al .[38] | USA | skin fibroblast cultures | IP3-mediated Ca 2+ release | High-throughput Ca2+ imaging | The study found a significantly depressed IP3-mediated Ca2+ signals in ASD. | ||
| O'Roak et al .[31] | USA | 209 trios |
(1)CACNA1D (2)CACNA1E |
CNVs | Exome read-depth CNV analysis | The result showed that rare de novo alleles of CACNA1D and CACNA1E contributed to the genetic etiology of ASD. | |
| De Rubeis et al .[11] | USA |
(1) 3871 ASD (2)9937 controls |
(1)CACNA2D3 (2) CACNA1D |
de novo variant | loss of function | Exome sequencing | The result identified two de novo CACNA2D3 loss of function mutations in ASD cases and none in controls. |
| Girirajan et al .[17] | USA |
(1) 2588 ASD (2) 580 controls |
CACNA2D3 | CNVs | Microarray analyses | The study reported an enrichment of CACNA2D3 deletion in ASD subjects compared to controls | |
| Jiang et al. [22] | USA | 32 families with ASD | CACNA1C | rare missense variant | Whole-genome sequencin | It found a rare missense mutation in CACNA1C (R1522Q) in a proband with ASD and an unaffected sibling. | |
| Yuen et al. [51] | Canada |
85 quartet families (parents and two ASD-affected siblings) |
CACNB2 | rare missense variant | Whole-genome sequencing | It found a CACNB2 (V2D) mutation in two affected siblings. | |
| Yatsenko et al .[50] | USA | 20 ASD | CACNA1B | duplication |
(1) whole genome sequencing (2) custom 9q34 microarray |
The study found a duplication of the chromosomal region 9q43.3, comprising the gene CACNA1B, in 12 out of 20 patients. | |
| Prasad et al .[36] | USA |
(1)696 unrelated ASD (2)1000 controls |
CACNA2D4 | CNVs | CGH microarray | The study identified multiple novel CNVs in ASD subjects, including the loss of CACNA2D4. |
These included articles were published between 2004 and 2017, and nine of them were published in the last 5 years. Several early studies contained relatively small numbers of subjects, therefore, they might not have adequate statistical power to detect true risk alleles [26]. These included studies mainly originated in the USA (n = 19), followed by China, Germany, Australia and Canada (n = each 2) and New Zealand (n = 1). A series of genetic methods has been adopted to identify VGCC genetic variants associated with ASD, the most commonly used of which included next-generation sequencing, gene network analysis, and microarrays.
The search generated a total of 18 studies regarding the genetic association between Cav gene variants and ASD, 8 studies regarding the genetic association between auxiliary subunit gene variants and ASD, and 4 studies reported associations between calcium signaling pathways and ASD.
Genetic associations between Cav genes and ASD
The search generated a total of 18 studies regarding the genetic association between Cav gene variants and ASD. Evidence generated from this review consistently implicated the role of Cav gene variants in the etiology of ASD.
The search yielded 3 studies that rendered CACNA1A a promising etiological candidate gene for ASD. Li et al. [23] examined the genetic relationship between CACNA1A and ASD in a Chinese Han population and reported an association between rs12609735 and rs7249246 in CACNA1A with ASD in a total of 553 trios, although this association would not survive after Bonferroni correction. Damaj et al. [10] identified 16 individuals carrying CACNA1A loss-of-function variants and presented ASD-like behavioral deficits by investigating four non-consanguineous families. The sequencing results revealed one CACNA1A gene deletion as well as two deleterious CACNA1A point mutations, including one known stop-gain and one new frameshift variant and a new splice site variant, which pointed to an association between CACNA1A variants and ASD symptoms. The study from Skafidas et al. [40] generated a predictive genetic classifier based on a linear function of 237 SNPs that distinguished ASD from controls and found that a SNP (rs10409541) in CACNA1A was among the top 15 most contributory SNPs for ASD diagnosis prediction.
The search returned only one study regarding the role of CACNA1B variants in the etiology of ASD. Yatsenko et al. [50] analyzed 20 patients with copy number gains involving the subtelomeric 9q34 region and reported a monogenic duplication of the CACNA1B gene in 12 out of 20 patients with a phenotype including ASD.
The search generated 7 studies that implicated CACNA1C as a susceptibility gene for ASD. The Cross-Disorder Group of the Psychiatric Genomics Consortium [9] compiled data on five major psychiatric disorders in a large genome-wide meta-analysis with 33,332 cases and 27,888 controls, rendering SNPs in CACNA1C a promising etiological candidate gene for five major psychiatric disorders, including ASD. Wang et al. [47] reported findings from a genetic analysis in a large number of ASD cases, which provided suggestive evidence of a relationship between SNPs in CACNA1C and ASD. The study of Splawski et al. [42] described the phenotypic characterization of Timothy syndrome (TS) and identified two analogous mutations, p.G406R and p.G402S in CACNA1C, which led to significantly impaired current inactivation of the Cav1.2 splice form, raised the possibility that the ASD phenotypes associated with TS may result from the CACNA1C gain-of-function mutation. Smith et al. [41] conducted a detailed microarray analysis of 69 ASD probands and 35 parents and identified a rare homozygous deletion in a male proband that removed one copy of the CACNA1C calcium channel genes (12p13.33). Li et al. [23] examined the relationship between CACNA1C variants and ASD in 553 nuclear families of Chinese Han ancestry and found a nominal significant association between two SNPs (rs1006737 and rs4765905) in CACNA1C and ASD, suggesting that CACNA1C might play a role in the genetic etiology of ASD. A whole-genome sequencing study from Jiang et al. [22], who examined rare inherited genetic variants in 32 families with ASD, identified a rare missense mutation in CACNA1C in a proband with ASD and an unaffected sibling; therefore, it was unclear whether the mutation in CACNA1C was causative of ASD in this family. A genome-wide association study from Lu et al. [25] examined the potential role of VGCC variants in the etiology of ASD, rendering SNPs in CACNA1C a predisposing risk factor contributing to ASD.
Evidence from 4 studies proposed the role of CACNA1D variants in the genetic etiology of ASD. Pinggera et al. [33] functionally expressed two de novo mutations, p.A749G and p.G407R, in CACNA1D in tsA-201 cells to study their functional consequences using whole-cell patch-clamp analysis. The novel functional data strongly argued for an important role of CACNA1D gain-of-function mutations in the pathophysiology of ASD. Pinggera et al. [34] reported a de novo missense mutation in CACNA1D in a patient with ASD and examined the function of this mutation using whole-cell patch-clamp analysis, strengthening the evidence for CACNA1D gain-of-function mutations in the pathophysiology of ASD. O'Roak et al. [31] sequenced the exomes of 209 parent-child trios and identified rare de novo alleles of CACNA1D as top de novo risk mutations for ASD. De Rubeis et al. [11] conducted the largest ASD WES study to date to identify rare coding variants in 3871 ASD subjects and 9937 controls and identified five mutations located in CACNA1D, including G407R and A749G, in ASD subjects.
The search yielded only one study regarding the role of CACNA1E variants in the etiology of ASD. The study by O'Roak et al. [31] sequenced the exomes of 209 parent-child trios and identified de novo mutations in CACNA1E that contributed to the genetic etiology of ASD.
Two studies reported the potential role of CACNA1F variants in the etiology of ASD. Myers et al. [27] presented results from a large-sample resequencing study of candidate genes coupled with population genetics to identify rare variants associated with ASD and found a significant excess of rare missense CACNA1F variants in the cohort of ASD patients. The study of Hemara-Wahanui et al. [18] identified a CACNA1F mutation in a family with inherited night blindness, with some of the male members being affected with ASD. The mutation significantly affected the gating properties of the Cav 1.4 channel when exogenously expressed in tsA-201 cells, indicating that the molecular mechanism of the pathology was likely to involve a gain, rather than loss, of Cav 1.4 channel function.
The search yielded 2 studies examining the association between CACNA1G variants and ASD. Strom et al. [45] examined the association between variants in the chromosomal interval 17q11-q21 and ASD in 284 independent trios and identified the calcium channel subunit gene CACNA1G as a novel candidate gene for ASD. Lu et al. [25] examined the potential role of calcium channel genes in ASD by focusing on 10 genes that encode α 1 subunits in a cohort of 2781 parent/affected child trios and identified SNPs in CACNA1G as predisposing risk factors contributing to ASD.
The search returned one study regarding the role of CACNA1H variants in the genetic etiology of ASD. Splawski et al. [43] examined the role of calcium channel gene mutations in the pathogenesis of ASD and identified missense mutations in CACNA1H in 6 of 461 subjects with ASD that resulted in decreased activity of the Cav3.2 channel, implicating the role of CACNA1H loss-of-function mutations in the development of ASD. However, some of the mutations were also present in unaffected family members, indicating that the mutations were not fully penetrant.
The search yielded only one study regarding the role of CACNA1I variants in the development of ASD. Lu et al. [25] examined the role of VGCC variants in the development of ASD by focusing on 10 genes that encode α1 subunits and identified SNPs in CACNA1I as genetic risks for ASD.
Genetic associations between auxiliary subunit genes and ASD
Given that the auxiliary subunits Cavα2δ and Cavβ have profound effects on the biophysical properties and membrane targeting of the Cavα1 subunit, genes encoding these subunits may also be linked to ASD. The search generated a total of 8 studies regarding the genetic association between auxiliary subunit gene variants and ASD. Existing evidence suggests that variants in the gene encode the auxiliary subunits Cavα2δ and Cavβ, which also contribute to the pathogenesis of ASD.
The search yielded 3 studies that reported an association between CACNB2 variants and ASD. Breitenkamp et al. [5] sequenced the exons and flanking introns of CACNB2 in 155 ASD subjects and 259 unaffected controls and identified three missense variants in the coding region of CACNB2 (G167S, S197F, and F240L) in ASD probands that resulted in decelerated inactivation of the Cav1.2 subunit, supporting the role of CACNB2 gain-of-function mutations in the pathophysiology of ASD. In line with these results, a study from the Psychiatric Genomics Consortium (2013), which analyzed genome-wide single-nucleotide polymorphism (SNP) data for the five disorders, identified CACNB2 as a risk locus for these five major psychiatric disorders, including ASD. An exome sequencing study by Yuen et al. [51] investigating 85 quartet families in which two siblings were affected with ASD to identify ASD-related gene variants found a CACNB2 (V2D) mutation in two affected siblings.
The search generated 3 studies regarding the role of CACNA2D3 variants in the development of ASD. Iossifov et al. [21] reported on the sequence analysis of whole exomes from 343 families, each with a single ASD child and at least one unaffected sibling. The results from exome sequencing showed an excess of de novo splice site mutations in CACNA2D3, one of many identified gene-disrupting mutations, in ASD subjects compared to unaffected siblings. De Rubeis et al. [11] identified rare coding variants in 3871 ASD subjects and 9937 ancestry-matched or parental controls and implicated CACNA2D3 as a risk gene following the identification of two de novo loss-of-function mutations in cases and none in controls. A copy number variation (CNV) study from Girirajan et al. [17], who comprehensively characterized recurrent CNVs for both large and putative smaller hotspots in 2588 ASD subjects and 580 controls, reported an enrichment of CACNA2D3 deletion in ASD subjects compared to controls.
The search returned 2 studies regarding the association between CACNA2D4 variants and ASD. Smith et al. [41] conducted a detailed microarray analysis of 69 ASD probands and 35 parents and identified a rare homozygous deletion in a male proband that removed one copy of the CACNA2D4 calcium channel genes (12p13.33). Prasad et al. [36] examined de novo copy number variations (CNVs) through combined analysis of CGH and SNP array data sets in a cohort of 696 unrelated ASD subjects and 1000 controls. The high-resolution CGH data identified multiple novel CNVs in ASD subjects, including the loss of CACNA2D4.
Associations between calcium signaling pathways and ASD
The genetic contributions of VGCC variants to the pathogenesis of ASD may arise from functional disturbances in a variety of signaling cascades in which these genes are involved, since it is the combination of these factors that ultimately shapes the phenotype. Calcium signaling pathways may serve as scaffolds that unite genetic lesions into a consensus etiology of ASD. Identification of variants in calcium channel genes led to the discovery of molecular signaling pathways involved in ASD, including the IP3-Ca2+ pathway and the MAPK pathway, providing further insights into the etiology of ASD.
The inositol triphosphate receptor (IP3R) forms a calcium-permeable ion channel located in the membrane of the endoplasmic reticulum (ER) [29]. The IP3 pathway changes intracellular Ca2+ concentration by releasing calcium stored in the ER, which is critical for synaptic plasticity, neuronal excitability, neurotransmitter release, and axonal growth, demonstrating the central integration position of IP3R in neurons [19, 46]. Suppressed IP3-mediated calcium signaling has been linked to ASD etiology in two studies. Schmunk et al. [38] evaluated functional deficits in IP3-mediated Ca2+ signaling from three monogenic models of ASD and identified significantly depressed IP3-mediated Ca2+ signals in ASD. Schmunk et al. [39] further extended these findings to a cohort of sporadic ASD patients and reported a significant reduction in IP3-mediated Ca2+ release from the ER. Recent research has implicated the abnormal MAPK pathway as a possible molecular mechanism of ASD. Wen et al. [48] performed a systematic analysis of the ASD pathway network and reported a high level of involvement of the MAPK signaling pathway together with calcium channel genes in the ASD pathway network. One study proposed a role of abnormal calcium homeostasis in the etiology of ASD. Palmieri et al. [32] examined the Ca2+ concentrations in the postmortem temporocortical gray matter of six matched patient-control pairs and reported a significantly higher level of Ca2+ in ASD patients than in healthy controls, supporting the involvement of altered Ca2+ homeostasis in the etiological pathway of ASD.
Discussion
The identification of VGCC genetic variants as risk factors for ASD may not only be extracted from the ability of calcium channels to mediate Ca2+ into neurons, which can trigger many calcium-modulated functions, but also derive from the role of calcium channels as signaling hubs, which can link together different cellular signaling pathways.
The present review summarized recent works regarding the genetic association between VGCCs and ASD. The most prominent VGCC variants found in ASD were those falling within loci encoding the α subunits CACNA1A, CACNA1B, CACNA1C, CACNA1D, CACNA1E, CACNA1F, CACNA1G, CACNA1H, and CACNA1I as well as those of their accessory subunits CACNB2, CACNA2D3, and CACNA2D4. Two signaling pathways, the IP3-Ca2+ pathway and the MAPK pathway, were identified as scaffolds that united genetic lesions into a consensus etiology of ASD.
Structure-function analyses involving the introduction of gene variants into cloned VGCCs have provided important clues concerning gain-of-function and loss-of-function properties of these calcium channels. Variants located in CACNA1C, CACNA1D, CACNA1F and CACNB2 cause gain of function by preventing voltage-dependent inactivation of Cav1.2, Cav1.3, Cav1.4, and Cavβ2, leading to excessive influx of Ca2+. Variants located in CACNA1A and CACNA1H turned out to rather cause loss of function in Cav2.1 and Cav3.2 due to reduced conductance and shifted voltage dependence of activation, resulting in decreased channel activity. It is difficult to form a clear consensus concerning gain or loss of function of the VGCC variants associated with ASD, which forms the basis of this review. It is conceivable that dysregulation of these calcium channels in either direction could result in functional and developmental abnormalities considering the requirement for precise regulation of internal Ca2+ concentrations for normal cell signaling and gene transcription during development.
Cytosolic calcium signals originate from either the influx of extracellular Ca2+ or the release of intracellular Ca2+ stored in the ER. The mitochondria actively communicate with the ER calcium signaling apparatus in the generation of rapid calcium signals, forming a bidirectional link between energy metabolism and transmitted cellular signals. The interaction of ER-mitochondria in the control of calcium homeostasis implies that calcium signaling may serve as a scaffold that integrates VGCC genetic lesions and organelle dysfunction, including the ER and mitochondria, into a consensus pathophysiology of ASD.
Given the consistent evidence generated from this review supporting the role of VGCC genetic variants in the pathophysiology of ASD, it is worthwhile to consider targeting Cavα1, Cavα2δ, and Cavβ subunits as a potential therapeutic strategy to treat this disorder. Despite the existence of several drugs targeting Cavα1 and Cavα2δ subunits, they are mainly used in the treatment of stroke, epilepsy, and cardiovascular disease [52]. Lamotrigine, a drug that blocks Cav2.3 channels, has been used to treat bipolar disorder (BD) [37]. Z160, a drug that blocks the Cav2.2 channel, has shown some promise in treating anxiety [53]. Calcium channel blockers are promising for the treatment of ASD, but gain-of-function mutations are in many cases less sensitive to blocking because of the loss of inactivation [20, 42].
Conclusion
Findings generated from this review proposed that the genetic component of ASD may involve a combination of multiple common alleles in the VGCC gene, represented by SNPs, each with a relatively small impact, together with a few rare alleles in the VGCC gene, represented by CNVs and deleterious mutations, which might produce a relatively large increased risk. The phenotype of ASD may be conferred by the sum of genetic risks with regard to both rare and common alleles in the VGCC gene together with risks from environmental stimuli.
Given the consistent evidence generated from this review supporting A total of 1163 records were generated from the initial search of these three databases. After eliminating 269 duplicates, 894 articles considered as potentially relevant for subsequent analysis. The titles and abstracts of all non-duplicated papers were preliminary checked by two authors, 803 articles were excluded as lacking of relevance. The full-text of the leaving 91 citations were further appraised independently for eligibility by two authors, 63 articles were excluded as failing to meet the selection criteria. A final total of 28 articles were selected as source documents for this review. Genetic variants in the pathophysiology of ASD, it is of great value to examine whether existing FDA-approved drugs modulate Ca2+ signaling function in the treatment of ASD. Although these findings have confirmed the genetic associations between VGCCs and ASD, the underlying mechanisms by which these variants produce the ASD phenotypes have not been characterized. Existing evidence implicated Ca2+ signaling as the most relevant node of an integrative network model for gene-environment interactions in the ASD context [54]. Further studies should shift attention from the role of individual variants to the compounded impacts of different variants as they interact with other genes and the environment.
Acknowledgements
This research did not receive any specific grant from agencies in the public, commercial, or not-for-profit sectors.
Statement of interest
All authors of this paper declare that there is no conflict of interest related to the content of this manuscript.
Abbreviations
- ASD
Autism Spectrum Disorder
- VGCCs
Voltage-Gated Calcium Channels
- HVA
High voltage-activated channels (HVA)
- LVA
Low voltage-activated channels
- SNPs
Single nucleotide polymorphisms
- IP3R
Inositol triphosphate receptor
- ER
Endoplasmic reticulum
- VDI
Voltage- dependent inactivation
- CDI
Calcium-dependent inactivation
- BD
Bipolar Disorder
- CNVs
Copy number variations
Authors’ contributions
All authors have read and agree to the published version of the manuscript. Xiaoli Liao conceived the manuscript idea, searched database, identified eligible studies, extracted data, drafted the manuscript, and critically reviewed the manuscript. Yamin Li conceived the manuscript idea, identified eligible studies, extracted data, responsible for funding acquisition, and critically reviewed the manuscript.
Funding
The research was funded by the National Natural Science Foundation of China (No: 81873806) and the National Natural Science Foundation of Hunan Province (No. 2019JJ40437).
Availability of data and materials
Not applicable.
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Footnotes
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Andrade A, Brennecke A, Mallat S, et al. Genetic Associations between Voltage-Gated Calcium Channels and Psychiatric Disorders. Int J Mol Sci. 2019;20(14):3537–3546. doi: 10.3390/ijms20143537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Arguello PA, Gogos JA. Modeling madness in mice: one piece at a time. Neuron. 2006;52(1):179–196. doi: 10.1016/j.neuron.2006.09.023. [DOI] [PubMed] [Google Scholar]
- 3.Baio J. Prevalence of autism spectrum disorder among children aged 8 years-autism and developmental disabilities monitoring network, 11 sites, United States, 2010. Morb Mortal Wkly Rep. 2014;63:1–2. [PubMed] [Google Scholar]
- 4.Blumberg SJ, Bramlett MD, Kogan MD, et al. Changes in prevalence of parent-reported autism Spectrum disorder in school-aged US children: 2007 to 2011-2012. Natl Center Health Stat Rep. 2013;65:1–11. [PubMed] [Google Scholar]
- 5.Breitenkamp AFS, Matthes J, Nass RD, et al. Rare mutations of CACNB2 found in autism spectrum disease-affected families alter calcium channel function. PLoS One. 2014;9(4):e95579. doi: 10.1371/journal.pone.0095579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Catterall WA. Structure and regulation of voltage-gated Ca2+ channels. Annu Rev Cell Dev Biol. 2000;16:521–555. doi: 10.1146/annurev.cellbio.16.1.521. [DOI] [PubMed] [Google Scholar]
- 7.Catterall WA, Striessnig J, Snutch TP, et al. International Union of Pharmacology. XL Compendium of voltage-gated ion channels: calcium channels. Pharmacol Rev. 2003;55:579–581. doi: 10.1124/pr.55.4.8. [DOI] [PubMed] [Google Scholar]
- 8.Catterall WA, Perez-Reyes E, Snutch TP, et al. International Union of Pharmacology. XLVIII Nomenclature and structure-function relationships of voltage-gated calcium channels. Pharmacol Rev. 2005;57:411–425. doi: 10.1124/pr.57.4.5. [DOI] [PubMed] [Google Scholar]
- 9.Cross-Disorder Group of the Psychiatric Genomics Consortium Identification of risk loci with shared effects on five major psychiatric disorders: a genome-wide analysis. Lancet. 2013;381(9875):1371–1379. doi: 10.1016/S0140-6736(12)62129-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Damaj L, Lupien-Meilleur A, Lortie A, et al. CACNA1A haploinsufficiency causes cognitive impairment, autism and epileptic encephalopathy with mild cerebellar symptoms. Eur J Hum Genet. 2015;23(11):1505. doi: 10.1038/ejhg.2015.21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.De Rubeis S, He X, Goldberg AP, et al. Synaptic, transcriptional and chromatin genes disrupted in autism. Nature. 2014;515(7526):209–215. doi: 10.1038/nature13772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dolmetsch RE, Pajvani U, Fife K, et al. Signaling to the nucleus by an L-type calcium channel-calmodulin complex through the MAP kinase pathway. Science. 2001;294(5541):333–339. doi: 10.1126/science.1063395. [DOI] [PubMed] [Google Scholar]
- 13.Dolphin AC. A short history of voltage-gated calcium channels. Br J Pharmacol. 2006;147(Suppl 1):S56–S62. doi: 10.1038/sj.bjp.0706442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dolphin AC. Calcium channel diversity: multiple roles of calcium channel subunits. Curr Opin Neurobiol. 2009;19:237–244. doi: 10.1016/j.conb.2009.06.006. [DOI] [PubMed] [Google Scholar]
- 15.Ebert DH, Greenberg ME. Activity-dependent neuronal signaling and autism spectrum disorder. Nature. 2013;493:327–337. doi: 10.1038/nature11860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Felix R, Calderón-Rivera A, Andrade A. Regulation of high-voltage-activated Ca2+ channel function,trafficking, and membrane stability by auxiliary subunits. Wiley Interdiscip Rev Membr Transp Signal. 2013;2:207–220. doi: 10.1002/wmts.93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Girirajan S, Dennis MY, Baker C, et al. Refinement and discovery of new hotspots of copy-number variation associated with autism spectrum disorder. Am J Hum Genet. 2013;92:221–237. doi: 10.1016/j.ajhg.2012.12.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hemara-Wahanui A, Berjukow S, Hope C, et al. A CACNA1F mutation identified in an X-linked retinal disorder shifts the voltage dependence of Cav1.4 channel activation. Proc Natl Acad Sci U S A. 2005;102:7553–7558. doi: 10.1073/pnas.0501907102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hernández-López S, Tkatch T, Perez-Garci E, et al. D2 dopamine receptors in striatal medium spiny neurons reduce L-type Ca2+ currents and excitability via a novel PLCβ1-IP3-calcineurin-signaling cascade. J Neurosci. 2000;20(24):8987–8995. doi: 10.1523/JNEUROSCI.20-24-08987.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Heyes S, Pratt WS, Rees E, et al. Genetic disruption of voltage-gated calcium channels in psychiatric and neurological disorders. Prog Neurobiol. 2015;134:36–54. doi: 10.1016/j.pneurobio.2015.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Iossifov I, Ronemus M, Levy D, et al. De novo gene disruptions in children on the autistic Spectrum. Neuron. 2012;74:285–299. doi: 10.1016/j.neuron.2012.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jiang YH, Yuen RK, Jin X, et al. Detection of clinically relevant genetic variants in autism spectrum disorder by whole-genome sequencing. Am J Hum Genet. 2013;93:249–263. doi: 10.1016/j.ajhg.2013.06.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Li J, Zhao L, You Y, et al. Schizophrenia related variants in CACNA1C also confer risk of autism. PLoS One. 2015;10(7):e0133247. doi: 10.1371/journal.pone.0133247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lord C, Cook EH, Leventhal BL, et al. Autism spectrum disorders. Neuron. 2000;28:355–363. doi: 10.1016/s0896-6273(00)00115-x. [DOI] [PubMed] [Google Scholar]
- 25.Lu ATH, Dai X, Martinez-Agosto JA, et al. Support for calcium channel gene defects in autism spectrum disorders. Mol Autism. 2012;3(1):18. doi: 10.1186/2040-2392-3-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mitchell KJ. The genetics of neurodevelopmental disease. Curr Opin Neurobiol. 2011;21:197–203. doi: 10.1016/j.conb.2010.08.009. [DOI] [PubMed] [Google Scholar]
- 27.Myers RA, Casals F, Gauthier J, et al. A population genetic approach to mapping neurological disorder genes using deep resequencing. PLoS Genet. 2011;7:e1001318. doi: 10.1371/journal.pgen.1001318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Neelands TR, King APJ, Macdonald RL. Functional expression of L-, N-, P/Q-, and R-type calcium channels in the human NT2-N cell line. J Neurophysiol. 2000;84(6):2933–2944. doi: 10.1152/jn.2000.84.6.2933. [DOI] [PubMed] [Google Scholar]
- 29.Nguyen RL, Medvedeva YV, Ayyagari TE, et al. Intracellular calcium dysregulation in autism spectrum disorder: an analysis of converging organelle signaling pathways. BBA-Mol Cell Res. 2018;1865(11):1718–1732. doi: 10.1016/j.bbamcr.2018.08.003. [DOI] [PubMed] [Google Scholar]
- 30.Oddi D, Crusio WE, D'Amato FR, et al. Monogenic mouse models of social dysfunction: implications for autism. Behav Brain Res. 2013;251:75–84. doi: 10.1016/j.bbr.2013.01.002. [DOI] [PubMed] [Google Scholar]
- 31.O'Roak BJ, Vives L, Girirajan S, et al. Sporadic autism exomes reveal a highly interconnected protein network of de novo mutations. Nature. 2012;485:246–250. doi: 10.1038/nature10989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Palmieri L, Papaleo V, Porcelli V, et al. Altered calcium homeostasis in autism-spectrum disorders: evidence from biochemical and genetic studies of the mitochondrial aspartate/glutamate carrier AGC1. Mol Psychiatry. 2010;15(1):38. doi: 10.1038/mp.2008.63. [DOI] [PubMed] [Google Scholar]
- 33.Pinggera A, Lieb A, Benedetti B, et al. CACNA1D de novo mutations in autism spectrum disorders activate Cav1.3 L-type calcium channels. Biol Psychiatry. 2015;77(9):816–822. doi: 10.1016/j.biopsych.2014.11.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Pinggera A, Mackenroth L, Rump A, et al. New gain-of-function mutation shows CACNA1D as recurrently mutated gene in autism spectrum disorders and epilepsy. Hum Mol Genet. 2017;26(15):2923–2932. doi: 10.1093/hmg/ddx175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Perez-Reyes E. Molecular physiology of low-voltage-activated t-type calcium channels. Physiol Rev. 2003;83:117–161. doi: 10.1152/physrev.00018.2002. [DOI] [PubMed] [Google Scholar]
- 36.Prasad A, Merico D, Thiruvahindrapuram B, et al. A discovery resource of rare copy number variations in individuals with autism spectrum disorder. G3-Genes Genom Genet. 2012;2(12):1665–1685. doi: 10.1534/g3.112.004689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Prabhavalkar KS, Poovanpallil NB, Bhatt LK. Management of bipolar depression with lamotrigine: an antiepileptic mood stabilizer. Front Pharmacol. 2015;6:242. doi: 10.3389/fphar.2015.00242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Schmunk G, Boubion BJ, Smith IF, et al. Shared functional defect in IP3R-mediated calcium signaling in diverse monogenic autism syndromes. Transl Psychiat. 2015;5(9):e643. doi: 10.1038/tp.2015.123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Schmunk G, Nguyen RL, Ferguson DL, et al. High-throughput screen detects calcium signaling dysfunction in typical sporadic autism spectrum disorder. Sci Rep. 2017;7:40740. doi: 10.1038/srep40740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Skafidas E, Testa R, Zantomio D, et al. Predicting the diagnosis of autism spectrum disorder using gene pathway analysis. Mol Psychiatry. 2014;19(4):504–510. doi: 10.1038/mp.2012.126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Smith M, Flodman PL, Gargus JJ, et al. Mitochondrial and ion channel gene alterations in autism. BBA-Bioenergetics. 2012;1817(10):1796–1802. doi: 10.1016/j.bbabio.2012.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Splawski I, Timothy KW, Sharpe LM, et al. Cav1.2 calcium channel dysfunction causes a multisystem disorder including arrhythmia and autism. Cell. 2004;119(1):19–31. doi: 10.1016/j.cell.2004.09.011. [DOI] [PubMed] [Google Scholar]
- 43.Splawski I, Yoo DS, Stotz SC, et al. CACNA1H mutations in autism spectrum disorders. J Biol Chem. 2006;281(31):22085–22091. doi: 10.1074/jbc.M603316200. [DOI] [PubMed] [Google Scholar]
- 44.Stotz SC, Zamponi GW. Structural determinants of fast inactivation of high voltage-activated Ca2+ channels. Trends Neurosci. 2001;24(3):176–182. doi: 10.1016/s0166-2236(00)01738-0. [DOI] [PubMed] [Google Scholar]
- 45.Strom SP, Stone JL, Ten Bosch JR, et al. High-density SNP association study of the 17q21 chromosomal region linked to autism identifies CACNA1G as a novel candidate gene. Mol Psychiatry. 2010;15(10):996. doi: 10.1038/mp.2009.41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Stutzmann GE, LaFerla FM, Parker I. Ca2+ signaling in mouse cortical neurons studied by two-photon imaging and photoreleased inositol triphosphate. J Neurosci. 2003;23(3):758–765. doi: 10.1523/JNEUROSCI.23-03-00758.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wang K, Zhang H, Ma D, et al. Common genetic variants on 5p14.1 associate with autism spectrum disorders. Nature. 2009;459:528–533. doi: 10.1038/nature07999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wen Y, Alshikho MJ, Herbert MR. Pathway network analyses for autism reveal multisystem involvement, major overlaps with other diseases and convergence upon MAPK and calcium signaling. PLoS One. 2016;11(4):e0153329. doi: 10.1371/journal.pone.0153329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wolfe JT, Wang H, Howard J, et al. T-type calcium channel regulation by specific G-protein βγ subunits. Nature. 2003;424(6945):209–213. doi: 10.1038/nature01772. [DOI] [PubMed] [Google Scholar]
- 50.Yatsenko SA, Hixson P, Roney EK, et al. Human subtelomeric copy number gains suggest a DNA replication mechanism for formation: beyond breakage-fusion-bridge for telomere stabilization. Hum Genet. 2012;131(12):1895–1910. doi: 10.1007/s00439-012-1216-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Yuen RK, Thiruvahindrapuram B, Merico D, et al. Whole-genome sequencing of quartet families with autism spectrum disorder. Nat Med. 2015;21:185–191. doi: 10.1038/nm.3792. [DOI] [PubMed] [Google Scholar]
- 52.Zamponi GW. Targeting voltage-gated calcium channels in neurological and psychiatric diseases. Nat Rev Drug Discov. 2016;15:19–34. doi: 10.1038/nrd.2015.5. [DOI] [PubMed] [Google Scholar]
- 53.Zamponi GW, Feng ZP, Zhang L, et al. Scaffold-based design and synthesis of potent N-type calcium channel blockers. Bioorg Med Chem Lett. 2009;19(22):6467–6472. doi: 10.1016/j.bmcl.2009.09.008. [DOI] [PubMed] [Google Scholar]
- 54.Zeidan-Chulia F, Rybarczyk-Filho JL, Salmina AB, et al. Exploring the multifactorial nature of autism through computational systems biology: calcium and the rho GTPase RAC1 under the spotlight. NeuroMolecular Med. 2013;15(2):364–383. doi: 10.1007/s12017-013-8224-3. [DOI] [PubMed] [Google Scholar]
- 55.Zhang Y, Mori M, Burgess DL, et al. Mutations in high-voltage-activated calcium channel genes stimulate low-voltage-activated currents in mouse thalamic relay neurons. J Neurosci. 2002;22(15):6362–6371. doi: 10.1523/JNEUROSCI.22-15-06362.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Not applicable.