Abstract
Background
Aberrant hedgehog (HH) signaling is implicated in the development of various cancer entities such as medulloblastoma. Activation of GLI transcription factors was revealed as the driving force upon pathway activation. Increased phosphorylation of essential effectors such as Smoothened (SMO) and GLI proteins by kinases including Protein Kinase A, Casein Kinase 1, and Glycogen Synthase Kinase 3 β controls effector activity, stability and processing. However, a deeper and more comprehensive understanding of phosphorylation in the signal transduction remains unclear, particularly during early response processes involved in SMO activation and preceding GLI target gene regulation.
Methods
We applied temporal quantitative phosphoproteomics to reveal phosphorylation dynamics underlying the short-term chemical activation and inhibition of early hedgehog signaling in HH responsive human medulloblastoma cells. Medulloblastoma cells were treated for 5.0 and 15 min with Smoothened Agonist (SAG) to induce and with vismodegib to inhibit the HH pathway.
Results
Our phosphoproteomic profiling resulted in the quantification of 7700 and 10,000 phosphosites after 5.0 and 15 min treatment, respectively. The data suggest a central role of phosphorylation in the regulation of ciliary assembly, trafficking, and signal transduction already after 5.0 min treatment. ERK/MAPK signaling, besides Protein Kinase A signaling and mTOR signaling, were differentially regulated after short-term treatment. Activation of Polo-like Kinase 1 and inhibition of Casein Kinase 2A1 were characteristic for vismodegib treatment, while SAG treatment induced Aurora Kinase A activity. Distinctive phosphorylation of central players of HH signaling such as SMO, SUFU, GLI2 and GLI3 was observed only after 15 min treatment.
Conclusions
This study provides evidence that phosphorylation triggered in response to SMO modulation dictates the localization of hedgehog pathway components within the primary cilium and affects the regulation of the SMO-SUFU-GLI axis. The data are relevant for the development of targeted therapies of HH-associated cancers including sonic HH-type medulloblastoma. A deeper understanding of the mechanisms of action of SMO inhibitors such as vismodegib may lead to the development of compounds causing fewer adverse effects and lower frequencies of drug resistance.
Video Abstract
Keywords: Oncogenic signaling, Hedgehog signaling, Phosphorylation, Phosphoproteomics, Medulloblastoma, Kinases, DAOY cells
Background
The role of protein phosphorylation in the control of cellular behavior has been well appreciated and intensely studied for many years. Phosphorylation is one of the most important post translational modifications (PTMs) and regulated through a well-controlled interplay of kinases and phosphatases to regulate cellular signaling [1]. Thus, protein phosphorylation affects processes such as cellular growth, cell division, and metabolism. Kinases are the driving force of phosphorylation cascades, and kinase dysfunction/dysregulation has been associated with oncogenesis [2–5], as for instance demonstrated for the Epidermal Growth Factor Receptor (EGFR) in various cancers. With its unique potential to monitor thousands of phosphorylation sites in parallel, phosphoproteomics has become one of the key methods to monitor kinase activity in cancer cells and oncogenic pathways [6–8]. One of the first phosphoproteomics studies intensively monitored phosphorylation dynamics in HeLa cells upon treatment with Epidermal Growth Factor (EGF) already in 2006 [9]. Phosphoproteomics has also been used to study various oncogenic signaling pathways such as Ras signaling [10] and PI3K signaling [11]. It has been applied to study lung cancer [12, 13], ovarian cancer [14], neuroblastoma [15, 16], and many other cancer entities.
In vertebrates, the hedgehog (HH) pathway plays a central role in the control of embryonic development, cell proliferation, and survival. The HH pathway is induced by the release of HH ligands such as sonic HH. Upon binding of HH protein to Patched 1 (PTCH-1), the G-protein coupled-like receptor Smoothened (SMO) is released from inhibition by PTCH1, allowing its translocation to and accumulation in the primary cilium. The primary cilium is a microtubule-based surface projection essential for canonical HH signal transduction in vertebrates [17, 18]. After HH pathway activation, SMO induces downstream signaling by inhibition of the central GLI inhibitory molecule SUFU, thereby allowing unprocessed full-length- GLI-transcription factors to translocate into the nucleus to induce HH target gene expression (reviewed in ref. [19]). The movement of hedgehog pathway components into and within the primary cilium is orchestrated by intraflagellar transport proteins as parts of the intraflagellar transport machinery (IFT) [20].
Persistent and irreversible activation of HH signaling has been implicated in the development of various cancers such as basal cell carcinoma and medulloblastoma [21–23]. In order to study phosphorylation dynamics in the HH pathway, Varjosalo et al. screened for kinases implicated in HH signaling by studying the kinome collection [24]. Marumoto et al. used comprehensive phosphoproteomics to reveal a critical role of the HH pathway in osteoblast transitions [25]. Recently, comprehensive phosphoproteomics elucidated Casein Kinase 2 as a key driver of the development of sonic HH-driven human medulloblastoma [26]. Besides Casein Kinase 1, other kinases such as Protein Kinase A and Glykogen Synthase Kinase 3β (GSK3β) are known as antagonistic players in the HH signaling cascade [27, 28]. Phosphorylation of the GLI transcription factors by these kinases promotes their ubiquitination and proteasomal processing or degradation [29]. Kinases were also reported to have dual roles in the pathway regulation. Casein Kinase I α (CSKIα), for instance, contributes to GLI transcription factor processing [30] but has also been implicated in the positive regulation of the HH pathway via phosphorylation of SMO [31]. Furthermore, it has been reported that PKA and GSK3β phosphorylate the inhibitory molecule SUFU, thereby stabilizing its molecular structure and promoting its localization in the primary cilium [32], thus impeding pathway activation.
Phosphatases are the major counterparts of kinases and represent an important class of enzymes involved in the regulation of the HH pathway, and their role in sonic HH signaling has been reviewed recently [33]. However, the knowledge about the function of phosphatases is limited. PP2A has been shown to influence GLI3 localization and activity [34] and to be involved in the dephosphorylation of the ciliary trafficking protein Kif7 [35]. Furthermore, PP4 was shown to be implicated in SMO dephosphorylation [36], and lipid phosphatases such as inositol polyphosphate 5-phosphatase INPP5E are critical for proper ciliary trafficking [37]. PP2C family member phosphatase Wip1, also known as PPM1D, influences the stability and localization of GLI1, indicating a direct modulation of transcriptional activity of GLI1 by dephosphorylation [38].
Despite the examples discussed above, the complex role of protein phosphorylation in the early phase of HH pathway activation and inhibition has not been elucidated so far. It is essential to better understand SMO modifying drugs, particularly in light of resistance development and the severe adverse effects such as muscle spasms caused by a number of, but not all, SMO inhibitors [39]. Hence, our study aims at the extensive analysis of phosphorylation patterns after short-term activation and inhibition of the HH pathway in human medulloblastoma cells using comprehensive, HPLC-MS/MS-based phosphoproteome analysis upon enrichment of phosphopeptides. As a cellular model, we used the HH responsive medulloblastoma cell line DAOY [40] to analyze phosphorylation changes in early HH signaling. Smoothened Agonist (SAG) [41] was utilized to activate the HH pathway in vitro, whereas vismodegib (Vismo) – an FDA-approved drug for the treatment of advanced and metastatic basal cell carcinoma [42] - served as inhibitor of the HH pathway. Taking advantage of comprehensive databases of biological pathways and interactions, as well as their function in cancer, such as the Ingenuity Database, we shed light on the complex global phosphorylation patterns in the context of activation and inhibition of the HH pathway in human cancer cells.
Methods
Experimental design and statistical rationale
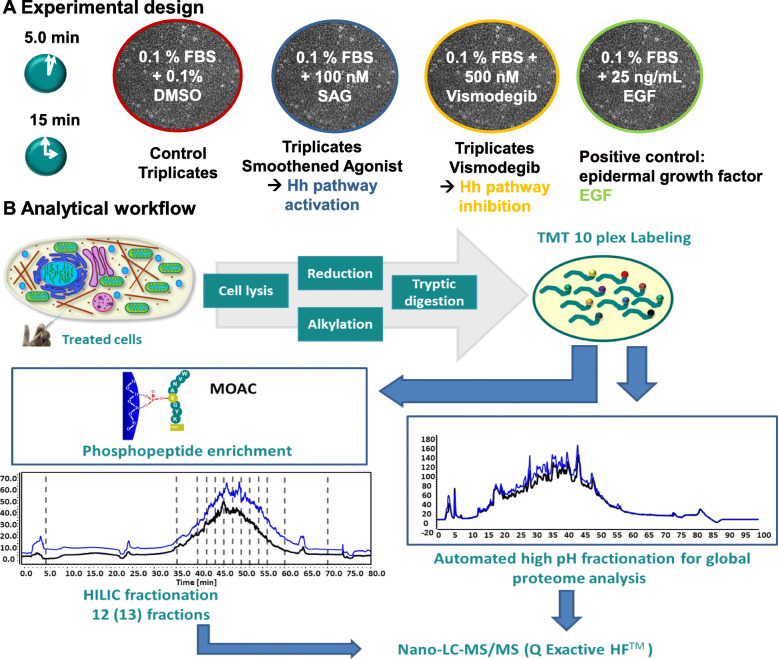
In order to systematically study the role of phosphorylation in hedgehog-driven cancer signaling, we used the human medulloblastoma cell line DAOY for our studies, which we have previously shown to allow manipulation of canonical pathway activity by (partial) agonist and antagonist treatment [40]. We confirmed the HH responsiveness of DAOY cells by treatment with SAG and vismodegib followed by qPCR analysis of the HH target genes GLI1, HHIP and PTCH1 (Figure S-1). To demonstrate comparable activities of natural and synthetic HH pathway inducers (i.e. natural Sonic Hh protein and the synthetic Smoothened agonist compound SAG, respectively), we performed time course analysis of HH target gene expression by qPCR (Figure S-2). For global phosphoproteomics, we performed short-term treatment of DAOY cells with SAG and vismodegib. For both time points, i.e. 5.0 and 15 min, we stimulated the cells with DMSO, SAG and vismodegib in biological triplicates (Fig. 1a). We used DMSO treatment for 5.0 and 15 min, respectively, in biological triplicates as controls, thus allowing the analysis of control, SAG and vismodegib treatment for a single time point within a single TMT 10-plex experiment. This ensures statistical power and comparability independent of instrumental variability. EGF treatment, known to induce extensive phosphorylation, served as (a single) positive control, by mixing DAOY cells treated for 5.0 and 15 min to a single control. This sample was used as common 10th channel in both TMT 10-plex sample sets (5.0 min and 15 min) to allow the quantitative comparison of the different time points of treatment. The labelling scheme for each TMT 10-plex sample is provided in Table 1.
Fig. 1.
Scheme of experimental design (a) and analytical workflow. a Three biological replicates of human medulloblastoma cells were treated with DMSO, SAG, vismodegib or EGF in starving medium for 5.0 and 15 min. b Analytical workflow: After treatment, sample preparation, and TMT 10-plex labeling, peptides of each time point and treatment were pooled. Aliquots were utilized for pH 8 fractionation and deep proteome profiling or phosphopeptide enrichment by metal oxide affinity chromatography. Measurements of peptides and phosphopeptides were conducted with reversed-phase HPLC coupled to high-resolution mass spectrometry
Table 1.
TMT Labeling Scheme for Sample 1a and Sample 2b
| Sample 1a | Sample 2b | TMT label |
|---|---|---|
| DMSO_5.0 min | DMSO_15min | 126 |
| DMSO_5.0 min | DMSO_15min | 127n |
| DMSO_5.0 min | DMSO_15min | 127c |
| SAG_5.0 min | SAG_15min | 128n |
| SAG_5.0 min | SAG_15min | 128c |
| SAG_5.0 min | SAG_15min | 129n |
| Vismo_5.0 min | Vismo_15min | 129c |
| Vismo_5.0 min | Vismo_15min | 130n |
| Vismo_5.0 min | Vismo_15min | 130c |
| EGF 5.0 + 15 min | EGF 5.0 + 15 min | 131 |
a5.0 min treatment; b15 min treatment
Scheme of experimental design (A) and analytical workflow
(A) Three biological replicates of human medulloblastoma cells were treated with DMSO, SAG, vismodegib or EGF in starving medium. (B) Analytical workflow: After treatment, sample preparation, and TMT 10-plex labeling, peptides of each time point and treatment were pooled. Aliquots were utilized for pH 8 fractionation and deep proteome profiling or phosphopeptide enrichment by metal oxide affinity chromatography. Measurements of peptides and phosphopeptides were conducted with reversed-phase HPLC coupled to high-resolution mass spectrometry.
After cell lysis, reduction and alkylation, tryptic digestion, and TMT labeling for relative quantification, phosphopeptides were enriched by metal oxide affinity chromatography (MOAC) using titanium dioxide (TiO2) beads. Following enrichment, phosphopeptides were fractionated using hydrophilic interaction liquid chromatography (HILIC) prior to nano-HPLC-MS/MS analysis for detection and relative quantification of phosphopeptides (Fig. 1b). In parallel, deep proteome profiling was performed with 50 μg of the sample using high pH fractionation to normalize the phosphoproteome data and to detect potential changes in protein abundance. For each phosphopeptide (TiO2) or protein (global), the average of the three normalized intensity values of each treatment or control was used to calculate the ratio of each particular treatment versus control. All three biological replicates were considered for ratio calculation. To determine p-values and statistical significance of more than 7000 phosphopeptides in an unbiased manner, LIMMA (Linear Models for Microarray) statistical testing was applied.
Chemicals and samples
Hyperconfluent HH responsive DAOY cells [40] (ATCC: HTB-186 American Type Culture Collection, ATCC, Manassas, VA, USA) pre-starved for 24 h in serum-reduced MEM medium containing 0.1% (v/v) FBS were treated with 0.10% (v/v) DMSO, 100 nM Smoothened Agonist (SAG, Sigma-Aldrich, Steinheim, Germany), 500 nM vismodegib (vismodegib, LC Laboratories, Woburn, USA), or 25 ng/mL Epidermal Growth Factor (EGF, Sigma-Aldrich, Steinheim, Germany) for 5.0 and 15 min, respectively, in triplicates. After treatment, the cells were washed with PBS prior to lysis in a solution containing 1.0% SDS (w/v), 50 mM Tris-HCl pH 7.80, 150 mM NaCl, supplemented with protease inhibitor cocktail Complete Mini and phosphatase inhibitor cocktail PhosSTOP (Roche, Basel, Switzerland). To reduce viscosity and remove nucleic acid, samples were incubated with 5.0 μL benzonase nuclease (25 units/μl) (Merck, Darmstadt, Germany) for 30 min at 37 °C, followed by centrifugation at 18,000 g for 30 min at 4.0 °C. Pellets were discarded. To determine protein concentration, a bicinchoninic acid (BCA) assay (Thermo Scientific, USA) was performed according to the manufacturer’s protocol. Cysteines were reduced with DTT (10 mM final concentration, Roche Diagnostics, Mannheim, Germany) for 30 min at 56 °C and alkylated with iodoacetamide (30 mM final concentration, Sigma Aldrich, Steinheim, Germany) for 20 min at room temperature in the dark. For each sample, 150 μg of protein were loaded onto a 30 kDa molecular weight cut-off spin filter (Pall Nanosep®, Sigma-Aldrich, Germany) for filter-aided sample preparation [43, 44] with a few modifications. Protein digestion occurred in 50 mM triethylammonium bicarbonate (TEAB, Sigma Aldrich, Steinheim, Germany), 2.0 mM CaCl2 (Merck, Darmstadt, Germany), 200 mM guanidine hydrochloride and trypsin (ratio 1:20 enzyme:protein, w/w) (Promega, USA) for 14 h at 37 °C. Later, spin filters were transferred to new 2 mL protein LoBind Eppendorf tubes (Sigma Aldrich, Steinheim, Germany), centrifuged at 13,500 g for 25 min, and followed by two other 15 min centrifugation steps with the addition of 100 μL 50 mM TEAB and 50 μL water on top of the membrane. To control digestion efficiency, 1.0 μg of each digest was injected into a monolithic HPLC system as described in Burkhart et al. [45]. All samples were dried under vacuum.
TMT 10-plex labeling
Samples were individually solubilized in 100 mM TEAB buffer and incubated with 0.80 mg TMT label (Thermo Fischer Scientific, USA) (dissolved in 41 μL acetonitrile) for 60 min at 25 °C with slight agitation. Reaction was stopped upon incubating for 15 min at 25 °C with 0.27% hydroxylamine (v/v). Samples were pooled together in a ratio of 1:1 and dried under vacuum.
Sample desalting
TMT pooled samples were solubilized in 300 μL 0.10% aqueous TFA and desalted with SPEC C18 AR 15 mg cartridge (Agilent, Germany). The cartridge was first washed with 400 μL 100% acetonitrile (ACN), equilibrated twice with 400 μL 0.10% (v/v) TFA, loaded twice with TMT sample, washed twice with 400 μL 0.10% (v/v) aqueous TFA, and the sample was eluted twice with 200 μL 70% (v/v) ACN into a protein LoBind Eppendorf tube (Sigma-Aldrich, Steinheim, Germany). After vortexing, 50 μg of each pool were removed for high pH reversed-phase fractionation. Samples were dried under vacuum.
High-pH reversed-phase fractionation
The aliquots were solubilized in 15 μL10 mM ammonium acetate, pH 8.0 (buffer A) and fractionated on an HPLC System (Ultimate 3000, Thermo Scientific). Peptides were separated on a 150 mm × 1.0 mm i.d. C18 column (ZORBAX 300SB, Agilent Technologies, Germany) with a 55 min gradient ranging from 2.5 to 42% ACN in 10 mM ammonium acetate, pH 8.0, at a flow rate of 12.5 μL/min. In total, 20 fractions were collected in 60 s intervals in concatenation mode. Each fraction was dried under vacuum and resuspended in 15 μL of 0.10% (v/v) aqueous TFA for nano-HPLC–MS/MS analysis.
Phosphopeptide enrichment and HILIC fractionation
TMT pooled samples were solubilized in 80% (v/v) ACN, 5.0% (v/v) TFA and 1.0 M glycolic acid. Based on Engholm-Keller’s et al. [46] protocol, phosphopeptide enrichment was applied as described in Gonczarowska-Jorge et al. [47]. Phosphopeptides were HILIC fractionated on an HPLC system (Ultimate 3000 nano RSLC, Thermo Scientific) coupled with a self-made 150 mm × 0.250 mm i.d. TSKgel Amide-80 (Tosoh, Japan) column. After injection, sample was loaded for 20 min in 98% ACN, 0.10% TFA (eluent A) and fractionated in 40 min gradient ranging from 1 to 35% eluent B (0.10% TFA). At 10% eluent B, flow rate decreased linearly from 3.0 μL/min to 2.5 μL/min at 40% eluent B. A total of 12 fractions were collected for each sample. Each fraction was dried under vacuum and resuspended in 15 μL of 0.10% aqueous TFA for nano-HPLC–MS/MS analysis.
Nano-HPLC-MS/MS analysis
Samples were analyzed using a nano HPLC system (Ultimate 3000 nano RSLC, Thermo Scientific) coupled to a quadrupole-Orbitrap mass spectrometer (Q Exactive HF, Thermo Scientific) in data dependent acquisition mode. All samples were preconcentrated on a 20 mm × 75 μm i.d. trapping column (C18 Acclaim Pepmap, Thermo Scientific, Germering, Germany) in 0.1% aqueous TFA for 10 min using a flow rate of 20 μL/min, followed by separation in a 50 cm × 75 μm i.d. separation column (Acclaim Pepmap C18, Thermo Scientific) at a flow rate of 250 nL/min using a binary gradient (A: 0.10% aqueous formic acid, B: 84% acetonitrile, 0.1% formic acid) as indicated in Table 1. MS survey scans were acquired in the Orbitrap from m/z 300 to 1500 at a resolution of 60,000 using the polysiloxane ion at 371.1012 m/z as lock mass [48]. The AGC target value was 2 × 105 and the maximum injection time 120 ms. For MS/MS, precursors were selected using the top 15 ions with 30 s dynamic exclusion. Peptides were fragmented using higher-energy collisional dissociation (HCD) at 33% relative collision energy, and fragment ion spectra were acquired in the Orbitrap with a resolution of 60,000, an AGC target value of 2 × 105 ions, a maximum injection time of 200 ms, and an isolation window of 0.4 m/z for the high pH fractions and isolation window of 0.8 m/z for the HILIC fractions (phosphoproteome). A Table with an overview of applied gradients is added in the supplementary section in Table S-1.
Data analysis
MS raw files were converted to the open standard format mzML and centroided using msconvert (3.0.11781). Further processing was performed using the KNIME Analytics platform (Version 3.7.0) [49] and OpenMS 2.4.0 [50, 51].
Identification was performed using the MSGFPlusAdapter with MS-GF+ [52](v2018.01.30). A Swiss-Prot human (# of entries: 20410, downloaded 17th of October 2018), cRAP (# of entries: 116) decoy database was created using the DecoyDatabase tool (# of entries: 41050). For database search, enzyme specificity was set to trypsin; Carbamidomethyl (C) (+ 57.021 Da), TMT6plex (N-term, K) (+ 229.163 Da) were set as fixed modifications. Oxidation (M) (+ 15.995 Da) was set as variable modification. For the HILIC fractions, phosphorylation of Ser, Thr, and Tyr (+ 79.966 Da) was included in the variable modifications. MS and MS/MS tolerances were set to 10 ppm and 0.02 Da, respectively. Afterwards, target decoy annotation was added, posterior error probability estimation was performed using percolator (v 3.02.0) [53]. Peptides with a false discovery rate below 0.01 were retained. For the dataset with phospho modifications LuciphorAdapter was used to run LuciPHOr2 (v 1.2014Oct10) [54] for phosphosite localization. Here, phosphopeptides with a False Localization Rate (FLR) smaller 0.05 were considered for further analysis. Quantification was performed using the IsobaricAnalyzer with the TMT10plex method and the correction matrix corresponding to the used chemicals. Afterwards, identification and quantitative information were mapped, unannotated and unassigned identifications filtered and exported in the mztab format.
Further location of the phosphosite position in the protein was assessed by extracting the position based on the annotated protein in the fasta database and the information of the peptide phosphosite localization.
For the proteomics dataset, the peptide level analysis was performed as described above, apart from phospho modifications and phosphosite scoring. Additionally, the unfiltered identifications were used for protein inference performed with the FidoAdapter using Fido (2012) [55]. Protein level quantification was performed using the Fido results with a protein FDR < 0.01.
The normalization of the phosphoproteome was performed as described by Shema et al. [56]. Median normalization of each channel was performed for the proteome. In order to facilitate reproduction of the computational analysis workflows and R scripts are included in the supplementary material.
LIMMA statistical testing (R package limma v. 3.36.5) [57] was applied on proteome normalized log2 intensity values of the phosphoproteomics dataset and used to generate volcano plots using R version 3.5.1. Scripts are attached in the online supplementary and all applied R packages with corresponding versions and citations are provided in Table S-2. Kinase Substrate Enrichment [58] was performed using KSEA App (casecpb.shinyapps.io/ksea/) [59] using the R package “KSEAapp” on CRAN using R version 3.5.1. Pathway enrichment was performed using Ingenuity pathway Analysis IPA (QIAGEN Inc., https://apps.ingenuity.com/).
Results and discussion
Comprehensive phosphopeptide profiling detects almost 10.000 phosphosites in stimulated medulloblastoma cells
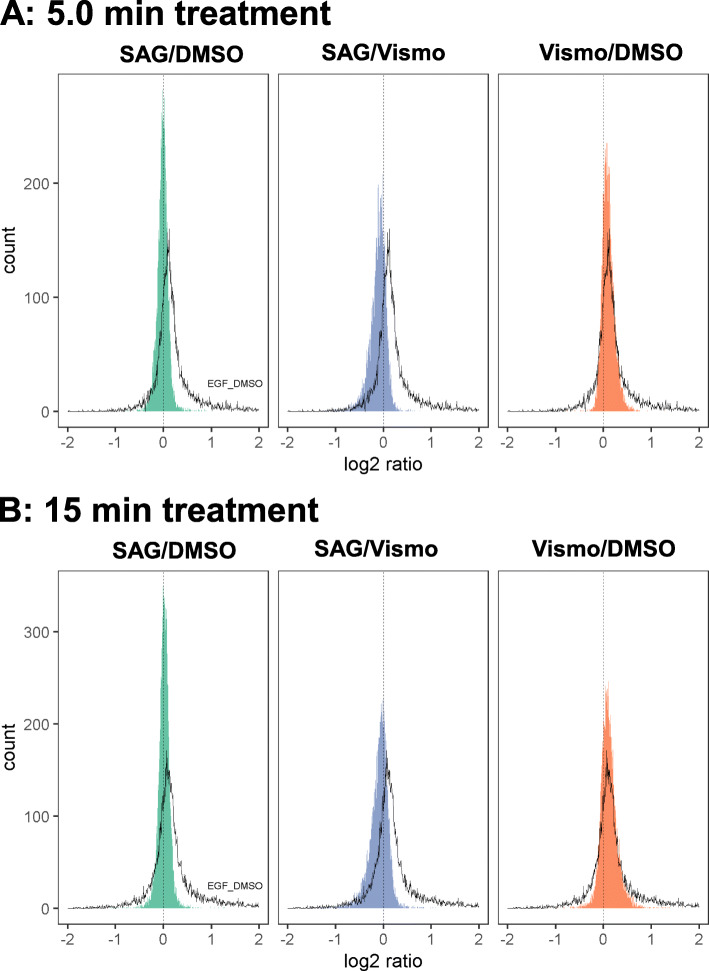
Following the analytical strategy described above, we identified and quantified a total of 7696 highly confident phosphopeptides after 5.0 min and 9976 phosphopeptides after 15 min treatment with a false localization rate (FLR) below 0.05. We further quantified 7547 protein groups after 5.0 min and 7763 protein groups after 15 min using extensive pH 8 fractionation. As expected, the changes in the global proteome after short-term treatment were negligible. We found a normal and narrow distribution of the ratios SAG/DMSO, Vismo/DMSO and SAG/Vismo, similar to the distribution of the positive control EGF as depicted in Figure S-3. Interestingly, the total number of identified phosphosites was higher after 15 min treatment, whereas the total number of identified protein groups was not increased after 15 min treatment. This can be due to technical variation in the sample preparation for the phosphoproteomics analysis or more phosphorylation events upon 15 min stimulation due to the activation of downstream phosphorylation cascades.
Strikingly, there were several distinctive features in the distributions of the phosphopeptide ratios SAG/DMSO, Vismo/DMSO and EGF/DMSO. Figure 2a illustrates the distribution of phosphopeptide ratios determined from proteome-normalized data for 5.0 min treatment and Fig. 2b for 15 min, respectively. The illustration of the distribution of the phosphopeptide ratios of the pooled EGF treated cells to DMSO treatment (EGF/DMSO) was taken as a reference distribution (black line). EGF is known to induce various phosphorylation events in the DAOY cell line by activation of EGFR signaling [60]. Hence, the reference distribution is skewed to positive ratios. In contrast, only minor changes in the total phosphoproteome were found after SAG treatment. The distribution of SAG/DMSO (green graph) after 5.0 min is quite narrow and almost normally distributed. The broader distribution of vismodegib (Vismo/DMSO) (orange graph) is skewed to positive ratios similar to the reference distribution of EGF/DMSO. This indicates an upregulation of phosphorylation events after 5.0 min of vismodegib treatment. These effects are maintained after 15 min treatment. The distribution of the ratio of Vismo/DMSO is even broader and more prominently skewed to positive ratios. This indicates that in particular inhibition of the Hedgehog pathway simulated by vismodegib treatment in DAOY cells is driven by a global increase in phosphorylation.
Fig. 2.
Distribution of proteome normalized ratios of phosphopeptides identified after 5.0 min treatment (a) and 15 min treatment (b). Intensity values of each channel of the phosphoproteome were normalized by correction factors derived from respective channels of the measured proteome. Ratios were determined by the mean of the treatment SAG or vismodegib (N = 3) divided by the mean of DMSO (N = 3) of the respective time point. SAG/DMSO (green graph) represents the distribution of the phosphopeptide ratios of SAG treatment compared with DMSO treatment, whereas Vismo/DMSO (orange graph) describes the distribution of phosphopeptide ratio of vismodegib treatment compared to control (DMSO) treatment. The ratio was determined by division of the proteome-normalized intensity by the mean of DMSO treatment. SAG/Vismo (blue graph) describes the phosphopeptide ratio generated by division of SAG/DMSO by Vismo/DMSO
Ingenuity pathway analysis reveals differences in regulation of various cancer related pathways after 5.0 min treatment
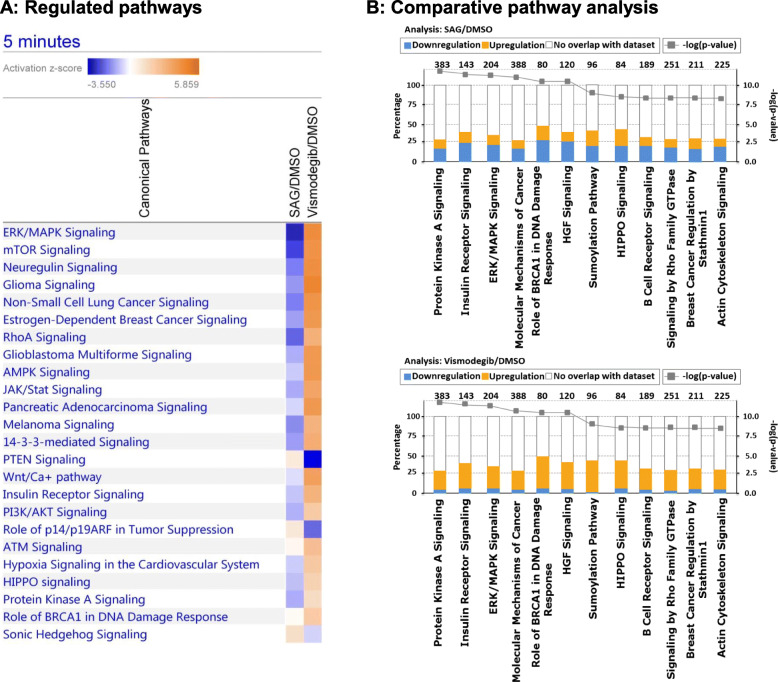
Ingenuity pathway analysis (IPA) was performed with all quantified phosphopeptides for both time points. All proteome-normalized phosphopeptide ratios of respective treatment relative to DMSO as control treatment were uploaded and analyzed using the phosphorylation analysis tool in the IPA software platform. Notably, differences in global phosphorylation dynamics between activation and inhibition of the HH pathway were more pronounced after the short-time treatment time point of 5.0 min as depicted in Fig. 3.
Fig. 3.
Ingenuity pathway analysis of all phosphopeptides identified after 5.0 min. a Comparative analysis of regulated pathways after SAG and vismodegib treatment. Pathways were manually filtered for cancer relevance and the log10 p-value cut-off was set to 1.3. Activated pathways are shown in orange, inhibited pathways in blue. b Top 12 significantly enriched pathways after 5.0 min SAG (upper bar chart) and vismodegib treatment (lower bar chart). The total number of phosphoproteins which are assigned to a particular pathway are displayed on top of the particular bar, share of those phosphoproteins identified in the experiment is represented by the primary x-axis, while the –log p-value determined by Fisher’s exact testing is represented by the secondary y-axis. The proportion of upregulated phosphoproteins is shown in orange, downregulation is indicated by blue bars
Protein phosphorylation is a fast and highly dynamic process, which can transmit intracellular signaling within minutes and even seconds [9, 61, 62]. Ingenuity pathway analysis of our data revealed various cancer-related pathways as differentially regulated already after 5.0 min of vismodegib or SAG treatment (Fig. 3a). For instance, ERK/MAPK signaling, mTOR signaling, JAK/STAT, or 14–3-3-mediated signaling were found to be significantly inhibited after 5.0 min SAG treatment but activated after treatment with vismodegib. We observed activation of 14–3-3-mediated signaling and Protein Kinase A signaling after 5.0 min of vismodegib treatment, consistent with a previous study showing that 14–3-3 epsilon binds to GLI after phosphorylation by Protein Kinase A, resulting in the inhibition of HH pathway activity [63].
The most significantly enriched pathways after 5.0 min treatment were Protein Kinase A signaling, Insulin Receptor signaling, as well as ERK/MAPK signaling (Fig. 3b). Interestingly, the majority of the phosphosites in the respective pathways was found to be downregulated after 5.0 min SAG treatment, whereas upregulation was predominantly observed after vismodegib treatment.
Protein Kinase A has been recognized to negatively modulate the hedgehog pathway and thus influence cell fate and proliferation [64]. In accordance with the literature we determined PKA signaling as activated after vismodegib treatment while inhibited after SAG treatment (Fig. 3). Intriguingly, this initial activation of PKA signaling after 5.0 min vismodegib treatment is not maintained after 15 min (Figure S-4A). The initial phosphorylation of PKA substrates after 5.0 min might induce downstream processes and appears to be balanced with time. PKA signaling was clearly inactivated after 15 min vismodegib treatment compared to 5.0 min as shown in Figure S-5. We confirmed activation and inactivation of PKA by vismodegib and SAG, respectively by Western blot analysis using activation-specific anti-phopho-PKA antibodies (see suppl. Figure S-6). Furthermore, Insulin Receptor Signaling is known to synergize with sonic HH in medulloblastoma formation [65] and was significantly enriched in our data set. Surprisingly, 5.0 min of vismodegib treatment activated Insulin Receptor signaling, while SAG treatment impeded the pathway, which may be explained by the partial agonistic effect reported for therapeutic SMO inhibitors [39]. However, SAG activates Insulin Receptor Signaling over time (Figure S-5) which was clearly induced after 15 min SAG treatment compared to 5.0 min treatment.
Additionally, we find ERK/MAPK signaling both enriched and activated after 5.0 min vismodegib treatment (Fig. 3). Extensive research has been conducted to decipher the interactions between sonic HH and ERK/MAPK signaling, however with contradictory results [40, 66, 67]. The regulation of HH signaling by ERK/MAPK signaling has been validated biochemically [68] and observed in multiple cancer entities such as non-melanoma and melanoma skin cancer [69–72], and gastric cancer [73], and hepatocellular carcinoma [74]. This has led to the suggestion to combine MEK or ERK1/2 inhibitors and HH pathway inhibitors to synergistically fight human cancers [75–78]. However, our data supports a negative crosstalk between HH and ERK/MAPK signaling since an activation of ERK/MAPK signaling was observed after vismodegib treatment. Of note, a negative interaction of HH and MEK/ERK signaling has also been reported by Götschel et al. [40] and by Neill et al. [79] for epidermal cells, suggesting context-dependent pathway interactions. Furthermore, we find increased activation of ERK/MAPK signaling after 15 min compared to 5.0 min SAG treatment (Figure S-5). Hence, SAG treatment induced ERK/MAPK signaling in a time-dependent manner indicating a slower kinetics compared to vismodegib treatment.
Research on the crosstalk of sonic HH signaling and the mTOR/S6K1 pathway revealed an activation of GLI1 in a SMO-independent manner indicating a positive crosstalk via non-canonical HH pathway activation [80]. In contrast, in our short-term pathway activation study, we find mTOR signaling to be activated after vismodegib but inhibited after SAG treatment, implying a negative crosstalk of mTOR signaling and the HH pathway.
Moreover, our data indicates that SAG and vismodegib treatment differentially influences JAK/STAT signaling (Fig. 3a). A positive crosstalk of sonic HH signaling with STAT3 was found in basal cell carcinoma [81, 82] and in human papillary thyroid cancer [83], but to our knowledge has not yet been reported for medulloblastoma. We find JAK/STAT signaling activated after 5.0 min of vismodegib treatment, while being inhibited after SAG treatment.
Interestingly, PTEN signaling was strongly inhibited after 5.0 min of vismodegib treatment. In contrast to our results, Hartmann et al. reported a link between PTEN loss and proliferation of medulloblastoma cells on genomic and epigenetic levels [84]. However, our data indicates that short-term vismodegib treatment can inhibit PTEN signaling in human medulloblastoma cells via phosphorylation cascades, demonstrating an additional level of crosstalk distinct from the previously reported. Likewise, Metcalfe et al. showed that allografts with PTEN deficient medulloblastoma do respond to vismodegib treatment, and therefore, still depend on sonic HH signaling [85]. Interestingly, SAG treatment influenced PTEN signaling in a time-dependent manner as shown in Figure S-5. We find decreased activation of PTEN signaling after 15 min compared to 5.0 min SAG treatment. This indicates initial activation of PTEN signaling by SAG which diminishes with time. Similarly, increased activation of PI3K/AKT signaling, the antagonist of PTEN signaling, is observed after 15 min compared to 5.0 min SAG treatment (Figure S-5). The crosstalk of PI3K/AKT/mTOR signaling and HH signaling has been reported frequently [67, 80]. Our data points to an initial induction of PTEN signaling by SAG, which is then balanced out by activation of PI3K/AKT signaling. This time-dependent activation of PI3K/AKT pathway after initial activation of PTEN signaling represents a potential mechanism of action of non-canonical HH pathway activation. These findings go along with the study of Chaturvedi et al. who combined PI3K-mTOR inhibition with vismodegib treatment, showing therapeutic effects in medulloblastoma in vitro and in vivo [86].
These time-dependent dynamics in pathway activation and inhibition explain that the differences between SAG and vismodegib treatment on pathway level found after 5.0 min were not observable after 15 min treatment (Figure S-4A). Vismodegib treatment might induce faster phosphorylation dynamics than SAG, meaning both drugs show different kinetics. However, we identified an overlap of 4960 phosphopeptides after both time points as shown in Figure S-7. In total, we found even more phosphopeptides after 15 min treatment compared to 5.0 min, which can be explained by enhanced signaling cascades downstream of initial phosphorylation events. These downstream signaling cascades result in similar pathway activation and phosphorylation patterns after 15 min both for SAG and vismodegib treatment as depicted in Figure S-4B.
BRCA1 phosphorylation and IFT172 phosphorylation may be involved in HH pathway inhibition and ciliary assembly
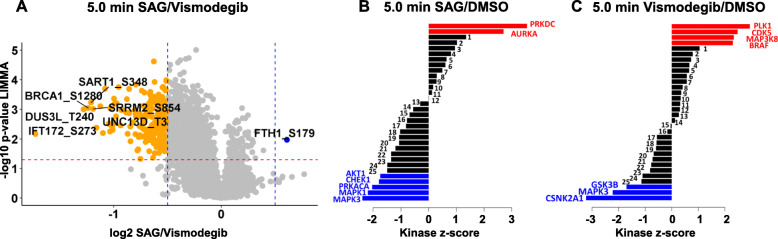
LIMMA significance testing was applied on the proteome-normalized intensity values of the three biological replicates of respective treatments. P-values were determined to evaluate the statistical significance of up- and downregulated phosphopeptides after 5.0 and 15 min treatment. Statistical significance was evaluated for each treatment relative to DMSO as control treatment and between both treatments SAG and vismodegib. Whereas only one phosphopeptide was found to be significantly upregulated after 5.0 min SAG treatment compared to DMSO as control (Figure S-8A), 18 phosphopeptides were significantly upregulated after vismodegib treatment (Figure S-8B). Even 216 phosphopeptides were significantly regulated between both treatments after 5.0 min with a significance threshold of p < 0.05. The corresponding volcano plot is shown in Fig. 4a. Here, the ratio of SAG/vismodegib reflects the ratio of SAG/DMSO divided by vismodegib/DMSO, hence negative ratios represent an upregulation after vismodegib treatment compared to SAG treatment.
Fig. 4.
Volcano plot and kinase substrate enrichment analysis for phosphopeptides identified after 5.0 min treatment. a Distribution and significantly regulated phosphopeptides between SAG and vismodegib treatment. Ratios were determined by division of the ratio SAG/DMSO by the ratio vismodegib/DMSO: SAG/vismodegib. LIMMA statistical testing was applied to determine p-values and statistically significant regulated phosphopeptides. Orange and blue dots, significantly down- or upregulated phosphopeptides, respectively (threshold of p < 0.05.0); log2-fold-change cutoff was set to 0.5 and – 0.5 (b) and (c) Kinase Substrate Enrichment Analysis. Proteome normalized ratios of SAG/DMSO (b) and Vismo/DMSO (c) were taken to infer the kinase activation score with a p-value cutoff of 0.05. Significantly activated kinases are presented in red, significantly attenuated kinases in blue. A table with enriched kinases with corresponding numbers is attached in the supplementary material
Interestingly, phosphorylation of Breast cancer type 1 susceptibility protein (BRCA1) at serine 1280 was significantly upregulated after HH pathway inhibition (Fig. 4a). BRCA1 plays important roles in the repair of DNA damage. ATM-dependent phosphorylation of BRCA1 in response to DNA damage has been found by Cortez et al. [87]. ATM-related kinase ATR has been described to phosphorylate BRCA1 at serine 1280, promoting its delocalization into the nucleus [88, 89]. Furthermore, phosphorylation of BRCA1 by AURKA is known to inhibit BRCA1 activity [90]. In accordance with these findings, kinase substrate enrichment revealed AURKA as significantly activated after 5.0 min SAG treatment, i.e. HH pathway induction (Fig. 4b). Furthermore, AURKA was the most significantly activated kinase after kinase substrate enrichment of 5.0 min SAG compared to vismodegib treatment (Figure S-9). AURKA has been implicated in regulating the proliferation and growth of human glioma cells [91], and in the (dis) assembly of the primary cilium. In line with its role in tumor cell proliferation, inhibitors of AURKA have successfully been applied in the treatment of tumor-propagating cells in medulloblastoma mouse models [92].
Furthermore, Intraflagellar transport protein 172 homolog (IFT172) was significantly higher phosphorylated at serine 273 after vismodegib treatment compared to SAG treatment. IFT172 is part of Intraflagellar transport complex B (IFT B) and important for ciliary assembly and essential for brain development [93]. Our point to a possible role of Ser273 phosphorylation of IFT172 in the regulation of ciliary transport of effector proteins involved in HH signaling. It will be interesting to address in future studies whether this phosphorylation event influences the regulation and coordination of HH signaling in the primary cilium.
Polo-like-kinase 1 activation and casein kinase 2 A1 inhibition by vismodegib treatment
Kinase substrate enrichment analysis of phosphopeptides identified after 5.0 min treatment revealed activation of Polo-like kinase 1 (PLK1) after vismodegib treatment along with CDK5, MAP 3 K8 and BRAF (Fig. 4c). Like AURKA, Polo-like Kinase-1 (PLK1) has been implicated in the control of cilia formation. It will therefore be important to address the role of these kinases in the regulation of HH signaling at the level of the primary cilium [94, 95] and in the context of therapeutic interventions using SMO modifiers. PLK1 is best known for its positive function in G2/M phase progression [96] and therefore has been intensely investigated as an attractive drug target for cancer therapy [97]. In accordance with our data, Zhang et al. found PLK1 as a negative regulator of Hh signaling by phosphorylation of GLI1 [98]. In contrast to our findings, Polo-like kinase-1 inhibition by small molecules was found to be beneficial in the treatment of HH dependent medulloblastoma cell lines such as DAOY cells [99].
Furthermore, Casein Kinase 2A1 (CSNK2A1) was predicted to be significantly downregulated after 5.0 min vismodegib treatment (Fig. 4c). Casein Kinase 2 is known as an important positive regulator of Hh signaling in Drosophila [100] and lung cancer cell lines [101]. In accordance with our data, Casein Kinase 2 was recently identified as a major regulator of medulloblastoma [102] and described as a key driver of HH signaling in murine granule neuron precursors [102].
Significant changes in phosphorylation of HH pathway components are likely to drive ciliary assembly after 5.0 min and signal transduction in the primary cilium after 15 min
We filtered the phosphoproteome of each time point for proteins, which are involved in or linked to HH signaling. Interestingly, the quantitative changes in the phosphoproteome of HH pathway components were rather low. However, various differences in the phosphorylation pattern of particular components of the Hedgehog pathway were observable. Pathway maps relying on identified phosphopeptides and the corresponding heatmap of HH pathway phosphoproteins are presented in Fig. 5 for 5.0 min and Fig. 6 for 15 min treatment.
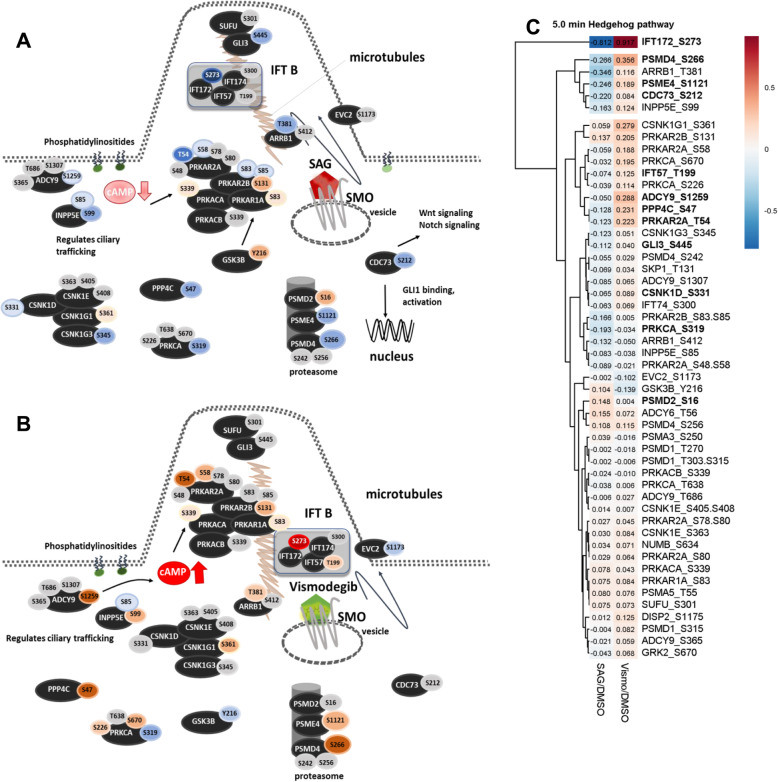
Fig. 5.
Phosphorylation of HH pathway components after 5.0 min treatment. a SAG binding to Smoothened induces ciliary trafficking after 5.0 min stimulation. b Vismodegib treatment induces phosphorylation and increases cAMP levels after 5.0 min treatment. c Heatmap of regulated and identified phosphosites after 5.0 min SAG and vismodegib treatment. Blue and red ellipses represent phosphosites down- or upregulated compared to control. All quantified phosphopeptides of phosphoproteins after 5.0 min treatment assigned to sonic hedgehog signaling were considered for the heatmap without applying any cut-off value. SAG/DMSO represents the ratio determined by the mean of the proteome-normalized intensities detected after SAG treatment (N = 3) divided by the mean of proteome-normalized intensities after DMSO (N = 3). Vismo/DMSO represents the ratio determined by the mean of proteome-normalized intensities detected after vismodegib treatment (N = 3) divided by the mean of proteome-normalized intensities after DMSO (N = 3). Significantly regulated phosphopeptides between SAG and vismodegib treatment are highlighted in bold. Significantly regulated phosphopeptides between SAG and vismodegib treatment are highlighted in bold
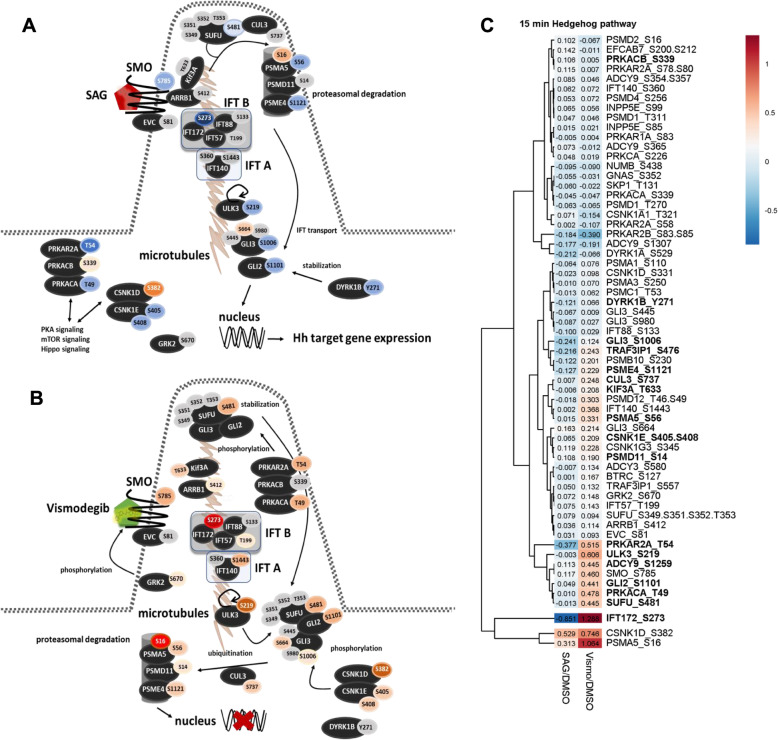
Fig. 6.
Phosphorylation of HH pathway components after 15 min treatment. a SAG impairs GLI phosphorylation after 15 min stimulation. b Vismodegib binding induces delocalization of Smoothened in the primary cilium. c Heatmap of regulated and identified phosphosites after 15 min SAG and vismodegib treatment. Blue and red ellipses, phosphosites down- or upregulated compared to control. All quantified phosphopeptides of phosphoproteins after 15 min treatment assigned to sonic hedgehog signaling search were considered for the heatmap without applying any cutoff value. SAG/DMSO represents the ratio determined by the mean of the proteome-normalized intensities detected after SAG treatment (N = 3) divided by the mean of proteome-normalized intensities after DMSO (N = 3). Vismo/DMSO represents the ratio determined by the mean of proteome-normalized intensities detected after vismodegib treatment (N = 3) divided by the mean of proteome-normalized intensities after DMSO (N = 3). Significantly regulated phosphopeptides between SAG and vismodegib treatment are highlighted in bold
In total, we identified 51 phosphopeptides involved in HH signaling after 5.0 min treatment, of which 12 phosphopeptides were significantly different between SAG and vismodegib treatment (corresponding proteins highlighted in bold in Fig. 5c). Various phosphosites of Protein Kinase A subunits, Casein Kinase 1 subunits, and also Protein Kinase C were identified after 5.0 min treatment and indicated in Fig. 5a and b. In particular threonine 54 of cAMP-dependent Protein Kinase type II-alpha regulatory subunit (PRKAR2A) was significantly downregulated after SAG and highly upregulated after vismodegib treatment. GSK3β was found to be phosphorylated and thus activated at tyrosine 216 after SAG treatment. Both kinases play central regulatory roles in sonic HH signaling [27, 32, 103].
Moreover, we found phosphorylation of the catalytic subunit of the phosphatase PPP4C, which is involved in activation and deactivation of PP4 for SMO dephosphorylation after 5.0 min treatment. Ser47 phosphorylation of PPP4C was significantly downregulated after SAG treatment and upregulated after vismodegib treatment. In accordance with our observation, Jia et al. reported a negative regulatory function of PP4 in SMO phosphorylation and HH pathway transduction [36]. However, PP4 controls microtubule organization and is implicated in various cellular processes [104]. Hence, the differences in PP4 phosphorylation could also reflect its functional role in microtubular coordination for signal transduction, e.g. in the primary cilium.
Interestingly, proteasomal subunits such as PSMD4 and PSME4 were differentially phosphorylated after both treatments, suggesting that post-translational modification of the proteasome machinery is an early response to SMO modulation. Moreover, various Adenylylcyclases and 72 kDa Inositol polyphosphate 5.0-phosphatase INPP5E were differentially phosphorylated after 5.0 min SAG and vismodegib treatment. Ciliary phosphoinositides control ciliary trafficking and thus contribute to the regulation of HH signaling [105]. INPP5E in particular regulates the level of inhibitors of Hh signaling in the primary cilium [106]. Moore et al. showed that cilia have a high cAMP level which is controlled by PKA and phosphoinositide dependent signaling [107]. In accordance with these findings we observe inactivation of Adenylylcyclases, INPP5E and PKA via dephosphorylation in response to activation of the HH pathway by 5.0 min SAG treatment. This observation is in line with the current model of reduced PKA activity in response to SMO activation and thus provides a basis for future studies to unravel in detail the elusive molecular link between SMO, PKA, SUFU and GLI.
Intriguingly and consistent with the effect of SAG, vismodegib treatment induced rapid phosphorylation of PKA subunits and the components of phosphoinositol signaling, indicating higher levels of cAMP and PKA activity in the cilium, which is required for the formation of GLI repressor forms. Our data also point to a role of IFT172 in the ciliary regulation and regulation of HH signaling. It represents a part of the IFT B complex and its phosphorylation was found to be highly downregulated after SAG and upregulated after vismodegib treatment at serine 273. Intraflagellar transport is crucial for ciliary trafficking and assembly, and thus for the control of HH signaling [108]. Corresponding to our findings, it has already been shown that disruption of IFT172 led to a complete lack of primary cilia [109]. Protein phosphorylation was reported to be important for flagellar disassembly by Pan et al. [110] but the importance of serine 273 phosphorylation at IFT172 was not demonstrated before. This phosphosite remains significantly regulated also after 15 min treatment.
Hence, we propose the following model of the HH pathway for the 5.0 min time point, visualized in Fig. 5a and b. SAG treatment induces dephosphorylation of central phosphoproteins of the Intraflagellar Transport complex (IFT) B and beta-Arrestin, which leads to the transport of Smoothened into the cilium. Furthermore, the ciliary cAMP level is reduced by dephosphorylation of ADCY9 and INPP5E resulting in inhibition of ciliary Protein Kinase A and other cytosolic kinases such as Casein kinases and Protein Kinase C. Protein Kinase A delocalizes into the cytoplasm and interacts with GSK3β which is activated by phosphorylation at Tyrosine 216. Dephosphorylation of CDC73 further activates sonic HH signaling by binding to GLI1 on gene level. In contrast, vismodegib binding to Smoothened causes phosphorylation of intraflagellar transport proteins in particular IFT172 (serine 273). ADCY9 and INPP5E are phosphorylated resulting in increased cAMP levels and activation of ciliary Protein Kinase A and other cytosolic kinases such as Casein kinases and Protein Kinase C. GSK3β is inhibited by dephosphorylation at tyrosine 216.
We could identify additional phosphopeptides with a possible role in HH signaling after 15 min treatment (Fig. 6). Out of 61 phosphopeptides, 17 were significantly regulated between both treatments. Vismodegib treatment induced phosphorylation of central HH pathway components such as SMO, SUFU, GLI2 and GLI3. GLI phosphorylation is likely to be a consequence of increased PKA activity (see above), which is known to promote the proteasomal processing and degradation, respectively [103]. Furthermore, SUFU phosphorylation stabilizes the inhibitory SUFU-GLI complex [32]. In contrast, SAG treatment induced dephosphorylation at these phosphosites possibly promoting proteasomal degradation of SUFU and activation of the GLI transcription factors.
Moreover, Serine/threonine-protein Kinase ULK3 (ULK3) was found to be significantly higher phosphorylated after 15 min of vismodegib treatment compared to SAG treatment (Fig. 6). A dual function of ULK3 in the regulation of the HH pathway was already reported 2010 by Maloverjan et al. [111, 112] They showed that ULK3 both interacts with SUFU and GLI2. Our data shows that 15 min vismodegib treatment induces ULK3 phosphorylation at serine 219 while SAG treatment reduces phosphorylation at this phosphosite compared to control treatment. The phosphorylation of ULK3 at serine 219 was also significantly upregulated after vismodegib treatment compared to DMSO treatment as visualized in the volcano plot in Figure S-10 B. Interestingly, Kinesin-like protein Kif3A phosphorylation at Thr633 was significantly increased after 15 min vismodegib treatment. Kif3A is essential for HH pathway activation [113] and required for ciliary trafficking of Smoothened [114]. Furthermore, the significant upregulation at Ser273 of IFT172 observed after 5.0 min vismodegib treatment persists after 15 min. An increase in the phosphorylation from 5.0 to 15 min can be observed only after vismodegib treatment. In contrast, IFT172 expression levels do not change over time and phosphorylation does not change after SAG treatment as shown in Figure S-11. Our data thus supports a model where phosphorylation serves as a regulatory mechanism to control ciliary trafficking of central components of the HH pathway, possibly via Kif3A and its function in the primary cilium. Phosphorylation events are also likely to impact on the stability of the SUFU-GLI complex, which may involve differential phosphorylation of ULK3 in response to SMO modulation.
In sum, we come up with the following effects on the phosphorylation dynamics within the HH pathway after 15 min SAG and vismodegib treatment as depicted in Fig. 6a and b. Upon SAG treatment, ARRB1 and Kif3A bind to dephosphorylated Smoothened and facilitate the delocalization into the primary cilium. SUFU gets dephosphorylated at Ser481 and Cul3 facilitates its proteasomal degradation. GLI2 and GLI3 are released from the SUFU complex and mostly dephosphorylated. SUFU is ubiquitinated and subjected to proteasomal degradation. GLI2 and GLI3 are transported by intraflagellar transport proteins of the IFT A and B complex. Dephosphorylation of DYRK1B (Y271) stabilizes GLI2 in its activator form, leading to Hh target gene expression. Protein Kinase A is mostly inactivated by dephosphorylation. On the other hand, Smoothened gets phosphorylated after 15 min vismodegib treatment by GRK2 at Ser785. SUFU is further phosphorylated at Ser481, which stabilizes the complex to GLI2 and GLI3. IFT B and IFT A proteins get phosphorylated and facilitate the transport of the SUFU-Gli complex into the cytosol. Ciliary protein Kinase A and cytoplasmic Casein kinases contribute to phosphorylation of GLI2 and GLI3. ULK3 is phosphorylated and interacts with SUFU promoting the CUL3 driven ubiquitination of GLI2 and GLI3 and their subsequent proteasomal degradation.
Conclusions
Monitoring of close to 10,000 phosphopeptides of human medulloblastoma upon short-term stimulation with SMO activators and inhibitors, respectively, allowed us to investigate the complex early phosphosignaling during HH pathway regulation. Changes in the phosphoproteome in response to activation or inhibition of the HH pathway are already observed after 5.0 min of treatment and mainly affect signaling pathways involved in cellular growth and proliferation. Of note, rapid changes in phosphorylation patterns of HH pathway components mainly affect components of the primary cilium as well as signaling via the SUFU-GLI axis. Intraflagellar transport proteins such as IFT172 were differentially phosphorylated indicating a crucial role of phosphorylation in the regulation of the intraflagellar transport machinery and hence the transport of HH pathway components in the primary cilium. Furthermore, phosphorylation influences Protein Kinase A signaling besides PI3K/AKT/mTOR and PTEN signaling as we find these pathways differentially regulated by phosphorylation dynamics upon short-term activation and inhibition of the HH pathway.
Our data suggests that several kinases play important roles in the regulation of short-term activation or inhibition of HH signaling. Aurora Kinase A may be involved in the activation of HH signaling upon SAG treatment. El-Sheikh et al. already showed that inhibition of Aurora Kinase A enhances the chemosensitivity of medulloblastoma [115], and targeting Aurora Kinase A in combination with HH pathway inhibitors was reported as novel therapeutic strategy in the treatment of human medulloblastoma [92]. However, inhibition of HH signaling simulated by vismodegib treatment was dominated by Polo-like Kinase-1 (PLK1). PLK1 was only recently shown to negatively regulate sonic HH signaling [98], which goes along with our findings. Furthermore, inhibition of Casein Kinase 2A1 after short-term vismodegib treatment underlines the discovery of Casein Kinase 2 as a key driver of hedgehog signaling by Purzner et al. [26]. However, our data highlights the involvement of subunit alpha in this functional role.
Elucidation of these deep phosphorylation cascades in HH signaling paves the way to a profound understanding of the basis of the immediate events of HH pathway activation important for the development of targeted therapies of sonic HH-type medulloblastoma patients. The new insights into changes in the phosphorylation landscape upon vismodegib treatment clears up its mechanism of action and will be beneficial for the prevention of adverse effects and therapeutic resistance.
Endnote
1The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
Supplementary information
Additional file 1: Table S-1. Eluent B (B) gradient range and gradient duration for the different analyzed fractions. Table S-2. Applied R-packages. Responsiveness of DAOY cells to SAG and vismodegib treatment. RNA isolation and quantitative PCR (qPCR). Table S-3. Primer Sequences used for qPCR. Figure S-1. Hedgehog target gene expression in SAG and vismodegib treated DAOY cells. Figure S-2. Comparable Hedgehog pathway induction by natural and synthetic HH pathway activators. Figure S-3. Distribution of protein groups identified after 5.0 min treatment (A) and 15 min treatment (B). Figure S-4. Ingenuity pathway analysis of all phosphopeptides identified after 15 min using a phosphorylation analysis. Figure S-5. Time dependent Ingenuity pathway analysis of cancer associated pathways. Figure S-6. Western blot validation of PKA activity modulation in response to SMO agonist and antagonists. Table S-4. Antibodies used for Western blot analysis. Figure S-7. Overlap of quantified phosphopeptides after 5.0 and 15 min. Figure S-8. Volcano plots of phosphopeptides identified after 5.0 min treatment. Figure S-9. Kinase substrate enrichment analysis (KSEA) was performed for phosphopeptides identified after 5.0 min treatment for the ratio SAG/Vismo using the online platform KSEA App (https://casecpb.shinyapps.io/ksea/). Figure S-10. Volcano plots and kinase set enrichment analysis for phosphopeptides identified after 15 min treatment. Figure S-11. IFT172 phosphorylation and expression after 5.0 and 15 min.
Acknowledgements
We thank Wolfgang Esser-Skala for his help with the computational visualization and analysis of the data.
Abbreviations
- ADCY9
Adenylate cyclase type 9
- ARRB1
Beta-arrestin-1
- ATM
Serine protein kinase ATM
- ATR
ATM-related kinase
- AURKA
Aurora kinase A
- BRAF
Serine/threonine-protein kinase B-raf
- BRCA 1
Breast cancer type 1 susceptibility protein
- cAMP
Cyclic adenosine monophosphate
- CDC73
Parafibromin
- CDK5
Cyclin-dependent-like kinase 5
- CSNK2A1
Casein Kinase 2A1
- Cul3
Cullin-3
- DTT
Dithiothreitol
- DYRK1B
Dual specificity tyrosine-phosphorylation-regulated kinase 1B
- EGF
Epidermal Growth Factor
- ERK
Extracellular Signal regulated kinase
- GLI
Glioma-associated oncogene
- GRK2
Beta-adrenergic receptor kinase 1
- GSK3β
Glycogen synthase kinase 3β
- HH
Hedgehog
- HILIC
Hydrophilic interaction liquid chromatography
- HPLC
High performance liquid chromatography
- IFT B
Intraflagellar transport complex B
- IFT
Intraflagellar Transport
- IFT172
Intraflagellar transport protein 172 homolog
- INPP5E
72 kDa inositol Polyphosphate 5-phosphatase
- IPA
Ingenuity Pathway Analysis
- JAK
Januskinase
- KIF 3A
Kinesin-like protein KIF3A
- Kif7
Kinesin-like protein KIF7
- LIMMA
Linear Models for Microarray
- MAP 3K8
Mitogen-activated protein kinase kinase kinase 8
- MAPK
Mitogen-activated protein kinase
- MS
Mass spectrometry
- mTOR
Mechanistic Target of Rapamycin
- PI3K
Phosphatidylinositol 3-kinases
- PKA
Protein Kinase A
- PLK1
Polo-like kinase 1
- PLK1
Polo-like kinase-1
- PP2A
Protein phosphatase 2
- PP4
Protein phosphatase 4
- PPP4C
Serine/threonine-protein phosphatase 4 catalytic subunit
- PRKAR2A
cAMP-dependent Protein Kinase type II-alpha regulatory subunit
- PSMD4
26S proteasome non-ATPase regulatory subunit 4
- PSME4
Proteasome activator complex subunit 4
- PTCH 1
Patched 1
- PTEN
Phosphatidylinositol 3,4,5-trisphosphate 3-phosphatase and dual-specificity protein phosphatase
- PTM
Post-translational Modification
- Ras
Rat Sarcoma
- S6K1
Ribosomal protein S6 kinase beta-1
- SAG
Smoothened Agonist
- SMO
Smoothened
- STAT
Signal Transducers and Activators of Transcription
- SUFU
Suppressor of Fused
- TEAB
Triethylammonium bicarbonate
- TFA
Trifluoroacetic acid
- ULK3
Serine/threonine-protein kinase ULK3
- Vismo
Vismodegib
Authors’ contributions
T. S., H. G.-J., M. D. A, W. G., R. Z., F. A., and C. G. H. conceived the study and experimental design. T. S., H. G.-J., M. D. A., W. G, and F. R. conducted sample preparation and measurements, T. S., H. G.-J., F. R., O. A., M. R., and O. K. performed data evaluation. O. A. developed the Knime Workflow and performed the optimization of data evaluation and statistical analysis. T.S. and O.A. coded the R-Script for data visualization. The manuscript was written through contributions of all authors. All authors have given approval to the final version of the manuscript.
Funding
This work was supported by the International PhD Program “Immunity in Cancer and Allergy” (project W1213) of the Austrian Science Fund, the Cancer Cluster Salzburg, the Allergy-Cancer-Bionano Research Center of the Paris-Lodron University of Salzburg. OA and OK acknowledge funding from the German Ministry of Research and Education (BMBF) under grant (project 031A5.035.0A, de. NBI/CIBI).
Availability of data and materials
The mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium via the PRIDE partner repository with the dataset identifier PXD013408.
Reviewer account details:
Username: reviewer62442@ebi.ac.uk
Password: NPewuMva
Further data has been attached:
Proteomics data:
Protein lists with all quantified proteins and corresponding ratios (5min_pH8_Ratios.csv; 15min_pH8_Ratios.csv)
Phosphoproteomics data:
Phosphopeptidelists with all confident phosphopeptides with normalized raw intensity values (FLR < 0.05.0), ratios SAG_DMSO, Vismo_DMSO and EGF_DMSO and p-values according to LIMMA statistical testing for SAG vs DMSO (SD), Vismo vs DMSO (VD) and SAG vs Vismo (SV): 5min_HILIC_final.csv, 15min_HILIC_final.csv
Supplementary figures (pdf)
List of kinases of Fig. 4 (KSEA_kinases_Figure4.xlsx)
KNIME workflows applied for proteomics and phosphoproteomics raw data analysis and R scripts applied for further data filtering and visualization can be accessed here: Phosphoproteomics Workflows:
5 min: http://www.myexperiment.org/workflows/5113.html
15 min: http://www.myexperiment.org/workflows/5114.html
Proteomics Workflows:
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests1.
Footnotes
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Tamara Scheidt, Email: tamara.scheidt@sbg.ac.at.
Oliver Alka, Email: alka@informatik.uni-tuebingen.de.
Humberto Gonczarowska-Jorge, Email: humgj87@gmail.com.
Wolfgang Gruber, Email: gruber.wolfi@gmail.com.
Florian Rathje, Email: florian.rathje@sbg.ac.at.
Margherita Dell’Aica, Email: margherita.dellaica@gmail.com.
Marc Rurik, Email: rurik@informatik.uni-tuebingen.de.
Oliver Kohlbacher, Email: oliver.kohlbacher@uni-tuebingen.de.
René P. Zahedi, Email: Rene.Zahedi@ladydavis.ca
Fritz Aberger, Email: fritz.aberger@sbg.ac.at.
Christian G. Huber, Email: c.huber@sbg.ac.at
Supplementary information
Supplementary information accompanies this paper at 10.1186/s12964-020-00591-0.
References
- 1.Graves JD, Krebs EG. Protein phosphorylation and signal transduction. Pharmacol Ther. 1999;82:111–121. doi: 10.1016/s0163-7258(98)00056-4. [DOI] [PubMed] [Google Scholar]
- 2.Hanahan D, Weinberg RA. The hallmarks of cancer. Cell. 2000;100:57–70. doi: 10.1016/s0092-8674(00)81683-9. [DOI] [PubMed] [Google Scholar]
- 3.Blume-Jensen P, Hunter T. Oncogenic kinase signalling. Nature. 2001;411:355–365. doi: 10.1038/35077225. [DOI] [PubMed] [Google Scholar]
- 4.Zhang J, Yang PL, Gray NS. Targeting cancer with small molecule kinase inhibitors. Nat Rev Cancer. 2009;9:28–39. doi: 10.1038/nrc2559. [DOI] [PubMed] [Google Scholar]
- 5.Fleuren ED, Zhang L, Wu J, Daly RJ. The kinome 'at large' in cancer. Nat Rev Cancer. 2016;16:83–98. doi: 10.1038/nrc.2015.18. [DOI] [PubMed] [Google Scholar]
- 6.Harsha HC, Pandey A. Phosphoproteomics in cancer. Mol Oncol. 2010;4:482–495. doi: 10.1016/j.molonc.2010.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wu X, Xing X, Dowlut D, Zeng Y, Liu J, Liu X. Integrating phosphoproteomics into kinase-targeted cancer therapies in precision medicine. J Proteome. 2019;191:68–79. doi: 10.1016/j.jprot.2018.03.033. [DOI] [PubMed] [Google Scholar]
- 8.von Stechow L, Francavilla C, Olsen JV. Recent findings and technological advances in phosphoproteomics for cells and tissues. Expert Rev Proteomics. 2015;12:469–487. doi: 10.1586/14789450.2015.1078730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Olsen JV, Blagoev B, Gnad F, Macek B, Kumar C, Mortensen P, Mann M. Global, in vivo, and site-specific phosphorylation dynamics in signaling networks. Cell. 2006;127:635–648. doi: 10.1016/j.cell.2006.09.026. [DOI] [PubMed] [Google Scholar]
- 10.Sudhir PR, Hsu CL, Wang MJ, Wang YT, Chen YJ, Sung TY, Hsu WL, Yang UC, Chen JY. Phosphoproteomics identifies oncogenic ras signaling targets and their involvement in lung adenocarcinomas. PLoS One. 2011;6:e20199. doi: 10.1371/journal.pone.0020199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gnad F, Wallin J, Edgar K, Doll S, Arnott D, Robillard L, Kirkpatrick DS, Stokes MP, Vijapurkar U, Hatzivassiliou G, et al. Quantitative phosphoproteomic analysis of the pi3k-regulated signaling network. Proteomics. 2016;16:1992–1997. doi: 10.1002/pmic.201600118. [DOI] [PubMed] [Google Scholar]
- 12.Wiredja DD, Ayati M, Mazhar S, Sangodkar J, Maxwell S, Schlatzer D, Narla G, Koyutürk M, Chance MR. Proteomics. 2017;17, 1700214. 10.1002/pmic.201700214. [DOI] [PMC free article] [PubMed]
- 13.Wang Y-T, Pan S-H, Tsai C-F, Kuo T-C, Hsu Y-L, Yen H-Y, Choong W-K, Wu H-Y, Liao Y-C, Hong T-M, et al. Phosphoproteomics reveals hmga1, a ck2 substrate, as a drug-resistant target in non-small cell lung cancer. Sci Rep. 2017;7:44021. doi: 10.1038/srep44021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Francavilla C, Lupia M, Tsafou K, Villa A, Kowalczyk K, Rakownikow Jersie-Christensen R, Bertalot G, Confalonieri S, Brunak S, Jensen LJ, et al. Phosphoproteomics of primary cells reveals druggable kinase signatures in ovarian cancer. Cell Rep. 2017;18:3242–3256. doi: 10.1016/j.celrep.2017.03.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chen K, Lv F, Xu G, Zhang M, Wu Y, Wu Z. Phosphoproteomics reveals alk promote cell progress via ras/ jnk pathway in neuroblastoma. Oncotarget. 2016;7:75968–75980. doi: 10.18632/oncotarget.12513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.DeNardo BD, Holloway MP, Ji Q, Nguyen KT, Cheng Y, Valentine MB, Salomon A, Altura RA. Quantitative phosphoproteomic analysis identifies activation of the ret and igf-1r/ir signaling pathways in neuroblastoma. PLoS One. 2013;8:e82513. doi: 10.1371/journal.pone.0082513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wong SY, Reiter JF. The primary cilium at the crossroads of mammalian hedgehog signaling. Curr Top Dev Biol. 2008;85:225–260. doi: 10.1016/S0070-2153(08)00809-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Goetz SC, Ocbina PJ, Anderson KV. The primary cilium as a hedgehog signal transduction machine. Methods Cell Biol. 2009;94:199–222. doi: 10.1016/S0091-679X(08)94010-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kasper M, Regl G, Frischauf AM, Aberger F. Gli transcription factors: mediators of oncogenic hedgehog signalling. Eur J Cancer. 2006;42:437–445. doi: 10.1016/j.ejca.2005.08.039. [DOI] [PubMed] [Google Scholar]
- 20.Huangfu D, Anderson KV. Cilia and hedgehog responsiveness in the mouse. Proc Natl Acad Sci U S A. 2005;102:11325–11330. doi: 10.1073/pnas.0505328102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ruiz i Altaba A, Sanchez P, Dahmane N. Gli and hedgehog in cancer: Tumours, embryos and stem cells. Nat Rev Cancer. 2002;2:361–372. doi: 10.1038/nrc796. [DOI] [PubMed] [Google Scholar]
- 22.Mullor JL, Sanchez P, Ruiz i Altaba A. Pathways and consequences: hedgehog signaling in human disease. Trends Cell Biol. 2002;12:562–569. doi: 10.1016/s0962-8924(02)02405-4. [DOI] [PubMed] [Google Scholar]
- 23.Aberger F, Ruiz IAA. Context-dependent signal integration by the gli code: The oncogenic load, pathways, modifiers and implications for cancer therapy. Semin Cell Dev Biol. 2014;33:93–104. doi: 10.1016/j.semcdb.2014.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Varjosalo M, Bjorklund M, Cheng F, Syvanen H, Kivioja T, Kilpinen S, Sun Z, Kallioniemi O, Stunnenberg HG, He WW, et al. Application of active and kinase-deficient kinome collection for identification of kinases regulating hedgehog signaling. Cell. 2008;133:537–548. doi: 10.1016/j.cell.2008.02.047. [DOI] [PubMed] [Google Scholar]
- 25.Marumoto A, Milani R, da Silva RA, da Costa Fernandes CJ, Granjeiro JM, Ferreira CV, Peppelenbosch MP, Zambuzzi WF. Phosphoproteome analysis reveals a critical role for hedgehog signalling in osteoblast morphological transitions. Bone. 2017;103:55–63. doi: 10.1016/j.bone.2017.06.012. [DOI] [PubMed] [Google Scholar]
- 26.Purzner T, Purzner J, Buckstaff T, et al. Developmental phosphoproteomics identifies the kinase CK2 as a driver of Hedgehog signaling and atherapeutic target in medulloblastoma. Sci Signal. 2018;11(547):eaau5147. 10.1126/scisignal.aau5147. [DOI] [PMC free article] [PubMed]
- 27.Aikin RA, Ayers KL, Thérond PP. The role of kinases in the hedgehog signalling pathway. EMBO Rep. 2008;9:330–336. doi: 10.1038/embor.2008.38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hammerschmidt M, Bitgood MJ, McMahon AP. Protein kinase a is a common negative regulator of hedgehog signaling in the vertebrate embryo. Genes Dev. 1996;10:647–658. doi: 10.1101/gad.10.6.647. [DOI] [PubMed] [Google Scholar]
- 29.Niewiadomski P, Kong JH, Ahrends R, Ma Y, Humke EW, Khan S, Teruel MN, Novitch BG, Rohatgi R. Gli protein activity is controlled by multisite phosphorylation in vertebrate hedgehog signaling. Cell Rep. 2014;6:168–181. doi: 10.1016/j.celrep.2013.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Price MA. Cki, there's more than one: casein kinase i family members in wnt and hedgehog signaling. Genes Dev. 2006;20:399–410. doi: 10.1101/gad.1394306. [DOI] [PubMed] [Google Scholar]
- 31.Zhang C, Williams EH, Guo Y, Lum L, Beachy PA. Extensive phosphorylation of smoothened in hedgehog pathway activation. Proc Natl Acad Sci U S A. 2004;101:17900–17907. doi: 10.1073/pnas.0408093101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chen Y, Yue S, Xie L, Pu XH, Jin T, Cheng SY. Dual phosphorylation of suppressor of fused (sufu) by pka and gsk3beta regulates its stability and localization in the primary cilium. J Biol Chem. 2011;286:13502–13511. doi: 10.1074/jbc.M110.217604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zhao L, Wang L, Chi C, Lan W, Su Y. The emerging roles of phosphatases in hedgehog pathway. Cell Commun Signal. 2017;15:35. doi: 10.1186/s12964-017-0191-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Krauss S, Foerster J, Schneider R, Schweiger S. Protein phosphatase 2a and rapamycin regulate the nuclear localization and activity of the transcription factor gli3. Cancer Res. 2008;68:4658–4665. doi: 10.1158/0008-5472.CAN-07-6174. [DOI] [PubMed] [Google Scholar]
- 35.Liu YC, Couzens AL, Deshwar AR, McBroom-Cerajewski LDB, Zhang X, Puviindran V, Scott IC, Gingras AC, Hui CC, Angers S. The ppfia1-pp2a protein complex promotes trafficking of kif7 to the ciliary tip and hedgehog signaling. Sci Signal. 2014;7:ra117. doi: 10.1126/scisignal.2005608. [DOI] [PubMed] [Google Scholar]
- 36.Jia H, Liu Y, Yan W, Jia J. Pp4 and pp2a regulate hedgehog signaling by controlling smo and ci phosphorylation. Development (Cambridge, England) 2009;136:307–316. doi: 10.1242/dev.030015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Dyson JM, Conduit SE, Feeney SJ, Hakim S, DiTommaso T, Fulcher AJ, Sriratana A, Ramm G, Horan KA, Gurung R, et al. Inpp5e regulates phosphoinositide-dependent cilia transition zone function. J Cell Biol. 2017;216:247–263. doi: 10.1083/jcb.201511055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Pandolfi S, Montagnani V, Penachioni JY, Vinci MC, Olivito B, Borgognoni L, Stecca B. Wip1 phosphatase modulates the hedgehog signaling by enhancing gli1 function. Oncogene. 2013;32:4737–4747. doi: 10.1038/onc.2012.502. [DOI] [PubMed] [Google Scholar]
- 39.Teperino R, Amann S, Bayer M, McGee SL, Loipetzberger A, Connor T, Jaeger C, Kammerer B, Winter L, Wiche G, et al. Hedgehog partial agonism drives Warburg-like metabolism in muscle and brown fat. Cell. 2012;151:414–426. doi: 10.1016/j.cell.2012.09.021. [DOI] [PubMed] [Google Scholar]
- 40.Götschel F, Berg D, Gruber W, Bender C, Eberl M, Friedel M, Sonntag J, Rüngeler E, Hache H, Wierling C, et al. Synergism between hedgehog-gli and egfr signaling in hedgehog-responsive human medulloblastoma cells induces downregulation of canonical hedgehog-target genes and stabilized expression of gli1. PLoS One. 2013;8:e65403. doi: 10.1371/journal.pone.0065403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Chen JK, Taipale J, Young KE, Maiti T, Beachy PA. Small molecule modulation of Smoothened activity. Proc Natl Acad Sci U S A. 2002;99(22):14071–6. 10.1073/pnas.182542899. [DOI] [PMC free article] [PubMed]
- 42.Aditya S, Rattan A. Vismodegib: A smoothened inhibitor for the treatment of advanced basal cell carcinoma. Indian Dermatol Online J. 2013;4:365–368. doi: 10.4103/2229-5178.120685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Manza LL, Stamer SL, Ham AJ, Codreanu SG, Liebler DC. Sample preparation and digestion for proteomic analyses using spin filters. Proteomics. 2005;5:1742–1745. doi: 10.1002/pmic.200401063. [DOI] [PubMed] [Google Scholar]
- 44.Wiśniewski JR, Zougman A, Mann M. Combination of fasp and stagetip-based fractionation allows in-depth analysis of the hippocampal membrane proteome. J Proteome Res. 2009;8:5674–5678. doi: 10.1021/pr900748n. [DOI] [PubMed] [Google Scholar]
- 45.Burkhart JM, Schumbrutzki C, Wortelkamp S, Sickmann A, Zahedi RP. Systematic and quantitative comparison of digest efficiency and specificity reveals the impact of trypsin quality on ms-based proteomics. J Proteome. 2012;75:1454–1462. doi: 10.1016/j.jprot.2011.11.016. [DOI] [PubMed] [Google Scholar]
- 46.Engholm-Keller K, Birck P, Storling J, Pociot F, Mandrup-Poulsen T, Larsen MR. Tish--a robust and sensitive global phosphoproteomics strategy employing a combination of tio2, simac, and hilic. J Proteome. 2012;75:5749–5761. doi: 10.1016/j.jprot.2012.08.007. [DOI] [PubMed] [Google Scholar]
- 47.Gonczarowska-Jorge H, Dell’Aica M, Dickhut C, Zahedi RP. Variable digestion strategies for phosphoproteomics analysis. In: von Stechow L, editor. Phospho-proteomics: Methods and protocols. New York: Springer New York; 2016. pp. 225–239. [DOI] [PubMed] [Google Scholar]
- 48.Olsen JV, de Godoy LM, Li G, Macek B, Mortensen P, Pesch R, Makarov A, Lange O, Horning S, Mann M. Parts per million mass accuracy on an orbitrap mass spectrometer via lock mass injection into a c-trap. Mol Cell Proteomics. 2005;4:2010–2021. doi: 10.1074/mcp.T500030-MCP200. [DOI] [PubMed] [Google Scholar]
- 49.Fillbrunn A, Dietz C, Pfeuffer J, Rahn R, Landrum GA, Berthold MR. Knime for reproducible cross-domain analysis of life science data. J Biotechnol. 2017;261:149–156. doi: 10.1016/j.jbiotec.2017.07.028. [DOI] [PubMed] [Google Scholar]
- 50.Rost HL, Sachsenberg T, Aiche S, Bielow C, Weisser H, Aicheler F, Andreotti S, Ehrlich HC, Gutenbrunner P, Kenar E, et al. Openms: A flexible open-source software platform for mass spectrometry data analysis. Nat Methods. 2016;13:741–748. doi: 10.1038/nmeth.3959. [DOI] [PubMed] [Google Scholar]
- 51.Pfeuffer J, Sachsenberg T, Alka O, Walzer M, Fillbrunn A, Nilse L, Schilling O, Reinert K, Kohlbacher O. Openms - a platform for reproducible analysis of mass spectrometry data. J Biotechnol. 2017;261:142–148. doi: 10.1016/j.jbiotec.2017.05.016. [DOI] [PubMed] [Google Scholar]
- 52.Kim S, Pevzner PA. Ms-gf+ makes progress towards a universal database search tool for proteomics. Nat Commun. 2014;5:5277. doi: 10.1038/ncomms6277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.The M, MacCoss MJ, Noble WS, Kall L. Fast and accurate protein false discovery rates on large-scale proteomics data sets with percolator 3.0. J Am Soc Mass Spectrom. 2016;27:1719–1727. doi: 10.1007/s13361-016-1460-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Fermin D, Avtonomov D, Choi H, Nesvizhskii AI. Luciphor2: site localization of generic post-translational modifications from tandem mass spectrometry data. Bioinformatics. 2015;31:1141–1143. doi: 10.1093/bioinformatics/btu788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Serang O, MacCoss MJ, Noble WS. Efficient marginalization to compute protein posterior probabilities from shotgun mass spectrometry data. J Proteome Res. 2010;9:5346–5357. doi: 10.1021/pr100594k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Shema G, Nguyen MTN, Solari FA, Loroch S, Venne AS, Kollipara L, Sickmann A, Verhelst SHL, Zahedi RP. Simple, scalable, and ultrasensitive tip-based identification of protease substrates. Mol Cell Proteomics. 2018;17:826–834. doi: 10.1074/mcp.TIR117.000302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Ritchie ME, Phipson B, Wu D, Hu Y, Law CW, Shi W, Smyth GK. Limma powers differential expression analyses for rna-sequencing and microarray studies. Nucl Acids Res. 2015;43:e47. doi: 10.1093/nar/gkv007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Casado P, Rodriguez-Prados JC, Cosulich SC, Guichard S, Vanhaesebroeck B, Joel S, Cutillas PR. Kinase-substrate enrichment analysis provides insights into the heterogeneity of signaling pathway activation in leukemia cells. Sci Signal. 2013;6:rs6. doi: 10.1126/scisignal.2003573. [DOI] [PubMed] [Google Scholar]
- 59.Wiredja DD, Koyutürk M, Chance MR. The KSEA App: a web-based tool for kinase activity inference from quantitative phosphoproteomics.Bioinformatics. 2017;33(21):3489–91. 10.1093/bioinformatics/btx415. [DOI] [PMC free article] [PubMed]
- 60.Dudu V, Able RA, Jr, Rotari V, Kong Q, Vazquez M. Role of epidermal growth factor-triggered pi3k/akt signaling in the migration of medulloblastoma-derived cells. Cell Mol Bioeng. 2012;5:502–413. doi: 10.1007/s12195-012-0253-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Beck F, Geiger J, Gambaryan S, Solari FA, Dell'Aica M, Loroch S, Mattheij NJ, Mindukshev I, Potz O, Jurk K, et al. Temporal quantitative phosphoproteomics of adp stimulation reveals novel central nodes in platelet activation and inhibition. Blood. 2017;129:e1–e12. doi: 10.1182/blood-2016-05-714048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Beck F, Geiger J, Gambaryan S, Veit J, Vaudel M, Nollau P, Kohlbacher O, Martens L, Walter U, Sickmann A, et al. Time-resolved characterization of camp/pka-dependent signaling reveals that platelet inhibition is a concerted process involving multiple signaling pathways. Blood. 2014;123:e1–e10. doi: 10.1182/blood-2013-07-512384. [DOI] [PubMed] [Google Scholar]
- 63.Asaoka Y, Kanai F, Ichimura T, Tateishi K, Tanaka Y, Ohta M, Seto M, Tada M, Ijichi H, Ikenoue T, et al. Identification of a suppressive mechanism for hedgehog signaling through a novel interaction of gli with 14-3-3. J Biol Chem. 2010;285:4185–4194. doi: 10.1074/jbc.M109.038232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Kotani T, Chapter twelve - Protein Kinase A Activity and Hedgehog Signaling Pathway, publisher’s location: Vitamins & Hormones, Academic Press. 2012;88:273-91. ISSN 0083-6729, ISBN 9780123946225. 10.1016/B978-0-12-394622-5.00012-2. [DOI] [PubMed]
- 65.Rao G, Pedone CA, Del Valle L, Reiss K, Holland EC, Fults DW. Sonic hedgehog and insulin-like growth factor signaling synergize to induce medulloblastoma formation from nestin-expressing neural progenitors in mice. Oncogene. 2004;23:6156–6162. doi: 10.1038/sj.onc.1207818. [DOI] [PubMed] [Google Scholar]
- 66.Zhao X, Ponomaryov T, Ornell KJ, Zhou P, Dabral SK, Pak E, Li W, Atwood SX, Whitson RJ, Chang ALS, et al. Ras/mapk activation drives resistance to smo inhibition, metastasis, and tumor evolution in shh pathway-dependent tumors. Cancer Res. 2015;75:3623–3635. doi: 10.1158/0008-5472.CAN-14-2999-T. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Brechbiel J, Miller-Moslin K, Adjei AA. Crosstalk between hedgehog and other signaling pathways as a basis for combination therapies in cancer. Cancer Treat Rev. 2014;40:750–759. doi: 10.1016/j.ctrv.2014.02.003. [DOI] [PubMed] [Google Scholar]
- 68.Whisenant TC, Ho DT, Benz RW, et al. Computational prediction and experimental verification of new MAP kinase docking sites and substrates including Gli transcription factors. PLoS Comput Biol. 2010;6(8):e1000908. 10.1371/journal.pcbi.1000908. [DOI] [PMC free article] [PubMed]
- 69.Stecca B, Mas C, Clement V, Zbinden M, Correa R, Piguet V, Beermann F, Ruiz IAA. Melanomas require hedgehog-gli signaling regulated by interactions between gli1 and the ras-mek/akt pathways. Proc Natl Acad Sci U S A. 2007;104:5895–5900. doi: 10.1073/pnas.0700776104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Kasper M, Schnidar H, Neill GW, Hanneder M, Klingler S, Blaas L, Schmid C, Hauser-Kronberger C, Regl G, Philpott MP, et al. Selective modulation of hedgehog/gli target gene expression by epidermal growth factor signaling in human keratinocytes. Mol Cell Biol. 2006;26:6283–6298. doi: 10.1128/MCB.02317-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Schnidar H, Eberl M, Klingler S, Mangelberger D, Kasper M, Hauser-Kronberger C, Regl G, Kroismayr R, Moriggl R, Sibilia M, et al. Epidermal growth factor receptor signaling synergizes with hedgehog/gli in oncogenic transformation via activation of the mek/erk/Jun pathway. Cancer Res. 2009;69:1284–1292. doi: 10.1158/0008-5472.CAN-08-2331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Eberl M, Klingler S, Mangelberger D, Loipetzberger A, Damhofer H, Zoidl K, Schnidar H, Hache H, Bauer HC, Solca F, et al. Hedgehog-egfr cooperation response genes determine the oncogenic phenotype of basal cell carcinoma and tumour-initiating pancreatic cancer cells. EMBO Mol Med. 2012;4:218–233. doi: 10.1002/emmm.201100201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Seto M, Ohta M, Asaoka Y, Ikenoue T, Tada M, Miyabayashi K, Mohri D, Tanaka Y, Ijichi H, Tateishi K, et al. Regulation of the hedgehog signaling by the mitogen-activated protein kinase cascade in gastric cancer. Mol Carcinog. 2009;48:703–712. doi: 10.1002/mc.20516. [DOI] [PubMed] [Google Scholar]
- 74.Lu J-T, Zhao W-D, He W, Wei W. Hedgehog signaling pathway mediates invasion and metastasis of hepatocellular carcinoma via erk pathway. Acta Pharmacol Sin. 2012;33:691–700. doi: 10.1038/aps.2012.24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Rovida E, Stecca B. Mitogen-activated protein kinases and hedgehog-gli signaling in cancer: A crosstalk providing therapeutic opportunities? Semin Cancer Biol. 2015;35:154–167. doi: 10.1016/j.semcancer.2015.08.003. [DOI] [PubMed] [Google Scholar]
- 76.Stecca B, Ruiz IAA. Context-dependent regulation of the gli code in cancer by hedgehog and non-hedgehog signals. J Mol Cell Biol. 2010;2:84–95. doi: 10.1093/jmcb/mjp052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Mangelberger D, Kern D, Loipetzberger A, Eberl M, Aberger F. Cooperative hedgehog-egfr signaling. Front Biosci (Landmark Ed) 2012;17:90–99. doi: 10.2741/3917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Teglund S, Toftgard R. Hedgehog beyond medulloblastoma and basal cell carcinoma. Biochim Biophys Acta. 1805;2010:181–208. doi: 10.1016/j.bbcan.2010.01.003. [DOI] [PubMed] [Google Scholar]
- 79.Neill GW, Harrison WJ, Ikram MS, Williams TD, Bianchi LS, Nadendla SK, Green JL, Ghali L, Frischauf AM, O'Toole EA, et al. Gli1 repression of erk activity correlates with colony formation and impaired migration in human epidermal keratinocytes. Carcinogenesis. 2008;29:738–746. doi: 10.1093/carcin/bgn037. [DOI] [PubMed] [Google Scholar]
- 80.Wang Y, Ding Q, Yen C-J, Xia W, Izzo JG, Lang J-Y, Li C-W, Hsu JL, Miller SA, Wang X, et al. The crosstalk of mtor/s6k1 and hedgehog pathways. Cancer Cell. 2012;21:374–387. doi: 10.1016/j.ccr.2011.12.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Sternberg C, Gruber W, Eberl M, Tesanovic S, Stadler M, Elmer DP, Schlederer M, Grund S, Roos S, Wolff F, et al. Synergistic cross-talk of hedgehog and interleukin-6 signaling drives growth of basal cell carcinoma. Int J Cancer. 2018;143:2943–2954. doi: 10.1002/ijc.31724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Gu D, Fan Q, Zhang X, Xie J. A role for transcription factor stat3 signaling in oncogene smoothened-driven carcinogenesis. J Biol Chem. 2012;287:38356–38366. doi: 10.1074/jbc.M112.377382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Dong W, Cui J, Tian X, He L, Wang Z, Zhang P, Zhang H. Aberrant sonic hedgehog signaling pathway and stat3 activation in papillary thyroid cancer. Int J Clin Exp Med. 2014;7:1786–1793. [PMC free article] [PubMed] [Google Scholar]
- 84.Hartmann W, Digon-Sontgerath B, Koch A, Waha A, Endl E, Dani I, Denkhaus D, Goodyer CG, Sorensen N, Wiestler OD, et al. Phosphatidylinositol 3′-kinase/akt signaling is activated in medulloblastoma cell proliferation and is associated with reduced expression of pten. Clin Cancer Res. 2006;12:3019–3027. doi: 10.1158/1078-0432.CCR-05-2187. [DOI] [PubMed] [Google Scholar]
- 85.Metcalfe C, Alicke B, Crow A, Lamoureux M, Dijkgraaf GJ, Peale F, Gould SE, de Sauvage FJ. Pten loss mitigates the response of medulloblastoma to hedgehog pathway inhibition. Cancer Res. 2013;73:7034–7042. doi: 10.1158/0008-5472.CAN-13-1222. [DOI] [PubMed] [Google Scholar]
- 86.Chaturvedi NK, Kling MJ, Coulter DW, McGuire TR, Ray S, Kesherwani V, Joshi SS, Sharp JG. Improved therapy for medulloblastoma: targeting hedgehog and pi3k-mtor signaling pathways in combination with chemotherapy. Oncotarget. 2018;9:16619–16633. doi: 10.18632/oncotarget.24618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Cortez D, Wang Y, Qin J, Elledge SJ. Requirement of atm-dependent phosphorylation of brca1 in the DNA damage response to double-strand breaks. Science. 1999;286:1162–1166. doi: 10.1126/science.286.5442.1162. [DOI] [PubMed] [Google Scholar]
- 88.Au WW, Henderson BR. Identification of sequences that target brca1 to nuclear foci following alkylative DNA damage. Cell Signal. 2007;19:1879–1892. doi: 10.1016/j.cellsig.2007.04.010. [DOI] [PubMed] [Google Scholar]
- 89.Tibbetts RS, Cortez D, Brumbaugh KM, Scully R, Livingston D, Elledge SJ, Abraham RT. Functional interactions between brca1 and the checkpoint kinase atr during genotoxic stress. Genes Dev. 2000;14:2989–3002. doi: 10.1101/gad.851000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Sankaran S, Crone DE, Palazzo RE, Parvin JD. Aurora-a kinase regulates breast cancer associated gene 1 inhibition of centrosome-dependent microtubule nucleation. Cancer Res. 2007;67:11186–11194. doi: 10.1158/0008-5472.CAN-07-2578. [DOI] [PubMed] [Google Scholar]
- 91.Xu L, Liu H, Yan Z, Sun Z, Luo S, Lu Q. Inhibition of the hedgehog signaling pathway suppresses cell proliferation by regulating the gli2/mir-124/aurka axis in human glioma cells. Int J Oncol. 2017;50:1868–1878. doi: 10.3892/ijo.2017.3946. [DOI] [PubMed] [Google Scholar]
- 92.Markant SL, Esparza LA, Sun J, Barton KL, McCoig LM, Grant GA, Crawford JR, Levy ML, Northcott PA, Shih D, et al. Targeting sonic hedgehog-associated medulloblastoma through inhibition of aurora and polo-like kinases. Cancer Res. 2013;73:6310–6322. doi: 10.1158/0008-5472.CAN-12-4258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Gorivodsky M, Mukhopadhyay M, Wilsch-Braeuninger M, Phillips M, Teufel A, Kim C, Malik N, Huttner W, Westphal H. Intraflagellar transport protein 172 is essential for primary cilia formation and plays a vital role in patterning the mammalian brain. Dev Biol. 2009;325:24–32. doi: 10.1016/j.ydbio.2008.09.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Seeger-Nukpezah T, Liebau MC, Hopker K, Lamkemeyer T, Benzing T, Golemis EA, Schermer B. The centrosomal kinase plk1 localizes to the transition zone of primary cilia and induces phosphorylation of nephrocystin-1. PLoS One. 2012;7:e38838. doi: 10.1371/journal.pone.0038838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Wang G, Chen Q, Zhang X, Zhang B, Zhuo X, Liu J, Jiang Q, Zhang C. Pcm1 recruits plk1 to the pericentriolar matrix to promote primary cilia disassembly before mitotic entry. J Cell Sci. 2013;126:1355–1365. doi: 10.1242/jcs.114918. [DOI] [PubMed] [Google Scholar]
- 96.Cholewa BD, Liu X, Ahmad N. The role of polo-like kinase 1 in carcinogenesis: cause or consequence? Cancer Res. 2013;73:6848–6855. doi: 10.1158/0008-5472.CAN-13-2197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Liu Z, Sun Q, Wang X. Plk1, a potential target for cancer therapy. Transl Oncol. 2017;10:22–32. doi: 10.1016/j.tranon.2016.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Zhang T, Xin G, Jia M, et al. The Plk1 kinase negatively regulates the Hedgehog signaling pathway by phosphorylating Gli1. J Cell Sci.2019;132(2):jcs220384. 10.1242/jcs.220384. [DOI] [PubMed]
- 99.Harris PS, Venkataraman S, Alimova I, Birks DK, Donson AM, Knipstein J, Dubuc A, Taylor MD, Handler MH, Foreman NK, et al. Polo-like kinase 1 (plk1) inhibition suppresses cell growth and enhances radiation sensitivity in medulloblastoma cells. BMC Cancer. 2012;12:80. doi: 10.1186/1471-2407-12-80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Jia H, Liu Y, Xia R, Tong C, Yue T, Jiang J, Jia J. Casein kinase 2 promotes hedgehog signaling by regulating both smoothened and cubitus interruptus. J Biol Chem. 2010;285:37218–37226. doi: 10.1074/jbc.M110.174565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Zhang S, Wang Y, Mao JH, Hsieh D, Kim IJ, Hu LM, Xu Z, Long H, Jablons DM, You L. Inhibition of ck2alpha down-regulates hedgehog/gli signaling leading to a reduction of a stem-like side population in human lung cancer cells. PLoS One. 2012;7:e38996. doi: 10.1371/journal.pone.0038996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Nitta R, Bolin SM, Nwagbo G, Purzner T, Kahn S, Cho Y-J, Li G. Abstract lb-322: Casein kinase 2 is a major regulator of medulloblastoma growth. Cancer Res. 2018;78:LB-322. [Google Scholar]
- 103.Chen Y, Jiang J. Decoding the phosphorylation code in hedgehog signal transduction. Cell Res. 2013;23:186–200. doi: 10.1038/cr.2013.10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Toyo-oka K, Mori D, Yano Y, Shiota M, Iwao H, Goto H, Inagaki M, Hiraiwa N, Muramatsu M, Wynshaw-Boris A, et al. Protein phosphatase 4 catalytic subunit regulates cdk1 activity and microtubule organization via ndel1 dephosphorylation. J Cell Biol. 2008;180:1133–1147. doi: 10.1083/jcb.200705148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Chavez M, Ena S, Van Sande J, de Kerchove d'Exaerde A, Schurmans S, Schiffmann SN. Modulation of ciliary phosphoinositide content regulates trafficking and sonic hedgehog signaling output. Dev Cell. 2015;34:338–350. doi: 10.1016/j.devcel.2015.06.016. [DOI] [PubMed] [Google Scholar]
- 106.Garcia-Gonzalo FR, Phua SC, Roberson EC, Garcia G, 3rd, Abedin M, Schurmans S, Inoue T, Reiter JF. Phosphoinositides regulate ciliary protein trafficking to modulate hedgehog signaling. Dev Cell. 2015;34:400–409. doi: 10.1016/j.devcel.2015.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Moore BS, Stepanchick AN, Tewson PH, Hartle CM, Zhang J, Quinn AM, Hughes TE, Mirshahi T. Cilia have high camp levels that are inhibited by sonic hedgehog-regulated calcium dynamics. Proc Natl Acad Sci U S A. 2016;113:13069–13074. doi: 10.1073/pnas.1602393113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Goetz SC, Anderson KV. The primary cilium: A signalling Centre during vertebrate development. Nat Rev Genet. 2010;11:331–344. doi: 10.1038/nrg2774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Bangs F, Anderson KV. Primary cilia and mammalian hedgehog signaling. Cold Spring Harb Perspect Biol. 2017;9:a028175. doi: 10.1101/cshperspect.a028175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Pan J, Naumann-Busch B, Wang L, Specht M, Scholz M, Trompelt K, Hippler M. Protein phosphorylation is a key event of flagellar disassembly revealed by analysis of flagellar phosphoproteins during flagellar shortening in chlamydomonas. J Proteome Res. 2011;10:3830–3839. doi: 10.1021/pr200428n. [DOI] [PubMed] [Google Scholar]
- 111.Maloverjan A, Piirsoo M, Kasak L, Peil L, Osterlund T, Kogerman P. Dual function of unc-51-like kinase 3 (ulk3) in the sonic hedgehog signaling pathway. J Biol Chem. 2010;285:30079–30090. doi: 10.1074/jbc.M110.133991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Maloverjan A, Piirsoo M, Michelson P, Kogerman P, Osterlund T. Identification of a novel serine/threonine kinase ulk3 as a positive regulator of hedgehog pathway. Exp Cell Res. 2010;316:627–637. doi: 10.1016/j.yexcr.2009.10.018. [DOI] [PubMed] [Google Scholar]
- 113.Humke EW, Dorn KV, Milenkovic L, Scott MP, Rohatgi R. The output of hedgehog signaling is controlled by the dynamic association between suppressor of fused and the gli proteins. Genes Dev. 2010;24:670–682. doi: 10.1101/gad.1902910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Philipp M, Caron MG. Hedgehog signaling: is smo a g protein-coupled receptor? Curr Biol. 2009;19:R125–R127. doi: 10.1016/j.cub.2008.12.010. [DOI] [PubMed] [Google Scholar]
- 115.El-Sheikh A, Fan R, Birks D, Donson A, Foreman NK, Vibhakar R. Inhibition of aurora kinase a enhances chemosensitivity of medulloblastoma cell lines. Pediatr Blood Cancer. 2010;55:35–41. doi: 10.1002/pbc.22465. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional file 1: Table S-1. Eluent B (B) gradient range and gradient duration for the different analyzed fractions. Table S-2. Applied R-packages. Responsiveness of DAOY cells to SAG and vismodegib treatment. RNA isolation and quantitative PCR (qPCR). Table S-3. Primer Sequences used for qPCR. Figure S-1. Hedgehog target gene expression in SAG and vismodegib treated DAOY cells. Figure S-2. Comparable Hedgehog pathway induction by natural and synthetic HH pathway activators. Figure S-3. Distribution of protein groups identified after 5.0 min treatment (A) and 15 min treatment (B). Figure S-4. Ingenuity pathway analysis of all phosphopeptides identified after 15 min using a phosphorylation analysis. Figure S-5. Time dependent Ingenuity pathway analysis of cancer associated pathways. Figure S-6. Western blot validation of PKA activity modulation in response to SMO agonist and antagonists. Table S-4. Antibodies used for Western blot analysis. Figure S-7. Overlap of quantified phosphopeptides after 5.0 and 15 min. Figure S-8. Volcano plots of phosphopeptides identified after 5.0 min treatment. Figure S-9. Kinase substrate enrichment analysis (KSEA) was performed for phosphopeptides identified after 5.0 min treatment for the ratio SAG/Vismo using the online platform KSEA App (https://casecpb.shinyapps.io/ksea/). Figure S-10. Volcano plots and kinase set enrichment analysis for phosphopeptides identified after 15 min treatment. Figure S-11. IFT172 phosphorylation and expression after 5.0 and 15 min.
Data Availability Statement
The mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium via the PRIDE partner repository with the dataset identifier PXD013408.
Reviewer account details:
Username: reviewer62442@ebi.ac.uk
Password: NPewuMva
Further data has been attached:
Proteomics data:
Protein lists with all quantified proteins and corresponding ratios (5min_pH8_Ratios.csv; 15min_pH8_Ratios.csv)
Phosphoproteomics data:
Phosphopeptidelists with all confident phosphopeptides with normalized raw intensity values (FLR < 0.05.0), ratios SAG_DMSO, Vismo_DMSO and EGF_DMSO and p-values according to LIMMA statistical testing for SAG vs DMSO (SD), Vismo vs DMSO (VD) and SAG vs Vismo (SV): 5min_HILIC_final.csv, 15min_HILIC_final.csv
Supplementary figures (pdf)
List of kinases of Fig. 4 (KSEA_kinases_Figure4.xlsx)
KNIME workflows applied for proteomics and phosphoproteomics raw data analysis and R scripts applied for further data filtering and visualization can be accessed here: Phosphoproteomics Workflows:
5 min: http://www.myexperiment.org/workflows/5113.html
15 min: http://www.myexperiment.org/workflows/5114.html
Proteomics Workflows: